Abstract
Early in mitosis, the mammalian Golgi apparatus disassembles, and fluorescence microscopy reveals Golgi clusters and an extensive, nonresolvable haze that either represents scattered vesicles or a merged endoplasmic reticulum (ER)-Golgi compartment. To help decide between these alternatives, we have carried out a combined microscopic and pharmacological analysis, by using a BS-C-1 cell line stably coexpressing ER and Golgi markers. Video fluorescence microscopy showed that these two organelles were morphologically distinguishable at all stages of mitosis, and photobleaching experiments showed that diffusion of the Golgi marker was unaffected by the presence of the ER. Fragmentation of the ER by using filipin III completely blocked diffusion of the ER marker but had no effect on the Golgi marker, unless it was first relocated to the ER by using brefeldin A. The Golgi haze was also studied using BODIPY ceramide. Its diffusion was slower in mitotic Golgi than in mitotic ER, but similar to that of a Golgi enzyme marker in the mitotic Golgi haze or in Golgi vesicles generated by ilimaquinone. Together, these results support the idea that the Golgi and the ER remain separate during mitosis and strongly suggest that Golgi markers move by vesicle diffusion, as opposed to lateral diffusion in continuous membranes.
INTRODUCTION
The status of the Golgi apparatus as an organelle has been the subject of a long-running debate that focused initially on the route taken by transiting cargo (cisternal maturation or vesicle-mediated transport; Glick and Malhotra, 1998; Pelham and Rothman, 2000) and, more recently, on the mechanisms of duplication and partitioning that underlie its biogenesis (Marsh and Howell, 2002; Munro, 2002). If the Golgi apparatus is responsible for its biogenesis, then it can be viewed as an autonomous organelle (Pelletier et al., 2002). If, on the other hand, it depends on the endoplasmic reticulum, then it can be viewed as a dependent organelle, which exists only as a consequence of ER functioning (Zaal et al., 1999; Bevis et al., 2002; Munro, 2002).
Golgi partitioning during mitosis in animal cells has been particularly controversial, with models ranging from partitioning by Golgi elements themselves (Lucocq et al., 1989; Misteli and Warren, 1995; Jesch and Linstedt, 1998; Shima et al., 1998; Jesch et al., 2001; Jokitalo et al., 2001) to the partial or even complete merger of the Golgi with the ER, which then mediates the partitioning process (Thyberg and Moskalewski, 1992; Zaal et al., 1999; Kano et al., 2000; Terasaki, 2000). Although most biochemical experiments have yielded consistent results, suggesting a separation of the ER and Golgi during mitosis (Jesch and Linstedt, 1998; Farmaki et al., 1999; Jesch et al., 2001), microscopic experiments have often yielded contradictory results (Thyberg and Moskalewski, 1992; Shima et al., 1998; Zaal et al., 1999; Jokitalo et al., 2001).
A particular issue has been the Golgi haze observed by fluorescence microscopy of mitotic cells. Breakdown and fragmentation of the Golgi ribbon at the onset of mitosis is accompanied by the appearance of a Golgi haze, which pervades the entire mitotic cell cytoplasm. The amount depends on the extent to which the Golgi breaks down, and this varies from cell to cell and even worker to worker. Some of these differences can be attributed to the quality of the antibodies used, particularly in early studies (Warren, 1985), but careful studies of both fixed and live cells show that the haze is a genuine product of the Golgi fragmentation process (Jesch and Linstedt, 1998; Zaal et al., 1999; Jesch et al., 2001). The problem, however, is that the haze cannot be resolved by fluorescence microscopy and has been attributed to two different structures, depending on the partitioning mechanism.
Experiments that favor the separation of the Golgi and the ER during mitosis attribute the haze to free vesicles formed by vesiculation of Golgi stacks during the early phases of mitosis. In vitro and in vivo approaches suggest that these vesicles are derived by continued budding of COPI vesicles in the absence of fusion (Warren, 1985; Misteli and Warren, 1994, 1995; Sönnichsen et al., 1996; Nakamura et al., 1997; Lowe et al., 1998). Mitotic Golgi vesicles have been identified using several electron microscopy (EM) techniques and shown to be distinct from the mitotic ER (Lucocq et al., 1989; Misteli and Warren, 1995; Jokitalo et al., 2001). Fractionation of mitotic cells by velocity gradient centrifugation has been used to separate mitotic Golgi vesicles from the mitotic ER (Jesch and Linstedt, 1998; Jesch et al., 2001). In all cases, these vesicles are ∼60 nm in diameter, which is below the resolution level of fluorescence microscopy (300 nm), and so they should be visualized as a haze. The spindle region of mitotic cells lacks ER but contains the haze, further suggesting that it is independent of the ER (Jesch and Linstedt, 1998; Jesch et al., 2001).
Experiments that favor the merger of the Golgi and the ER during mitosis attribute the haze to a merged ER-Golgi compartment. Early data suggested the presence of mannosidase II in the mitotic ER, although the amounts could not be assessed because the immunocytochemical method used cannot provide quantitation (Thyberg and Moskalewski, 1992). Later experiments suggested that the enzymes found in the mitotic ER were the result of new enzyme synthesis, rather than relocation from the Golgi (Farmaki et al., 1999). Zaal et al. (1999) put forward several lines of evidence for a merged ER-Golgi compartment. A β1,4 galactosyltransferase (GalT)-green fluorescent protein (GFP) construct was localized to the ER by immuno-EM though this probably reflected, at least in part, newly synthesized protein undergoing slow folding (Jokitalo et al., 2001). Endogenous GalT was also localized to ER-like structures in mitotic cells, but the lack of quantitation and double-labeling to identify the ER precludes any firm conclusions. Blocking ER exit at the end of mitosis prevented Golgi reassembly arguing that critical Golgi components were relocated to the ER during the mitosis (Zaal et al., 1999).
Perhaps the most supportive line of evidence came from experiments exploiting the properties of the lipid dye BODIPY ceramide, which has been used as a marker for the Golgi apparatus in interphase cells (Pagano et al., 1991). Lipids move much faster in membranes than proteins but should move at the same rate if both are present in common carriers such as small vesicles. Fluorescence recovery after photobleaching (FRAP) analysis in mitotic cells showed that the rate of diffusion of this dye was 10 times higher than that of a GalT-GFP construct, arguing that both were present in a continuous membrane system rather than small vesicles. These membranes were identified as the ER because the diffusion coefficients for the GalT-GFP construct were the same in mitotic cells as in interphase cells treated with brefeldin A (BFA) (Zaal et al., 1999).
Another model-related issue is the status of mitotic Golgi clusters (MGCs), which are visualized as bright resolvable puncta by fluorescence microscopy, and tubulo-vesicular clusters by EM (Lucocq et al., 1987, 1989; Misteli and Warren, 1995; Shima et al., 1998; Jokitalo et al., 2001). In the autonomous partitioning model, they are viewed as the mediators of Golgi partitioning (Shima et al., 1998). They are formed by the vesiculation of Golgi stacks and shed vesicles to a variable extent, forming the mitotic Golgi haze. There is evidence that the MGCs have an underlying matrix (Seemann et al., 2000) that is normally populated by Golgi enzymes, but these do not have to accompany the matrix during the partitioning process (Seemann et al., 2002). This might help explain the variability in the amount of Golgi haze that is seen in different cell types. The status of the matrix is, however, still unclear, and its existence has been disputed (Zaal et al., 1999; Miles et al., 2001; Ward et al., 2001; Stroud et al., 2003). In the merged ER-Golgi model of partitioning, the MGCs are viewed as intermediates on the pathway leading to merger of the Golgi and the ER.
In this article, we have used quantitative fluorescence microscopy to address the independence of the Golgi and the ER during mitosis. We have focused on the Golgi haze, exploiting its presence in the spindle region to discriminate it from the ER. Quantitative FRAP analysis and pharmacological approaches have been applied to a cell line stably expressing both a Golgi and an ER marker, each tagged with a different variant of GFP. The diffusion of BODIPY ceramide has also been reinvestigated. Together, our data strongly support the idea that the Golgi remains distinct from the ER throughout mitosis and that the Golgi haze consists of vesicles moving throughout the mitotic cytoplasm.
MATERIALS AND METHODS
Chemicals
The following working concentrations (and stock solutions) were used: BFA (Epicenter Technologies, Madison, WI), 5 μg/ml (5 mg/ml in ethanol); filipin III (Sigma-Aldrich, St. Louis, MO), 5 μg/ml (10 mg/ml in methanol); BODIPY FL ceramide (Molecular Probes, Eugene, OR), 5 μM (5 mM in methanol), and ilimaquinone (IQ; Sigma-Aldrich), 25 μM (5 mg/ml in dimethyl sulfoxide). BODIPY ceramide was used as described by Sciaky et al. (1997) and ilimaquinone as described by Takizawa et al. (1993).
Plasmid Constructs
The plasmid encoding GalNAc-T2-YFP, a yellow fluorescent protein (YFP) version of GalNAc-T2-fluorescent protein (Storrie et al., 1998; White et al., 2001), was obtained from Dr. J. White (European Molecular Biology Laboratory, Heidelberg, Germany). It consists of the stalk region of UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2), sufficient for correct Golgi localization (Storrie et al., 1998, and references therein), inserted into the vector pEYFP-N1 (BD Biosciences Clontech, Palo Alto, CA). The cyan fluorescent protein (CFP) version of Sec61β (VLP 25; Rolls et al., 1999) was obtained from Dr. M. Rolls (Harvard Medical School, Boston, MA). CFPSec61β-Hygro was generated by the ligation of a SnaBI-NotI fragment from this construct into the SnaBI-NotI digested vector pcDNA3.1/Hygro(+) (Invitrogen, Carlsbad, CA).
Cell Culture and Transfection
BS-C-1 cells (ATCC CCL 26) were routinely cultured in minimum essential medium (Invitrogen), supplemented with 10% fetal bovine serum (Gemini, Irvine, CA), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen), and 2 mM l-glutamine (Invitrogen). Transfection (plasmids prepared using EndoFree maxi kit; QIAGEN, Valencia, CA) was performed using the Super-Fect transfection reagent (QIAGEN), according to the manufacturer's protocol. Cells were transferred to selective medium after 24 h, and after 2-3 wk, colonies were picked by repetitive pipetting of trypsin and expanded. Single stable GalNAc-T2-YFP transfectants were first selected in geneticin (G-418 sulfate, 800 μg/ml; Invitrogen), an appropriate clone was further transfected with CFP-Sec61β-Hygro, and double-stable transfectants were then selected and maintained using Hygromycin B (130 μg/ml; Calbiochem, San Diego, CA) in addition to geneticin.
Confocal Microscopy and Quantitative FRAP
Microscopy was performed in phenol red-free minimum essential medium (Invitrogen) with 10% fetal bovine serum, 2 mM l-glutamine, and 25 mM HEPES buffer, pH 7.3, on 60% confluent cells in 0.17-mm glass bottom petri dishes (MatTek, Ashland, MA). A 37°C environment was maintained using a stage heater (Carl Zeiss, Thornwood, NY), except during filipin III treatment and the following imaging or FRAP, which were performed at room temperature. Single confocal planes were collected with an inverted laser scanning confocal microscope (LSM510; Carl Zeiss), by using 458-nm laser excitation for CFP and 514 nm for YFP and BODIPY ceramide, a C-Apochromat 40× objective lens, 6× zoom, and a pinhole equivalent to one Airy disk diameter. CFP, YFP, and BODIPY ceramide were imaged together by using 458- and 514-nm laser excitations and the META detector system in the range 467-606 nm with 10.7-nm steps. Signals were separated based on reference spectra from live cells expressing only CFP or YFP, or wild-type cells stained with BODIPY ceramide. All imaging and FRAP of BODIPY ceramide was performed within 30-60 min after the dye was washed away, and the cells were returned to 37°C, as described by Sciaky et al. (1997).
Photobleaching was performed using high laser intensity (100% transmission, two scans for BODIPY ceramide, 20 for YFP, and 50 for CFP) on a 2.5- by 2.5-μm region (selected to be free of resolvable MGCs when viewing Golgi). The FRAP was monitored by continuous scanning of the whole cell at low transmission, and values were obtained from the bleached area and an unbleached control area by using the LSM510 software (Carl Zeiss). Values were corrected for background using a cell-free region of the field of view, and values from the bleached area were compensated for bleaching during monitoring, by using values from the control area. As for BODIPY ceramide, the monitoring affected ER and Golgi differently, and bleached and control areas were selected to represent the same organelle, as judged by positional criteria. Values are given as percentage of the prebleach values, which are averages of two measurements.
RESULTS
The Generation of a Cell Line Expressing Fluorescent Golgi and ER Markers
The O-glycosylation enzyme GalNAc-T2 is a marker of the whole Golgi stack, but with a slight preference for trans cisternae (Rottger et al., 1998). Substitution of the catalytic domain with GFP does not affect its location (Storrie et al., 1998), and a BS-C-1 cell clone stably expressing a YFP analogue of this chimera was selected. Both by YFP fluorescence and by immunofluorescence with anti-GFP antibody, the construct localized to the juxtanuclear Golgi ribbon (our unpublished data). A trace level of ER staining could be distinguished in some cells, probably representing the slow cycling of Golgi enzymes through the ER (Storrie et al., 1998). The ratio of Golgi-to-ER staining seemed similar both by YFP fluorescence and anti-GFP antibody, arguing against an ER pool of misfolded chimera. A CFP-labeled Sec61β (Rolls et al., 1999), a translocon component localized to the ER (Kalies et al., 1998; Rolls et al., 1999), was then expressed to obtain a double-stable cell line. This construct localized to the ER, including the nuclear envelope, as judged by CFP fluorescence.
The Mitotic Golgi and ER Are Morphologically Distinct
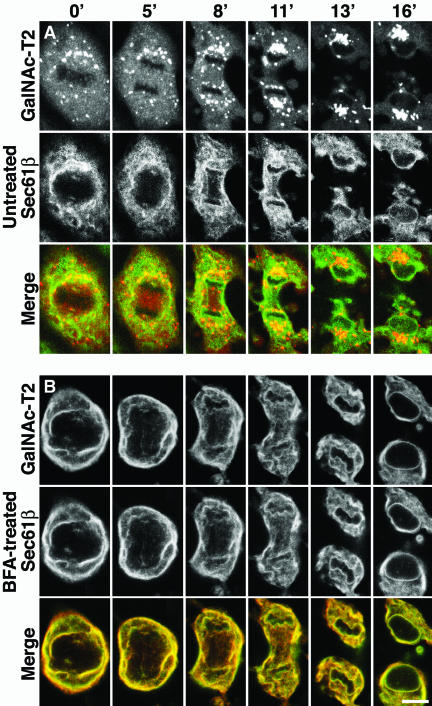
Unsynchronized cultures of GalNAc-T2-YFP/CFP-Sec61β cells were imaged live by using laser scanning confocal microscopy. Figure 1A shows a representative cell monitored from metaphase to cytokinesis. The ER looked like a continuous network, readily distinguishable from the Golgi, which instead looked like MGCs and a nonresolvable haze. As has been reported previously (Jesch and Linstedt, 1998; Jesch et al., 2001), the spindle region (or more correctly, the region delimited by polar microtubules) of the metaphase cell (0 min) lacked ER but contained the Golgi haze. This striking feature persisted through anaphase and telophase (5-8 min), and the ER did not invade this region until spindle disassembly at cytokinesis (11 min). The Golgi haze was homogenously distributed throughout the entire cell, except for the chromosomal regions, and there were no differences in intensity between spindle regions and other areas.
Figure 1.
Morphological differences between mitotic ER and Golgi in live cells. BSC-1 cells, stably expressing GalNAc-T2-YFP, a Golgi enzyme marker (red), and CFP-Sec61β, an ER marker (green), were monitored by laser scanning confocal microscopy at 37°C from metaphase to cytokinesis (lapsed time in minutes indicated at the top). (A) Untreated cell. (B) Cell possessing an artificial ER-Golgi hybrid, generated by addition of BFA before M-phase entry (3 h before the first image). Bar, 10 μm.
To illustrate how the Golgi enzyme marker would look if localized to the ER, cells were treated with BFA before M-phase entry. Figure 1B reveals an almost complete colocalization between the Golgi enzyme and the ER marker, in an ER-like structure, and both markers were absent from the spindle region. This shows that the morphological differences between the Golgi and the ER seen in Figure 1A are real and not due to differences between YFP and CFP, or other technical artifacts.
The Diffusion Rate of the Golgi Haze Is Unaffected by the Presence of ER
We next carried out FRAP experiments by using this cell line, because the diffusion rate of Golgi enzymes in mitotic cells has been taken as evidence for their presence in the ER (Zaal et al., 1999). Two approaches were used.
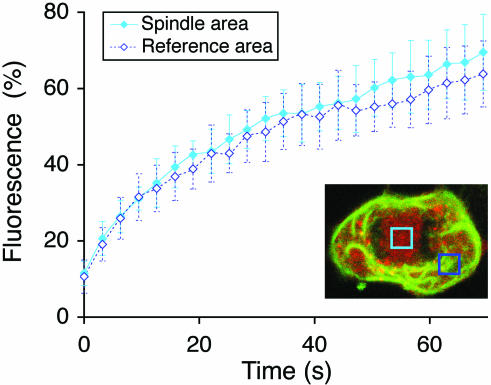
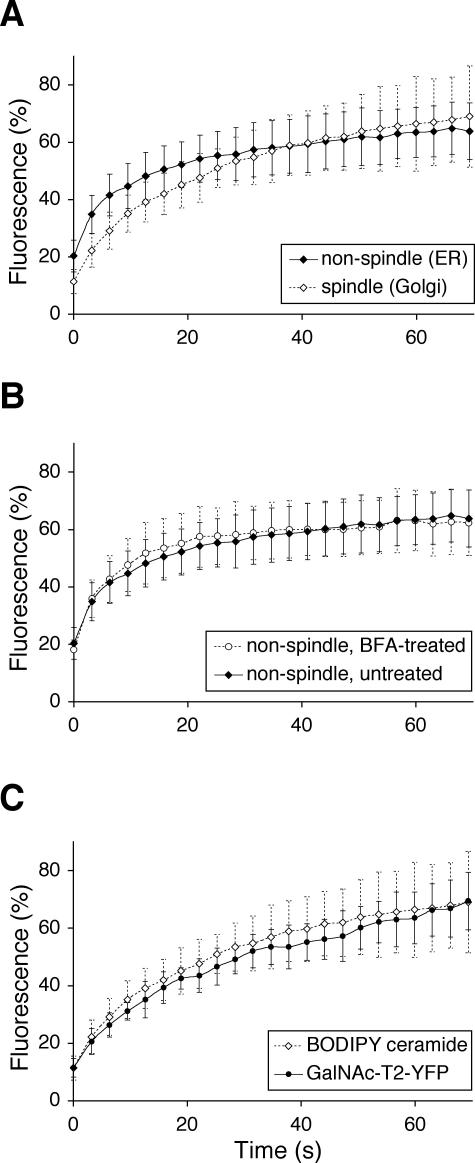
The first exploited the discovery by Linstedt and colleagues (Jesch and Linstedt, 1998; Jesch et al., 2001) that the spindle region is free of ER membranes. Quantitative FRAP of the Golgi haze (GalNAc-T2-YFP) in the spindle region and an ER-containing reference region were compared. Areas were selected to avoid resolvable Golgi puncta (MGCs). Anaphase cells were used, because these possess spindle regions that are both large enough for recovery studies and are free from chromosomes that might otherwise disturb the homogeneity and diffusion of the haze. Figure 2, inset, shows an example of the areas that were bleached in the spindle and reference regions. The ER marker (CFP-Sec61β) was monitored in all cells to confirm the absence of ER in the spindle regions and its presence in the reference regions. As shown in Figure 2, the FRAP kinetics was essentially the same in the ER-free spindle region and in an ER-containing reference region, with an apparent t1/2 of ≈12 s and a mobile fraction of ≈70% in both cases. This shows that the rate of Golgi marker diffusion is not dependent on the presence of the ER, which in turn suggests that the Golgi haze has its own ER-independent diffusion.
Figure 2.
Diffusion rate of the Golgi haze is unaffected by the presence of ER. Anaphase cells were subjected to quantitative FRAP of the Golgi marker GalNAc-T2-YFP (red) both over the ER-free spindle region (filled diamonds) and an ER-containing reference region outside the spindle region (open diamonds). Means ± SD, n = 10. As illustrated in the inset, the absence of ER in the spindle region (light blue box), or presence in the reference region (dark blue box), could be confirmed in every cell imaged by observing the ER marker, CFP-Sec61β.
Fragmentation of the ER Blocks Diffusion of the ER but Not the Golgi Marker
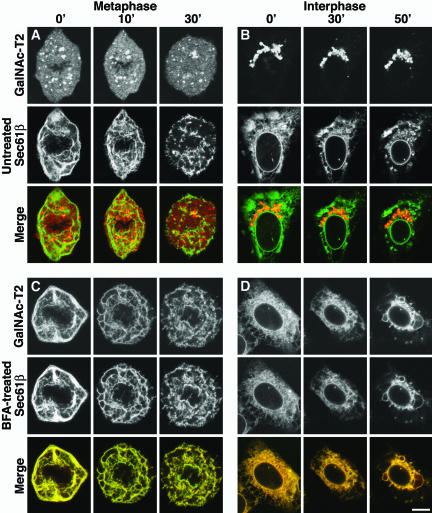
The second approach was pharmacological, exploiting the effect of filipin III, a drug that breaks down the ER in living cells (McEwen et al., 1987). The underlying mechanism is still unclear, but it may be secondary to the cholesterol sequestering action of this compound. The procedures were first optimized for BS-C-1 cells. The effect of filipin III seemed to be best if cells were not fully confluent, and at room temperature rather than 37°C. Metaphase cells, which have small spindle regions, were used so as to maximize the amount of ER available for analysis. The effect of filipin III was rapid, with breakdown of the mitotic ER readily visible by 10 min and complete by 30 min (Figure 3A). Breakdown of the ER in interphase cells took longer, ∼50 min, for reasons that are still unclear (Figure 3B).
Figure 3.
Mitotic and interphase ER can be broken down by using filipin III. Cells were monitored at room temperature (≈20°C) after the addition of filipin III (5 μg/ml) at time points indicated at the top. (A) Metaphase cell. (B) Interphase cell. (C) Metaphase cell, possessing an artificial ER-Golgi hybrid, obtained by addition of BFA before M-phase entry (3 h before the first picture). (D) Interphase cell, likewise treated with BFA for 3 h. Bar, 10 μm.
The interphase Golgi ribbon seemed to be unaffected by filipin III treatment (Figure 3B), at least over the time periods used in these experiments. So, too, were the Golgi haze and the MGCs in mitotic cells (Figure 3A). Filipin III was also tested on cells pretreated with BFA, and as expected, both markers labeled an ER that was broken down by filipin III in both metaphase (Figure 3C) and interphase (Figure 3D) cells. ER breakdown was, however, often less complete in cells pretreated with BFA than in untreated cells, perhaps because the ER-Golgi hybrid acquired some of the resistant properties of the Golgi.
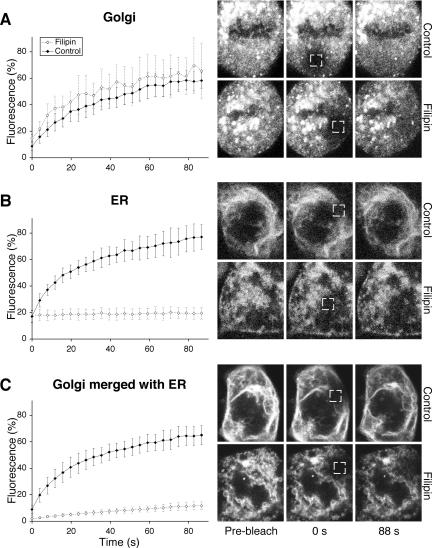
Figure 4A shows quantitative FRAP of the Golgi haze in metaphase cells. The kinetics was not significantly changed by filipin III treatment. If anything, the recovery seemed slightly faster in treated (apparent t1/2 of ≈10 s) than in control cells (apparent t1/2 of ≈15 s). This, and the greater fluctuations in the recovery curve of filipin III-treated cells, was probably due to increased movement of MGCs into the areas being monitored, which were initially selected to be free of these structures. One possibility is that the broken down ER offers less hindrance to movement of the MGCs.
Figure 4.
Breakdown of the mitotic ER blocks diffusion of the ER but not the Golgi marker. Metaphase cells were treated with 5 μg/ml filipin III (open diamonds) or left as untreated controls (filled diamonds) for 30-45 min at room temperature, and then subjected to quantitative FRAP (at room temperature). Means ± SD, n = 10. (A) Golgi marker, GalNAc-T2-YFP. (B) ER marker, CFPSec61β. (C) Golgi marker GalNAc-T2-YFP, relocalized to the ER by addition of BFA before M-phase entry (3 h before addition of filipin III). Shown at right are cells representative of each curve (time points at bottom). Boxes indicate bleaching areas (2.5 × 2.5 μm).
The ER marker of control cells showed a rapid recovery after photobleaching (apparent t1/2 of ≈10 s; Figure 4B). Strikingly, however, in filipin III-treated cells, the ER marker recovery seemed to be negligible. This probably reflects the lack of membrane continuity in ER membranes broken down by filipin III (Figure 3). Together, Figure 4, A and B, show that the diffusion of Golgi and ER markers are differentially affected by treatment with filipin III and provide strong evidence that they reside and diffuse in different structures.
As a control, we studied the effect of filipin III on the Golgi marker after it had been relocalized to the ER by BFA treatment before M-phase entry. The Golgi marker now responded as if it were an ER marker, showing fast recovery in the absence of filipin III (apparent t1/2 of ≈12 s) and very slow recovery in its presence (t1/2 and mobile fraction not measurable within the monitoring interval used). The little recovery that is seen might reflect the slightly less complete breakdown of a merged ER-Golgi discussed above. The fact that diffusion of the Golgi marker can be rendered filipin III sensitive further attests to it not being in the ER during mitosis.
BODIPY Ceramide Diffuses More Slowly in the Mitotic Golgi than in Mitotic ER
The lipid dye BODIPY ceramide has been used as a marker for the Golgi apparatus in interphase cells (Pagano et al., 1991). It has also been used to argue against the mitotic fragmentation of the Golgi into small vesicles that do not fuse with the ER. If this lipid (and its metabolites) shared mitotic Golgi vesicles with Golgi enzymes, then the diffusion rates should be the same, limited by the diffusion of the small vesicles. In fact, the diffusion rate of the lipid was about 10 times that of a GalT-GFP construct, arguing that both were present in the mitotic ER, which does not fragment extensively during mitosis (Zaal et al., 1999). In the light of our results, we decided to reinvestigate the diffusion of BODIPY ceramide and to determine the extent to which it colocalized with the Golgi marker GalNAc-T2 and the ER marker Sec61β during mitosis.
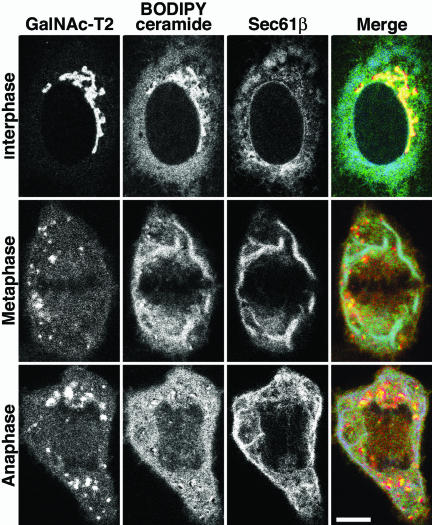
The GalNAc-T2-YFP/CFP-Sec61β cell line was stained with BODIPY ceramide in accordance with the protocol used by Sciaky et al. (1997), and BODIPY, YFP, and CFP were monitored in live cells by using the META system (Carl Zeiss) that allows fluorophores with overlapping spectra to be separated. The top row in Figure 5 shows an interphase cell, and as shown previously (Pagano et al., 1991), a significant amount of the BODIPY ceramide localized to the ER, irrespective of the amounts found in the Golgi ribbon. In metaphase cells (Figure 5, middle row), in contrast, most of the BODIPY signal seemed to colocalize with the ER marker in continuous tubuli. These were clearly distinct from the Golgi haze and MGCs. A faint haze in the spindle region suggested, however, that a small portion of the BODIPY ceramide was in the Golgi. The same phenomenon was seen in the anaphase cells (Figure 5, bottom row), where the ER-containing area outside the mitotic spindle had a much brighter BODIPY signal than the haze of the spindle. These observations were verified using regular YFP filters to visualize the BODIPY ceramide in cells not expressing overlapping fluorophores and were also confirmed in HeLa cells, the cell line used by Zaal et al. (1999; our unpublished data). The results suggest that most of the BODIPY ceramide localizes to the ER in mitotic cells, in accordance with the interpretations of previous kinetic data (Zaal et al., 1999). At the same time, the results further underline the structural differences between the mitotic ER and Golgi observed in Figure 1. The fact that BODIPY ceramide (and its metabolites) is mostly in the ER of mitotic cells does not mean that the Golgi is in the ER.
Figure 5.
BODIPY ceramide localizes mostly to the ER in mitotic cells. The GalNAc-T2-YFP (red)/CFP-Sec61β (blue) cell line was loaded with BODIPY ceramide (from a methanol stock; green) for 10 min in the cold, washed, and incubated in fat-free bovine serum albumin for 30 min at 37°C to remove excess dye. Images were sampled, at 37°C, 30-60 min after loading the dye, by using the META system (Carl Zeiss) to separate the spectra. Bar, 10 μm.
The relatively faint BODIPY signal within the anaphase spindle was subjected to FRAP, to study the kinetics of BODIPY ceramide in mitotic Golgi. Figure 6A shows the FRAP curve together with a corresponding curve from a nonspindle area. The curves are very different, revealing an apparent t1/2 of ≈10 s for the spindle region, compared with ≈2 s for the nonspindle region. The latter value seemed to reflect mostly ER diffusion, because the kinetics of nonspindle areas was essentially identical in cells pretreated with BFA for 2 h (Figure 6B; apparent t1/2 of ≈2 s). Apparent t1/2 values were also obtained for interphase ER (BFA, 2 h) and interphase Golgi ribbons, and were ≈1 and ≈2 s, respectively (our unpublished data). Together, the apparent t1/2 was in the range of ≈1-2 s in all structures where lateral diffusion in continuous membranes can be expected. The fivefold difference in t1/2 found for mitotic Golgi (≈10 s) therefore suggests a different mode of diffusion for this organelle. Figure 6C shows an overlay of the FRAP curves for BODIPY ceramide (Figure 6A) and GalNAc-T2-YFP (Figure 2) in the anaphase spindle region. These are similar, with apparent t1/2 of ≈10 and 12 s, respectively, suggesting that these two markers diffuse in common carriers in the anaphase spindle region.
Figure 6.
BODIPY ceramide diffuses more slowly in the mitotic Golgi than in the mitotic ER. Wild-type BS-C-1 cells were stained with BODIPY ceramide as in Figure 5, and 30-60 min after loading the dye, anaphase cells were subjected to quantitative FRAP. (A) Comparison between the ER-free spindle region (open diamonds) and an ER-containing region outside of the spindle (filled diamonds). (B) Comparison between the ER-containing regions of cells that were not (filled diamonds) or were (open circles) pretreated with BFA for 2 h before BODIPY ceramide addition. (C) Comparison between BODIPY ceramide (open diamonds) and GalNAc-T2-YFP (as in Figure 2; filled circles) over the ER-free spindle region. Means ± SD, n = 10.
The Recovery Rate of the Golgi Haze Is Similar to That of Golgi Vesicles in IQ-treated Cells
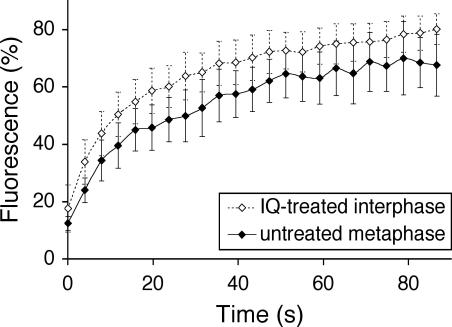
As a positive control for Golgi vesicles, we used the well-characterized drug IQ, which converts the interphase Golgi apparatus into collections of small vesicles, 90 nm in diameter (Takizawa et al., 1993). By using the GalNAc-T2-YFP/CFP-Sec61β cell line, the Golgi was converted into a perinuclear haze, but there was no apparent effect on the ER. Figure 7 shows an apparent t1/2 for recovery of ≈8 s, which is very similar to that for metaphase (≈8 s) and anaphase (≈12 s; Figure 2) cells. Together with the data in the previous section, these results argue strongly that the mitotic Golgi haze comprises free vesicles, corroborating earlier work with biochemical and EM methods (Lucocq et al., 1989; Misteli and Warren, 1995; Jesch and Linstedt, 1998; Jesch et al., 2001; Jokitalo et al., 2001).
Figure 7.
Golgi haze kinetics are consistent with the haze being free vesicles. Quantitative FRAP as for GalNAc-T2-YFP of interphase cells treated for 30-60 min in IQ (open diamonds) and untreated metaphase cells (filled diamonds). Means ± SD, n = 10.
DISCUSSION
In this study, quantitative FRAP analysis was used to examine the dynamics of Golgi and ER membranes during mitosis. To view both membranes simultaneously, a stable cell line was generated expressing two tagged proteins, the retention domain of a Golgi enzyme, GalNAc-T2, linked to YFP, and a component of the ER translocon, Sec61β, linked to CFP. Earlier studies had relied on the transient or stable expression of a single tagged marker, so that its pattern could only be compared with that for other organelles after fixation and labeling with antibodies. Using a cell line stably expressing ER and Golgi markers permitted the first real-time comparison of these two organelles in mitotic cells. This not only avoided the problems associated with cell fixation, background labeling, and antigen accessibility but also increased substantially the number of images that could be carefully compared and provided an unambiguous order of events. The results of these comparisons showed that the Golgi and the ER have distinct distributions throughout mitosis, unless the Golgi was first relocated to the ER by pretreatment of the cells with BFA before they entered mitosis. Then the patterns were virtually identical.
These morphological differences were then confirmed by exploiting two observations. The first was discovered by Linstedt and colleagues (Jesch and Linstedt, 1998; Jesch et al., 2001), who showed that the spindle region contains Golgi but not ER markers. The second was the selective effect of filipin III on ER but not Golgi membranes. Although the reasons are still unclear, the cholesterol-sequestering effect of filipin III has a dramatic effect on the ER. Over a 30-min time period, the mitotic ER undergoes extensive fragmentation. FRAP analysis showed that this was accompanied by a dramatic change in the movement of the Sec61β marker. Although the t1/2 for recovery of this marker in mitotic ER was ≈10 s, there was essentially no recovery in the presence of filipin III, presumably because the ER membrane fragments were no longer connected. In marked contrast, the t1/2 for recovery of the GalNAc-T2 marker was about the same in the absence or presence of filipin III. If anything, recovery was a little faster (≈10 vs. ≈15 s), perhaps because the fragmentation of the ER gave the Golgi more room to move around. Furthermore, when the cells were pretreated with BFA before they entered mitosis, the Golgi marker now behaved in exactly the same way as the ER marker. There was essentially no recovery after photobleaching in the presence of filipin III. Together, these data clearly show that during mitosis, the diffusion of the Golgi marker occurs in a different compartment from that in which the ER marker is diffusing.
Does this separate compartment, the mitotic Golgi haze, then comprise vesicles? The recovery rates for the Golgi marker in the Golgi and in the ER (after pretreatment with BFA) were surprisingly similar. The t1/2 values were ≈15 and ≈12 s, respectively. One might have expected Golgi enzymes to diffuse much more slowly in the Golgi than in the ER, because the diffusion rate in the Golgi would be limited by the diffusion of the vesicles. This argument was in fact used by Zaal et al. (1999), who observed that a GalT construct diffused at the same rate in interphase ER and mitotic cells. They therefore concluded that the GalT construct must be present in the mitotic ER, not Golgi vesicles.
To our knowledge, however, the diffusion rate of vesicles in mitotic cytosol has neither been estimated nor measured. It could simply be coincidence that Golgi enzymes diffuse at the same rate in Golgi vesicles as in the ER, and then FRAP could not be used to identify the membrane in which the Golgi enzyme is diffusing. To determine whether the recovery rate for GalNAc-T2-YFP is consistent with movement in vesicles, the cells were treated with IQ. This sponge metabolite has been extensively characterized by Malhotra and colleagues (Takizawa et al., 1993). They showed that the Golgi ribbon was converted into a collection of homogeneously sized vesicles, 90 nm in diameter. FRAP analysis showed that the apparent t1/2 for GalNAc-T2-YFP in these treated cells (≈8 s) was similar to that in both metaphase (≈8 s) and anaphase (≈12 s) cells, arguing that the recovery rates are entirely consistent with diffusion in vesicles. The similarity in diffusion rates of GalNAc-T2-YFP in the Golgi and the ER could then be explained in two ways. Either the mitotic Golgi vesicles diffuse much more rapidly than might be expected, or the GalNAc-T2-YFP in mitotic ER diffuses much more slowly. Golgi enzymes can exist as large oligomeric structures (Nilsson et al., 1994; Nilsson and Warren, 1994) and this might make them move more slowly in ER membranes. Alternatively, or in addition, movement through the ER might be obstructed by large oligomeric structures such as the ribosome/translocon complexes that translocate proteins across the ER membrane.
The presence of Golgi enzymes in small vesicles was also supported by a reinvestigation of the diffusion of BODIPY ceramide in mitotic cells. The rapid diffusion of this Golgi lipid marker compared with a Golgi enzyme marker was taken as evidence that both were in the ER (Zaal et al., 1999). Had they both been present in small vesicles, the diffusion rate would have been the same. Using exactly the same conditions as Lippincott-Schwartz and colleagues, BODIPY ceramide was found mostly in the Golgi ribbon in interphase BSC-1 cells and in the ER in mitotic cells. This assignment was facilitated by triple labeling experiments and the use of the Zeiss META detector to separate out the overlapping fluorescence signals. It was also confirmed in single labeling experiments and in other cell lines such as HeLa cells. There was, however, detectable BODIPY ceramide in the spindle region, which excluded ER but not the haze of the Golgi enzyme marker. Quantitative FRAP showed that the recovery rate of the lipid in this region was very similar to that of the Golgi enzyme marker (t1/2 of ≈10 and ≈12 s, respectively). This argues strongly that both the lipid and the Golgi enzyme are sharing the same carriers, most likely small vesicles. The fact that this lipid pool is small, because most goes back to the ER, could explain why it was overlooked by Lippincott-Schwartz and colleagues. It also raises the interesting possibility that there is movement of Golgi components back to the ER during mitosis, but it is lipids rather than proteins that move back. Movement could be mediated either indirectly by lipid-rich vesicles or directly by junctions that have been observed between the trans-Golgi and the ER (Ladinsky et al., 2002). However, the first step has to be a repetition of these experiments studying endogenous lipids, to show that they behave in the same manner as BODIPY ceramide.
Although the Golgi haze can clearly be equated with small, dispersed vesicles, this does not explain their physiological function. In the earliest experiments, they were thought to be the end product of Golgi vesiculation and therefore the Golgi partitioning unit. The predicted number of vesicles, ≈10,000, was sufficient to provide a theoretical accuracy of partitioning of a few percent (Birky, 1983; Lucocq et al., 1989). Later experiments suggested that MGCs were in fact the partitioning unit, and they mediated ordered partitioning in association with microtubules (Shima et al., 1998). Vesicles were relegated to a subordinate role with no obvious function, shed from the MGCs to a variable extent. But even shedding is not essential for partitioning to occur. Pretreatment of cells with BFA before their entry into mitosis relocates the enzymes to the ER yet has no effect of the partitioning of MGCs identified by matrix markers (Seemann et al., 2002). Microinjection of a mutant p47 seems to prevent vesiculation at the onset of mitosis but has no apparent effect on partitioning (Uchiyama et al., 2003). The mechanism is unclear because this cofactor is usually involved in the p97-mediated reassembly of the Golgi apparatus at the end of mitosis. Nevertheless, it does suggest that Golgi stacks themselves can mediate accurate partitioning, which would put mammalian cells into the same category as plants and fungi which also do not fragment their Golgi during mitosis (Warren, 1993).
This would leave us with the possibility that mitotic Golgi vesicles are simply the side-product of the inhibition of intra-Golgi transport that is a characteristic feature of mitotic mammalian cells (Warren, 1985). Nevertheless, it should be noted that all these experiments have been carried out on individual fibroblastic cells growing on plastic or glass surfaces. Cells growing in tissues have to coordinate mitosis and cell division with neighboring cells. In the future, it will be important to study the Golgi haze in systems that better approximate the multicellular state of mammals.
Acknowledgments
We thank Drs. Jamie White, Tommy Nilsson, Melissa Rolls, and Tom Rapoport for plasmid constructs. We are also grateful to Drs. Cecile Chalouni, Amy Chow, Cynthia He, and Derek Toomre for technical help with the confocal microscopy and critical reading of the manuscript. M.A. was supported by a postdoctoral fellowship from the Swedish Foundation for International Cooperation in Research and Higher Education. This work was supported by the National Institutes of Health.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-07-0459. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-07-0459.
Abbreviations used: BFA, brefeldin A; CFP, cyan fluorescent protein; EM, electron microscopy; ER, endoplasmic reticulum; FRAP, fluorescence recovery after photobleaching; GalNAc-T2, UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 2; GalT, β1,4 galactosyltransferase; GFP, green fluorescent protein; IQ, ilimaquinone; MGC, mitotic Golgi cluster; YFP, yellow fluorescent protein.
References
- Bevis, B.J., Hammond, A.T., Reinke, C.A., and Glick, B.S. (2002). De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol. 4, 750-756. [DOI] [PubMed] [Google Scholar]
- Birky, C.W. (1983). The partitioning of cytoplasmic organelles at cell division. Int. Rev. Cytol. 15, 49-89. [DOI] [PubMed] [Google Scholar]
- Farmaki, T., Ponnambalam, S., Prescott, A.R., Clausen, H., Tang, B.L., Hong, W., and Lucocq, J.M. (1999). Forward and retrograde trafficking in mitotic animal cells. ER-Golgi transport arrest restricts protein export from the E.R. into COPII-coated structures. J. Cell Sci. 112, 589-600. [DOI] [PubMed] [Google Scholar]
- Glick, B.S., and Malhotra, V. (1998). The curious status of the Golgi apparatus. Cell 95, 883-889. [DOI] [PubMed] [Google Scholar]
- Jesch, S.A., and Linstedt, A.D. (1998). The Golgi and endoplasmic-reticulum remain independent during mitosis in HeLa cells. Mol. Biol. Cell 9, 623-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch, S.A., Mehta, A.J., Velliste, M., Murphy, R.F., and Linstedt, A.D. (2001). Mitotic Golgi is in a dynamic equilibrium between clustered and free vesicles independent of the ER. Traffic 2, 873-884. [DOI] [PubMed] [Google Scholar]
- Jokitalo, E., Cabrera-Poch, N., Warren, G., and Shima, D.T. (2001). Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J. Cell Biol. 154, 317-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalies, K.U., Rapoport, T.A., and Hartmann, E. (1998). The beta subunit of the Sec61 complex facilitates cotranslational protein transport and interacts with the signal peptidase during translocation. J. Cell Biol. 141, 887-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano, F., Takenaka, K., Yamamoto, A., Nagayama, K., Nishida, E., and Murata, M. (2000). MEK and Cdc2 kinase are sequentially required for Golgi disassembly in MDCK cells by the mitotic Xenopus extracts. J. Cell Biol. 149, 357-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky, M.S., Wu, C.C., McIntosh, S., McIntosh, J.R., and Howell, K.E. (2002). Structure of the Golgi and distribution of reporter molecules at 20°C reveals the complexity of the exit compartments. Mol. Biol. Cell 13, 2810-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, M., Rabouille, C., Nakamura, N., Watson, R., Jackman, M., Jämsä, E., Rahman, D., Pappin, D.J.C., and Warren, G. (1998). Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell 94, 783-793. [DOI] [PubMed] [Google Scholar]
- Lucocq, J.M., Berger, E.G., and Warren, G. (1989). Mitotic Golgi fragments in HeLa cells and their role in the reassembly pathway. J. Cell Biol. 109, 463-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq, J.M., Pryde, J.G., Berger, E.G., and Warren, G. (1987). A mitotic form of the Golgi apparatus in HeLa cells. J. Cell Biol. 104, 865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, B.J., and Howell, K.E. (2002). The mammalian Golgi-complex debates. Nat. Rev. Mol. Cell. Biol. 3, 789-795. [DOI] [PubMed] [Google Scholar]
- McEwen, B.F., Telford, J.N., Handelman, C.T., and Arion, W.J. (1987). A critical evaluation of the use of filipin-permeabilized rat hepatocytes to study functions of the endoplasmic reticulum in situ. Cell. Biochem. Funct. 5, 263-272. [DOI] [PubMed] [Google Scholar]
- Miles, S., McManus, H., Forsten, K.E., and Storrie, B. (2001). Evidence that the entire Golgi apparatus cycles in interphase HeLa cells: sensitivity of Golgi matrix proteins to an ER exit block. J. Cell Biol. 155, 543-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., and Warren, G. (1994). COP-coated vesicles are involved in the mitotic fragmentation of Golgi stacks in a cell-free system. J. Cell Biol. 125, 269-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., and Warren, G. (1995). Mitotic disassembly of the Golgi apparatus in vivo. J. Cell Sci. 108, 2715-2727. [DOI] [PubMed] [Google Scholar]
- Munro, S. (2002). More than one way to replicate the Golgi apparatus. Nat. Cell Biol. 4, E223-224. [DOI] [PubMed] [Google Scholar]
- Nakamura, N., Lowe, M., Levine, T.P., Rabouille, C., and Warren, G. (1997). The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell 89, 445-455. [DOI] [PubMed] [Google Scholar]
- Nilsson, T., Hoe, M.H., Slusarewicz, P., Rabouille, C., Watson, R., Hunte, F., Watzele, G., Berger, E.G., and Warren, G. (1994). Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J. 13, 562-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, T., and Warren, G. (1994). Retention and retrieval in the endoplasmic-reticulum and the Golgi-apparatus. Curr. Opin. Cell Biol. 6, 517-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano, R.E., Martin, O.C., Kang, H.C., and Haugland, R.P. (1991). A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J. Cell Biol. 113, 1267-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham, H.R., and Rothman, J.E. (2000). The debate about transport in the Golgi-two sides of the same coin? Cell 102, 713-719. [DOI] [PubMed] [Google Scholar]
- Pelletier, L., et al. (2002). Golgi biogenesis in Toxoplasma gondii. Nature 418, 548-552. [DOI] [PubMed] [Google Scholar]
- Rolls, M.M., Stein, P.A., Taylor, S.S., Ha, E., McKeon, F., and Rapoport, T.A. (1999). A visual screen of a GFP-fusion library identifies a new type of nuclear envelope membrane protein. J. Cell Biol. 146, 29-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottger, S., White, J., Wandall, H.H., Olivo, J.C., Stark, A., Bennett, E.P., Whitehouse, C., Berger, E.G., Clausen, H., and Nilsson, T. (1998). Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. J. Cell Sci. 111, 45-60. [DOI] [PubMed] [Google Scholar]
- Sciaky, N., Presley, J., Smith, C., Zaal, K.J.M., Cole, N., Moreira, J.E., Terasaki, M., Siggia, E., and Lippincott-Schwartz, J. (1997). Golgi tubule traffic and the effects of brefeldin-A visualized in living cells. J. Cell Biol. 139, 1137-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann, J., Jokitalo, E., Pypaert, M., and Warren, G. (2000). Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407, 1022-1026. [DOI] [PubMed] [Google Scholar]
- Seemann, J., Pypaert, M., Taguchi, T., Malsam, J., and Warren, G. (2002). Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science 295, 848-851. [DOI] [PubMed] [Google Scholar]
- Shima, D.T., Cabrera-Poch, N., Pepperkok, R., and Warren, G. (1998). An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J. Cell Biol. 141, 955-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen, B., Watson, R., Clausen, H., Misteli, T., and Warren, G. (1996). Sorting by COP I-coated vesicles under interphase and mitotic conditions. J. Cell Biol. 134, 1411-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie, B., White, J., Rottger, S., Stelzer, E.H.K., Suganuma, T., and Nilsson, T. (1998). Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 143, 1505-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud, W.J., Jiang, S., Jack, G., and Storrie, B. (2003). Persistence of Golgi matrix distribution exhibits the same dependence on Sar1p activity as a Golgi glycosyltransferase. Traffic 4, 631-641. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A., Yucel, J.K., Veit, B., Faulkner, D.J., Deerinck, T., Soto, G., Ellisman, M., and Malhotra, V. (1993). Complete vesiculation of Golgi membranes and inhibition of protein-transport by a novel sea sponge metabolite, ilimaquinone. Cell 73, 1079-1090. [DOI] [PubMed] [Google Scholar]
- Terasaki, M. (2000). Dynamics of the endoplasmic reticulum and Golgi apparatus during early sea urchin development. Mol. Biol. Cell 11, 897-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyberg, J., and Moskalewski, S. (1992). Reorganization of the Golgi complex in association with mitosis: redistribution of mannosidase II to the endoplasmic reticulum and effects of brefeldin A. J. Submicrosc. Cytol. Pathol. 24, 495-508. [PubMed] [Google Scholar]
- Uchiyama, K., Jokitalo, E., Lindman, M., Jackman, M., Kano, F., Murata, M., Zhang, X., and Kondo, H. (2003). The localization and phosphorylation of p47 are important for Golgi disassembly-assembly during the cell cycle. J. Cell Biol. 161, 1067-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, T.H., Polishchuk, R.S., Caplan, S., Hirschberg, K., and Lippincott-Schwartz, J. (2001). Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155, 557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, G. (1985). Membrane traffic and organelle division. Trends Biochem. Sci. 10, 439-443. [Google Scholar]
- Warren, G. (1993). Membrane partitioning during cell division. Annu. Rev. Biochem. 62, 323-348. [DOI] [PubMed] [Google Scholar]
- White, J., Keller, P., and Stelzer, E.H. (2001). Spatial partitioning of secretory cargo from Golgi resident proteins in live cells. BMC Cell Biol. 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaal, K.J., et al. (1999). Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell 99, 589-601. [DOI] [PubMed] [Google Scholar]