Abstract
The control of muscle-specific expression is one of the principal mechanisms by which diversity is generated among muscle types. In an attempt to elucidate the regulatory mechanisms that control fiber diversity in any given muscle, we have focused our attention on the transcriptional regulation of the Drosophila Troponin T gene. Two, nonredundant, functionally identical, enhancer-like elements activate Troponin T transcription independently in all major muscles of the embryo and larvae as well as in adult somatic and visceral muscles. Here, we propose that the differential but concerted interaction of these two elements underlies the mechanism by which a particular muscle-type establish the correct levels of Troponin T expression, adapting these levels to their specific needs. This mechanism is not exclusive to the Troponin T gene, but is also relevant to the muscle-specific Troponin I gene. In conjunction with in vivo transgenic studies, an in silico analysis of the Troponin T enhancer-like sequences revealed that both these elements are organized in a modular manner. Extending this analysis to the Troponin I and Tropomyosin regulatory elements, the two other components of the muscle-regulatory complex, we have discovered a similar modular organization of phylogenetically conserved domains.
INTRODUCTION
The differentiation of the distinct types of muscle is a complex, multistep developmental process involving multiple gene regulatory mechanisms (Stockdale, 1997; Hughes and Salinas, 1999; McKinsey et al., 2002). In this process a large battery of genes encoding muscle-specific proteins are rapidly activated (Buckingham et al., 1992). The differential accumulation of these proteins in each muscle type, according to their specific contractile properties and functions (i.e., the rate of force generation, the relaxation rate and the fatigability of the myofibers) contributes to the generation of muscle diversity (Bernstein et al., 1993). Two of the most important aspects of this process are 1) the possibility of achieving very high levels of protein expression in a very short period of time and 2) the pattern of protein isoforms expressed that, together with the modulation of the quantities of protein in each individual fiber, serves to establish the correct stoichiometry necessary to maintain myofibril integrity and for the proper function of each particular fiber (Buckingham et al., 1992; Bernstein et al., 1993). As such, transcriptional regulation is a major mechanism for the generation of muscle diversity. This elaborated process requires specific interactions between genomic sequences and components of the transcriptional machinery, resulting in local structural changes of the chromatin (Davidson et al., 2002).
A number of genes encoding muscle-specific proteins have been shown to be regulated by two or more enhancer-like elements (Karlik and Fyrberg, 1986; Hanke and Storti, 1988; Gremke et al., 1993; Buonanno and Rosenthal, 1996; Schiaffino and Reggiani, 1996; Arnone and Davidson, 1997; Hughes and Salinas, 1999). Indeed, short-range enhancers or enhancer-like elements are often situated within a gene locus (Buonanno and Rosenthal, 1996; Schiaffino and Reggiani, 1996; Arnone and Davidson, 1997; Hughes and Salinas, 1999; Markstein and Levine, 2002) and in many of these genes, positive regulatory elements have been found within the first intron (Hess et al., 1989; Banerjee-Basu and Buonanno, 1993; Gremke et al., 1993; Meredith and Storti, 1993; Calvo et al., 2001; Hallauer and Hastings, 2002). The exact functional role of these enhancer elements has remained unclear although there is mounting evidence that they are involved in the control of gene expression at specific stages of development or in a specific set of cells and in some cases in achieving higher levels of gene expression (Blackwood and Kadonaga, 1998). In this context, it is important to understand why a gene needs two or more enhancer-like elements that often seem to perform the same function. However, the complexity of the mouse model coupled with the simplicity of cell culture systems to analyze the precise functions of these enhancers makes it difficult to obtain a clear answer to this question. In contrast to mammals, few specialized muscle types are generated in Drosophila and each muscle type is composed of only one fiber type (Bate, 1990; Baylies et al., 1998). Nevertheless, flies and vertebrates mostly share the evolutionary conserved molecular pathways controlling muscle formation. This makes Drosophila a good model system in which to study the processes that leads to the specification of muscles.
Troponin T, in combination with Troponin C, Troponin I, and Tropomyosin, participates in a complex that is involved in the regulation of calcium-mediated muscle contraction (Perry, 1998). The thin-filament contractile protein Troponin T appears to be the most important regulatory protein of the sarcomere, a key regulator of cardiac contraction (Javadpour et al., 2003; Schwartz and Mercadier, 2003). Interest in Troponin T is further enhanced by the fact that mutations of the TnT gene (TNNT2), encoding cardiac Troponin T, are responsible for 15% of all cases of familial hypertrophic cardiomyopathies (Thierfelder et al., 1994; Watkins et al., 1995; Maron et al., 1999; Roberts and Sigwart, 2001a, 2001b). The mutant proteins are thought to act as dominant-negative isoforms that impair the function of heart muscle (Roberts and Sigwart, 2001a, 2001b; Seidman and Seidman, 2001). Moreover, changes in cardiac TnT isoform expression have been correlated with heart disease (Perry, 1998; Roberts and Sigwart, 2001a, 2001b).
The Drosophila Troponin T gene is one of a group of more than 15 genes encoding contractile proteins that are coordinately activated during myogenesis. The gene contains an untranslated exon followed by a relatively large intron (Benoist et al., 1998), an organization that is shared by a number of genes encoding contractile proteins (Karlik and Fyrberg, 1986; Konieczny and Emerson, 1987; Bernstein et al., 1993; Hallauer et al., 1993). In Drosophila, four Troponin T isoforms are generated by developmentally regulated alternative splicing, these being specifically expressed in larval hypodermic, adult hypodermic, visceral, and heart (dorsal vessel) muscle, and in jump (TDT-tergal depressor of the trochanter) and flight (IFM-indirect flight muscles) muscles (Benoist et al., 1998).
The detailed characterization of cell-specific enhancers in transgenic embryos is a laborious process and as a consequence, comparatively few enhancers have been examined in this way. Although several computational approaches have been developed to identify cis-regulatory motifs and regions in silico, until recently very few of these predictions have been tested in multicellular animals (Frith et al., 2001; Pennachhio and Rubin, 2001). Here, we have analyzed the regulation of the Troponin T gene by linking a reporter gene to distinct genomic sequences and analyzing gene expression in vivo in germ line transformants of Drosophila. Our studies have revealed that the complex spatiotemporal pattern of Drosophila Troponin T expression depends on two nonredundant, functionally equivalent, enhancer-like elements found within the gene locus named the upstream regulatory element (URE) and intronic regulatory element (IRE). Cooperation between these two elements is essential to ensure the correct expression of the TnT gene. We demonstrate that the quantities of protein found in each particular muscle are established through a concerted interaction between both elements. Our data reveal a modular organization of these enhancer-like elements and we found two discrete, yet strongly conserved regions, that are transcriptionally active within each element. Within the URE, a proximal region governs expression in all the major muscles in embryos, larvae, and adult. However, the expression in flight and jump muscles, the specialized thoracic muscles, is directed from a distal region. This region also functions as an enhancer-like sequence in all muscle-types, directing robust expression. Furthermore, within the IRE, two distinct regulatory modules were found that independently control Troponin T gene expression.
MATERIALS AND METHODS
Isolation of Genomic Clones, Construction of P-transformation Plasmids, and Generation of Transformed Drosophila Lines
The Drosophila melanogaster and D. virilis genomic clones, containing both the 5′ region upstream from the transcription initiation sites and intron 1 of the TnT gene, were subcloned and sequenced as described (Benoist et al., 1998). The D. virilis genomic clones available contain only -2.1 kb upstream from the transcription initiation site of TnT gene. The genomic D. pseudoobscura sequence database was accessed through Drosophila Genome Project (http://www.hgsc.bcm.tmc.edu/projects/drosophila). Distinct fragments of these regions were cloned into P-transformation vectors respecting their native orientation relative to the basal promoters. The main transcription initiation starting point of the TnT gene in previously identified clones is referred to as +1 bp on our map (Benoist et al., 1998). Construct TnT URE/IRE contains 3.5 kb of the 5′upstream region, exon 1, intron 1, and part of exon 2 (until ATG). Construct TnT IRE contains 110 bp of the 5′upstream region, exon 1, intron 1, and part of exon 2 (until the ATG). Constructs TnT URE, TnT URE 2.5, TnT URE 1.4, TnT URE 1.1, TnT URE 0.8, and TnT URE 0.6 contain 3.5, 2.5, 1.4, 1.1, 0.8, and 0.6 kb of the 5′upstream region, respectively, exon 1 and part of exon 2, but do not contain intron 1. It was noteworthy that TnT URE and TnT URE 2.5 transgenic flies gave the same pattern of muscle transgene expression in both β-galactosidase staining and Northern blot analysis (unpublished data). Constructs TnT IREΔI, TnT IREΔII, and TnT IREΔ(I+II) contain 110 bp of the 5′upstream region, exon 1, the intronic fragment analyzed, and part of exon 2. These constructs try to maintain the internal organization of the endogenous TnT gene. In TnT IREΔI, TnT IREΔII, and TnT IREΔ(I+II), modules I, II, or both were deleted from intron 1. Module I comprises the sequence between +396 and +878 bp, and module II comprises the sequence between +1531 and +2241 bp. Construct TnT -/- contains 110 bp of the 5′upstream region, exon 1, and part of exon 2. Furthermore, constructs TnT pelican IREΔI and TnT pelican IREΔII contain the IRE element, except module I or module II, respectively, linked to a heterologous strong promoter, hsp70. Thus, the internal organization of the gene is not maintained. All of these constructs are in-phase with the β-galactosidase/reporter gene. Appropriate restriction enzyme sites were used to insert the fragments into the plasmids. All constructs were sequenced and cloned into pCaSpeR β-gal (Thummel et al., 1988), and one copy of the constructs was inserted into the lines analyzed. The transformation vector used for TnT pelican IREΔI and TnTpelican IREΔII was pH-Pelican (Barolo et al., 2000). Three or four transformant lines were analyzed for each construct. The generation of germ line transgenic flies using the P element-mediated transformation technique was essentially as earlier described (Spradling and Rubin, 1982).
Immunostaining of Embryos
Embryos were collected, dechorionized, and stained with anti-β-galactosidase (Cappel, Cochranville, PA) diluted 1:2000 and biotinylated goat anti-rabbit IgG diluted 1:400. The color reaction was developed with Vectasin ABC Kit (Vector Laboratories).
Embryo, Larvae, and Adult Staining
β-galactosidase enzyme activity was assayed in the larvae and adults of transgenic lines using standard protocols with minor modifications (Sullivan et al., 2000; Arredondo et al., 2001). At least, three different lines were analyzed in each case. Third instar larvae and newly emerged flies were microdissected, fixed, and stained as described previously (Meredith and Storti, 1993). The levels of β-galactosidase activity were used as a means of comparing the transcriptional efficiency between different constructs. In larval and adult muscles, a rough quantification of the expression levels between different constructs was achieved by visually monitoring the time required for the blue reaction products from the X-gal substrate/β-galactosidase reaction to appear. The complete expression patterns were visualized after 2 h of staining in larvae and microdissected abdomen and after 45 min in thoracic sections, if not otherwise specified.
Reverse Transcriptase Polymerase Chain Reactions and Sequencing
Reverse transcriptase polymerase chain reactions (RT-PCR) using RNA from TnT -/--transformed lines was performed according to standard protocols (Sambrook and Russell, 2001). Oligonucleotides covering TnT exon 1 and different sequences of the LacZ gene were used as PCR primers. Products were analyzed on 2% agarose gels and where necessary, bands were excised, cloned in pGEMT, and sequenced using Sp6 and T7 primers as described (Sanger et al., 1977).
RNA Purification and Northern Hybridization
Total RNA from late pupa and 16-24-h-old embryos was purified, and Northern blots were performed as described previously (Sambrook and Russell, 2001). Specific probes against β-galactosidase, TnT, or triose phosphate isomerase (TPI) were used to hybridize blots. β-galactosidase mRNA levels were determined and standardized against endogenous TnT or TPI mRNA levels through densitometric analysis of the x-ray films using the NIH image 1.6.1 program and Quantity One Program (Bio-Rad, Richmond, CA).
SDS-Polyacrylamide Gels and Immunoblot Analysis
Microdissected IFM, TDT muscles, and abdomens were homogenized in Laemmli buffer (Maroto et al., 1996), separated by electrophoresis, and immunoblotted as described previously (Benoist et al., 1998). The amount of protein loaded in each lane was ∼10 μg. A rabbit antiserum against TnT prepared in the laboratory against the purified D. melanogaster TnT protein (Domingo et al., 1998) and a commercial rabbit antiserum against β-galactosidase (Cappel, Cochranville, PA) were used to probe the blots. Immunostaining was visualized using the ECL system (Amersham Corp., Little Chalfont, United Kingdom).
Genomic Analyses
Sequence comparison between Drosophila species and searches for transcription factor binding sites in short genomic sequences near the transcription initiation site of the genes was performed with the Gene Jockey II program (Biosoft, Cambridge, United Kingdom). Consensus sequences used to search for binding sites were as follows: C/TTAWWWWTAA/G or CCAAAAATAA/G for MEF2 (Taylor et al., 1995), TCAAGTG for TINMAN (Gajewski et al., 1997), A/GTATATA/GTA for CF2 (Transfac databases at www.gene-regulation.com), and AAAA/GTGTTA/GTAA/T for PDP1 (Reddy et al., 2000). The genomic sequences around the TnT gene and the relative positions of the distinct genes were obtained from the GenBank at NCBI and FlyBase (The FlyBase Consortium, 2003). The search for MEF2, TINMAN, PDP1, and CF2 binding sites in large genomic sequences was performed using the DNA-pattern program at regulatory sequence analysis tools (www.rsat.ulb.ac.be/rsat) and CIS-ANALYST tool (http://rana.lbl.gov/cis-analyst; Berman et al., 2002).
Nucleotide Sequence Accession Numbers
The complete 5′ upstream and first intron sequences of the TnT gene from D. melanogaster and D. virilis have been submitted to GenBankTM/EMBL Data Bank with accession numbers AJ002261-AJ002262 and AJ002263-AJ002264, respectively.
RESULTS
Two Distinct Muscle Enhancer-like Elements Independently Promote TnT Expression in All Muscle Types
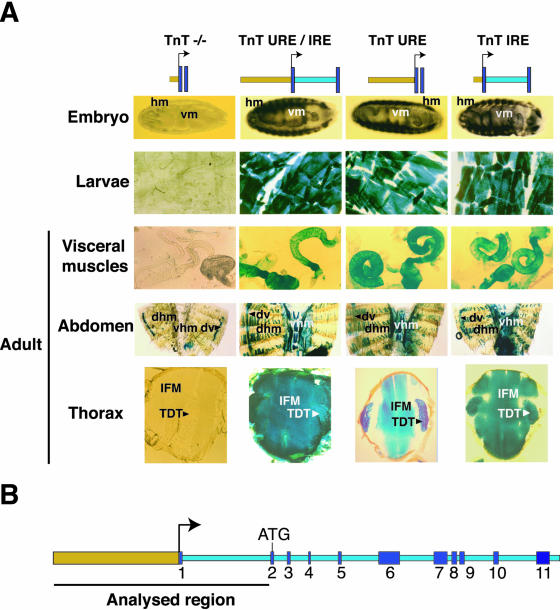
To define the minimal sequence required for muscle-specific transcription of the Troponin T gene, we generated transgenic lines with four genomic fragments. These four constructs differed in the presence or absence of the 5′upstream region or intron 1. Nevertheless, each construct maintained the endogenous structural organization of the gene with regards the relative positions of the TnT upstream sequences, intron, and transcription initiation site (see MATERIAL AND METHODS). The constructs were named TnT URE/IRE, TnT URE, TnT IRE, and TnT -/-, according to the presence or absence of the upstream (URE) or downstream (IRE) regions. The expression levels of the different constructs were tested after 2 h (Figure 1 and Table 1). The TnT -/- transgene did not drive reporter gene expression in larvae or adults. In stark contrast, flies harboring the TnT URE/IRE, TnT URE, or TnT IRE constructs showed high levels of transgene expression in all of the major muscle groups of the embryo and larva, as well as in adults. Indeed, the temporal and spatial expression patterns were similar to those obtained with the endogenous TnT protein (Fyrberg et al., 1990; Benoist et al., 1998). The only difference observed was in flight muscles where flies containing the TnT URE construct displayed less activity than the other constructs, TnT URE/IRE and TnT IRE (Figure 1, thorax panels, and Table 1).
Figure 1.
Upstream and downstream intron sequences independently drive TnT expression in all muscle types. (A) The β-galactosidase staining of embryos, third instar larvae, visceral muscles, dissected abdomens, and thin sections of the thorax transformed with distinct constructs. Constructs inserted in the pCaSper β-gal plasmid are identified by name. TnT URE/IRE contains 3.5 kb of the 5′ upstream region and intron 1; TnT URE contains 3.5 kb of the 5′ upstream region but not contain intron 1; TnT IRE contains 110 bp of the 5′upstream region and intron 1; and TnT -/- contains 110 bp of the 5′upstream region without intron 1. IFM, indirect flight muscles; TDT, tergal depressor of the trochanter; dhm, dorsal hypodermic muscles; vhm, ventral hypodermic muscles; dv, dorsal vessel. The staining patterns were visualized after 2 h, except for in thoracic cryosections that were visualized at 45 min.(B) A schematic representation of the D. melanogaster TnT gene and the region analyzed. Exons are shown as dark blue boxes and are specified by number.
Table 1.
β-galactosidase expression in URE and IRE lines
| Thorax
|
|||||||
|---|---|---|---|---|---|---|---|
| Analyzed lines | IFM | TDT | Abdomen | Visceral muscles | Larvae | Dorsal vessel | |
| TnT URE/IRE | 3 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
| TnT URE | 4 | + | ++ | ++ | ++ | ++ | ++ |
| TnT IRE | 3 | ++++ | +++ | +++ | ++ | ++ | ++ |
| TnT −/− | 3 | − | − | − | − | − | − |
Comparison of β-galactosidase reporter expression driven by selected sequences upstream of the transcription start site and intron 1 of the TnT transcription unit. The highest expressing line, TnT URE/IRE, produced the maximum intensity of staining for most muscles within approximately 1 h and was referred to as “+++++.” The level of intensity of the other lines, TnT URE, TnT URE, and TnT IRE was determined by comparison with this line.
Interestingly, these results suggested that both regulatory elements have equivalent roles in controlling TnT gene transcription, in all muscles except for flight muscles. Thus, we also cloned the IRE fragment upstream of a heterologous strong promoter (MATERIAL AND METHODS). Flies harboring this construct expressed very high levels of the transgene in all muscles. Thus, the TnT IRE fragment was able to confer muscle specificity irrespective of its position and spatial organization (unpublished data). These observations indicate that two distinct enhancer-like elements, URE and IRE, were independently capable of reproducing the spatiotemporal expression of TnT in a manner indistinguishable from the endogenous gene.
Cooperation between the URE and IRE Elements Is Required for Strong Muscle Transcription in Larval Muscles
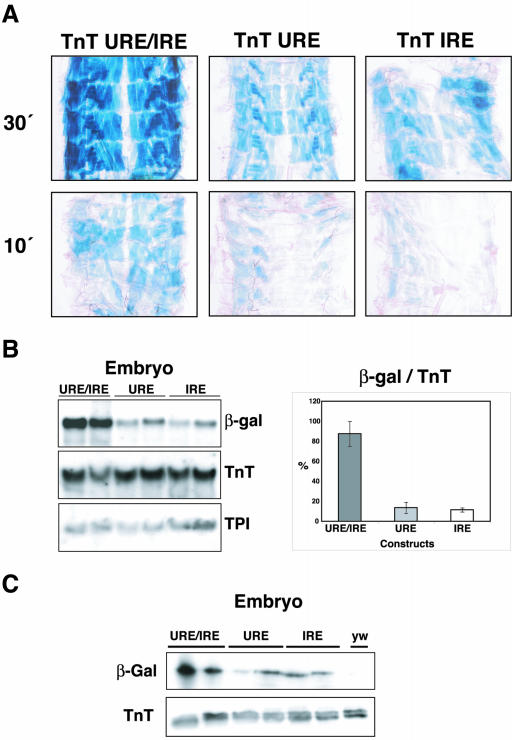
To detect variations in the levels of expression between the TnT URE/IRE, TnT URE, and TnT IRE lines, we monitored the appearance of the β-galactosidase reaction products within the linear phase of the assay, i.e., at 10 and 30 min. In body wall muscle of larvae under these conditions, the highest levels of expression were found in the lines carrying the reporter gene driven by both the URE and IRE (TnT URE/IRE). Lower levels of expression were seen in larval muscles carrying the reporter gene driven by either the URE or IRE than those detected in TnT URE/IRE lines (Figure 2A).
Figure 2.
The URE and IRE elements interact synergistically to produce higher levels of transcription in larval muscles. (A) The β-galactosidase staining of 3rd instar larvae transformed with TnT URE/IRE, TnT URE, and TnT IRE constructs. TnT URE/IRE construct carries the URE and IRE elements, and TnT URE and TnT IRE carry, either URE or IRE elements, respectively. A representative line of the three different lines analyzed is presented in each case. The measurement of β-galactosidase activity was performed after short reaction times (10-30 min). (B) Northern blot analysis of the total RNA from late embryos (16-24 h) of the TnT URE/IRE, TnT URE, and TnT IRE transgenic lines from two representative lines in each case (the lines with highest and lowest activity). On the right, the β-galactosidase mRNA data from the three lines analyzed are standardized to endogenous Troponin T. (C) Western blot analysis with β-galactosidase- and TnT-specific antibodies from two lines of the TnT URE/IRE, TnT URE, and TnT IRE transgenic lines.
The accumulation of RNA transcripts and protein in these lines was measured relative to the levels of endogenous Troponin T expression in late embryos (16-24 h) and first instar larvae (MATERIAL AND METHODS). As described previously, there is no posttranscriptional control of TnT expression; thus, TnT transcription and translation are closely coupled (Fyrberg et al., 1990; Benoist et al., 1998; Domingo et al., 1998). In both Northern and Western blots, the contribution of URE and IRE elements is approximately equal in both embryonic and larval musculature (Figure 2, B and C). Similar amounts of transgene transcripts and protein accumulated in larval muscles from the TnT URE and TnT IRE lines. However, synergism can be seen when the two elements work together in the TnT URE/IRE larvae where a four- to fivefold increase in expression was observed.
The contribution of URE and IRE Elements to Achieve Maximum Transgene Expression Varies between Adult Muscle Types
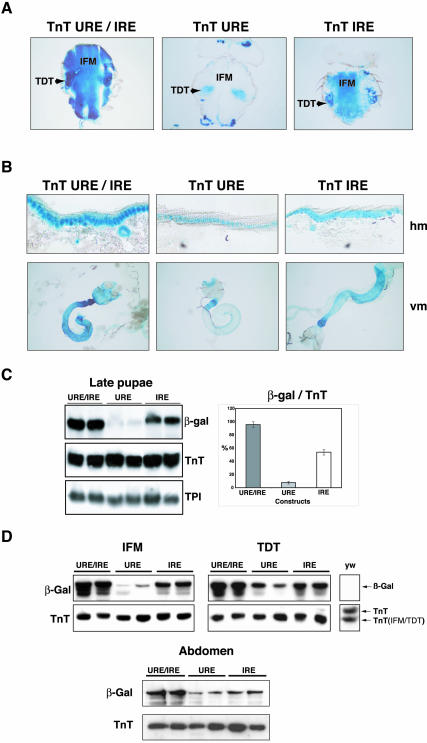
To quantify the levels of expression in specific adult muscle types that differ in their contractile properties, we examined the amount of protein produced by these lines in dissected flight muscles (IFM), dissected jump muscles (TDT), and abdominal muscles (the hypodermic and visceral muscles are the main muscle-types in the abdomen). As seen in larvae, the adult TnT URE/IRE muscles drove the highest levels of transgene expression and the absence of either the URE or IRE elements significantly diminished expression in the four muscle types analyzed (Figure 3). In the IFM muscles, a clear decrease was observed in TnT URE lines but transgene expression was not completely abolished (Figures 3D and 1A, thorax panels).
Figure 3.
The cooperative role of URE and IRE elements in the major adult muscle groups. Staining of a β-galactosidase reporter of four adult muscle types from TnT URE/IRE, TnT URE, and TnT IRE transformed lines is shown. A representative line of the three analyzed is presented in each case. β-galactosidase reaction time was 30 min. (A) β-galactosidase reporter staining in IFM and TDT thin sections. IFM transgene expression in TnT URE is not completely abolished. (B) β-galactosidase reporter staining in hypodermic and visceral muscles. (C) Northern blot analysis of total RNA from late pupae of the TnT URE/IRE, TnT URE, and TnT IRE transgenic lines. Two lines with each construct are shown. On the right, the β-galactosidase mRNA data are standardized against endogenous Troponin T, three lines were analyzed for each construct. (D) Western blot analysis with extracts from dissected IFM, TDT, and abdomen samples with β-galactosidase- and TnT-specific antibodies from two representative lines of the TnT URE/IRE, TnT URE, and TnT IRE lines are shown. Arrows indicate the localization of the β-galactosidase- and TnT-specific isoforms.
In contrast to larvae, the cooperation between the URE and IRE is distinct in the adult musculature (Figures 3, C and D), where the contribution of the IRE element in flight muscles, a fibrillar muscle-type, is nine times higher than that of the URE. Indeed, the protein content strongly correlates with the mRNA levels seen in Northern blots on total RNA from pupae (Figure 3D), confirming the high contribution of the flight muscles to the total muscle mass in Drosophila. In contrast, the contribution of the IRE is about two to three times higher in the synchronous tubular jump muscles, whereas in the hypodermic and visceral muscles of which the abdomen is mainly composed, the IRE contribution is ∼1.5-fold that of the URE. Moreover, the synergistic effect of the two elements observed in adult muscles is less than that seen in larval muscles.
A search for clusters of myogenic motifs in the genomic region of the TnT gene on the X chromosome using a regulatory sequence analysis tool revealed important clusters containing binding sites for myogenic factors upstream and downstream of the TnT transcription start site (see supplemental material). Indeed, DNase I digestion patterns confirmed that the two regulatory elements are simultaneously active in transcriptionally active 15-24 h embryos (see supplemental material).
Modular Organization of the URE Element: Two Conserved Regions Are Involved in TnT Gene Expression
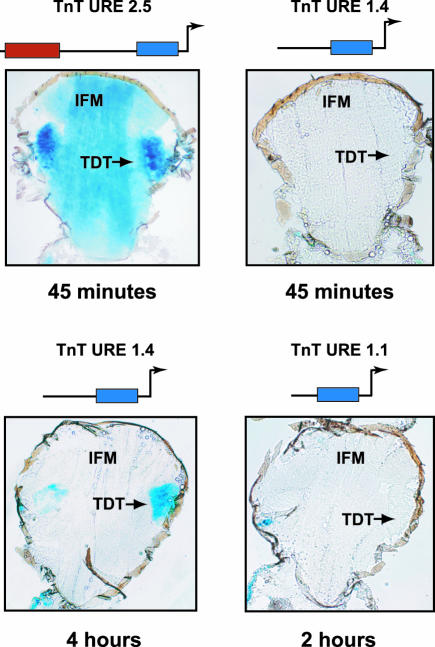
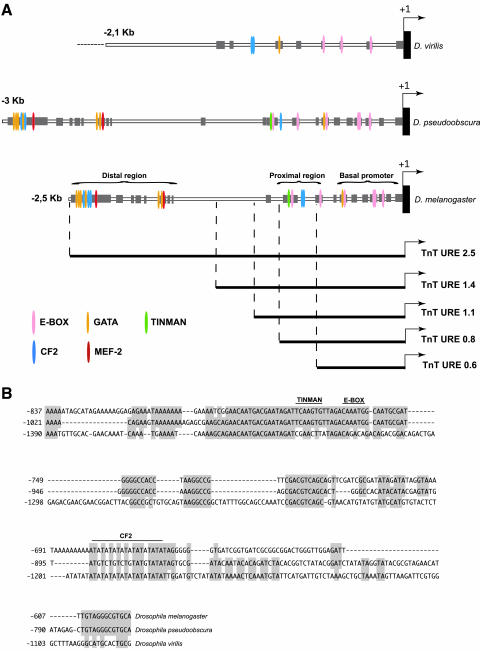
To identify the discrete sequences responsible for the muscle-type-specific TnT gene expression in the URE enhancer-like element, the region was further subdivided into smaller fragments that were tested for reporter activity. The resulting expression patterns are summarized in Figures 4, 5, and 6 and in Table 2. In TnT URE 2.5 flies, significant levels of β-galactosidase expression were observed in all muscles (Figures 4, 5, 6). In contrast, TnT URE 1.4 flies expressed the transgene in all larval and adult muscles, except in the flight and jump muscles (Figures 4, 5, 6). When dissected thoraces were stained longer (4 h), transgene expression driven by TnT URE 1.4 was still not detected in the flight muscles, although weak expression was observed in the jump muscles (bottom left panel in Figure 4). Two putative conserved MEF2 binding sites are located between -2.5 and -1.4 kb of the upstream sequence. Electrophoretic mobility shift assays using in vitro-transcribed/translated MEF-2, combined with antibody supershifts, indicate that the MEF2 gene product binds specifically to these sequences (unpublished data).
Figure 4.
Elements controlling expression in specialized adult muscles. A distal region situated in between -2.5 and -1.4 kb, upstream of the transcription start site of TnT gene is involved in controlling transgene expression in the adult specialized muscles. β-galactosidase staining of thin sections of thoraxes transformed with distinct constructs containing 2.5 kb (TnT URE 2.5), 1.4 kb (TnT URE 1.4), and 1.1 kb (TnT URE 1.1) of the 5′ upstream region linked to the β-galactosidase gene. IFM, indirect flight muscles and TDT, tergal depressor of the trochanter. The reaction time for the β-galactosidase staining is indicated in the lower part of each panel.
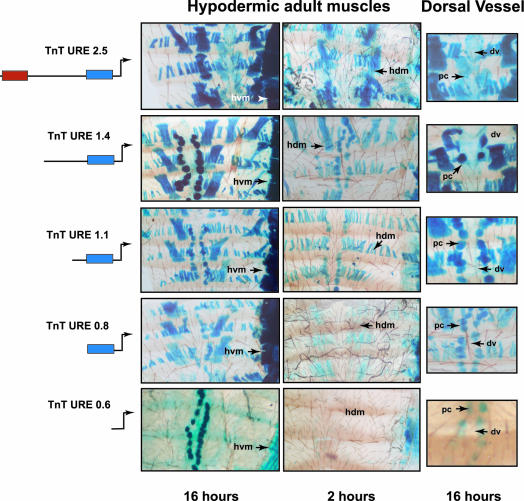
Figure 5.
Elements controlling expression in adult hypodermic muscles and the heart. The proximal region (-0.8 and -0.6 Kb upstream of the transcription start site of TnT gene) may be implicated in the control of transgene expression in adult hypodermic muscles and the heart. β-galactosidase staining of dissected abdomens and dorsal vessels in TnT URE 2.5, TnT URE 1.4, TnT URE 1.1, TnT URE 0.8, and TnT URE 0.6 transformed flies. β-galactosidase reaction time is indicated in the lower part of the panels. dv, dorsal vessel (heart); pc, pericardial cells; hvm, hypodermic ventral muscles; and hdm, hypodermic dorsal muscles. The strong staining in the pericardial cells in all lines is nonspecific.
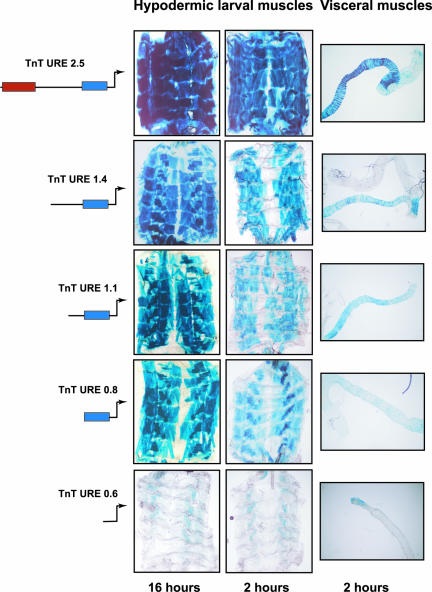
Figure 6.
The distal region is important to maintain the levels of transgene expression in larval hypodermic and visceral muscles. β-galactosidase staining of dissected larvae and visceral muscles in TnT URE 2.5, TnT URE 1.4, TnT URE 1.1, TnT URE 0.8, and TnT URE 0.6 transformed flies. β-galactosidase reaction time is indicated in the lower part of the panels.
Table 2.
β-galactosidase reporter expression
| Thorax
|
Abdomen
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Analyzed lines | IFM | TDT | Dorsal | Ventral | Visceral muscles | Larvae | Dorsal vessel | |
| TnT URE 2.5 | 3 | ++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| TnT URE 1.4 | 4 | − | + | +++ | +++ | +++ | +++ | +++ |
| TnT URE 1.1 | 3 | − | − | ++ | ++ | ++ | ++ | ++ |
| TnT URE 0.8 | 3 | − | − | + | + | + | + | − |
| TnT URE 0.6 | 3 | − | − | − | + | − | − | − |
| TnT IRE | 3 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
| TnT IRE ΔI | 5 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
| TnT IRE ΔII | 3 | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
| TnT IRE Δ(I+II) | 5 | − | − | − | + | + | − | − |
Comparison of β-galactosidase reporter expression driven by selected sequences upstream of the transcription start site and intron 1 of the TnT transcription unit. The highest expressing line, TnT IRE, produced the maximum intensity of staining for most muscles within approximately 1 h and were referred to as “+++++.” The level of intensity of the other lines, TnT URE 1.4, TnT URE 1.1, TnT URE 0.8, TnT 0.6, and TnT IREΔI and TnT IREΔII, and TnT IREΔ(I+II) lines was determined by comparison with these two lines.
TnT URE 0.8 flies expressed β-galactosidase in all hypodermic muscles as well as in visceral muscles in both larvae and adults (Figures 5 and 6, and Table 2). In all these lines, transgene expression was not observed in the dorsal vessel (heart). Although no significant levels of β-galactosidase expression were seen when the TnT URE 0.6 flies were analyzed (Figures 5 and 6, and Table 2), upon more detailed examination, small amounts of the transgene could be detected in the adult hypodermic ventral muscles (Figures 5). Three putative binding sites for TINMAN and CF2 were found in the region -0.8 and -0.6 kb. In vitro-transcribed/translated TINMAN and CF2 product specifically bound to these sequences (unpublished data) and band shift analysis with oligonucleotides corresponding to these regions revealed that binding to these sites was specific (unpublished data).
When β-galactosidase reactions were performed for 2 h, transgene expression in TnT URE 2.5, TnT URE 1.4, TnT URE 1.1, and TnT URE 0.8 larval and adult musculatures were compared (Figures 5 and 6). A sequential reduction in the levels of expression in larval and adult hypodermic muscles as well as in visceral muscles was observed (middle panels in Figures 5 and 6). However, when the reactions were left for 16 h, these differences disappeared. These results indicated the importance of the distal region to potentiate expression in hypodermic as well as visceral muscles.
Finally, the TnT URE elements from D. melanogaster, D. virilis, and D. pseudoobscura were analyzed in parallel using bioinformatic tools (see MATERIALS AND METHODS). In this way, annotated maps reflecting both the level of sequence conservation and the precise location of matches to transcription factor binding sites were established (Figure 7). The conservation of the spatial arrangement of binding sequences for myogenic factors such as MEF-2, TINMAN, GATA, and CF2 factors became evident through these maps. MEF2 and TINMAN have been described as being essential for the specification of muscle and cardiac lineages (Azpiazu and Frasch, 1993; Bodmer, 1993; Borkowski et al., 1995; Taylor et al., 1995; Black and Olson, 1998). Furthermore, both CF2 (Bagni et al., 2002) and PDP1 (Reddy et al., 2000) have been shown to have important roles in muscle development. Thus, within the URE element, we could identify two conserved regions, one located between approximately -0.55 and -0.85 kb and the other between -2.5 and -1.6 kb and coinciding with the transcriptionally active regions described above. These two regions of 300 and 900 bp were denominated as the proximal and distal regions, respectively (Figure 7). The distal region might be more closely related to the expression in the adult specialized muscles, the flight and jump muscles, and the proximal region in embryo/larval musculature, and visceral muscles as well as in adult hypodermic musculature.
Figure 7.
Conserved regions within the URE element. (A) The alignment of the sequences upstream of the transcriptional start sites of the TnT genes in the D. melanogaster, D. pseudoobscura, and D. virilis species. A schematic representation of conserved binding sites for myogenic motifs, including MEF2, TINMAN, GATA, E box, and CF2 is shown. In addition, several conserved sequences that are not targets for known myogenic factors were found (gray squares). Sequences situated -2.5/2.4 kb upstream in the D. virilis TnT gene are unknown. (B) The alignment of the conserved sequences situated in the proximal regions of the TnT genes in the D. melanogaster, D. pseudoobscura, and D. virilis species.
Two Distinct Regulatory Modules within the IRE Element Independently Control TnT Gene Expression
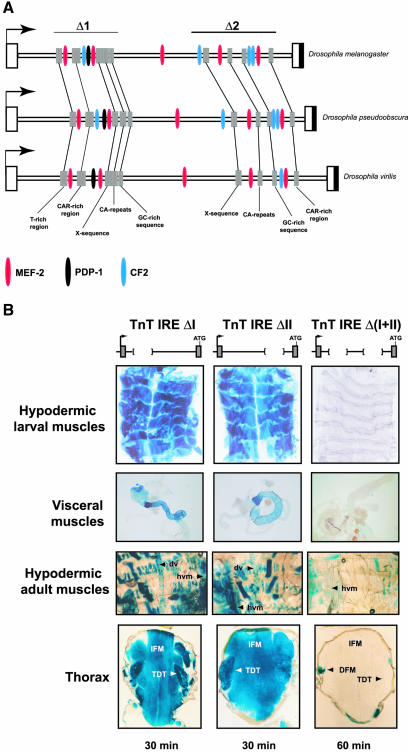
Phylogenetic footprinting was performed on intron 1, and a high degree of sequence homology was found at two sites of 504 and 711 bp, respectively, designated as modules I and II (Figure 8A). As in other genes encoding contractile proteins, the importance of the MEF2 sites in controlling TnT gene expression was confirmed in our in silico analysis, which revealed that the five MEF2 sites situated in the IRE element are >99% conserved. Four of them are located within modules I and II and specifically bind MEF2 protein (unpublished data). Moreover, each module contained one binding site for PDP1, two binding sites for CF2, and several conserved regions ranging in size from 20 to 60 bp.
Figure 8.
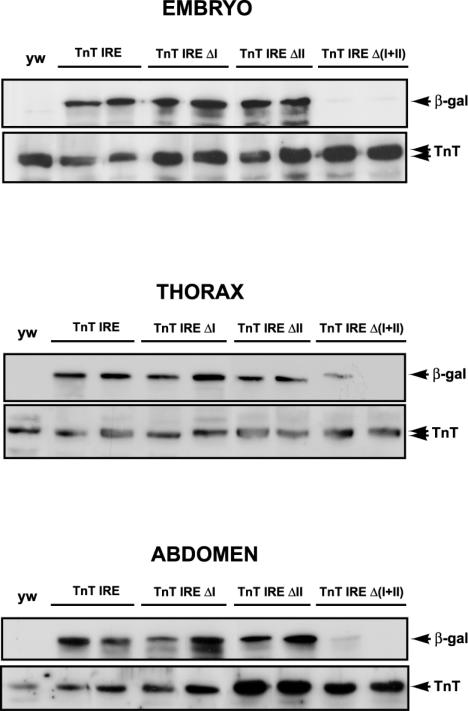
Two distinct regulatory modules within IRE element exhibit functional redundancy in Drosophila muscles. (A) The alignment of the intron sequences of the TnT genes in the D. melanogaster, D. pseudoobscura, and D. virilis species. A schematic representation of conserved binding sites for myogenic motifs, including MEF2, PDP1 and CF2, is shown. Several conserved sequences that are not targets for known myogenic factors were found (gray squares). (B) β-galactosidase expression in distinct adult and larval muscles in the TnT IRE, TnT IRE ΔI and TnT IREΔII, and TnT IREΔ(I+II) transgenic lines is presented. β-galactosidase reaction time is indicated in the lower part of the panels. Shorter reaction times give similar staining patterns. IFM, indirect flight muscles; TDT, tergal depressor of the trochanter; dv, dorsal vessel (heart); hvm, hypodermic ventral muscles; and hdm, hypodermic dorsal muscles.
To determine if the cis-acting sequences governing TnT transcription in the IRE enhancer-like element were localized in modules I or II, we transformed embryos with smaller IRE fragments and tested for reporter activity (MATERIALS AND METHODS). Within the intronic sequence, module I or II or both were deleted and the resulting fragments were cloned, maintaining the endogenous organization of the TnT gene. Accordingly, to indicate the absence of module I, II or both, the constructs were named TnT IREΔI, TnT IREΔII, or TnT IREΔ(I+II). Surprisingly, the temporal and spatial expression exhibited in the muscles by the TnT IREΔI and TnT IREΔII constructs mimic the expression of the TnT IRE flies (Figure 8 and Table 2). No significant differences in activity were observed, even when expression was analyzed after very short reaction times (2-5 min). However, when neither of the two modules was present in TnT IREΔ(I+II) flies, no activity was detected in the majority of muscles (Figure 8 and Table 2), and only very weak transgene expression in direct flight muscles (DFM) and ventral hypodermic muscles could be detected after 16 h of staining (Figure 8B). The fragments, TnT IREΔI and TnT IREΔII, were both cloned upstream of a heterologous promoter (MATERIALS AND METHODS), and these lines exhibited very high activity in all muscles. Thus, the TnT IREΔI and TnT IREΔII fragments were able to confer muscle-specificity, driving expression in all Drosophila muscles (unpublished data). Moreover, the control of TnT gene transcription in intron 1 seems to be independent of position and spatial organization.
The relative levels of expression in these lines were established by Western blotting (Figure 9 and unpublished data). Surprisingly, the protein levels in extracts from late embryos carrying either the deletion of module I or II, the TnT IREΔI or TnT IREΔII lines, respectively, were similar to that produced in those lines harboring both elements. However, in TnT IREΔ(I+II) lines only very low levels of activity were detected. A similar effect was observed in dissected thoraces, IFM, TDT, and abdomens of the same lines (unpublished data). These findings clearly indicated that the two regulatory modules within the IRE enhancer-line element are interchangeable and thus, the removal of either module alone does not produce changes in transgene transcription.
Figure 9.
Western blot analysis of the TnT IRE, TnT IREΔI, TnT IREΔII, and TnT IREΔ(I+II) lines. Western blot analysis with β-galactosidase and TnT-specific antibodies on extracts from embryos, dissected thoraxes, and abdomens from two representative lines of the of the TnT IRE, TnT IREΔI and TnT IREΔII, and TnT IREΔ(I+II) lines are shown. Arrows indicate the localization of the β-galactosidase and TnT-specific isoforms.
DISCUSSION
To discover more about the complex transcriptional control of muscle-specific genes, we have studied the regulation of the Drosophila TnT gene. One of the interesting features of the Drosophila TnT gene is that it contains an untranslated exon followed by a relatively large intron (Benoist et al., 1998), an organization that is shared by a number of genes encoding contractile proteins (Karlik and Fyrberg, 1986; Konieczny and Emerson, 1987; Bernstein et al., 1993; Hallauer and Hastings, 2002). The fact that this feature is so widely conserved suggests that this general structure might be important for the coordinated regulation of this group of genes (Gremke et al., 1993; Meredith and Storti, 1993; Hallauer and Hastings, 2002).
Expression from the TnT gene is regulated by two different elements, the upstream URE and the IRE that is located downstream of the transcription initiation site for this gene. We have shown that these two elements function as positive regulatory elements, because removal of either provokes a decrease in the rate of transcription of a reporter gene (Figures 2 and 3). Furthermore, each element is itself sufficient to achieve the appropriate spatiotemporal pattern of gene expression. Indeed, they both direct the expression of a transgene in a manner indistinguishable from the endogenous gene, and independently they are sufficient to activate transcription in all muscle types. Thus, the TnT gene has two positive regulatory elements, functionally equivalent but nonredundant, because the concerted action of the two elements is required to reach correct TnT levels associated with specific muscle-type differentiation. Nevertheless, as reflected by an IRE/URE ratio greater than 1, the contribution of the IRE to the total reporter gene expression was always higher in adult muscles than in the larval musculature. Our findings also reveal that the contributions of IRE and URE elements vary depending on the type of muscle, and that different contributions of IRE and URE elements are necessary to ensure the correct level of transcription in each muscle type. Thus, in the embryonic/larval musculature, a supercontractile fibrillar muscle type responsible for larval movements, the contribution of URE and IRE elements is approximately equal (Figure 2, B and C). However, synergism can be seen when the two elements work together in the TnT URE/IRE larvae where an important increase in expression was observed (Figure 2). This degree of cooperation changes in the adult musculature (Figure 3), where the contribution of the IRE element in flight muscles, an asynchronous fibrillar muscle-type, is much higher (9 times) than that of the URE. In contrast, the contribution of the IRE is two or three times higher in the synchronous tubular jump muscles, whereas in the somatic hypodermic and visceral muscles, of which the abdomen is mainly composed, the contribution of the IRE is 1.5-fold that of the URE. Thus, the synergistic effect observed in adult muscles is less than that seen in larvae muscles.
The application of bioinformatic tools is tremendously useful when attempting to define elements that play a role in transcriptional control. Indeed, more accurate and precise sequences have been identified using this approach (Konig et al., 2002). The importance of the URE and IRE enhancer-like elements was highlighted by the discovery of important clusters containing binding sites for MEF2 and CF2 myogenic factors upstream and downstream of the TnT transcription start site. Similar clusters were found in Troponin I, Tropomyosin, Myosin heavy chain, Actin 57B, and Actin 87E genes. These clusters were not found in nonmuscle genes (our unpublished data). These elements exhibited similar overall structures in which focally conserved sequences rich in putative muscle-specific transcription factor binding sites were distributed throughout more discrete regions. The conserved MEF2, CF2, E boxes, TINMAN, PDP1, and GATA binding sites present in the proximal and distal regions within the URE element and modules I and II within the IRE element in the TnT gene are contained within larger conserved regions that range from 20 to 30/40 bp (Figures 7 and 8). The degree of sequence conservation in these regions is >70%. Furthermore, four different sequences, a CAR-rich region, a CA repeat region, a CG rich region, and an X sequence, are present in both modules, I and II (Figure 8 and additional information). These studies suggest that the presence of factors other than MEF2, CF2, TINMAN, and bHLH factors may possibly participate in the regulatory complexes that control TnT expression.
Our in silico studies of the URE and IRE sequences in the TnT genes in conjunction with the in vivo analyses presented here have allowed us to better define the location of the discrete regions situated in these elements. Thus, in the URE element, the 150-bp sequence proximal to the TnT start site drives low levels of transgene expression (detected exclusively by RT-PCR), suggesting that they contain binding sites for the basal transcriptional machinery. Besides the proximal promoter, two other discrete elements have been identified: a 300-bp proximal region, approximately located between -0.55 and -0.85 kb, that may govern expression in all the major muscles of embryos, larvae, and adult; and a 0.9 kb conserved distal region, situated in between -1.6 kb and -2.5, that is active in flight and jump muscles, the specialized thoracic muscles. In addition, the presence of the distal region acting as an enhancer-like sequence in all larval and adult muscles is important to achieve optimal transgene transcription. Thus, its absence in the TnT URE 1.4, TnT URE 1.1, and TnT URE 0.8 lines results in a reduced activity in larval and adult somatic muscles as well as visceral muscles (Figures 5 and 6). In fact, the removal of the distal region, transforms the URE element into a weak tissue-specific enhancer. A direct interaction between these two proximal and distal regions may be needed to reach the optimal levels of transgene expression within the URE element in Drosophila muscles.
With respect to the IRE element, two discrete modules govern transgene expression in all the major muscles of embryos, larvae, and adult, including the specialized thoracic muscles (Figure 8). Modules I and II seem to direct expression in an independent manner and thus, the removal of either module in the TnT IREΔI or TnT IREΔII lines results in similar transgene expression. However, the simultaneous deletion of both modules in the IRE element severely diminishes activity. Using this approach we could not found a unique and specific role for either of the modules. Moreover, both constructs can drive reporter gene activity in all muscles independently of their position. These results are consistent with the idea that within the IRE element, the two regulatory elements, modules I and II, might be redundant. With respect to the exact role of the sequences in between the two modules, our results indicate that they may play a role governing expression in the direct flight muscles. The absence of the two modules in TnT IREΔ(I+II) transgenic lines completely abolished reporter activity in all but the direct flight muscles.
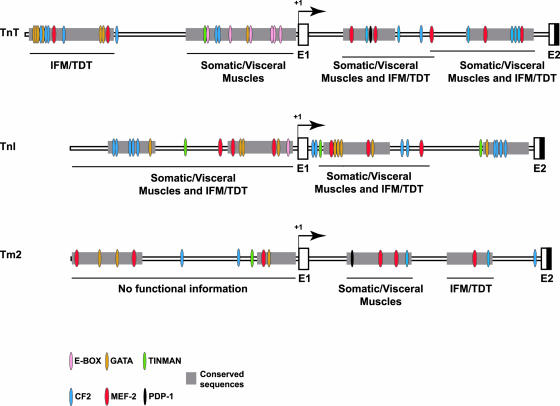
In summary, the structure of the active regions within URE and IRE reveals a modular organization of these regulatory elements. The size of these regions varies from 900 bp (distal) and 300 bp (proximal) in the URE elements to 500 bp (module I) and 700 bp (module II) in the IRE element. These patterns suggest that the order and/or maybe the physical distance between transcription factor binding sites spread over the URE and IRE elements may be crucial to the proper developmental regulation of TnT gene expression. To address the possibility of a shared core promoter structure, we extended the pairwise comparison to include the Troponin I and Tropomyosin promoters, two other components of the troponin complex for which additional functional expression results have been published (Figure 10). In combination with Troponin C, Troponin I, and Tropomyosin, TnT participates in the formation of a complex that is involved in the regulation of calcium-mediated muscle contraction (Perry, 1998). Recent studies have shown that Drosophila muscle Troponin I gene transcription is also controlled by two enhancer elements located upstream and downstream of the start site of the gene. Moreover, these two TnI regulatory elements also contain clustered binding sites for distinct combinations of myogenic factors that collaborate to achieve maximal levels of expression (Marín et al., 2004). In the case of the Drosophila Tropomyosin 2 gene, two enhancer-like elements were described within intron 1 that confer the appropriate temporal and tissue-specific expression. One of the enhancer-like elements governs transgene expression in somatic and visceral muscles and the other one governs transgene expression in the flight and jump muscles, the specialized thoracic muscles (Karlik and Fyrberg, 1986; Hanke and Storti, 1988; Gremke et al., 1993).
Figure 10.
Phylogenetically conserved domains within the URE and IRE elements of the Drosophila TnT, TnI, and Tropomyosin 2 genes together with in vivo analysis. A schematic representation of clusters of binding sites for myogenic factors, including MEF2, PDP1 and CF2, GATA, TINMAN, and E-boxes is shown. Several conserved sequences that are not targets for known myogenic factors were found (gray squares).
To complete our studies, we decided to compare the sequences of the two regulatory regions of Troponin I and Tropomyosin genes in D. melanogaster and D. pseudoobscura. These two species of Drosophila diverged more than 30 million years ago. A comparative analysis of the TnT, TnI, and Tm2 genes together with the in vivo analysis reveals a similar modular organization of phylogenetically conserved domains that control the expression of the Drosophila troponin/tropomyosin genes. Furthermore, these modules share a group of known and unknown binding sites for myogenic factors. Thus, all proximal modules within the intron contain two MEF2 binding sites and are involved in somatic and visceral muscle expression. Moreover, the three genes have a specific module controlling transgene expression for the adult specialized muscles. It is clear that additional studies in Tm2 genes and other muscle genes are necessary to fully explain such a complex situation. Our findings highlight the complex nature of the regulatory mechanisms directing muscle gene expression. However, the molecular mechanisms underlying the cooperation between the different discrete regions identified are currently unknown. Communication between these modules may be needed to ensure the proper temporal and tissue-specific expression (Blackwood and Kadonaga, 1998). Although the precise mechanisms remain unclear, the two distinct levels of regulation, namely cis-transcription and chromatin remodeling must be integrated. It is unclear how the RNA polymerase resolves the problem of read-through and whether this solution may provide a mechanism for regulating the levels of protein expression in individual muscles. However, the results we present here reveal a new level of complexity to explain the role of the distinct discrete elements present in the URE and IRE elements in regulating protein expression in genes encoding muscle proteins.
Supplementary Material
Acknowledgments
We thank Drs. M. Frach, R. Storti, and Kafatos for the generous gifts of antibodies; Drs. A. Cano, A. Ferrús, M. Manzanares, R. Marco, and J. Vinós for their critical reading of the manuscript; and Vanesa Santos for her expert technical assistance. Grants PB97-0014 and BMC2001-1454 from the DGICYT (Spanish Ministry of Education, Culture and Sports), ESP94-865 (Plan Nacional deInvestigación Científica) supported this research. E.G.-Z. is a pre-doctoral fellow of the DGICYT, and J.A.M. is a postdoctoral fellow at the Universidad Autónoma deMadrid.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-10-0729. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-10-0729.
Online version is available at www.molbiolcell.org.
Online version of this article contains supplemental figures.
References
- Arnone, M.I., and Davidson, E.H. (1997). The hardwiring of development: organization and function of genomic regulatory systems. Development 124, 1851-1864. [DOI] [PubMed] [Google Scholar]
- Arredondo, J.J., Ferreres, R.M., Maroto, M., Cripps, R.M., Marco, R., Bernstein, S.I., and Cervera, M. (2001). Control of Drosophila paramyosin/miniparamyosin gene expression. Differential regulatory mechanisms for muscle-specific transcription. J. Biol. Chem. 276, 8278-8287. [DOI] [PubMed] [Google Scholar]
- Azpiazu, N., and Frasch, M. (1993). tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 7, 1325-1340. [DOI] [PubMed] [Google Scholar]
- Bagni, C., Bray, S., Gogos, J.A., Kafatos, F.C., and Hsu, T. (2002). The Drosophila zinc finger transcription factor CF2 is a myogenic marker downstream of MEF2 during muscle development. Mech. Dev. 117, 265-268. [DOI] [PubMed] [Google Scholar]
- Banerjee-Basu, S., and Buonanno, A. (1993). cis-acting sequences of the rat troponin I slow gene confer tissue- and development-specific transcription in cultured muscle cells as well as fiber type specificity in transgenic mice. Mol. Cell Biol. 13, 7019-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo, S., Carver, L.A., and Posakony, J.W. (2000). GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29, 726, 728, 730, 732. [DOI] [PubMed] [Google Scholar]
- Bate, M. (1990). The embryonic development of larval muscles in Drosophila. Development 110, 791-804. [DOI] [PubMed] [Google Scholar]
- Baylies, M.K., Bate, M., and Ruiz Gomez, M. (1998). Myogenesis: a view from Drosophila. Cell 93, 921-927. [DOI] [PubMed] [Google Scholar]
- Benoist, P., Mas, J.A., Marco, R., and Cervera, M. (1998). Differential muscle-type expression of the Drosophila troponin T gene. A 3-base pair microexon is involved in visceral and adult hypodermic muscle specification. J. Biol. Chem. 273, 7538-7546. [DOI] [PubMed] [Google Scholar]
- Berman, B.P., Nibu, Y., Pfeiffer, B.D., Tomancak, P., Celniker, S.E., Levine, M., Rubin, G.M., and Eisen, M.B. (2002). Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99, 757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, S.I., O'Donnell, P.T., and Cripps, R.M. (1993). Molecular genetic analysis of muscle development, structure, and function in Drosophila. Int. Rev. Cytol. 143, 63-152. [DOI] [PubMed] [Google Scholar]
- Black, B.L., and Olson, E.N. (1998). Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14, 167-196. [DOI] [PubMed] [Google Scholar]
- Blackwood, E.M., and Kadonaga, J.T. (1998). Going the distance: a current view of enhancer action. Science 281, 61-63. [DOI] [PubMed] [Google Scholar]
- Bodmer, R. (1993). The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 118, 719-729. [DOI] [PubMed] [Google Scholar]
- Borkowski, O.M., Brown, N.H., and Bate, M. (1995). Anterior-posterior subdivision and the diversification of the mesoderm in Drosophila. Development 121, 4183-4193. [DOI] [PubMed] [Google Scholar]
- Buckingham, M., Houzelstein, D., Lyons, G., Ontell, M., Ott, M.O., and Sassoon, D. (1992). Expression of muscle genes in the mouse embryo. Symp. Soc. Exp. Biol. 46, 203-217. [PubMed] [Google Scholar]
- Buonanno, A., and Rosenthal, N. (1996). Molecular control of muscle diversity and plasticity. Dev. Genet. 19, 95-107. [DOI] [PubMed] [Google Scholar]
- Calvo, S., Vullhorst, D., Venepally, P., Cheng, J., Karavanova, I., and Buonanno, A. (2001). Molecular dissection of DNA sequences and factors involved in slow muscle-specific transcription. Mol. Cell Biol. 21, 8490-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, E.H. et al. (2002). A genomic regulatory network for development. Science 295, 1669-1678. [DOI] [PubMed] [Google Scholar]
- Domingo, A., Gonzalez-Jurado, J., Maroto, M., Diaz, C., Vinos, J., Carrasco, C., Cervera, M., and Marco, R. (1998). Troponin-T is a calcium-binding protein in insect muscle: in vivo phosphorylation, muscle-specific isoforms and developmental profile in Drosophila melanogaster. J. Muscle Res. Cell Motil. 19, 393-403. [DOI] [PubMed] [Google Scholar]
- Frith, M.C., Hansen, U., and Weng, Z. (2001). Detection of cis-element clusters in higher eukaryotic DNA. Bioinformatics 17, 878-889. [DOI] [PubMed] [Google Scholar]
- Fyrberg, E., Fyrberg, C.C., Beall, C., and Saville, D.L. (1990). Drosophila melanogaster troponin-T mutations engender three distinct syndromes of myofibrillar abnormalities. J. Mol. Biol. 216, 657-675. [DOI] [PubMed] [Google Scholar]
- Gajewski, K., Kim, Y., Lee, Y.M., Olson, E.N., and Schulz, R.A. (1997). D-mef2 is a target for Tinman activation during Drosophila heart development. EMBO J. 16, 515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremke, L., Lord, P.C., Sabacan, L., Lin, S.C., Wohlwill, A., and Storti, R.V. (1993). Coordinate regulation of Drosophila tropomyosin gene expression is controlled by multiple muscle-type-specific positive and negative enhancer elements. Dev. Biol. 159, 513-527. [DOI] [PubMed] [Google Scholar]
- Hallauer, P.L., Bradshaw, H.L., and Hastings, K.E. (1993). Complex fiber-type-specific expression of fast skeletal muscle troponin I gene constructs in transgenic mice. Development 119, 691-701. [DOI] [PubMed] [Google Scholar]
- Hallauer, P.L., and Hastings, K.E. (2002). TnIfast IRE enhancer: multistep developmental regulation during skeletal muscle fiber type differentiation. Dev. Dyn. 224, 422-431. [DOI] [PubMed] [Google Scholar]
- Hanke, P.D., and Storti, R.V. (1988). The Drosophila melanogaster tropomyosin II gene produces multiple proteins by use of alternative tissue-specific promoters and alternative splicing. Mol. Cell Biol. 8, 3591-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, N., Kronert, W.A., and Bernstein, S.I. (1989). Transcriptional and post-transcriptional regulation of Drosophila myosin heavy chain expression. Cell Mol. Biol. Muscle Dev. 621-631.
- Hughes, S.M., and Salinas, P.C. (1999). Control of muscle fibre and motoneuron diversification. Curr. Opin. Neurobiol. 9, 54-64. [DOI] [PubMed] [Google Scholar]
- Javadpour, M.M., Tardiff, J.C., Pinz, I., and Ingwall, J.S. (2003). Decreased energetics in murine hearts bearing the R92Q mutation in cardiac troponin T. J. Clin. Invest. 112, 768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlik, C.C., and Fyrberg, E.A. (1986). Two Drosophila melanogaster tropomyosin genes: structural and functional aspects. Mol. Cell Biol. 6, 1965-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, S.F., and Emerson, C.P., Jr. (1987). Complex regulation of the muscle-specific contractile protein (troponin I) gene. Mol. Cell Biol. 7, 3065-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig, S., Burkman, J., Fitzgerald, J., Mitchell, M., Su, L., and Stedman, H. (2002). Modular organization of phylogenetically conserved domains controlling developmental regulation of the human skeletal myosin heavy chain gene family. J. Biol. Chem. 277, 27593-27605. [DOI] [PubMed] [Google Scholar]
- Marín, M.C., Rodríguez, J.R., and Ferrús, A. (2004). Transcription of Drosophila Troponin I gene is regulated by two conserved, functionally identical, synergistic elements. Mol. Biol. Cell (in press). [DOI] [PMC free article] [PubMed]
- Markstein, M., and Levine, M. (2002). Decoding cis-regulatory DNAs in the Drosophila genome. Curr. Opin. Genet. Dev. 12, 601-606. [DOI] [PubMed] [Google Scholar]
- Maron, B.J., McKenna, W.J., Elliott, P., Spirito, P., Frenneaux, M.P., Keren, A., Cecchi, F., Borggrefe, M., and Williams, W.G. (1999). Hypertrophic cardiomyopathy. J. Am. Med. Assoc. 282, 2302-2303. [DOI] [PubMed] [Google Scholar]
- Maroto, M., Arredondo, J., Goulding, D., Marco, R., Bullard, B., and Cervera, M. (1996). Drosophila paramyosin/miniparamyosin gene products show a large diversity in quantity, localization, and isoform pattern: a possible role in muscle maturation and function. J. Cell Biol. 134, 81-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey, T.A., Zhang, C.L., and Olson, E.N. (2002). Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 14, 763-772. [DOI] [PubMed] [Google Scholar]
- Meredith, J., and Storti, R.V. (1993). Developmental regulation of the Drosophila tropomyosin II gene in different muscles is controlled by muscle-type-specific intron enhancer elements and distal and proximal promoter control elements. Dev. Biol. 159, 500-512. [DOI] [PubMed] [Google Scholar]
- Pennachhio, L.A., and Rubin, E.M. (2001). Genomic estrategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2, 100-109. [DOI] [PubMed] [Google Scholar]
- Perry, S.V. (1998). Troponin T: genetics, properties and function. J. Muscle Res. Cell Motil. 19, 575-602. [DOI] [PubMed] [Google Scholar]
- Reddy, K.L., Wohlwill, A., Dzitoeva, S., Lin, M.H., Holbrook, S., and Storti, R.V. (2000). The Drosophila PAR domain protein 1 (Pdp1) gene encodes multiple differentially expressed mRNAs and proteins through the use of multiple enhancers and promoters. Dev. Biol. 224, 401-414. [DOI] [PubMed] [Google Scholar]
- Roberts, R., and Sigwart, U. (2001a). New concepts in hypertrophic cardiomyopathies, part I. Circulation 104, 2113-2116. [DOI] [PubMed] [Google Scholar]
- Roberts, R., and Sigwart, U. (2001b). New concepts in hypertrophic cardiomyopathies, part II. Circulation 104, 2249-2252. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sanger, F., Nicklen, S., and Coulson, A.R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino, S., and Reggiani, C. (1996). Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol. Rev. 76, 371-423. [DOI] [PubMed] [Google Scholar]
- Schwartz, K., and Mercadier, J.J. (2003). Cardiac troponin T and familial hypertrophic cardiomyopathy: an energetic affair. J. Clin. Invest. 112, 652-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman, J.G., and Seidman, C. (2001). The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell 104, 557-567. [DOI] [PubMed] [Google Scholar]
- Spradling, A.C., and Rubin, G.M. (1982). Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218, 341-347. [DOI] [PubMed] [Google Scholar]
- Stockdale, F.E. (1997). Mechanisms of formation of muscle fiber types. Cell Struct. Funct. 22, 37-43. [DOI] [PubMed] [Google Scholar]
- Sullivan, W., Ashurner, M., and Scott Hawley, R. (2000). Drosophila Protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Taylor, M.V., Beatty, K.E., Hunter, H.K., and Baylies, M.K. (1995). Drosophila MEF2 is regulated by twist and is expressed in both the primordia and differentiated cells of the embryonic somatic, visceral and heart musculature. Mech. Dev. 50, 29-41. [DOI] [PubMed] [Google Scholar]
- Thierfelder, L., Watkins, H., MacRae, C., Lamas, R., McKenna, W., Vosberg, H.P., Seidman, J.G., and Seidman, C.E. (1994). Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell 77, 701-712. [DOI] [PubMed] [Google Scholar]
- Thummel, C.S., Boulet, A.M., and Lipshitz, H.D. (1988). Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene 74, 445-456. [DOI] [PubMed] [Google Scholar]
- Watkins, H. et al. (1995). Mutations in the genes for cardiac troponin T and alpha-tropomyosin in hypertrophic cardiomyopathy. N. Engl. J. Med. 332, 1058-1064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.