Abstract
In Saccharomyces cerevisiae, the developmentally regulated Soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) protein Spo20p mediates the fusion of vesicles with the prospore membrane, which is required for the formation of spores. Spo20p is subject to both positive and negative regulation by separate sequences in its aminoterminal domain. We report that the positive activity is conferred by a short, amphipathic helix that is sufficient to confer plasma membrane or prospore membrane localization to green fluorescent protein. In vitro, this helix binds to acidic phospholipids, and mutations that reduce or eliminate phospholipid binding in vitro inactivate Spo20p in vivo. Genetic manipulation of phospholipid pools indicates that the likely in vivo ligand of this domain is phosphatidic acid. The inhibitory activity is a nuclear targeting signal, which confers nuclear localization in vegetative cells and in cells entering meiosis. However, as cells initiate spore formation, fusions containing the inhibitory domain exit the nucleus and localize to the nascent prospore membrane. Thus, the SNARE Spo20p is both positively and negatively regulated by control of its intracellular localization.
INTRODUCTION
The secretory pathway of eukaryotic cells consists of a series of membrane-bound compartments between which proteins and lipids move in vesicle carriers. To maintain the organization of the endomembrane system, it is of particular importance that fusion between membranes be strictly controlled so that any particular vesicle fuses only with the appropriate compartment. Soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) proteins are thought to play a critical role in maintaining the specificity of vesicle fusion events (Protopopov et al., 1993; Sollner et al., 1993; Rothman, 1994; Rothman and Warren, 1994; Pelham, 1999; Jahn et al., 2003). SNAREs mediate membrane fusion by the formation of a complex between a membrane protein of the vesicle (v-SNARE) and related proteins on the acceptor compartment membrane (t-SNAREs) (Protopopov et al., 1993; Sollner et al., 1993; Rothman, 1994; Rothman and Warren, 1994; Pelham, 1999; Jahn et al., 2003). The v- and t-SNAREs oligomerize through related helices found in all SNARE proteins, and this oligomerization is thought to lead directly to fusion of the lipid bilayers (Weimbs et al., 1997; Sutton et al., 1998; Weber et al., 1998). Furthermore, specific interactions between a v-SNARE and its cognate t-SNARE are proposed to provide much of the specificity for vesicle fusion (Rothman, 1994; Rothman and Warren, 1994; Parlati et al., 2002). Thus, regulation of SNARE proteins would provide the cell an excellent means of regulating activity of the secretory pathway. One such regulated change in SNARE protein usage occurs during sporulation in Saccharomyces cerevisiae.
In response to starvation, diploid cells of S. cerevisiae enter a developmental program in which they undergo meiosis and differentiate into haploid spores (Byers, 1981). The process of sporulation involves the de novo generation of intracellular membranes, called prospore membranes, which encapsulate each of the daughter nuclei, forming daughter cells (spores) (Moens, 1971; Byers, 1981). Prospore membrane formation is driven by the redirection of post-Golgi secretory vesicles from the plasma membrane to intracellular sites where they coalesce to form prospore membranes (Moens and Rapport, 1971; Neiman, 1998).
In addition to a redirection of secretory vesicles, proper formation of the prospore membrane requires the induction of a sporulation-specific SNARE molecule, Spo20p (Neiman, 1998). Spo20p is a member of the soluble N-ethylmaleimide-sensitive factor attachment protein (SNAP)-25 SNARE sub-family. Unlike most SNARE proteins, SNAP-25 family members do not contain a transmembrane domain (Hess et al., 1992; Weimbs et al., 1997). In mitotically growing yeast cells, a second yeast SNAP-25 orthologue, Sec9p, binds to the t-SNAREs Sso1/2p and the v-SNAREs Snc1/2p to form a complex responsible for mediating vesicle fusion at the plasma membrane (Gerst et al., 1992; Aalto et al., 1993; Protopopov et al., 1993; Brennwald et al., 1994; Couve and Gerst, 1994). During sporulation, Spo20p replaces Sec9p to mediate fusion at the prospore membrane (Neiman, 1998). Despite sharing 40% sequence identity in their SNAP-25 domains and the ability to oligomerize with the same partner SNAREs Sec9p and Spo20p cannot efficiently mediate fusion at the other's membrane of action. That is, even though they mediate the fusion of the same vesicle population, the function of each protein is limited to a specific acceptor compartment (Neiman, 1998; Neiman et al., 2000). Thus, the switch from use of Sec9p to Spo20p provides a useful model system in which to study SNARE specificity.
In previous work, a series of chimeric SPO20/SEC9 genes were constructed in an attempt to define regions of the proteins conferring plasma membrane or prospore membrane-specific functions (Neiman et al., 2000). These studies defined an important role for the unique amino-terminal region of Spo20p in the specificity of function. Deletion studies identified two separable activities in the Spo20p amino terminus. First, residues 51-95 of the full-length protein defined a region essential for SNARE function at the prospore membrane. Second, residues 4-50 of the protein contained an activity that inhibited SNARE function in vegetative cells. This latter activity could inhibit the function of either the Spo20p or the Sec9p SNARE domain. Thus, the Spo20p amino terminus both positively and negatively regulated SNARE function.
Studies of other SNARE proteins have also identified regulatory domains outside of the core SNARE helical regions (Nicholson et al., 1998; Cheever et al., 2001; Laage and Ungermann, 2001; Tochio et al., 2001). The VAM7 gene encodes a SNARE related to Spo20p and Sec9p that functions in vesicle fusion at the yeast vacuole (Sato et al., 1998; Ungermann and Wickner, 1998). Like Spo20p and Sec9p, Vam7p contains no transmembrane domain. Rather, in its amino terminal region Vam7p has a PX domain that binds to phosphatidylinositol-3-phosphate (PI3P) (Cheever et al., 2001). Binding of PI3P by the PX domain is required to target Vam7p to the vacuolar membrane and mutation of the PX domain interferes with Vam7p function (Cheever et al., 2001).
Inhibitory domains have also been defined within SNARE proteins. In the plasma membrane t-SNARE Sso1p, three short helical regions form a four helix bundle in combination with the SNARE helix (Munson et al., 2000). Formation of this intramolecular bundle competitively inhibits SNARE complex formation by blocking interactions between the Sso1p SNARE helix and its partner SNAREs (Nicholson et al., 1998; Munson and Hughson, 2002). Override of this intrinsic negative activity is important for activity in vivo (Munson and Hughson, 2002). Several other SNAREs have also been shown to be autoinhibited by their amino-terminal domains (Dulubova et al., 2001; Laage and Ungermann, 2001; Tochio et al., 2001; Antonin et al., 2002).
In this study, we have examined the basis for positive and negative regulation of Spo20p by its amino-terminal region. Our results indicate that the positive regulatory region contains a lipid binding motif necessary for proper localization of the Spo20 protein to the prospore membrane. The inhibitory activity of the first 50 amino acids of Spo20p is due to the presence of a nuclear targeting activity that sequesters the protein in the nucleus. In wild-type cells this activity is inactivated as cells enter meiosis II and begin to form prospore membranes. Thus, Spo20p is both positively and negatively regulated by control of its intracellular localization.
MATERIALS AND METHODS
Yeast Strains and Media
Unless otherwise noted, standard media and genetic methods were used (Rose and Fink, 1990). S. cerevisiae strains used in this study are listed in Table 1. AN211 was made as follows. AN63-2C (Neiman, 1998) and W303-1B (Thomas and Rothstein, 1989) were crossed and dissected to generate AN123-4A. AN123-4A was then crossed to an AN146-1D (a parent of AN147; Neiman et al., 2000) and dissected to generate AN204-5D(MATα ura3 trp1 leu2 lys2 spo20Δ::his5+ sec9-4). An ade2 derivative of AN117-4B (Neiman et al., 2000) was crossed to AN146-4D and dissected to generate AN206-1C (MATa ura3 his3 trp1 leu2 ade2 spo20Δ::his5+). AN204-5D and AN206-1C were mated and segregants were crossed to generate AN211.
Table 1.
Yeast strains used in this study
| Name | Genotype | Source |
|---|---|---|
| AN120 | MATa/MATα his3/his3 ura3/ura3 trp1 :: hisG/trp1 :: hisG leu2/leu2 arg4-NspI/ARG4 lys2/lys2 hoΔ :: LYS2/hoΔ :: LYS2 rme1 :: LEU2/RME1 | Neiman et al., 2000 |
| AN147 | MATa/MATαhis3/his3 ura3/ura3 trp1 :: hisG/trp1 :: hisG leu2/leu2 arg4-NspI/ARG4 lys2/lys2 hoΔ :: LYS2/hoΔ :: LYS2 rme1 :: LEU2/RME1 spo20Δ :: his5+/spo20Δ :: his5+ | Neiman et al., 2000 |
| W303-1A | MATa Leu2 his3 ade2 ura3 trp1 can1 | Thomas and Rothstein, 1989 |
| AAY202 | MATαleu2 ura3 his3 trp1 lys2 suc2 mss4 :: his3 harboring YCplac111mss4ts-102 | Stefan et al., 2002 |
| AN211 | MATa/MATα his3/his3 ura3/ura3 trp1/trp1 leu2/leu2 LYS2/lys2 Ade2/ade2 sec9-4/sec9-4 spo20Δ :: his5+/spo20Δ :: his5+ | This study |
| AAY102 | MATαleu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 stt4 :: HIS3 harboring pRS415 stt4-4 | Audhya et al., 2002 |
| AAY104 | MATαleu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 pik1 :: HIS3 harboring pRS314-pik1-83 | Audhya et al., 2002 |
Plasmids
Plasmids and oligonucleotides used in this study are listed in Tables 2 and 3, respectively. To construct the centromere plasmid pRS316-SEC9pr-Δ3-51PSPS a KpnI-SacI fragment of pRS306-SEC9pr-Δ3-51PSPS (Neiman et al., 2000) was cloned into KpnI and SacI sites of pRS316 (Sikorski and Hieter, 1989). The integrating plasmids pRS306-SPO20pr-SPO20 and pRS306-SPO20pr-Δ3-51SPO20 (Neiman et al., 2000) were used to express Spo20p and Spo20pΔ3-51 under the control of SPO20 promoter, respectively. pRS426-SPO20pr-SPO20, pRS426-SPO20pr-Δ3-51SPO20, and pRS426-SPO20pr-Δ3-95SPO20 were constructed by cloning the KpnI-SacI fragment of pRS306-SPO20pr-SPO20, pRS306-SPO20pr-Δ3-51SPO20, and pRS306-SPO20pr-Δ3-95SPO20 (Neiman et al., 2000) into similarly digested pRS426 (Christianson et al., 1992), respectively. To make pRS306-SPO20pr-Δ3-51SPO20(L67P), the XhoI and SpeI fragment of pRS316-SEC9pr-Δ3-51PSPS(L67P) (see below) was cloned into XhoI and SpeI digested pRS306-SPO20pr-Δ3-51SPO20.
Table 2.
Plasmids used in this study
| Name | Yeast markers | Promoter | Cloned gene | Source |
|---|---|---|---|---|
| pRS306-SEC9pr-Δ3-51PSPS | URA3, Integration | SEC9 | Δ3-51 PSPS chimera | Neiman et al. 2000 |
| pRS306-SPO20pr-SPO20 | URA3, Integration | SPO20 | SPO20 | Neiman et al. 2000 |
| pRS306-SPO20pr-Δ3-51SP020 | URA3, Integration | SPO20 | Δ3-51, SPO20 | Neiman et al. 2000 |
| pRS306-SPO20pr-Δ3-95SPO20 | URA3, Integration | SPO20 | Δ3-95 SPO20 | Neiman et al. 2000 |
| pRS306-SPO20pr-Δ3-51SPO20(L63N) | URA3, Integration | SPO20 | Δ3-51 SPO20 with L63N mutation | This study |
| pRS306-SPO20pr-Δ3-51SPO20(L67P) | URA3, Integration | SPO20 | Δ3-51 SPO20 with L67P mutation | This study |
| pRS306-SPO20pr-Δ3-51SPO20(L67N) | URA3, Integration | SPO20 | Δ3-51 SPO20 with L67N mutation | This study |
| pRS306-SPO20pr-Δ3-51SPO20(L70N) | URA3, Integration | SPO20 | Δ3-51 SPO20 with L70N mutation | This study |
| pRS306-SPO20pr-Δ3-51SPO20(I74N) | URA3, Integration | SPO20 | Δ3-51 SPO20 with I74N mutation | This study |
| pRS306-SPO20pr-Δ3-51SPO20(2A) | URA3, Integration | SPO20 | Δ3-51 SPO20 with K68A,R71A mutations | This study |
| pRS306-SPO20pr-Δ3-51SPO20(3A) | URA3, Integration | SPO20 | Δ3-51 SPO20 with K68A,R71A,H75A mutations | This study |
| pRS306-SPO20pr-Δ3-51 SPO20(2E) | URA3, Integration | SPO20 | Δ3-51 SPO20 with K66E,K68E mutations | This study |
| pRS306-SPO20pr-Δ3-51SPO20(4E) | URA3, Integration | SPO20 | Δ3-51 SPO20 with K66E,K68E,R71E,K73E mutations | This study |
| pRS306-SPO20pr-Δ3-51SPO20(L78N) | URA3, Integration | SPO20 | Δ3-51 SPO20 with L78N mutation | This study |
| pRS306-SPO20pr-SPO20(L63N) | URA3, Integration | SPO20 | SPO20 with L63N mutation | This study |
| pRS306-SPO20pr-SPO20(L67P) | URA3, Integration | SPO20 | SPO20 with L67P mutation | This study |
| pRS306-SPO20pr-SPO20(I74N) | URA3, Integration | SPO20 | SPO20 with I74N mutation | This study |
| pRS306-SPO20pr-SPO20(2E) | URA3, Integration | SPO20 | SPO20 with K66E,K68E mutations | This study |
| pRS306-SPO20pr-SPO20(4E) | URA3, Integration | SPO20 | SPO20 with K66E,K68E,R71E,K73E mutations | This study |
| pRS316-SEC9pr-Δ3-51PSPS | URA3, CEN | SEC9 | Δ3-51 PSPS chimera | This study |
| pRS316-SEC9pr-Δ3-51PSPS(L67P) | URA3, CEN | SEC9 | Δ3-51 PSPS chimera with L67P mutation | This study |
| pRS426-GFP | URA3, 2μ | TEF2 | GFP | This study |
| pRS426-SPO20pr-SPO20 | URA3, 2μ | SPO20 | SPO20 | This study |
| pRS426-SPO20pr-Δ3-51SPO20 | URA3, 2μ | SPO20 | Δ3-51 SPO20 | This study |
| pRS426-SPO20pr-Δ3-95SPO20 | URA3, 2μ | SPO20 | Δ3-95 SPO20 | This study |
| pRS426-SPO20pr-SPO20(L67P) | URA3, 2μ | SPO20 | SPO20 with L67P mutation | This study |
| pRS426-SPO20pr-SPO20(4E) | URA3, 2μ | SPO20 | SPO20 with K66E,K68E,R71E,K73E mutations | This study |
| pRS426-G1-50 | URA3, 2μ | TEF2 | GFP-SPO201-50 | This study |
| pRS426-G1-91 | URA3, 2μ | TEF2 | GFP-SPO201-91 | This study |
| pRS426-G20 | URA3, 2μ | TEF2 | GFP-SPO2051-91 | This study |
| pRS426-G51-80 | URA3, 2μ | TEF2 | GFP-SPO2051-80 | This study |
| pRS426-G61-91 | URA3, 2μ | TEF2 | GFP-SPO2061-91 | This study |
| pRS426-G61-78 | URA3, 2μ | TEF2 | GFP-SPO2061-78 | This study |
| pRS426-G61-80 | URA3, 2μ | TEF2 | GFP-SPO2061-80 | This study |
| pRS426-G68-91 | URA3, 2μ | TEF2 | GFP-SPO2068-91 | This study |
| pRS426-G20(L67P) | URA3, 2μ | TEF2 | GFP-SPO2051-91 with L67P mutation | This study |
| pRS426-G20(4E) | URA3, 2μ | TEF2 | GFP-SPO2051-91 with K66E,K68E,R71E,K73E mutations | This study |
| pRS426SPO20pr-G1-91 | URA3, 2μ | SPO20 | GFP-SPO201-91 | This study |
| pRS426GFP-2XPH(PLCδ) | URA3, 2μ | 2XGFP-PLC-δ1 PH domain | Stefan et al. 2002 | |
| YEp352GAP-SPO20 | URA3, 2μ | TDH3 | SPO20 | This study |
| YEp352GAP-Δ3-51SPO20 | URA3, 2μ | TDH3 | Δ3-51 SPO20 | This study |
| pRS424GAL1pr | TRP1, 2μ | GAL1 | This study | |
| pGFP-N-FUS | URA3, CEN | MET25 | Used to make GFP fusion gene | Niedenthal et al., 1996 |
| pGFP-C-FUS | URA3, CEN | MET25 | Used to make pRS426-GFP | Niedenthal et al., 1996 |
| pRS426TEF | URA3, 2μ | TEF2 | Yest expression plasmid with TEF2 promoter | Mumberg et al., 1995 |
| pFA6a-kanMX6-PGAL1 | Used to amplify the GAL1 promoter | Longtine et al., 1998 | ||
| pGEX-20R | GST-SPO2061-91 | This study | ||
| pGEX-20RL67P | GST-SPO2061-91 with L67P mutation | This study | ||
| pGEX-20R4E | GST-SPO2061-91 with K66E,K68E,R71E,K73E mutations | This study | ||
| pGEX-PLCδPH | GST-rat PLC-δ1 domain (aa 11-142) | This study | ||
| pGEX5X-1 | GST | Amersham Biosciences |
Table 3.
Primers used in this study
| Name | Sequence |
|---|---|
| ANO95 | 5′-GTGCTGAATTCTATATAATGGGGTTCAG-3′ |
| ANO111 | 5′-GGCTAGAATTCATATATCTAAAAATGGC-3′ |
| ANO195 | 5′-CCTTGAGATCTAAGTCTAGGCGCTTTCAAC-3′ |
| ANO205 | 5′-CATGTGGGGTGCGACTCAG-3′ |
| ANO208 | 5′-TCAGGCTGGACATTCTC-3′ |
| ANO227 | 5′-CTTGTTGAATTCATGGACAATTGTTCAGGAAGC-3′ |
| ANO230 | 5′-CATGTGAAGCTTGCATCCTTGGCGAATAAAATCCAC-3′ |
| ANO231 | 5′-GTGGATTTTATTCGCCAAGGATGCAAGCTTCACATG-3′ |
| ANO234 | 5′-GCAGAAGACGTGATAGGAACCATGTGAAGCTTAAATCC-3′ |
| ANO235 | 5′-GGATTTAAGCTTCACATGGTTCCTATCACGTCTTCTGC-3′ |
| ANO236 | 5′-GATAGGCTACATGTGAAGAACAAATCCTTGAGGAATAAAATCC-3′ |
| ANO237 | 5′-GGATTTTATTCCTCAAGGATTTGTTCTTCACATGTAGCCTATC-3′ |
| ANO238 | 5′-GGCTACATGTGAAGCTTAAATCCAACAGGAATAAAATCCACAAAC-3′ |
| ANO239 | 5′-GTTTGTGGATTTTATTCCTGTTGGATTTAAGCTTCACATGTAGCC-3′ |
| ANO240 | 5′-CCTTGAGGAATAAAAACCACAAACAACTTCACCC-3′ |
| ANO241 | 5′-GGGTGAAGTTGTTTGTGGTTTTTATTCCTCAAGG-3′ |
| BBO5 | 5′-CTTGTTTCTAGAATGGACAATTGTTCAGGAAGC-3′ |
| BBO6 | 5′-GTGCTTCTAGATATATAATGGGGTTCAG-3′ |
| DGK-F | 5′-GTTCTTACTAGTATGGCCAATAATACCACTGG-3′ |
| HNO141 | 5′-GTTCTTTCTAGAGATAGGCTACATGTGAAGCTT-3′ |
| HNO142 | 5′-GTTCTTCTCGAGTTATTGTTTGTGGATTTTATTCCT-3′ |
| HNO143 | 5′-GTTCTTGAATTCGATAGGCTACATGTGAAGCTT-3′ |
| HNO144 | 5′-GTTCTTGAATTCGATAGGCTACATGTGGAGCTT-3′ |
| HNO201 | 5′-GTTCTTCTCGAGCTACCTGAAGGGTGATAAATATT-3′ |
| HNO222 | 5′-GTTCTTGCGGCCGCAAAATCCTTGAGGAATAAAATC-3′ |
| HNO223 | 5′-GTTCTTGCGGCCGCTTTGTATAGTTCATCCATGCC-3′ |
| HNO271 | 5′-GTTCTTAGATCTGGCTCCGGCTCCGGCTCCGGCTCCGGCTCCGGCTCCTCCAAAATGCGACCATAACA-3′ |
| HNO273 | 5′-GTTCTTCTCGAGCTAGTTAACAGCAGCGTAAT-3′ |
| NHO274 | 5′-GTTCTTCAATTCGGATCCATGTCGCGATACCC-3′ |
| HNO321 | 5′-GTTCTTGAGCTCTGTAAAGAGCCCCATTATCT-3′ |
| HNO323 | 5′-GTTCTTACTAGTTTTGAGATCCGGGTTTTTTC-3′ |
| HNO333 | 5′-GTTCTTCTCGAGTTAACTAGTCTTAGTGGCGTCATCGAACCGACAGTTTGGGTGATTTTGTTTGTGGATTTTATTCC-3′ |
| HNO351 | 5′-TTGAGGAATAAAATCGCCAAACAACTTCACCC-3′ |
| HNO352 | 5′-GGGTGAAGTTGTTTGGCGATTTTATTCCTCAA-3′ |
| HNO361 | 5′-GTTCTTCTCGAGTCACCATCTTTTCCCGATCA-3′ |
| HNO371 | 5′-GATAGGCTACATGTGGAGCTTGAATCCTTGAGGAATA-3′ |
| HNO372 | 5′-TATTCCTCAAGGATTCAAGCTCCACATGTAGCCTATC-3′ |
| HNO381 | 5′-GAGCTTGAATCCTTGGAGAATGAAATCCACAAACAAC-3′ |
| HNO382 | 5′-GTTGTTTGTGGATTTCATTCTCCAAGGATTCAAGCTC-3′ |
| HNO391 | 5′-CCCCCGGGCTGCAGGAATTC-3′ |
| HNO392 | 5′-GTTCTTCTCGAGTTATGGGTGAAGTTGTTTGTGGA-3′ |
| HNO452 | 5′-GTTCTTCTCGAGTTACTGCAGCTTCTGCCGCTGGTCCATG-3′ |
| HNO453 | 5′-GTTCTTGAATTC CACGGGCTCCAGGATGACCCGGACCTTCAGG CCCTTCTGAAGGGCAGCCAGCTTCTGAAGGTGAAGTC-3′ |
| M13 | 5′-ACTGGCCGTCGTTTTAC-3′ |
| PDO2 | 5′-CTTGTTCTCGAGTTAACTAGTCTTAGTGGCGTC-3′ |
pRS426-G20, used to express GFP-Spo20p51-91 under the TEF2 promoter, was constructed in two steps. First, the coding sequence of Spo20p amino acids 51-91 was amplified by polymerase chain reaction (PCR) by using BBO5 and PDO2 as primers, and pRS306-SPO20pr-SPO20 as a template. The product was digested by XbaI and XhoI, and cloned into the XbaI and XhoI sites of pGFP-N-FUS (Niedenthal et al., 1996). The resulting plasmid was digested with EcoRI and XhoI, and the GFP fusion gene was cloned into the EcoRI and XhoI sites of pRS426TEF (Mumberg et al., 1995). pRS426-G20(L67P), pRS426-G1-50, pRS426-G1-91, pRS426-G61-91 and pRS426-G61-78, used to express GFP-Spo20p51-91 with the L67P mutation, GFP-Spo20p1-50, GFP-Spo20p1-91, GFP-Spo20p61-91, and GFP-Spo20p61-78, respectively, were constructed in the same way. BBO5 and PDO2, BBO6 and HNO201, BBO6 and PDO2, HNO141 and PDO2, and HNO141 and HNO142 were used to amplify the SPO20 gene fragments, respectively. For pRS426-G20(L67P), pRS306-SPO20pr-Δ3-51SPO20(L67P) was used as a template. pRS426-G51-80 and pRS426-G61-80, used to express GFP-Spo20p51-80 and GFP-Spo20p61-80, respectively, were constructed as follows. HNO391 and HNO392 were used to amplify green fluorescent protein (GFP) fused to Spo20p amino acids 51-80 and amino acids 61-80. pRS426-G20 and pRS426-G61-91 were used as templates, respectively. The PCR fragments were digested by EcoRI and XhoI, and cloned into the EcoRI and XhoI sites of pRS426TEF. pRS426-G68-91 was used to express GFP-Spo20p68-91. To make the plasmid, PCR was performed using HNO222 and HNO223 as primers, and pRS426-G61-91 as a template. The resulting fragment was digested by NotI and self-ligated. pRS426-GFP was constructed by cloning the EcoRI-XhoI fragment with a GFP gene from pGFP-C-FUS (Niedenthal et al., 1996) into similarly digested pRS426TEF.
pGEX5X-1(Amersham Biosciences, Piscataway, NJ) was used to express glutathione S-transferase (GST) fusion proteins in Escherichia coli cells. PCR fragments encoding Spo20p amino acids 61-91 and the fragments with L67P or K66E,K68E,R71E,K73E mutations were cloned into the EcoRI and XhoI sites of pGEX5X-1 to make pGEX-20R, pGEX-20RL67P, and pGEX-20R4E, respectively. To amplify the wt or L67P fragments, PDO2 and HNO143 were used as primers, and pRS306-SPO20pr-SPO20 and pRS306-SPO20pr-Δ3-51SPO20(L67P) (See Mutagenesis) were used as templates, respectively. The fragment carrying the K66E,K68E,R71E,K73E mutations was amplified by PDO2 and HNO144 as primers and pRS426-G20(4E) as a template.
pRS424GAL1pr-DGK, used to express E. coli diacylglycerol (DAG) kinase under the GAL1 promoter in yeast cells, was made as follows. First, the GAL1 promoter was amplified by PCR by using HNO321 and HNO323 as primers, and pFA6a-kanMX6-PGAL1 (Longtine et al., 1998) as a template and cloned between the SacI and SpeI sites of pRS424 (Christianson et al., 1992) to create pRS424GAL1pr. The E. coli DAG kinase gene was then amplified by PCR by using DGK-F and HNO271 as primers and pCTY85 (Kearns et al., 1997) as a template, and digested by SpeI and BglII. Three copies of the influenza hemagglutinin (HA) epitope were amplified by PCR by using HNO273 and NHO274 as primers and HAp316 (a gift of T. Odani and Y. Jigami, AIST, Tsukuba, Japan) as a template, and digested with EcoRI and XhoI. The two PCR fragments were cloned into the SpeI and BamHI sites, and the EcoRI and XhoI sites of pRS426TEF, respectively, to create pRS426TEF-DGK. Then, a SpeI and XhoI fragment containing HA-tagged DAG kinase gene was cloned between the SpeI and XhoI sites of pRS424GAL1pr.
YEp352GAP-SPO20 and YEp352GAP-Δ3-51SPO20 were constructed as follows. SPO20 and SPO20Δ3-51 genes were amplified by PCR by using ANO95 and HNO361, and ANO227 and HNO361, respectively. These PCR fragments were digested by EcoRI and XhoI, and cloned into the EcoRI and XhoI sites of YEp352GAP (Gao et al., 1999).
pRS426SPO20pr-G1-91 was constructed as follows. The SPO20 promoter was amplified by PCR by using ANO195 and ANO111 as primers, and pRS306-SPO20pr-SPO20 as a template. The PCR fragment was digested by EcoRI and BglII, and cloned into EcoRI and BamHI sites of pRS426 to create pRS426SPO20pr. A GFP fusion to 1-91SPO20 from pRS426-G1-91 was then cloned into EcoRI and XhoI sites of pRS426SPO20pr. All sequences of genes cloned by PCR were confirmed by sequencing.
pRS426GFP-2 × PH(PLCδ) was provided by S. Emr (University of California, San Diego, CA). To construct PGEX-PLCδPH, a fragment of the rat PLCδ-1 PH domain (aa 11-142) was amplified using HNO452 and HNO453 as primers and pRS426GFP-2XPH(PLCδ) (Stefan et al., 2002) as a template.
Mutagenesis
Random mutagenesis was performed by error-prone PCR mutagenesis (Muhlrad et al., 1992). Briefly, fragments containing 200 bp of the SEC9 promoter region and nucleotides 154-497 of SPO20 sequence were generated by mutagenic PCR and cotransformed into AN211 with plasmid pRS316-SEC9pr-Δ3-51PSPS that had first been linearized by digestion with XhoI and SpeI. Digestion with these two enzymes removes the region encoding amino acids 51-95 of the Spo20p amino terminus. Site-directed mutagenesis was performed with QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All sequences of genes cloned by PCR were confirmed by sequencing.
Sporulation Assays
Sporulation assays were performed essentially as described previously (Neiman, 1998). After overnight incubation, the percentage of sporulation was determined by observation in the light microscope. For ether test, strains were sporulated at 30°C and 5 × 103 cells were spotted onto YPD plate. The plate was inverted over a paper filter soaked with 2 ml of ethyl ether for 15 min and then removed and incubated at 30°C for 2 d.
Expression and Purification of GST Fusion Proteins
For each GST fusion protein, an expression plasmid was transformed into E. coli strain BL21, 1l of culture was grown to OD600 = 0.8 in LB + ampicillin medium, and expression of the fusion protein was induced by adding 1 mM isopropyl β-d-thiogalactoside. Cells were then frozen at -80°C, resuspended in phosphate-buffered saline (130 mM NaCl, 7 mM Na2HPO4, and 3 mM NaH2PO4) containing 200 μg/ml lysozyme, left on ice for 30 min, and sonicated for 3 × 10 s to lyse the cells. The GST fusion proteins were purified on glutathione Sepharose 4B (Amersham Biosciences), eluted with 50 mM Tris (pH 8.0) containing 0.25 M KCl and 5 mM reduced glutathione, and then dialyzed against 20 mM HEPES (pH. 7.5) containing 100 mM NaCl. Samples were mixed with an equal volume of 100% glycerol and stored at -20°C.
Liposome Binding Assay
Phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL). Sucrose-laden liposomes were generated essentially as described previously (Sciorra et al., 1999). The appropriate phospholipid mixtures (80 nmol/reaction) were air-dried, resuspended in 200 μl of 20 mM HEPES (pH 7.5) with 20 mM KCl and 0.2 M sucrose, and sonicated for 4 × 15 s. The solution with sucrose-laden vesicles was mixed with 700 μl of 20 mM HEPES (pH 7.5) containing 100 mM NaCl, and centrifuged at 200,000 × g for 15 min at 4°C. Vesicles were resuspended in 200 μl of 20 mM HEPES (pH 7.5) containing 100 mM NaCl and 10 μg of bovine serum albumin, and incubated with 2 μg of purified protein for 30 min on ice. Vesicles were pelleted as before, and coprecipitated protein was analyzed with SDS-PAGE and Coomassie Blue staining. Protein in supernatant was precipitated with trichloroacetic acid and analyzed, as well. A white/UV transilluminator (UVP, Upland, CA) was used to take images of gels and the intensity of the stained bands was quantitatively determined by using LabWork 4.0 software (UVP). For the comparison of binding of GST-PLC-δ1-PH and GST-Spo20p61-91 to liposomes 20 mM NaPO4 buffer (pH 7.5) containing 100 mM NaCl was used instead of HEPES buffer.
Microscopy
Indirect immunofluorescence was performed essentially as described previously (Tachikawa et al., 2001). The anti-GFP monoclonal antibody (BD Biosciences, Franklin Lakes, NJ) was used at a 1:100 dilution and anti-Spo20p antibodies (Neiman et al., 2000) were used at a 1:50 dilution. For direct detection of GFP fluorescence, either living cells or cells fixed with 3.7% formaldehyde for 10 min and then placed in mounting media containing 4,6-diamidino-2-phenylindole (DAPI) were used. Images were acquired using an Axioplan2 microscope (Carl Zeiss, Thornwood, NY) with a Zeiss mRM Axiocam and deconvolved using Zeiss Axiovision 3.1 software.
RESULTS
Isolation of a Point Mutation That Specifically Affects SNARE Activity during Sporulation
To elucidate the role of the Spo20p amino terminus in regulating SNARE function, we took advantage of a Spo20p/Sec9p chimera, Δ3-51 PSPS, that is capable of rescuing both the growth defect of a sec9-4 temperature-sensitive (ts) mutant and the sporulation defect of a spo20Δ mutant (Neiman et al., 2000). In this chimera, the helical regions of Spo20p have been replaced with those of Sec9p, and, in addition, the inhibitory domain of the Spo20p amino terminus has been removed. The chimeric protein can mediate vesicle fusion at the prospore membrane due to sequences in the 50-95 region of the Spo20p amino terminus, but this region is dispensable for the chimera to promote fusion at the plasma membrane (Neiman et al., 2000).
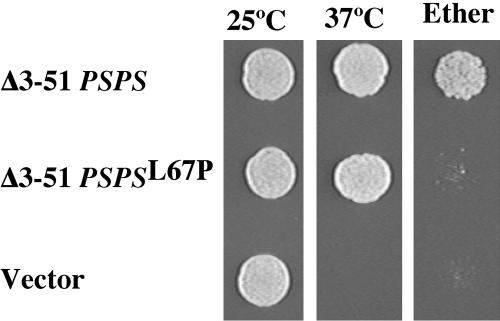
The Δ3-51 PSPS chimera was placed into a centromeric plasmid under control of the SEC9 promoter, and PCR mutagenesis was used to introduce mutations into the amino terminus between residues 50 and 95. The mutagenized pool was introduced into a diploid strain (AN211) that was homozygous for both the sec9-4 mutation and spo20Δ. Transformants were initially screened for growth at 37°C to identify plasmids expressing chimeras still capable of rescuing sec9-4. Transformants that grew at 37°C were then replica plated to SPO medium and then screened by exposure to ether vapor, which is lethal to vegetative cells but not spores, for sporulation. From 7435 transformants screened, three plasmids were identified that reproducibly rescued ts growth but not the sporulation defect of AN211 (Figure 1). The chimeric genes in each of the three plasmids were sequenced and all three were found to contain an identical T-to-C transition at nucleotide 200. This mutation changes a leucine at position 67 (in the native Spo20p sequence) to proline. Introduction of this same mutation into Δ3-51 SPO20 also greatly decreased rescue of the spo20Δ sporulation defect (Table 4). Thus, the L67P mutation perturbs the positive regulatory activity of the Spo20p amino terminus
Figure 1.
L67P mutation causes a specific sporulation defect in a Spo20p/Sec9p chimera. Centromeric plasmids carrying Δ3-51 PSPS, Δ3-51 PSPS gene with the L67P mutation, or no insert were transformed into AN211 (sec9-4/sec9-4 spo20Δ/spo20Δ). Cultures were grown to saturation at 25°C, and 5 × 103 cells of each culture were spotted onto a YPD plate and grown at or 37°C. As an assay of sporulation, 5 × 103 sporulated cells of each strain were spotted onto a YPD plate. The plate was exposed to ether vapor and then incubated at 25°C.
Table 4.
Effect of mutations in the amino-terminal helix on SPO20 function
| %Ascia
|
||
|---|---|---|
| Mutation | Δ3-51SPO20 | SPO20 |
| wt | 86.9 | 63.7 |
| L63N | 66 | 30.5 |
| L67P | 0.1 | 5.8 |
| L67N | <0.1 | n.d |
| L70N | 1 | n.d |
| 174N | 69 | 24.6 |
| L78N | 87 | n.d |
| K68A, R71A | 74 | n.d |
| K68A, R71A, H75A | 81 | n.d |
| K66E, K68E | 6 | 57.9 |
| K66E, K68E, R71E, K73E | <0.1 | 0.6 |
n.d., not determined.
Integrating plasmids carrying the indicated mutations in the context of Δ3-51 SPO20 or SPO20 were transformed into AN147 (spo20Δ/spo20Δ). Cultures were sporulated and then analyzed in the light microscope to determine the percentage sporulation; 1000 cells were counted per culture.
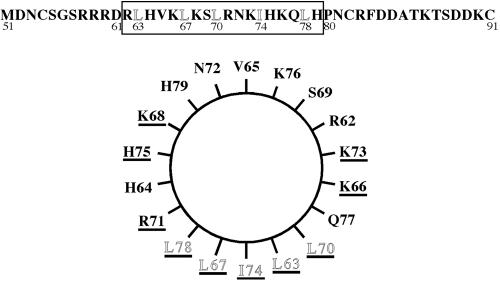
The Positive Regulatory Domain Is Predicted to Encode an Amphipathic Helix
The identification of leucine 67 as a critical residue for Spo20p function led us to examine more closely the primary sequence of this region of the protein. Although no known motifs are present, several different secondary structure prediction algorithms (available at http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_seccons.html) predict that residues 62-79 form a short amphipathic helix with hydrophobic and positively charged faces (Figure 2). Intriguingly, the L67P mutation would break this helix, suggesting a basis for the mutant phenotype. To test the importance of this putative helix, site-directed mutations in this region were introduced into Δ3-51 SPO20 (Table 4). Each mutant was cloned into an integrating plasmid, transformed into a spo20Δ strain, and the ability of the mutants to rescue sporulation was examined.
Figure 2.
Positive regulatory domain of Spo20p is predicted to contain an amphipathic α-helix. Top, primary sequence of the positive regulatory region is shown. The boxed region represents the predicted α-helix, the residues composing the hydrophobic face are outlined. Bottom, helical wheel representation of residues 62-79 (boxed amino acids in the top) of Spo20p. Amino acids changed by site-directed mutagenesis (Tables 4 and 5) are underlined.
Each of the residues of the hydrophobic face were changed, individually to asparagine residues. Though mutation of L63, I74, or L78 had, at most, modest effects on sporulation, mutation of L67 or L70 produced a dramatic reduction in complementation. Thus, the predicted hydrophobic surface, and in particular the center of that surface, is important for function.
The positively charged face was somewhat more refractory to mutation because changing two or three of the positive residues to alanine displayed no phenotype (Table 4). However, when these residues were changed to the negatively charged glutamic acid, the function of the Spo20 protein was affected. Indeed, introduction of four glutamates at positions 66, 68, 71, and 73 reduced function to the level seen in the original L67P allele. Therefore, the predicted positively charged surface is also important for function.
Several of the site-directed mutations were also introduced into the full length SPO20 gene. Some differences were observed from assays of Δ3-50 SPO20, in particular the phenotypes of L63N and I74N become more pronounced, whereas the phenotypes of L67P and K66E K68E became less so (Table 4). However, overall the same general pattern of effects was observed.
Because many of these mutations reduce but do not completely eliminate SPO20 activity when expressed from integrating plasmids, the ability of the SPO20L67P and SPO20K66E, K68E, R71E, K73E genes to rescue spo20Δ when overexpressed was examined. When present on multicopy plasmids both mutant alleles rescued the sporulation defect nearly as well as wild type (Table 5). By contrast Δ3-95 SPO20, which lacks the entire positive regulatory region, complemented very poorly when overexpressed. These results indicate that the point mutations reduce but do not completely inactivate the positive function of the Spo20p amino terminus.
Table 5.
Rescue of spo20Δ by overexpression of Spo20p mutants
| Gene Expressed | %Ascia |
|---|---|
| SPO20 | 48 |
| Δ3-51SPO20 | 51 |
| Δ3-95SPO20 | 3 |
| SPO20L67P | 53 |
| SPO20K66E, K68E, R71E, K73E | 37 |
High copy plasmids carrying the indicated SPO20 allele were transformed into AN147 (spo20Δ/spo20Δ). Cultures were sporulated and then analyzed in the light microscope to determine the percentage sporulation; 1000 cells were counted per culture.
A Short Peptide of the Spo20p Amino Terminus Is Sufficient to localize GFP to the Plasma Membrane in Vegetative Cells and the Prospore Membrane in Sporulating Cells
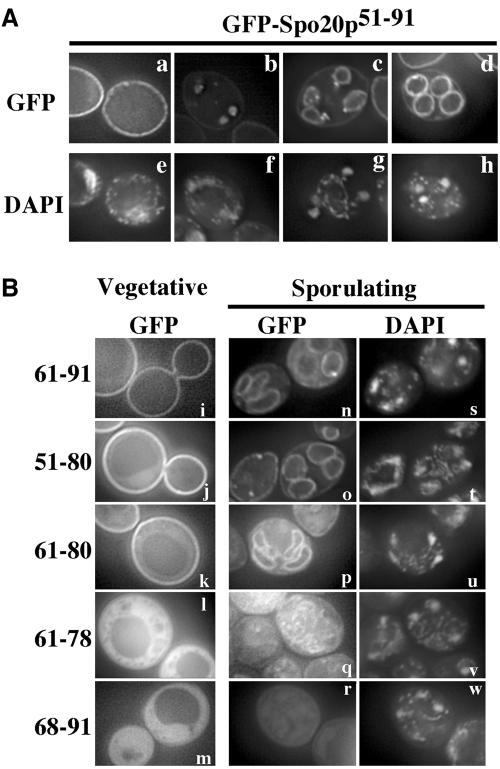
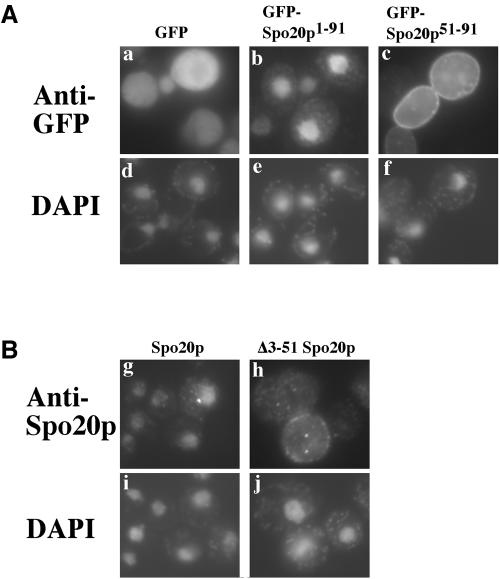
In Vam7p, the amino-terminal domain of the protein serves to localize the SNARE to its proper site of action (Cheever et al., 2001). To address whether the activation domain of Spo20p plays a similar role, a fusion of GFP to amino acid residues 51-91 of Spo20p (GFP-Spo20p51-91) was expressed in a wild-type strain, AN120, and its localization was observed. In vegetatively growing cells, GFP-Spo20p51-91 localized predominantly to the plasma membrane (Figure 3A, a). In sporulating cells, fluorescence accumulated intracellularly near the spindle poles in early meiosis II and subsequently resolved into a membrane pattern surrounding the nucleus (Figure 3A, b). As meiosis II progressed, the membranes extended along the nucleus and eventually captured each daughter nucleus at end of the meiosis (Figure 3A, c and d). This fluorescence pattern is characteristic of that of prospore membranes. Also, as meiosis II progressed, the intensity of fluorescence from the intracellular membranes became stronger than that of the plasma membrane. Therefore, GFP-Spo20p51-91 binds to the plasma membrane in vegetative cells and in meiotic cells it binds both the plasma membrane and the prospore membrane with some preference for the prospore membrane.
Figure 3.
Small region of the Spo20p amino terminus is sufficient to localize GFP to the plasma membrane and the prospore membrane. (A) Localization of GFP-Spo20p51-91 in vegetative and sporulating cells. A plasmid expressing GFP-Spo20p51-91 was introduced into AN120 and GFP fluorescence was observed in vegetative growing cells (a) and sporulating cells (b-d). DAPI staining of cells in a-d (e-h) (B) Deletion mapping of the membrane localization activity. Sequences encoding the indicated fragments of Spo20p were fused to GFP and expressed in AN120. GFP fluorescence was observed in vegetatively growing cells (i-m) and sporulating cells (n-r). (a-d and i-r) GFP fluorescence. (e-h and s-w), DAPI staining of corresponding cells in a-d and n-r, respectively.
To further define the region required to confer membrane localization, a series of deletions within GFP-Spo20p51-91 were constructed (Figure 3B). GFP-Spo20p61-91, GFPSpo20p51-80, and GFP-Spo2061-80 were observed to localize to the plasma membrane in vegetatively growing cells and the prospore membrane in sporulating cells (Figure 3B, i-k and n-p). By contrast, GFP-Spo20p61-78 and GFP-Spo20p68-91 were dispersed in the cytosol in both vegetative and sporulating cells (Figure 3B, l, m, q, and r). In addition, a fraction of the cells expressing GFP-Spo20p61-78 displayed an apparent mitochondrial localization of the fusion (our unpublished observations). Although this result might indicate that GFP-Spo20p61-78 has some affinity for mitochondrial membranes, this fusion was not observed at the plasma membrane or the prospore membranes, indicating that it lost the ability to bind to these compartments. In sum, these deletion studies indicate that amino acid residues 61-80 of the Spo20p amino terminus, the same region that is predicted to form an amphipathic α-helix, are necessary and sufficient to bind to both the plasma membrane and the prospore membrane.
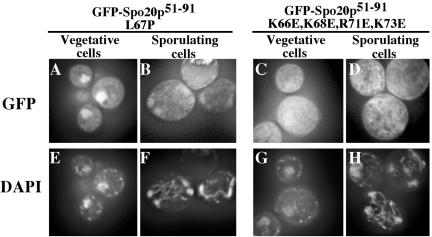
These data suggest that the positive regulatory region of the Spo20p amino terminus promotes SNARE function by localizing the protein to the membrane. If true, then the L67P and K66E, K68E, R71E, K73E mutations, which strongly perturb Spo20p function, should also perturb the localization of GFP-Spo20p51-91. To examine this possibility, the L67P and K66E, K68E, R71E, K73E mutations were introduced into GFP-Spo20p51-91. GFP-Spo20p51-91 with the L67P mutation did not show plasma membrane or prospore membrane localization but rather was concentrated in the nucleus in both vegetative and sporulating cells (Figure 4, A and B). GFP-Spo20p51-91 carrying the K66E, K68E, R71E, K73E mutations also failed to localize to either the prospore or plasma membrane, although in this case the GFP fusion was dispersed throughout the cytosol in both vegetative and sporulating cells rather than in the nucleus (Figure 4, C and D). Thus, as expected, both mutants delocalize the protein from the membrane, although perhaps by different mechanisms. The K66E, K68E, R71E, K73E mutations cause complete delocalization of the protein, whereas the L67P mutation may reveal a cryptic nuclear localization signal.
Figure 4.
L67P and K66E, K68E, R71E, K73E mutations perturb the membrane localization of GFP-Spo20p51-91. GFP-Spo20p51-91 with L67P or the K66E, K68E, R71E, K73E mutations was expressed in AN120 and GFP fluorescence was observed in vegetatively growing cells (A and C) and sporulating cells (B and D). (A-D), GFP fluorescence; (E-H), DAPI staining of corresponding cells in A-D.
The Positive Regulatory Region Is Necessary to Localize Spo20p to the Prospore Membrane during Sporulation
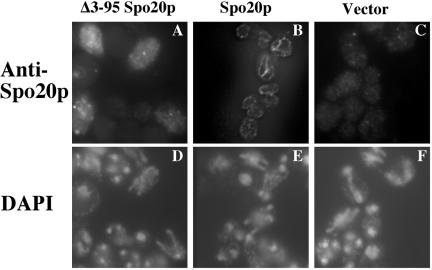
If the role of the positive regulatory region is to localize Spo20p properly, then Spo20p carrying the L67P or K66E, K68E, R71E, K73E mutations should be mislocalized during sporulation. Unfortunately, the direct test of this prediction is problematic as intact Spo20p can only be visualized when overexpressed (Neiman et al., 2000), but overexpression of the mutant SPO20 alleles suppresses their phenotype (Table 5). However, cells overexpressing of Δ3-95 SPO20, which lacks the entire positive regulatory region, still sporulate poorly (Table 5). Therefore, we examined the localization of Δ3-95 Spo20p when overexpressed in sporulating cells. High copy plasmids expressing either full-length Spo20p or Δ3-95 Spo20p were introduced into a wild-type strain. Chromosomally expressed SPO20 is present in this strain to ensure that the cells expressing Δ3-95 Spo20p sporulate well, but, as reported previously, the protein expressed from the chromosomal locus is not seen by immunofluorescence (Figure 5C). Unlike the full-length Spo20p, which was localized to the prospore membrane in meiosis II cells (Figure 5B), Δ3-95 Spo20p was found distributed in a punctate pattern in the cytosol throughout meiosis (Figure 5A). Thus, the positive regulatory region is required for proper targeting of Spo20p to the prospore membrane.
Figure 5.
Positive regulatory region is required for localization of Spo20p to the prospore membrane. Strain AN120 (wt) was transformed with high copy plasmids expressing Δ3-95 Spo20p, Spo20p, or containing no insert and the localization of the Spo20 protein was examined by indirect immunofluorescence. (A-C) Staining with anti-Spo20p antibodies. (D-F) Corresponding DAPI staining for the cells in A-C.
The Positive Regulatory Region Binds to Acidic Phospholipids In Vitro
The positive regulatory region of Spo20p is sufficient to provide a specific localization in vivo. Presumably, this region binds to a component of the plasma or prospore membrane to confer localization. Two-hybrid screens with the 51-91 region of Spo20p failed to identify good candidates for a protein ligand (our unpublished observations). An alternative possibility is that, analogously to Vam7p, this region confers localization by binding to a lipid that is uniquely enriched in the plasma membrane and prospore membrane.
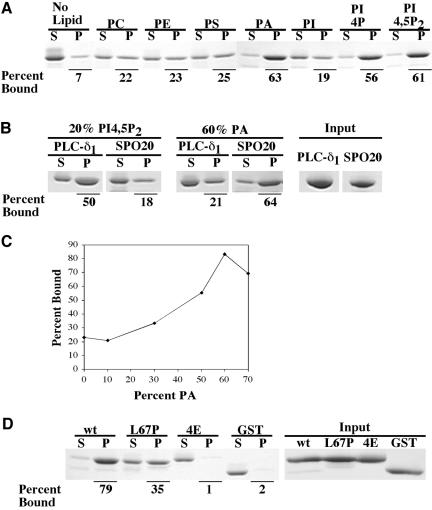
To examine this possibility, amino acid residues 61-91 of Spo20p were fused to the carboxy terminus of GST, the resulting fusion protein was purified from E. coli, and the ability of this fusion to bind to liposomes of varying composition was tested (Figure 6A). The GST fusion bound weakly to liposomes containing phosphatidylcholine (PC), phosphatidlylserine (PS), phosphatidylethanolamine (PE), or phosphatidylinositol (PI). However, when phosphatidic acid (PA), phosphatidylinositol-4-phosphate (PI4P), or phosphatidylinositol-4,5-bisphosphate (PI4,5P2) were included, >50% of the fusion protein cosedimented with the liposomes. GST alone did not precipitate to any extent (Figure 6D). Thus, amino acids 61-91 of Spo20p can confer binding to acidic phospholipids in vitro.
Figure 6.
GST-Spo20p61-91 binds to acidic liposome in vitro. (A) Binding of GSTSpo20p61-91 to phospholipids. Purified GSTSpo20p61-91 (2 μg) was mixed with sucrose laden liposomes containing 50% (40 nmol) of PC and 50% (40 nmol) of PC, PE, PS, PA, PI, PI4P, or PI4,5P2. After centrifugation, the protein precipitated with the liposomes (P) or remaining in the supernatant (S) was analyzed by SDS-PAGE and Coomassie staining. Percentage of the input protein that bound to the liposomes is given. (B) Comparison of binding of GST-PLC-δ1-PH and GSTSpo20p61-91 to PI4,5P2 or PA containing liposomes. Binding of 66 pmol of GST-PLC-δ1-PH (PLC-δ1) and GST-Spo20p61-91 (SPO20) to liposomes containing 20% PI4,5P2 or 60% PA was assayed as described in A. (C) Binding of GST-Spo20p61-91 to liposomes with increasing concentrations of PA. The curve shows the percentage of input GST-Spo20p61-91 (2 μg) in the pellet with PC-based liposomes containing 0,10, 30, 50, 60, or 70% of PA. The data shown are means of three determinations. (D) Binding of wt and mutant GST-Spo20p61-91 to PA. Binding of GST-Spo20p61-91 (wt), GST-Spo20p61-91 L67P (L67P), GST-Spo20p61-91 K66E, K68E, R71E, K73E (4E), or GST alone (GST) to liposomes containing 60% PA and 40% PC was assayed as described in A.
The apparent lack of specificity of GST-Spo2061-91 for a particular phospholipid led us to compare the binding of this fusion to a bona fide PI4,5P2 binding moiety, the PH domain from PLC-δ1 (Kavran et al., 1998). A fusion of PLC-δ1-PH to GST was purified from E. coli and comparable molar amounts of GST-Spo2061-91 and GST-PLC-δ1-PH were tested for their ability to bind either PI4,5P2 or PA containing liposomes (Figure 6B). Relative to GST-PLC-δ1-PH, GSTSpo2061-91 displayed a lower affinity for PI4,5P2 and a higher affinity for PA. The comparable binding of GST-Spo2061-91 to the different lipids is thus not an artifact of the assay system but a property of the protein in vitro.
Efficient coprecipitation in vitro required a relatively high concentration of the acidic phospholipid in the liposomes (Figure 6C). Maximal binding was seen when the liposomes were composed of 60% PA. This raises some concern about the physiological significance of the observed binding. If lipid binding is relevant to the in vivo function of this domain, then mutations that inactivate function in vivo should affect lipid binding in vitro. To test this, the L67P and K66E, K68E, R71E, K73E mutations were introduced into the GST-Spo20p61-91 fusion. These mutant fusion proteins were also purified from E. coli and their ability to bind to liposomes was assayed (Figure 6D). Under maximal binding conditions, in which 79% of the GST-Spo20p61-91 protein bound liposomes, only the 35% of the GST-Spo20p61-91 L67P fusion was precipitated. More strikingly, binding to lipids was completely lost in the GST-Spo20p61-91 K66E, K68E, R71E, K73E fusion. That these mutant forms show reduced binding to lipids suggests that their mutant phenotype may be due to an inability to bind lipids in vivo.
Localization of GFP-Spo20p51-91 Is Not Dependent on PI4,5P2
Binding to acidic phospholipids could provide an explanation for how the positive regulatory domain binds membranes in vivo. However, the lack of specificity for an individual lipid makes it unclear how specific targeting to the plasma and prospore membrane is achieved. Within the cell, each membrane compartment has a unique lipid composition and may be enriched in specific lipid types (Schneiter et al., 1999). For example, in S. cerevisiae fusion of GFP to a PI4,5P2 binding domain, GFP-2 × PH(PLCδ), will target the fusion protein to the plasma membrane, whereas fusion to a PI4P binding domain will target GFP to the Golgi compartment (Stefan et al., 2002). It may be that the positive regulatory region of Spo20p has greater specificity in vivo than in vitro, and in vivo targeting is achieved by binding to a particular lipid.
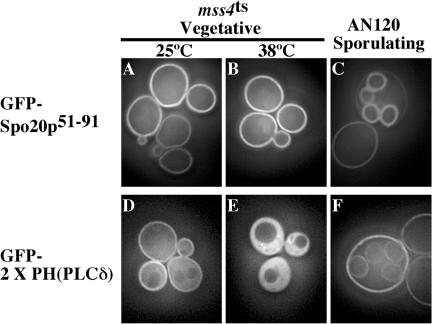
Because both GFP-Spo20p51-91 and GFP-2 × PH(PLCδ) are localized to the plasma membrane in vegetative cells and GST-Spo20p61-91 has a strong affinity for PI4,5P2 in vitro, PI4,5P2 seemed to be a good candidate for the in vivo GFP-Spo20p51-91 ligand. To test this possibility, the localization of GFP-Spo20p51-91 and GFP-2 × PH(PLCδ) were compared in strains carrying a mutation in the MSS4 gene. MSS4 encodes the sole phosphatidyl-inositol-4-phosphate-5-kinase in S. cerevisiae, and PI4,5P2 pools are rapidly depleted when cells carrying a ts allele of MSS4 are shifted to the nonpermissive temperature (Stefan et al., 2002). After 1-h incubation of the mss4-ts strain at 37°C, depletion of PI4,5P2 resulted in a redistribution of GFP-2 × PH(PLCδ) from the plasma membrane to the cytosol (Figure 7E), as reported previously (Stefan et al., 2002). By contrast, GFP-Spo20p51-91 retained a plasma membrane localization even after prolonged incubation of the strain at the restrictive temperature (Figure 7B). This results might be explained if GFP-2 × PH(PLCδ) a lower affinity for PI4,5P2 than GFP-Spo20p51-91; however, this does not seem to be the case in vitro (Figure 6B). Thus, this result suggests that PI4,5P2 is not the in vivo ligand of GFP-Spo20p51-91.
Figure 7.
Comparison of subcellular localization of GFP-Spo20p51-91 and GFP-2 × PH(PLCδ). GFP fluorescence of GFPSpo20p51-91 (A-C) and GFP-2 × PH(PLCδ) (D-F) was observed in vegetatively growing mss4ts cells (A, B, D, and E) and sporulating AN120 (C and F).
Moreover, in sporulating cells, GFP-2 × PH(PLCδ) was not observed at prospore membranes during meiosis II. A weak prospore membrane signal was eventually observed at late stages of spore formation, but in these cells a strong plasma membrane signal remained (Figure 7F). This pattern is very different from that of GFP-Spo20p51-91; in which prospore membrane localization occurs at the beginning of membrane formation, and the intensity of the prospore membrane signal is stronger than that of the plasma membrane (Figures 3A and 7C). Taken with the results in the mss4-ts strain, these results strongly indicate that the localization of GFP-Spo20p51-91 is not dependent on PI4,5P2.
Overexpression of DAG Kinase Alters GFP-Spo20p51-91 Localization
The phosphatidylinsoitol-4-kinase protein Stt4p produces a plasma membrane localized pool of PI4P, which serves as a substrate for Mss4p (Audhya and Emr, 2002). In principle, GFP-Spo20p51-91 could bind to this plasma membrane PI4P pool. To test this possibility, analogous experiments to those described for mss4-ts were performed with an stt4-ts strain. No change in the localization of GFP-Spo20p51-91 was seen even after incubations sufficient to affect the localization of GFP-2 × PH(PLCδ) (our unpublished observations). Similar results were also obtained with mutations of PIK1, which encodes a Golgi-localized phosphatidylinsoitol-4-kinase (Walch-Solimena and Novick, 1999). Thus, PI4P does not seem to be the in vivo ligand of GFP-Spo20p51-91.
Besides PI4P and PI4,5P2, the other phosphatidylinositol phosphate species in the yeast cell are found predominantly in intracellular membranes (Gillooly et al., 2000; Stefan et al., 2002) and are therefore not good candidates for in vivo ligands of the Spo20p positive regulatory domain. By contrast, the plasma membrane is enriched in PA relative to other membrane compartments of the cell (Schneiter et al., 1999). Analogous to the experiments with mss4-ts, we sought to examine the effect of altered intracellullar PA levels on the localization of GFP-Spo20p51-91. Mutation of the phospholipase D encoded by SPO14, which hydrolyzes PC to make PA, had no effect on the distribution of GFP-Spo20p51-91 (our unpublished observations); however, this mutation does not cause significant changes in the intracellular PA concentration in vegetative cells (Rudge et al., 2001). No other PA-generating enzymes have been defined in S. cerevisiae. Therefore, to alter intracellular PA levels the E. coli DAG kinase, which synthesizes PA from DAG, was used.
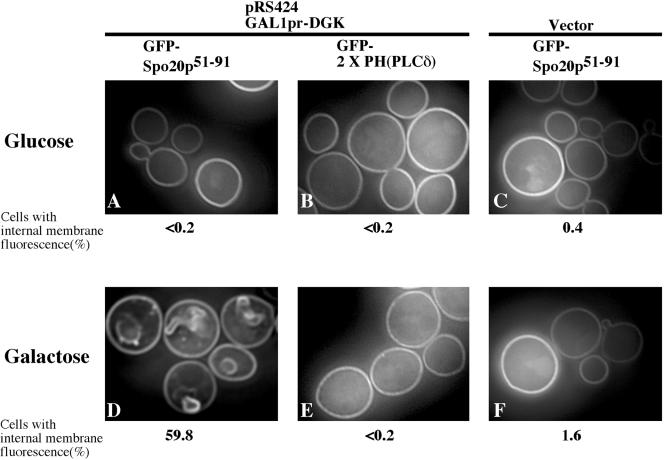
Expression of DAG kinase in yeast is reported to result in elevation of cellular PA levels (Kearns et al., 1997), although which membranes contain the PA is not known. A plasmid was constructed carrying the DAG kinase gene under the control of the GAL1 promoter, and cotransformed into cells along with either GFP-Spo20p51-91 or GFP-2 × PH(PLCδ). When the cells were grown on glucose medium, both GFP fusions were found on the plasma membrane. The cultures were then shifted to galactose medium to induce the expression of DAG kinase. The localization of GFP-2 × PH(PLCδ) was unaltered by induction of the DAG kinase (Figure 8, B and E). By contrast, after 8 h in galactose medium, ∼60% of the cells expressing GFP-Spo20p51-91 displayed fluorescence from intracellular membrane structures in addition to fluorescence from the plasma membrane (Figure 8D). Approximately 40% of these structures could be costained with the dye FM 4-64, indicating that some of them are endosomal structures. These data indicate that the localization of GFPSpo20p51-91 is responsive to PA levels in vivo.
Figure 8.
Effects of expression of DAG kinase on GFP-Spo20p51-91 and GFP-2 × PH(PLCδ) localization. GFP fluorescence of GFP-Spo2051-91 (A, C, D, and F) and GFP-2 × PH(PLCδ) (B and E) was observed in W303-1A harboring pRS424GAL1pr-DGK. DAG kinase was repressed in glucose medium (A-C) or expressed in galactose medium (D-F). Localization of GFP-Spo20p51-91 in W303-1A with empty vector (pRS424GAL1pr) in glucose medium (C) and galactose medium (F) was examined as a control. The percentage of cells with internal membrane fluorescence is given; 500 cells were counted for each culture.
The Inhibitory Region of the Spo20p Amino Terminus Sequesters the Protein in the Nucleus
As noted, the Δ3-51PSPS chimera expressed under the control of the SEC9 promoter complements both sec9-4 and spo20Δ mutants. However, the full-length PSPS chimera rescues a spo20Δ mutant poorly and a sec9-4 mutant not at all (Neiman et al., 2000). These observations define an inhibitory function in the 4-50 region of the Spo20p amino terminus. In constructing fusions of GFP to Spo20p amino-terminal fragments, we found that fusions that contained amino acid residues 1-50 of Spo20p never displayed plasma membrane fluorescence. For example, in cells expressing GFPSpo20p1-51 or GFP-Spo20p1-91 very little fluorescence was visible; only 2-6% of the cells displayed a fluorescence signal, and this was in a punctate, nuclear pattern (our unpublished observations). The reason for this loss of fluorescence is unclear, because both GFP-Spo20p1-51 and GFP-Spo20p1-91 were present in comparable abundance to GFP-Spo20p51-91 as determined by Western blotting with anti-GFP antibodies (our unpublished observations).
To determine the intracellular localization of the fusions including the 1-50 region, indirect immunofluorescence by using an anti-GFP antibody was performed. When detected by immunofluorescence, a majority of the cells displayed staining, and GFP-Spo20p1-91 localized to the nucleus in these cells (Figure 9A, b). By contrast, immunostaining of cells expressing GFP-Spo20p51-91 displayed the same plasma membrane localization seen with direct GFP fluorescence (Figure 9A, c). Thus, the addition of amino acids 1-50 relocalized the fusion protein from the plasma membrane to the nucleus.
Figure 9.
Intracellular localization of GFP-Spo20p1-51, GFPSpo20p1-91, Δ3-51 Spo20p, and full-length Spo20p in vegetative cells. (A) Localizations of GFP alone (a), GFP-Spo20p1-91 (b), and GFPSpo20p51-91 (c) were determined by indirect immunofluorescence by using anti-GFP antibodies. (d-f) DAPI staining of corresponding cells in a-c. (B) Localizations of full-length Spo20p (g) and Δ3-51 Spo20p (h) expressed from the TDH3 promoter in vegetative cells were determined by indirect immunofluorescence by using anti-Spo20p antibodies. (i and j) DAPI staining of corresponding cells in g and h.
Similarly, we observed that full-length Spo20p, when expressed from the TDH3 promoter in vegetative cells was concentrated in the nucleus, but Δ3-51 Spo20p expressed in the same way was localized either diffusely throughout the cytosol or concentrated at the plasma membrane (Figure 9B). The inhibitory region, therefore, also functions as a nuclear targeting domain in the context of the full-length protein.
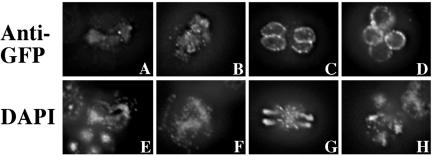
If the inhibitory activity of the Spo20p amino terminus sequesters the protein into the nucleus, then this activity must somehow be overcome during sporulation to allow Spo20p to reach the prospore membrane. To examine the behavior of the protein during sporulation, we expressed GFP-Spo20p1-91 in sporulating cells under control of native SPO20 promoter. Consistent with the sporulation-specific regulation of this promoter, the fusion protein was first detectable in cells during meiosis I and at that time the fusion was localized to the nucleus, as it was when expressed in vegetative cells (Figure 10A). As cells proceeded into meiosis II, however, localization to nascent prospore membranes became apparent (Figure 10B) and by late meiosis II nuclear localization was lost and the fusion was found exclusively on the prospore membrane where it remained in mature spores (Figure 10, C and D). Exit from the nucleus must be cued by a developmental signal and not simply the appearance of a PA-rich membrane in the cytosol because expression of DAG kinase does not cause exit of GFPSpo20p1-91 from the nucleus in vegetative cells (our unpublished observations). These results indicate that the inhibitory activity of the amino terminus sequesters Spo20p to the nucleus when it is first synthesized, but nuclear targeting is lost as cells enter meiosis II and Spo20p then localizes to the prospore membrane by virtue of its lipid binding domain.
Figure 10.
Intracellular localization of GFPSpo20p1-91 in sporulating cells. Sporulating cells expressing GFP-Spo20p1-91 under control of the SPO20 promoter were examined by indirect immunofluorescence by using anti-GFP antibodies. (A-D) Anti-GFP staining. (E-H), DAPI staining of corresponding cells in A-D.
DISCUSSION
In this study, we have investigated the basis for the positive and negative regulation of SNARE function by the Spo20p amino terminus. A genetic screen was used to identify point mutations in the Spo20p amino terminus that specifically interfere with the fusion of vesicles at the prospore membrane. The original mutation and further, targeted mutagenesis identified a probable amphipathic helix as the critical feature of the amino terminus. A region as small as 20 amino acids encompassing this helix was sufficient to target GFP to the plasma membrane in vegetative cells and the prospore membrane in sporulating cells. In vitro binding studies demonstrated that this region binds to acidic phospholipids. Mutations in the helical domain that eliminate membrane targeting of GFP also eliminate lipid binding in vitro and create a nonfunctional protein when introduced into full-length Spo20p. Thus, positive regulation of Spo20p by the amino terminus is mediated by localization of Spo20p to its site of action.
An important unresolved issue is the identity of the in vivo ligand for this domain. In the in vitro binding studies, no strong preference for PA over PI4,5P2 or PI4P was seen. In vivo GFP-Spo2051-91 localized to the plasma membrane, similar to a GFP fused to a known PI4,5P2 binding domain and distinct from the intracellular Golgi pattern of GFP fused to a PI4P binding domain (Figure 6; Stefan et al., 2002). However, during sporulation the pattern of localization of the PI4,5P2-binding fusion was distinct from that of GFPSpo2051-91. More strikingly, when PI4,5P2 was depleted using an mss4-ts mutant, localization of the PI4,5P2-binding fusion to the plasma membrane was lost, whereas GFPSpo2051-91 localization was unaffected. By contrast, overexpression of E. coli diacylglycerol kinase, which leads to an increase in intracellular PA concentration, disturbed GFPSpo2051-91 localization without greatly affecting the PI4,5P2 binding protein, suggesting that GFP-Spo2051-91 might bind PA in vivo. Analysis of the lipid composition of different membrane compartments has shown that the plasma membrane is enriched in PA relative to other intracellular compartments (Schneiter et al., 1999). Thus, binding to PA might be reasonably expected to provide specific targeting to the plasma membrane.
The phospholipase D enzyme Spo14p, which produces PA, is localized to the prospore membrane during sporulation (Rudge et al., 1998). Localization of Spo14p to the plasma membrane has been shown to enhance the function of the Spo20p SNARE domain in vegetative cells (Coluccio et al., 2004), suggesting that Spo20p might operate most efficiently in a PA-rich membrane environment. Thus, for a variety of reasons PA is an attractive candidate for the in vivo ligand. However, final demonstration of this will have to await more sensitive genetic tools for specifically manipulating intracellular PA pools.
An alternative possibility is that Spo2051-91 does not have a specific ligand, but rather binds to acidic phospholipids only when they are sufficiently concentrated. If so, then the membrane preference of Spo20p51-91 would not be defined by one specific phospholipid, but rather clustering or spatial concentration of acidic phospholipids might be the critical determinant of membrane association. Such a model would explain why GST-Spo2061-91 only bound to liposomes in vitro containing relatively high concentrations of acidic phospholipid.
Regardless of the specific lipid bound, Spo20p resembles the vacuolar SNARE Vam7p in that it contains a lipid binding motif in its amino-terminal region. In the case of Vam7p, binding of PI3P by its amino-terminal PX domain is important for localization to the vacuolar membrane and function and, as with Spo20p, overexpression of Vam7p can compensate for the mutation of the lipid binding site (Cheever et al., 2001). This similarity to Vam7p raises the possibility that the lipid binding domain of Spo20p plays a role in vesicle fusion beyond simply recruiting Spo20p to its appropriate membrane compartment. For Vam7p, biochemical reconstitution of vacuolar fusion indicates that the PX domain is required for an on/off cycle of localization necessary for the complete fusion reaction (Boeddinghaus et al., 2002). For Spo20p, the K66E, K68E, R71E, K73E mutations that abolish lipid binding and membrane localization can still suppress spo20Δ when overexpressed. By contrast, Δ3-95 Spo20p that completely lacks the activating region cannot rescue sporulation well, even when overexpressed. This result suggests that the activating region might play some role in the fusion process beyond simply localizing Spo20p to the membrane.
When expressed in vegetative cells all GFP fusions to Spo20p that included residues 1-50 of the amino terminus were found to localize to the nucleus rather than the plasma membrane. Strikingly, a fusion of residues 1-91 of Spo20p was localized to the nucleus in vegetative and early meiotic cells, but left the nucleus and localized to the prospore membrane at the onset of membrane formation in meiosis II. These results indicate that inhibition by the amino terminus is due to sequestration of the protein into the nucleus and that the protein is released from the nucleus when it is needed for prospore membrane formation.
One remaining question is why Spo20p contains an inhibitory region. Although this region serves to inhibit Spo20p in vegetative cells, the gene itself is expressed specifically in meiotic cells, so the need for inhibition in mitotic growth is not clear. During sporulation, however, induction of SPO20 expression precedes the onset of prospore membrane formation and retargeting of secretory vesicles away from the plasma membrane (Figure 10; our unpublished observations). It may, therefore, be important to keep Spo20p inactive until the appropriate moment. Although this is an attractive hypothesis, cells expressing Δ3-50 SPO20 that lacks the inhibitory domain, sporulate normally (Neiman et al., 2000). Thus, at the moment the significance of Spo20p sequestration remains unclear.
Previous studies have identified autoinhibitory regions in a number of different SNARE molecules (Nicholson et al., 1998; Dulubova et al., 2001; Tochio et al., 2001; Antonin et al., 2002). In these instances, however, the inhibitory region seems to interfere directly with the ability of the inhibited SNARE to oligomerize with partner proteins. Although there are many examples of regulated nuclear localization (Schwoebel and Moore, 2000; Schuller and Ruis, 2002), Spo20p is the first SNARE protein reported to be regulated in this manner.
At least one other secretory pathway protein has been shown to exhibit similar behavior. The Schizosaccharomyces pombe homologue of S. cerevisiae SEC14 (confusingly named spo20+) encodes a PI transfer protein that localizes to the cell periphery in vegetatively growing cells and to the forespore membrane, the S. pombe equivalent of the prospore membrane, during sporulation (Nakase et al., 2001). On transfer of cells to sporulation medium the protein disappears from the cell periphery and accumulates in the nucleus. After meiosis, the S. pombe Spo20p exits the nucleus and localizes to the forespore membrane. The significance of the nuclear localization of S. pombe Spo20p is not yet known, but these observations suggest that sequestration to the nucleus could be a common regulatory mechanism for secretory pathway functions during sporulation, and perhaps under other conditions as well.
Could control of intracellular localization be a more general strategy for the regulation of SNARE molecules? Certainly, it seems likely to be an effective strategy only for those SNAREs, such as SNAP-25 family members, that lack transmembrane domains. As noted, localization of Vam7p to the vacuolar membrane requires a PX domain (Cheever et al., 2001). Furthermore, localization of SNAP-25 to the plasma membrane requires palmitoylation and an associated, conserved motif (Schwoebel and Moore, 2000; Schuller and Ruis, 2002). Recently, the prenylated SNARE Ykt6p has been shown to localize predominantly to an unidentified membrane compartment in rat neurons (Hasegawa et al., 2003). This localization is conferred by an amino-terminal region of Ykt6p. Thus, targeting of nontransmembrane domain SNAREs to their site of action by cis-sequence motifs seems to be a common strategy. Given our results with Spo20p, whether the localization of any of these other SNARE proteins is regulated bears closer examination.
Acknowledgments
We thank Y. Jigami (AIST, Tsukuba, Japan) J. Engebrecht (University of California, Davis), and S. Emr (University of California, San Diego) for strains and plasmids. We also thank J. Engebrecht and P. Brennwald (University of North Carolina, Chapel Hill) for comments on the manuscript and V. Bankaitis (University of North Carolina, Chapel Hill) for advice on lipid experiments. This work was supported by National Institutes of Health grant GM-62184 and National Science Foundation grant MC-B0095309 to A.M.N.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-11-0798. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-11-0798.
References
- Aalto, M.K., Ronne, H., and Keranen, S. (1993). Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 12, 4095-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin, W., Dulubova, I., Arac, D., Pabst, S., Plitzner, J., Rizo, J., and Jahn, R. (2002). The N-terminal domains of syntaxin 7 and vti1b form three-helix bundles that differ in their ability to regulate SNARE complex assembly. J. Biol. Chem. 277, 36449-36456. [DOI] [PubMed] [Google Scholar]
- Audhya, A., and Emr, S.D. (2002). Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell. 2, 593-605. [DOI] [PubMed] [Google Scholar]
- Boeddinghaus, C., Merz, A.J., Laage, R., and Ungermann, C. (2002). A cycle of Vam7p release from and PtdIns 3-P-dependent rebinding to the yeast vacuole is required for homotypic vacuole fusion. J. Cell Biol. 157, 79- 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald, P., Kearns, B., Champion, K., Keranen, S., Bankaitis, V., and Novick, P. (1994). Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell 79, 245-258. [DOI] [PubMed] [Google Scholar]
- Byers, B. (1981). Cytology of the yeast life cycle. In: The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance, ed. J. N. Strathern, E. W. Jones, and J. R. Broach, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 59-96.
- Cheever, M.L., Sato, T.K., de Beer, T., Kutateladze, T.G., Emr, S.D., and Overduin, M. (2001). Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 3, 613-618. [DOI] [PubMed] [Google Scholar]
- Christianson, T.W., Sikorski, R.S., Dante, M., Shero, J.H., and Hieter, P. (1992). Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119-122. [DOI] [PubMed] [Google Scholar]
- Coluccio, A., Malzone, M., and Neiman, A.M. (2004). Genetic evidence of a role for membrane lipid composition in the regulation of SNARE function in Saccharomyces cerevisiae. Genetics (in press). [DOI] [PMC free article] [PubMed]
- Couve, A., and Gerst, J.E. (1994). Yeast Snc proteins complex with Sec9. Functional interactions between putative SNARE proteins. J. Biol. Chem. 269, 23391-23394. [PubMed] [Google Scholar]
- Dulubova, I., Yamaguchi, T., Wang, Y., Sudhof, T.C., and Rizo, J. (2001). Vam3p structure reveals conserved and divergent properties of syntaxins. Nat. Struct. Biol. 8, 258-264. [DOI] [PubMed] [Google Scholar]
- Gao, X.D., Kaigorodov, V., and Jigami, Y. (1999). YND1, a homologue of GDA1, encodes membrane-bound apyrase required for Golgi N- and O- glycosylation in Saccharomyces cerevisiae. J. Biol. Chem. 274, 21450-21456. [DOI] [PubMed] [Google Scholar]
- Gerst, J.E., Rodgers, L., Riggs, M., and Wigler, M. (1992). SNC1, a yeast homolog of the synaptic vesicle-associated membrane protein/synaptobrevin gene family: genetic interactions with the RAS and CAP genes. Proc. Natl. Acad. Sci. USA 89, 4338- 4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly, D.J., Morrow, I.C., Lindsay, M., Gould, R., Bryant, N.J., Gaullier, J.M., Parton, R.G., and Stenmark, H. (2000). Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, H., Zinsser, S., Rhee, Y., Vik-Mo, E.O., Davanger, S., and Hay, J.C. (2003). Mammalian Ykt6 is a neuronal SNARE targeted to a specialized compartment by its profilin-like amino terminal domain. Mol. Biol. Cell 14, 698-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, D.T., Slater, T.M., Wilson, M.C., and Skene, J.H. (1992). The 25 kDa synaptosomal-associated protein SNAP-25 is the major methionine-rich polypeptide in rapid axonal transport and a major substrate for palmitoylation in adult CNS. J. Neurosci. 12, 4634-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, R., Lang, T., and Sudhof, T.C. (2003). Membrane fusion. Cell 112, 519-533. [DOI] [PubMed] [Google Scholar]
- Kavran, J.M., Klein, D.E., Lee, A., Falasca, M., Isakoff, S.J., Skolnik, E.Y., and Lemmon, M.A. (1998). Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 273, 30497-30508. [DOI] [PubMed] [Google Scholar]
- Kearns, B.G., McGee, T.P., Mayinger, P., Gedvilaite, A., Phillips, S.E., Kagiwada, S., and Bankaitis, V.A. (1997). Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature 387, 101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laage, R., and Ungermann, C. (2001). The N-terminal domain of the t-SNARE Vam3p coordinates priming and docking in yeast vacuole fusion. Mol. Biol. Cell 12, 3375-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., 3rd, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Moens, P.B. (1971). Fine structure of ascospore development in the yeast Saccharomyces cerevisiae. Can. J. Microbiol. 17, 507-510. [DOI] [PubMed] [Google Scholar]
- Moens, P.B., and Rapport, E. (1971). Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen). J. Cell Biol. 50, 344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., Hunter, R., and Parker, R. (1992). A rapid method for localized mutagenesis of yeast genes. Yeast 8, 79- 82. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., Muller, R., and Funk, M. (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119-122. [DOI] [PubMed] [Google Scholar]
- Munson, M., Chen, X., Cocina, A.E., Schultz, S.M., and Hughson, F.M. (2000). Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly. Nat. Struct. Biol. 7, 894-902. [DOI] [PubMed] [Google Scholar]
- Munson, M., and Hughson, F.M. (2002). Conformational regulation of SNARE assembly and disassembly in vivo. J. Biol. Chem. 277, 9375-9381. [DOI] [PubMed] [Google Scholar]
- Nakase, Y., Nakamura, T., Hirata, A., Routt, S.M., Skinner, H.B., Bankaitis, V.A., and Shimoda, C. (2001). The Schizosaccharomyces pombe spo20(+) gene encoding a homologue of Saccharomyces cerevisiae Sec14 plays an important role in forespore membrane formation. Mol. Biol. Cell 12, 901-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman, A.M. (1998). Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140, 29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman, A.M., Katz, L., and Brennwald, P.J. (2000). Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics 155, 1643-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, K.L., Munson, M., Miller, R.B., Filip, T.J., Fairman, R., and Hugh- son, F.M. (1998). Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p. Nat. Struct. Biol. 5, 793-802. [DOI] [PubMed] [Google Scholar]
- Niedenthal, R.K., Riles, L., Johnston, M., and Hegemann, J.H. (1996). Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast 12, 773-786. [DOI] [PubMed] [Google Scholar]
- Parlati, F., Varlamov, O., Paz, K., McNew, J.A., Hurtado, D., Sollner, T.H., and Rothman, J.E. (2002). Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity. Proc. Natl. Acad. Sci. USA 99, 5424-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham, H.R. (1999). SNAREs and the secretory pathway-lessons from yeast. Exp. Cell Res. 247, 1-8. [DOI] [PubMed] [Google Scholar]
- Protopopov, V., Govindan, B., Novick, P., and Gerst, J.E. (1993). Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell 74, 855-861. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., and Fink, G.R. (1990). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Rothman, J.E. (1994). Intracellular membrane fusion. Adv. Second Messenger Phosphoprotein Res. 29, 81-96. [DOI] [PubMed] [Google Scholar]
- Rothman, J.E., and Warren, G. (1994). Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr. Biol. 4, 220-233. [DOI] [PubMed] [Google Scholar]
- Rudge, S.A., Morris, A.J., and Engebrecht, J. (1998). Relocalization of phospholipase D activity mediates membrane formation during meiosis. J. Cell Biol. 140, 81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge, S.A., Pettitt, T.R., Zhou, C., Wakelam, M.J., and Engebrecht, J.A. (2001). SPO14 separation-of-function mutations define unique roles for phospholipase D in secretion and cellular differentiation in Saccharomyces cerevisiae. Genetics 158, 1431-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T.K., Darsow, T., and Emr, S.D. (1998). Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol. Cell. Biol. 18, 5308-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter, R., et al. (1999). Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 146, 741-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller, C., and Ruis, H. (2002). Regulated nuclear transport. Results Probl. Cell Differ. 35, 169-189. [DOI] [PubMed] [Google Scholar]
- Schwoebel, E.D., and Moore, M.S. (2000). The control of gene expression by regulated nuclear transport. Essays Biochem. 36, 105-113. [DOI] [PubMed] [Google Scholar]
- Sciorra, V.A., Rudge, S.A., Prestwich, G.D., Frohman, M.A., Engebrecht, J., and Morris, A.J. (1999). Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. EMBO J. 18, 5911-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner, T., Whiteheart, S.W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P., and Rothman, J.E. (1993). SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318-324. [DOI] [PubMed] [Google Scholar]
- Stefan, C.J., Audhya, A., and Emr, S.D. (2002). The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)bisphosphate. Mol. Biol. Cell 13, 542-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, R.B., Fasshauer, D., Jahn, R., and Brunger, A.T. (1998). Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution [see comments]. Nature 395, 347-353. [DOI] [PubMed] [Google Scholar]
- Tachikawa, H., Bloecher, A., Tatchell, K., and Neiman A.M. (2001). A Giplp- Glc7p phosphatase complex regulates septin organization and spore wall formation. J. Cell. Biol. 155, 797-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, B.J., and Rothstein, R. (1989). Elevated recombination rates in transcriptionally active DNA. Cell 56, 619- 630. [DOI] [PubMed] [Google Scholar]
- Tochio, H., Tsui, M.M., Banfield, D.K., and Zhang, M. (2001). An autoinhibitory mechanism for nonsyntaxin SNARE proteins revealed by the structure of Ykt6p. Science 293, 698-702. [DOI] [PubMed] [Google Scholar]
- Ungermann, C., and Wickner, W. (1998). Vam7p, a vacuolar SNAP-25 homolog, is required for SNARE complex integrity and vacuole docking and fusion. EMBO J. 17, 3269-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena, C., and Novick, P. (1999). The yeast phosphatidylinositol- 4-OH kinase pik1 regulates secretion at the Golgi. Nat. Cell. Biol. 1, 523-525. [DOI] [PubMed] [Google Scholar]
- Weber, T., Zemelman, B.V., McNew, J.A., Westermann, B., Gmachl, M., Parlati, F., Sollner, T.H., and Rothman, J.E. (1998). SNAREpins: minimal machinery for membrane fusion. Cell 92, 759-772. [DOI] [PubMed] [Google Scholar]
- Weimbs, T., Low, S.H., Chapin, S.J., Mostov, K.E., Bucher, P., and Hofmann, K. (1997). A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc. Natl. Acad. Sci. USA 94, 3046-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]