Abstract
The soybean cyst nematode (SCN), Heterodera glycines, can cause significant reductions in soybean yield and quality in many parts of the world. Natural biological control may play an important role in regulating SCN population. In this study the bacterial communities associated with SCN cysts obtained from fields under different lengths of soybean monoculture were explored. Soil samples were collected in 2010 and 2011 from six fields that had been used for soybean monoculture for 2 to 41 yr. SCN population densities were determined and bacterial communities from SCN cysts were investigated by Biolog and PCR-DGGE methods. SCN population densities initially increased in the first 5 yr of soybean monoculture but then declined steeply as years of soybean monoculture increased. Catabolic diversity of bacterial communities associated with cysts tended to decline as number of years of monoculture increased. Some specific PCR-DGGE bands, mainly representing Streptomyces and Rhizobium, were obtained from the cysts collected from the long-term monoculture fields. Principal component analysis of Biolog and PCR-DGGE data revealed that bacterial communities associated with cysts could be divided into two groups: those from cysts obtained from shorter (< 8 yr) vs. longer (> 8 yr) monoculture. This research demonstrates that the composition of the bacterial communities obtained from SCN cysts changes with length of soybean monoculture; the suppressive impact of these bacterial communities to SCN is yet to be determined.
Keywords: bacterial community, biodiversity, biological control, cyst, Heterodera glycines, monoculture, soybean, soybean cyst nematode
The soybean cyst nematode (SCN, Heterodera glycines) is the most serious pathogen of soybean (Glycine max) in northeast China and other parts of the world (Ma et al., 2005; Alkharouf et al., 2006). Management of this nematode is difficult, in part because effective nematicides are either unavailable or expensive. Long-term rotations with nonhost crops can control SCN but are not usually economically feasible (Opperman and Bird, 1998). While SCN-resistant cultivars have been widely used in some soybean-growing regions in the world, especially in the United States, they are not commercially available in most parts in China. Moreover, use of resistant cultivars can select virulent SCN populations that eventually overcome the resistance (Zheng and Chen, 2011). Biological control with fungal and bacterial antagonists of nematodes is a potential alternative (Atibalentja et al., 2000; Liu and Chen, 2001).
One kind of biological control is manifested in suppressive soils, i.e., soils that contain pathogen but in which the pathogen causes little damage because of the natural presence of biological control agents. A number of nematode-suppressive soils have been reported (Chen and Dickson, 2012) including SCN-suppressive soils in China (Liu and Wu, 1992; Sun and Liu, 2000) and in the United States (Chen, 2007). In these soils, SCN population densities declined as monoculture of a susceptible soybean cultivar was continued for more than 5 yr.
Although the mechanisms that result in a nematode suppressive soil are complex and not well understood, the presence of fungi and bacteria are often suspected. Parasitism of SCN eggs by microorganisms was higher in a suppressive than in nonsuppressive soils in China (Sun and Liu, 2000). In some soils, the nematophagous fungi Hirsutella spp. were considered to be a main factor contributing to SCN suppression because parasitism of SCN second-stage juveniles by these fungi was greater in suppressive than in nonsuppressive soils (Xiang et al., 2010; Kidane et al., 2012). Several Rhizobium spp. and uncultured α-proteobacterial clones associated with cysts of Heterodera schachtii were found to be involved in the suppression of the nematode (Yin et al., 2003). Researchers have reported differences in the rhizosphere microbial community of healthy and SCN-infected soybean roots, and such differences might explain SCN suppression (Carris et al., 1989; Liu and Wu, 1992; Sun and Liu, 2000; Chen, 2007). A survey of soybean fields in Canada indicated that an average of 104 to 105 bacteria were associated with each SCN cyst (Nour et al., 2003). A recent study concerning sugar beet root rot caused by Rhizoctonia solani demonstrated that a number of rhizosphere microorganisms were involved in suppression of this disease (Mendes et al., 2011). These results indicate that a better understanding of the interactions of microorganisms and SCN in soil should yield new insights into the mechanisms of microbial ecology and perhaps new SCN management strategies using microbial consortium in suppressive soil.
However, the identities and functional diversity of microbes associated with SCN cysts under different numbers of years of soybean monoculture have not been investigated. With the development of molecular techniques, new tools have been available for monitoring changes in the microbial community associated with SCN. For example, denaturing gradient gel electrophoresis (DGGE) has been used to determine the diversity of bacteria colonizing SCN cysts (Nour et al., 2003). A culture-independent method termed “oligonucleotide fingerprinting of rRNA genes” has been used to identify bacterial rDNA associated with H. schachtii cysts (Yin et al., 2003). Application of these molecular methods might be useful for understanding the mechanism of microbial ecology and has revealed the existence of formerly unknown microorganisms in the agricultural ecosystems.
The objective of the present study was to investigate the change in bacterial communities associated with SCN cysts in response to the number of years (2 to 40 yr) of soybean monoculture, particularly in comparing short-term with long-term soybean monoculture, using culture-dependent and culture-independent approaches.
Materials And Methods
Field sites and soil sampling:
Six fields, located at Harbin County (45°40′–45°46′N, 126°35′–126°45′E), Heilongjiang Province, China, with different lengths of soybean monoculture were selected for this study. The major physical and chemical characteristics of the soil in the fields were similar to the following: 9.1% to 10.6% sand; 34.2% to 34.7% clay; 26.1 to 28.9 g/kg organic matter; 5.9 to 7.1 pH; 1.57 to 1.75 g/kg total N; 0.41 to 0.44 g/kg total P; and 19.5 to 20.3 g/kg total K. Although the soybean cultivars planted in the fields were unknown, no SCN-resistant cultivars are known to have been used in the region. Therefore, we assumed that the soybean cultivars in all fields were susceptible to SCN. Three soil samples were taken from each field in July 2010 and again in July 2011. Each sample consisted of five to seven soil cores (2-cm-diam. × 20-cm depth) collected in a zigzag pattern from an area about 50 m2. The distance between two samples in a field was 30 to 50 m. The soil samples were shipped immediately to the lab and stored at 4°C before processing within 14 d.
SCN extraction and population density determination:
Each soil sample was thoroughly mixed. A subsample of 100 g soil was used to determine SCN egg population density, and another 100-g subsample was used to determine SCN cyst population density. Cysts were extracted by decanting the soil suspension through nested 800- and 250-μm-aperture sieves. Cysts caught on the 250-μm-aperture sieve were separated from soil particles with centrifugation in 75% (w/v) sucrose solution at 1,795 g for 5 min. Eggs were released from extracted cysts by breaking the cysts in a 5-ml glass tissue grinder; released eggs were collected on a 25-μm-aperture sieve (Liu and Chen, 2001). The cysts and eggs were stored in distilled water at 4°C before being counted within 24 h. The cysts were examined under a dissect microscope for eggs; and the cysts containing eggs or without eggs (empty cysts) were counted separately. The extracted eggs were counted under an inverted compound microscope.
Approximately 150 cysts were extracted from another 0.5 to 5-kg soil subsample for DNA extraction and metabolic profile determination. The cysts were surface sterilized by immersion for 5 min in 0.3% NaOCl bleach, followed by five rinses in sterile distilled water. Half of the cysts were frozen at -80°C for DNA extraction and the other half of the cysts were used directly for metabolic profile determination.
Biolog analyses:
Twenty-five cysts from each sample were placed in sterile distilled water and macerated with a tissue homogenizer. Water was then added to increase the volume of the suspension to 1 ml. Serial dilutions were made in 10-ml suspensions and a 200-μl aliquot of the solution was spread on a petri plate containing 0.1-strength beef extract tryptone agar. Each dilution was replicated on five plates. The plates were incubated at 28°C for 48 h before the colonies were counted. The dilution that resulted in 103 CFU/ml was selected for the Biolog analysis. An aliquot of 150 μl of this dilution was added to each well of a Biolog EcoPlate (Biolog Inc., Hayward, CA). The plates were then incubated at 28°C and read every 24 h for 7 d with an automated Biolog Microplate Reader at a wavelength of 590 nm. The data were collected with MicroLog 4.01 software (Biolog Inc., Hayward, CA).
Microbial activity in each microplate, expressed as average well-color development (AWCD) was calculated for each of different measurement times using the formula:
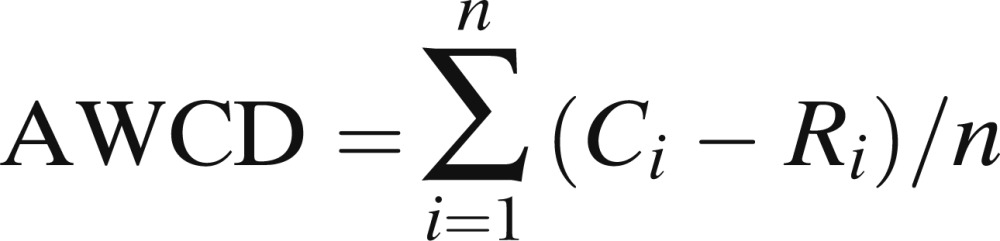 |
where Ci is the value of optical density at OD590 of the ith substrate, R is the optical density of the control well, and n is the number of substrates, equal to 31 in this study. Substrate richness (S) was calculated by counting total number of C substrates oxidized by individual treatments on each microplate (counting all positive OD readings), considering as “position”—that is, the substrate was metabolized-those wells with OD>0.10 (Garland, 1997).
The Shannon diversity index (H′) (Zak et al., 1994; Derry et al., 1998) was calculated as follows:
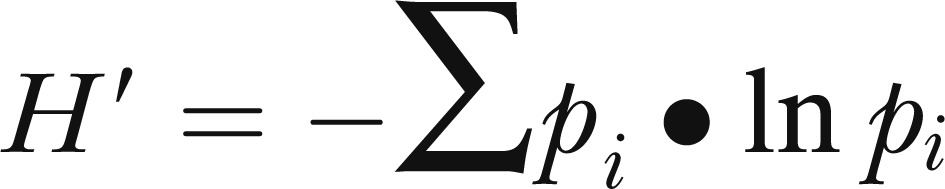 |
where pi is the proportion of members that a particular species contributes to the total in the sample.
DNA profiling:
DNA was released from cysts by placing the cysts in a PowerSoil DNA (MoBio, Carlsbad, CA) isolation kit and subjecting the cysts to 4,200 rpm for 30 sec in a Mini-Bead Beater (BioSpec Product Inc., Bartlesville, OK). DNA was subsequently extracted from this solution with the PowerCleanTM DNA Clean-UP kit (MoBio, Carlsbad, CA) as described by the manufacturer. The extracted DNA was dissolved in 50 μl of TE buffer (10 mmol/l Tris-HCl, 1 mmol/l EDTA, pH 8.0) and used as template for PCR.
PCR amplification for bacterial 16S rDNA was performed with the primer set of GC-338f (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGACTCCTACGGGAGGCAGCAG-3′) and 518r (5′-ATTACCGCGGCTGCTGG-3′) (Muyzer et al., 1993). The reaction mixture (50 μl) contained 0.5 μl of forward and reverse primers (50 pmol each), 5.0 μl of 10 × ExTaq Buffer (TaKaRa Biotechnology, Dalian, China), 5.0 μl of dNTP (2.5 mM each, TaKaRa), 2.0 μl (10 ng) of DNA template, and 0.5 μl of Taq DNA polymerase (TaKaRa). The PCR program consisted of 5 min at 94°C; followed by 30 cycles of 94°C for 40 sec, 52°C for 40 sec, and 72°C for 60 sec; and a final extension at 72°C for 10 min. PCR products were checked for the expected size in a 1.0% agarose gel with GoldViewTM nucleic acid stain (Beijing Solarbio Science & Technology Co. Ltd., Beijing, China) under ultraviolet light.
DGGE was performed by using 8% acrylamide gel with a 40% to 70% denaturant gradient, where 100% denaturant was defined as 7 mol/l urea plus 40% formamide. For each PCR sample, 20 μl containing 200-ng DNA was loaded on to the denaturing gradient gel, and DGGE was performed with a DCodeTM Universal Mutation Detection System (Bio-Rad Laboratories, Los Angeles, CA). The gel electrophoresis was run in 1 × TAE buffer (40 mM Tris–acetate, 1 mM EDTA, pH 8.0) for 16 h at 100V and 60°C. The gel was stained in 1:10000 (v/v) SYBRTM Green I (CAMBREX, Rockland, MD) nucleic acid staining solution for 20 min. An image of the gel was obtained with the Bio-Rad transilluminator (BIO-RAD Laboratories, Segrate, Italy) under UV light.
The specific DGGE bands were carefully excised with a razor blade under UV illumination and then placed in 50 μl of TE buffer. DNA was extracted from the gel by overnight incubation at 4°C (Sekiguchi et al., 2001), and then 2 μl of supernatant containing 10-ng DNA was used as template DNA for PCR amplification using 338f (5′-ACTCCTACGGGAGGCAGCAG-3′) and 518r (5′-ATTACCGCGGCTGCTGG-3′) primers without GC clamp. After purification by electrophoresis through 1.0% agarose and recovery by a DNA Recovery Kit, the DNA fragments were ligated to the pMD18-T cloning vector (TaKaRa) and transformed into Escherichia coli DH5a (TaKaRa). One or two positive clones from each band were selected randomly for DNA sequencing with an ABI 377 apparatus (SinoGenoMax Co., Ltd., Beijing, China). Sequence identification was performed by using BLAST search (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) (Altschul et al., 1990).
Banding patterns of the DGGE profile were analyzed by QuantityOne-1-D software (version 4.5; BIO-RAD Laboratories). The position and intensity of each band were determined automatically. The density value of each band was divided by the average band density in each sample to minimize the influence of differences DNA density between samples (Garland and Mills, 1991; Graham and Haynes, 2005). Sequences for DGGE bands A to F have been deposited in GenBank and given accession numbers JQ180206 to JQ180211.
Statistical analysis:
All data were examined for normality. SCN egg population density data was ln(x+1)-transformed, and AWCD, substrate richness, and diversity data were normalized before being subjected to analysis of variance (ANOVA) with the SPSS 13.0 for Windows. The ANOVA were performed for the data of the two years separately because the lengths of the monoculture were different between the two years. Based on examination of kinetic curves of AWCD, optical density (OD) values obtained 72 h after inoculation were used for statistical analysis. Means were compared with Tukey’s Test at α = 0.05. Regression was performed to determine the relationship between SCN egg population density and number of years of soybean monoculture using a quadratic model with ln-transformed average egg population density and year (2 to 13 yr). In addition, linear regression was performed to determine the relationship between the microbial activity (AWCD, Shannon index, and substrate richness) and the number of years of monoculture; and between eggs in cysts (eggs/cyst and percentage of cysts without eggs) and the number of years of monoculture. Sampling year and its interactions with number of years of monoculture were initially included in the regressions; however, sampling year and the interaction were not significant in any of the regression and subsequently removed from the regression models. Normalized data of DGGE band densities were used for principal component analysis (PCA) using SPSS 13.0 software (SPSS Inc., Chicago, IL) to determine relationships between bacterial community as represented by the DGGE profiles and the soybean monoculture. Redundancy analysis (RDA) as a direct ordination technique based on PCA, was used to explore the relationship between cyst microbial community (31 C-sources) and environmental variables, and performed using CANOCO for Windows 4.5 (Microcomputer Power, Ithaca, NY).
Results
SCN population density:
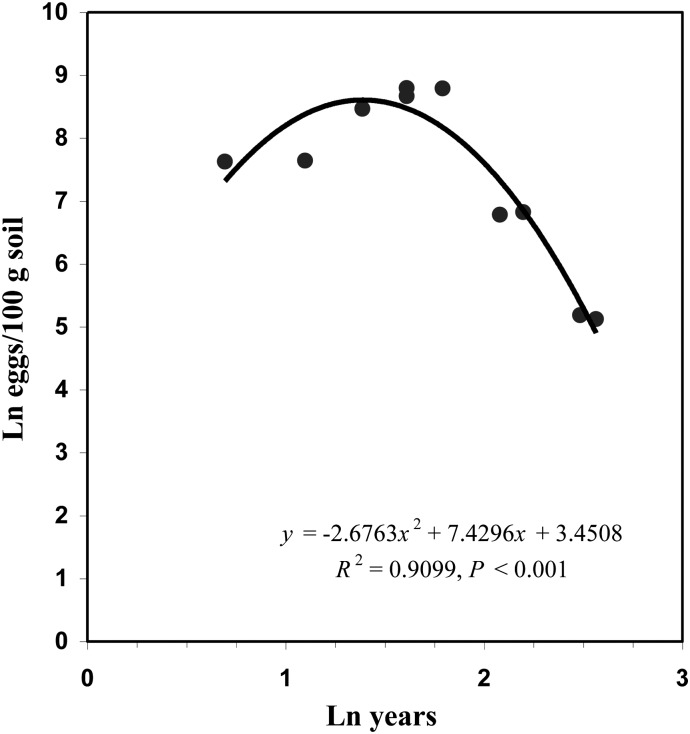
SCN egg population densities differed with years of monoculture (Table 1). Based on the regression analysis, egg densities increased as years of monoculture increased from 2 to about 5 yr and then declined sharply until 12 yr (Fig. 1). After 12 or more yr of soybean monoculture, fewer than 200 eggs/100 g of soil were detected (Table 1). Interestingly, egg numbers decreased to fewer than 50 eggs/cyst after 8 yr of monoculture. In addition, more than 40% of the cysts contained no eggs after 8 yr, and more than 70% of the cysts were empty after 40 to 41 yr of monoculture.
Table 1.
Numbers of cysts and eggs of Heterodera glycines in fields that were continuously cropped with soybean for 2 to 41 yr (the same fields were sampled in 2010 and 2011).
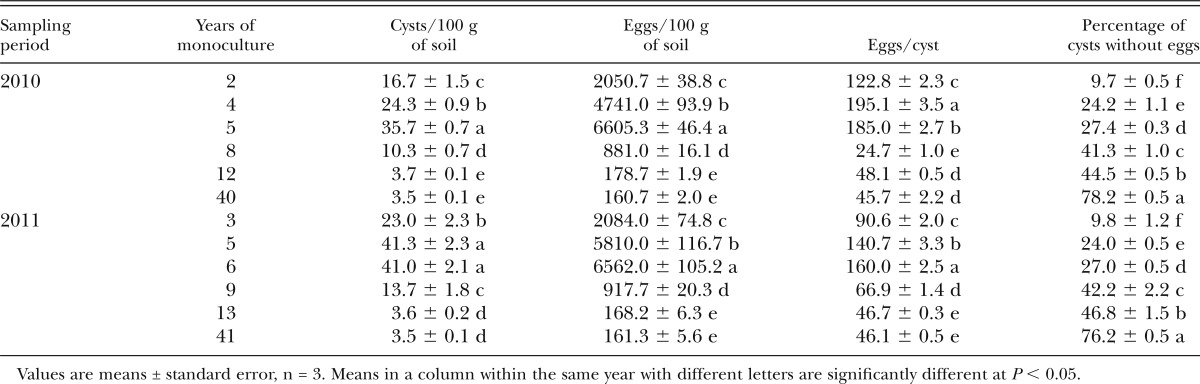
Fig. 1.
Relationship between egg population density (Ln eggs/100-g soil) of Heterodera glycines and number of years of soybean monoculture (Ln yr).
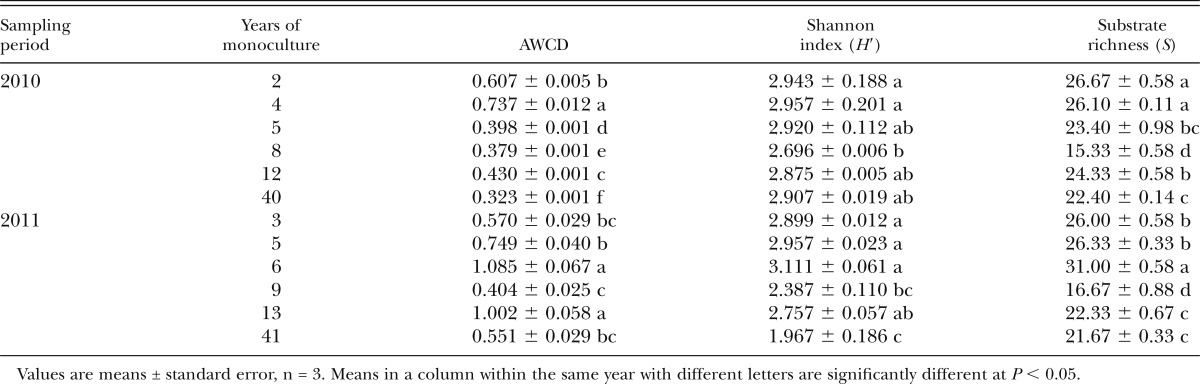
Metabolic activity and population densities of bacteria residing in SCN cysts:
There was no clear pattern of AWCD values among the fields, although differences were observed (Table 2). The lowest substrate richness was observed at 8 and 9 yr of monoculture in the same field, but linear regression analysis indicated no significant relationship of the variable with number of years of soybean monoculture (data not shown). However, linear regression analysis showed that the Shannon index declined with number of years of soybean monoculture (P < 0.05), although significant reduction was observed only in 8 yr in 2010, and 9 and 41 yr of monoculture based on ANOVA (Table 2).
Table 2.
Catabolic diversity (average well-color development (AWCD)), Shannon index, and substrate richness determined with Biolog EcoPlates) of the bacterial communities associated with Heterodera glycines cysts obtained from soybean (Glycine max) fields with different lengths of monoculture.
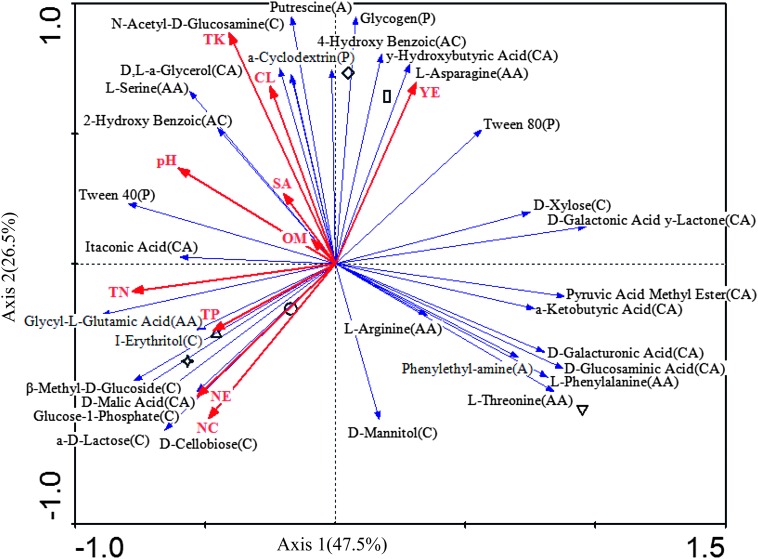
The Biolog EcoPlates contained 31 carbon sources, including nine carboxylic acids, six amino acids, four polymers, two amines, eight carbohydrates, and two aromatic compounds. RDA demonstrated that substrate utilization patterns of the cyst bacterial communities in 2010 differed with number of years of monoculture (Fig. 2). The main carbon substrates utilized by the cyst bacterial communities were carbohydrate (C), carboxylic acid (CA), and polymers (P) in monocultures of 2, 4 and 5 yr, 8 yr, and 12, 40 yr, respectively (Fig. 2). RDA analysis showed that the utilization of L-asparagine, γ-hydroxybutyric, and glycongen were greatly influenced by 12 and 40 yr of monoculture. Utilization of I-erythritol, β-methy-D-glucoside, D-malic acid, glycose-6-phosphate, α-D-lactose, and D-cellobiose were influenced by total phosphorus and nitrogen, cyst number and egg number in 2, 4, and 5 yr of monoculture. Monoculture years also affected the numbers of bacteria associated with cysts as determined by a culture-dependent method. In 2010, the bacterial population density after 2, 4, 5, 8, 12, and 40 yr of monoculture was 1.2 × 104, 7.9 × 104, 8.3 × 104, 0.03 × 104, 9.4 × 104, and 4.0 × 104 CFU/cyst, respectively. A similar pattern was found in 2011 (data not shown).
Fig. 2.
Redundancy analysis (RDA) of metabolic profiles for cyst samples used 31 carbon sources as species and environmental variables in 2010. Blue lines with arrows indicate the types of specific C sources (for capital letters in parentheses: C, carbohydrates; P, polymers; CA, carboxylic acids; AC, aromatic compounds; AA, amino acids and A, amines; the longer the line, the stronger the effect). Red lines with arrows indicate environmental variables: YE, years of soybean monoculture; OM, organic matter; SA, sand; CL, clay; TN, total nitrogen; TP; total P; TK, total K; pH, soil pH; NC, nematode cysts; NE, nematode eggs. Symbols indicate cysts from different years of continuous cropping (▵ 2 yr, ☆ 4 yr, ○ 5 yr, ▽ 8 yr, ◊ 12 yr, □ 40 yr).
Changes in the structure of the cyst bacterial community:
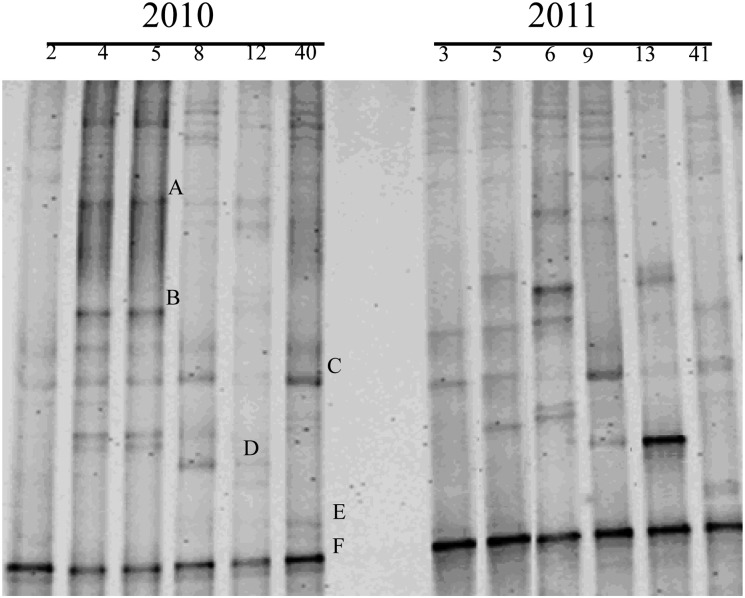
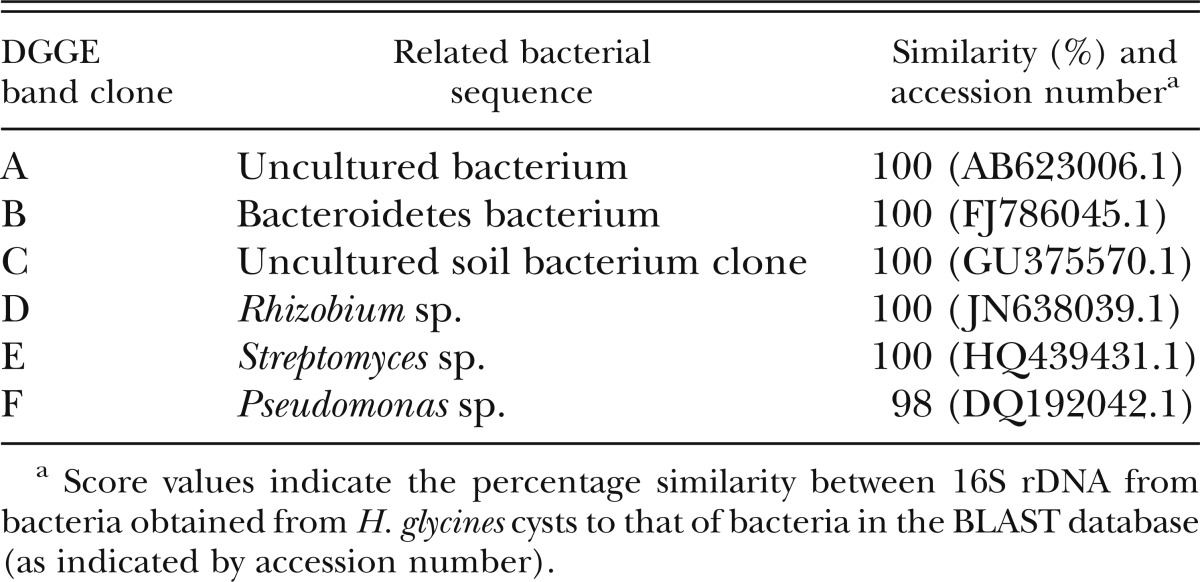
Each distinguishable band in the DGGE pattern represents an individual bacterial taxon generally at genus level. The DGGE patterns from cyst samples were reproducible in that the DGGE banding patterns generated by three samples from each particular field were similar (data not shown). The number of bands per lane ranged from 7 to 12 (Fig. 3). Some bands were common in almost all cyst samples (bands C and F), while others were present in only some of the cyst samples (bands A, B, D, and E). Although every cyst sample had a distinct community profile, and some bands were unique to some samples, the number of DGGE bands tended to decrease as the years of continuous monoculture increased. Moreover, some prominent bands (bands D and E) were found in the cyst samples from fields with ≥ 8 yr of monoculture but not in samples with < 8 yr of monoculture. The major bands were cloned, sequenced, and tentatively identified by comparison with sequences available in GenBank (Table 3). Band D was most closely related to the genus Rhizobium. Band E was related to the genus Streptomyces, and bands B and F to the genus Pseudomonas. Bands A to C represented unidentified bacterium clones.
Fig. 3.
DGGE analysis of DNA isolated from cysts and amplified with universal 16s rRNA gene primers. Each lane represents cysts from a single soybean soil. The bands marked with A to F were cloned and sequenced (Table 3). The numbers indicate number of years of monoculture.
Table 3.
Sequence analysis of bands excised from DGGE gels derived from bacterial 16S rDNA extracted from Heterodera glycines cysts.
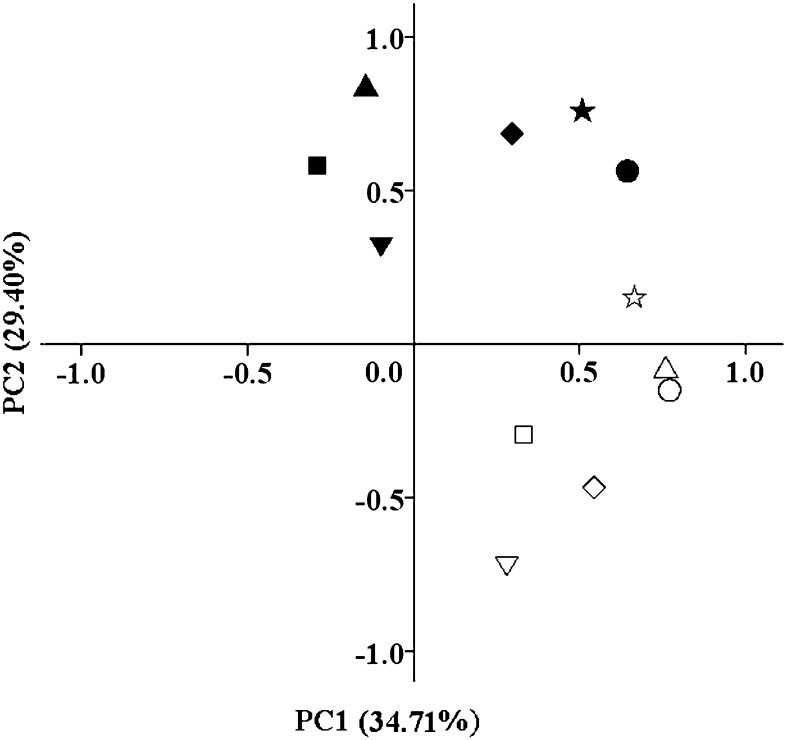
A PCA plot placed the bacteria from different cyst samples into two bacterial community groups (Fig. 4). One group contained samples from fields with 2 to 6 yr of monoculture, and the other group contained samples from fields with 8 to 41 yr of monoculture. When considered along with the data on egg density and carbon substrate utilization, these data on the structure of the cyst bacterial community indicate that the effect of soybean monoculture on SCN and the cyst bacterial community can be divided into two stages: a short-term stage (evident with fewer than 6 yr of monoculture) and a long-term stage (evident with 8 or more yr of monoculture).
Fig. 4.
Principal component analysis (PCA) of DGGE banding patterns of cyst-associated bacteria obtained from cysts in fields subjected to soybean monoculture for different numbers of years (▴ 2 yr, ♦ 4 yr, • 5 yr, ▵ 8 yr, ◊ 12 yr, and ○ 40 yr sampled in 2010; ▪ 3 yr, ▾ 5 yr, ★ 6 yr, ▽ 9 yr, □ 13 yr, and ☆ 41 yr sampled in 2011).
Discussion
Naturally occurring nematode-suppressive soils have been detected throughout the world (Chen and Dickson, 2012). For example, the population densities of cereal cyst nematode, Heterodera avenae, declined under crop monoculture in Europe (Kerry and Crump, 1998). A long-term stationary study in China found that numbers of SCN cysts increased to 351/250 g of soil in the first 5 yr, started to decrease after 7 yr, and then declined to 260/250 g of soil after 14 yr of soybean monoculture (Jin et al., 2006). The present study provides additional evidence that monoculture resulted in decline of SCN population density.
The decline of number of eggs per cyst and increase of empty cysts were observed in this study agreeing with a previous report in which the high percentages of empty cysts were found in a monoculture field where the soil was considered to be suppressive to the SCN (Liu and Wu, 1992). However, the cause of the increased empty cysts in the long-term monoculture fields and its relationship to the SCN egg population density were unclear. It is possible that some microbes can enhance the juvenile hatching from eggs or/and parasitize the eggs within the cysts resulting in more empty cysts in the longer monoculture fields.
The Biolog analysis provided one of the first analyses of bacteria associated with SCN cysts obtained from fields with different years of soybean monoculture. Carbon utilization by the bacterial communities associated with cysts was affected by years of soybean monoculture. The catabolic activity of cyst-associated bacteria was significantly lower for fields with 8 to 9 yr of monoculture than for fields with shorter or longer histories of monoculture, suggesting that 8 to 9 yr of monoculture could be a turning point in bacterial functional diversity associated with SCN cysts. However, further study may be needed to ascertain the observation.
The DGGE band patterns were consistent with the Biolog analysis in showing that bacterial communities associated with cysts changed with years of soybean monoculture. The DGGE data also indicated that Streptomyces spp. and Rhizobium spp. were dominant in the long-term monoculture samples. Streptomyces sp. has previously been isolated from SCN cysts from fields under 7 yr of monoculture of soybean in China (Sun and Liu, 2000). Streptomyces spp. produce many secondary metabolites that have been reported to inhibit egg hatch or root penetration by plant-parasitic nematodes (Kloepper et al., 1992; Dicklow et al., 1993; Esnard et al., 1995; Chen et al., 2009; Jayakumar, 2009), and Rhizobium spp. have been found to suppress SCN juveniles (Zhao et al., 2009; Yin et al., 2010).
In another study, the ability of a soil to suppress cyst nematode (Heterodera schachtii) was transferred to a second soil by transferring cysts from the first soil to the second; this demonstrated that the suppression was caused by microorganisms associated with the cysts in the suppressive soil (Westphal and Becker, 2001). In the present study, both Biolog analysis and DGGE patterns showed clear differences between the cyst-associated bacteria from short-term and long-term soybean monocultures, and some specific bacteria were found in the long-term monoculture soils, which were evidently suppressive based on the small numbers of SCN present. DGGE cannot provide a complete and unbiased fingerprint of the microbial community (Miletto et al., 2007), and its sensitivity is insufficient to detect all microorganisms within cysts (Muyzer, 1999); however, Streptomyces and Rhizobium spp. were determined to be dominant in the cysts of long-term monoculture soils. Therefore, we speculate these specific cyst microbes could potentially affect SCN population density and be responsible for nematode population changes with increasing years of soybean monoculture.
During a study of the soil microbial community in different soybean cropping systems, Li et al. (2010) found that soil quality was better with long-term than with short-term monoculture and that the turning point often occurred after 5 or 6 yr of monoculture. In the current study, SCN numbers increased for the first 5 to 6 yr of monoculture and then declined. During long-term monoculture, plants can modify the rhizosphere microbial community and thereby protect themselves against pathogens (Mendes et al., 2011). However, some microorganisms could benefit parasitic nematodes (Taylor et al., 2005; Zhao, 2008).
Many fungi have been isolated from nematode cysts, and some of them contribute to nematode suppression (Chen and Dickson, 2012). The results of this study show that there is a complex interaction of the bacterial community and SCN under different lengths of soybean monoculture, and that the changes in bacterial community structure (as indicated by substrate utilization data and DGGE profiles) could potentially be involved in the decline of SCN numbers with long-term monoculture of soybean soil. However, the role of these bacterial communities in nematode suppression requires further study.
Literature Cited
- Alkharouf NW, Klink VP, Matthews BF. Identification of Heterodera glycines (soybean cyst nematode [SCN]) cDNA sequences with high identity to those of Caenorhabditis elegans having lethal mutant or RNAi phenotypes. Experimental Parasitology. 2006;115:247–258. doi: 10.1016/j.exppara.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Atibalentja N, Noel GR, Domier LL. Phylogenetic position of the North American isolate of Pasteuria that parasitizes the soybean cyst nematode, Heterodera glycines, as inferred from 16S rDNA sequence analysis. International Journal of Systematic and Evolutionary Microbiology. 2000;50:605–613. doi: 10.1099/00207713-50-2-605. [DOI] [PubMed] [Google Scholar]
- Carris LM, Glawe DA, Smyth CA, Edwards DI. Fungi associated with populations of Heterodera glycines in two Illinois soybean fields. Mycologia. 1989;81:66–75. [Google Scholar]
- Chen LJ, Chen JS, Zheng YN, Dong J, Yu BS, Duan YX. Identification of actinomycetes strain Snea253 and its activity against soybean cyst nematode. Chinese Journal of Biological Control. 2009;25:66–69. [Google Scholar]
- Chen SY. Suppression of Heterodera glycines in soils from fields with long-term soybean monoculture. Biocontrol Science and Technology. 2007;17:125–134. [Google Scholar]
- Chen S, Dickson DW. 2012 Biological control of plant-parasitic nematodes. Pp. 761–811 in R. H. Manzanilla-López and N. Marbán-Mendoza, eds. Practical plant nematology. Guadalajara, Jalisco, Mexico: Colegio de Postgraduados and Mundi-Prensa, Biblioteca Básica de Agricultura. [Google Scholar]
- Derry AM, Staddon WJ, Trevors JT. Functional diversity and community structure of microorganisms in uncontaminated and creosote-contaminated soils as determined by sole-carbon-source utilization. World Journal of Microbiology and Biotechnology. 1998;14:571–578. [Google Scholar]
- Dicklow MB, Acosta N, Zuckerman BM. A novel Streptomyces species for controlling plant-parasitic nematodes. Journal of Chemical Ecology. 1993;19:159–173. doi: 10.1007/BF00993686. [DOI] [PubMed] [Google Scholar]
- Esnard J, Potter TL, Zuckerman BM. Streptomyces costaricanus sp. nov., isolated from nematode-suppressive soil. International Journal of Systematic Bacteriology. 1995;10:775–779. doi: 10.1099/00207713-45-4-775. [DOI] [PubMed] [Google Scholar]
- Garland JL. Analysis and interpretation of community level physiological profiles in microbial ecology. FEMS Microbiology Ecology. 1997;24:289–300. [Google Scholar]
- Garland JL, Mills AL. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Applied and Environmental Microbiology. 1991;57:2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MH, Haynes RJ. Catabolic diversity of soil microbial communities under sugarcane and other land uses estimated by Biolog and substrate-induced respiration methods. Applied Soil Ecology. 2005;29:155–164. [Google Scholar]
- Jayakumar J. Streptomyces avermitilis as a biopesticide for the management of root knot nematode. Karnataka Journal of Agricultural Sciences. 2009;22:564–566. [Google Scholar]
- Jin XH, Xin HP, Zheng W, Tai LM, Zhang YL, Yan FY. The influence of the long-term rotation and continuous cultivation on soybean cyst nematode in soil. Chinese Journal of Oil Crop Sciences. 2006;28:189–193. [Google Scholar]
- Kerry BR, Crump DH. The dynamics of the decline of the cereal cyst nematode, Heterodera avenae, in four soils under intensive cereal production. Fundamental and Applied Nematology. 1998;21:617–625. [Google Scholar]
- Kidane E, Hu W, Chen S, Liu X, Neher DA. Ecology of soils suppressive to the soybean cyst nematode: II. Effect of tillage and crop-biocide treatments on nematophagous fungi. Journal of Nematology. 2012;44:472. (Abstr.). [Google Scholar]
- Kloepper JW, Rodríguez-Kábana R, McInroy JA, Young RW. Rhizosphere bacteria antagonistic to soybean cyst (Heterodera glycines) and root-knot (Meloidogyne incognita) nematodes: Identification by fatty acid analysis and frequency of biological control activity. Plant Soil. 1992;139:75–84. [Google Scholar]
- Li CG, Li XM, Kong WD, Wu Y, Wang JG. Effect of monoculture soybean on soil microbial community in the Northeast China. Plant Soil. 2010;330:423–433. [Google Scholar]
- Liu XZ, Chen SY. Screening isolates of Hirsutella species for biocontrol of Heterodera glycines. Biocontrol Science and Technology. 2001;11:151–160. [Google Scholar]
- Liu XZ, Wu XY. 1992 Decline of soybean cyst nematode: A preliminary result. Pp. 61–69 in Proceedings of 2nd International Workshop on Plant Nematology, University of Karachi, Pakistan. [Google Scholar]
- Ma R, Liu XZ, Jian H, Li SD. Detection of Hirsutella spp. and Pasteuria sp. parasitizing second-stage juveniles of Heterodera glycines in soybean fields in China. Biological Control. 2005;33:223–229. [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, Raaijmakers JM. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- Miletto M, Bodelier PLE, Laanbroek HJ. Improved PCR-DGGE for high resolution diversity screening of complex sulfate-reducing prokaryotic communities in soils and sediments. Journal of Microbiological Methods. 2007;70:103–111. doi: 10.1016/j.mimet.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Muyzer G. DGGE/TGGE a method for identifying genes from natural ecosystems. Current Opinion in Microbiology. 1999;2:317–322. doi: 10.1016/S1369-5274(99)80055-1. [DOI] [PubMed] [Google Scholar]
- Muyzer G, De Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Applied and Environmental Microbiology. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour SM, Lawrence JR, Zhu H, Swerhone GDW, Welsh M, Welacky TW, Topp E. Bacteria associated with cysts of the soybean cyst nematode (Heterodera glycines) Applied and Environmental Microbiology. 2003;69:607–615. doi: 10.1128/AEM.69.1.607-615.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman CH, Bird DM. The soybean cyst nematode, Heterodera glycines: A genetic model system for the study of plant-parasitic nematodes. Current Opinion in Plant Biology. 1998;1:342–346. doi: 10.1016/1369-5266(88)80057-8. [DOI] [PubMed] [Google Scholar]
- Sekiguchi H, Tomioka N, Nakahara T, Uchiyama H. A single band does not always represent single bacterial strains in denaturing gradient gel electrophoresis analysis. Biotechnology Letters. 2001;23:1205–1208. [Google Scholar]
- Sun MH, Liu XZ. Suppressive soils of soybean cyst nematode in China. Acta Phytopathologica Sinica. 2000;30:353–356. [Google Scholar]
- Taylor MJ, Bandi C, Hoerauf A. Wolbachia bacterial endosymbionts of filarial nematodes. Advances in Parasitology. 2005;60:245–284. doi: 10.1016/S0065-308X(05)60004-8. [DOI] [PubMed] [Google Scholar]
- Westphal A, Becker JO. Components of soil suppressiveness against Heterodera schachtii. Soil Biology and Biochemistry. 2001;33:9–16. [Google Scholar]
- Xiang MC, Xiang PA, Jiang XZ, Duan WJ, Liu XZ. Detection and quantification of the nematophagous fungus Hirsutella minnesotensis in soil with real-time PCR. Applied Soil Ecology. 2010;44:170–175. [Google Scholar]
- Yin B, Valinsky L, Gao XB, Becker JO, Borneman J. Bacterial rRNA genes associated with soil suppressiveness against the plant-parasitic Nematode Heterodera schachtii. Applied and Environmental Microbiology. 2003;69:1573–1580. doi: 10.1128/AEM.69.3.1573-1580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin LN, Duan YX, Wang YY, Luo ZQ, Chen LJ. Screening of Rhizobia against soybean cyst nematode. Soybean Science. 2010;29:276–279. [Google Scholar]
- Zak J, Willig M, Moorhead D, Wildman H. Functional diversity of microbial communities: A quantitative approach. Soil Biology and Biochemistry. 1994;26:1101–1108. [Google Scholar]
- Zhao BG. 2008 Bacteria carried by the pine wood nematode and their symbiotic relationship with the nematode. Pp. 264–273 in B.G. Zhao, K. Futai, J.R. Sutherland, and Y. Takeuch, eds. Pine wilt disease. Berlin, New York: Springer. [Google Scholar]
- Zhao YS, Duan YX, Wang YY, Chen LJ, Yin LN. Resistance and biocontrol potential of soybean Rhizobia resources isolated from Liaoning Province. Soybean Science. 2009;28:113–117. [Google Scholar]
- Zheng JW, Chen SY. Estimation of virulence type and level of soybean cyst nematode field populations in response to resistant cultivars. Journal of Entomology and Nematology. 2011;3:37–43. [Google Scholar]