Abstract
Background
The RAS association domain family protein 1a gene (RASSF1A) is one of the tumor suppressor genes (TSG). Inactivation of RASSF1A is critical to the pathogenesis of cancer. Aberrant TSG methylation was considered an important epigenetic silencing mechanism in the progression of ovarian cancer. A number of studies have discussed association between RASSF1A promoter methylation and ovarian cancer. However, they were mostly based on a small number of samples and showed inconsist results, Therefore, we conducted a meta-analysis to better identify the association.
Methods
Eligible studies were identified by searching the PubMed, EMBASE, Web of Science, and CNKI databases using a systematic searching strategy. We pooled the odds ratio (ORs) from individual studies using a fixed-effects model. We performed heterogeneity and publication bias analysis simultaneously.
Results
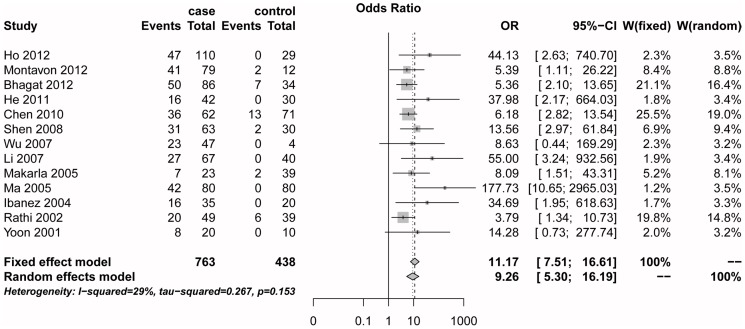
Thirteen studies, with 763 ovarian cancer patients and 438 controls were included in the meta-analysis. The frequencies of RASSF1A promoter methylation ranged from 30% to 58% (median is 48%) in the cancer group and 0 to 21% (median is 0) in the control group. The frequencies of RASSF1A promoter methylation in the cancer group were significantly higher than those in the control group. The pooled odds ratio was 11.17 (95% CI = 7.51–16.61) in the cancer group versus the corresponding control group under the fixed-effects model.
Conclusion
The results suggested that RASSF1A promoter methylation had a strong association with ovarian cancer.
Introduction
Ovarian cancer is the fifth most common cause of cancer deaths in women and accounts for the highest tumor-related mortality of gynecologic malignancies. Approximately 1.5% of females suffer ovarian from cancer and most cases are diagnosed at late stage owing to the lack of effective early detection methods [1], [2]. Over 80% of patients with advanced ovarian cancer relapse [3]. While the 5-year survival rate is close to 30% [4].
Hypermethylation of the tumor suppressor gene (TSG) promoter can lead to gene inactivation, which is critical to the pathogenesis of cancers, and occurs always in the early stage of cancer development in many types of cancer including ovarian cancer [5], [6]. Some studies also showed that methylation of TSG was detected in tumor tissue and was associated with clinical features [7], [8]. The RAS association domain family protein 1A (RASSF1A) is a putative tumor suppressor gene located on 3 p21 spanning 11,000 bp, containing eight exons and two different promoters [9]. Epigenetic inactivation of the gene often resulted from the methylation of CpG islands in promoters [5]. An in vitro study showed that aberrant methylation was frequently observed in ovarian cancer cell lines [10]. These studies showed that the methylation of RASSF1A promoter may play an important role in the development of ovarian cancer.
Some studies have reported differences in the methylation frequencies of RASSF1A between cancer tissues and non-cancerous tissues. However, they were mostly based on a small number of samples and showed inconsist results. Therefore, we performed a meta-analysis to better identify the association between RASSF1A promoter methylation and ovarian cancer.
Materials and Methods
Search Strategy and Study Selection
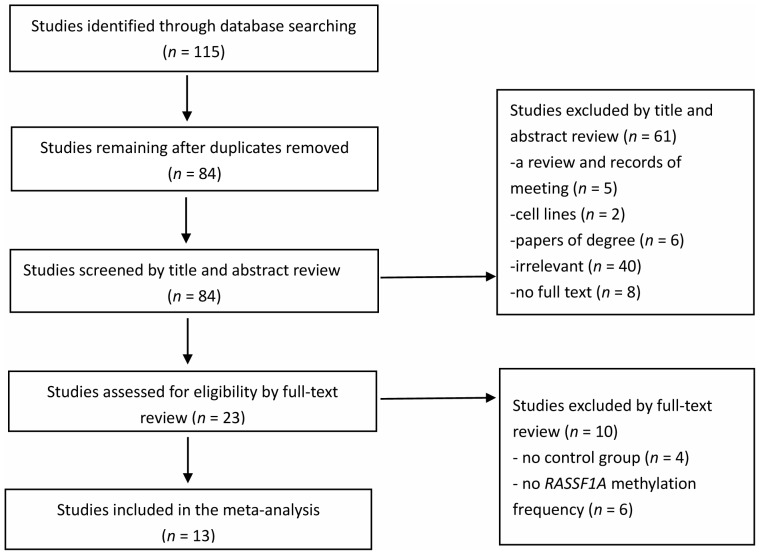
Online electronic databases (PubMed, EMBASE, Web of Science, and China National Knowledge Infrastructure (CNKI)) were searched for published studies up to June 1, 2013. The following search strategy was performed in PubMed: (ovarian OR ovary) AND (cancer OR carcinoma OR tumor) AND (RASSF1A methylation). The similar search strategy was used in other databases. The search was limited to human studies, without language restrictions. A study included in the meta-analysis had to meet the following criteria: 1) studies which evaluated the association of RASSF1A methylation with ovarian cancer, 2) a case-control study or one including case and control populations, 3) a study reporting the RASSF1A methylation frequency in case and control groups, 4) sample type limited to tissues. First, the title and abstract of studies from the initial search were evaluated according to the inclusion criteria. Then all potentially relevant studies were evaluated as full-text papers. If a study was published more than once, only the most complete and up-to-date information was included in the meta-analysis. Figure 1 presents detailed information on the study selection process. Finally, a total of 13 studies (PubMed 7, Web of Science 1, and CNKI 5) with 763 cases and 438 controls were included in our meta-analysis.
Figure 1. Selection of studies in the meta-analysis.
Data Extraction and Quality Assessment
Three reviewers (Hao Shi, Ya Li, and Changmei Gu) independently reviewed the selected studies. The following information was extracted from these studies: first author’s name, year of publication, study population, sample size, age of women in the case group, control type, the status of the individuals in the control group, the number of individuals in the case and control groups, the measuring methods of methylation, and modulation frequencies of RASSF1A in the case and control groups. All the detailed information in the included studies was checked by four reviewers (Meixia Lu, Shixuan Wang, Xiaozhong Wang, and Yangxin Huang) as prescribed by the Cochrane Handbook for systematic reviews.
The quality of those studies was evaluated according to the Newcastle Ottawa Scale (NOS) (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). The NOS has a maximum of nine ‘stars’ on items related to the selection of the study groups (four stars), the comparability of the groups (two stars) and the ascertainment of the outcome of interest (three stars). The studies were independently evaluated based on NOS by three reviewers (Cheng Lu, Lilan Yang, and Jiaqiang Xiong). Studies with quality scores greater than or equal to 5 were included.
Statistical Analysis
To assess the strength of the association between RASSF1A methylation and ovarian cancer the pooled odds ratios (ORs) and their 95% confidence intervals (CIs) were calculated. The x 2-based Cochran Q statistic test and I 2 statistics were used to test the heterogeneity among the included studies [11]. The selected studies were considered to have a severe heterogeneity when the I 2 was greater than 50%. When P<0.05 for the Q statistic, the heterogeneity was considered significant and a random-effects model (the DerSimonian-Laird estimate) was used to calculate the pooled ORs. Otherwise, a fixed-effects model (the Mantel-Haenszel method) was applied. A meta-regression (restricted maximum-likelihood estimator method) was employed to explore the source of heterogeneity. Furthermore, a subgroup analysis was performed to evaluate the source of heterogeneity. The contribution of each study to the final results of the meta-analysis was evaluated according to sensitivity analysis. We also used the Peters test, the Begg’s rank correlation method [12] and a funnel plot for Egger’s test [13] to assess publication bias. The fail-safe number [14] was also employed to assess publication bias. In our study, all the p values were two sides with a significant level at 0.05. When the individual studies have cells with zero counts, the default is to add 0.5 to all zero counts in the Meta package. All statistical analyses were performed with the Meta package [15] (version 2.2-1) in R (version 3.00; http://www.r-project.org/).
Results
Study Characteristics and Quality Assessment
A total of thirteen studies with 763 cases and 438 controls was included in the meta-analysis. 115 studies were initially identified by searching the electronic databases. 84 potentially relevant studies were retrieved for further evaluation after removing 31 duplicated articles. Based on their titles and abstracts, 61 studies were excluded (5 reviews or meeting reviews, 2 cell lines, 6 thesis papers, 40 irrelevant articles, and 8 papers that did not have full text versions). Four studies without a control group and six studies without RASSF1A methylation data were excluded from full-text review. The studies were published between 2001 and 2012, and covered from 20 to 110 cases. Finally, 13 studies were included in our meta- analysis. Eight studies were of Asian subjects and five studies were of Caucasian subjects. Six studies analyzed methylation of RASSF1A promoter CpG islands [16]–[21] and seven studies analyzed RASSF1A promoter methylation [22]–[28]. The case group consisted of cancer tissues form ovarian cancer patients. The control group included adjacent tissues (AT) from ovarian cancer patients, ovarian tissue from benign ovarian disease patients (BOT), and normal ovarian tissue from cancer-free patients or healthy people (NT). Among the 13 included studies, 11 studies used methylation-specific polymerase chain reaction (MSP), 1 study used methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) and 1 study used bisulfite sequencing PCR (BSP) to explore RASSF1A methylation in ovarian cancer and control. Characteristics of the 13 studies are presented in Table 1.
Table 1. Characteristics of studies included in the study characteristics of included studies.
| Case | Control | ||||||||
| Author | Year | Country | Age (y) | M | U | M | U | Method | Control type |
| Ho [22] | 2012 | Taiwan | NA | 47 | 63 | 0 | 29 | MS-MLPA | BOT |
| Montavon [16] | 2012 | Australia | 59.0 | 41 | 38 | 2 | 10 | MSP | BOT |
| Bhagat [23] | 2012 | India | 51.8 (22–71) | 50 | 36 | 7 | 12 | MSP | BOT |
| 0 | 15 | NT | |||||||
| He [24] | 2011 | China | NA | 16 | 26 | 0 | 19 | MSP | BOT |
| 0 | 11 | NT | |||||||
| Chen [17] | 2010 | China | 44.3 (23–68) | 36 | 26 | 13 | 38 | MSP | BOT |
| 0 | 20 | NT | |||||||
| Shen [25] | 2008 | China | 52.8 (33–76) | 31 | 32 | 1 | 19 | MSP | NT |
| 1 | 9 | AT | |||||||
| Wu [18] | 2007 | Norway | NA | 23 | 24 | 0 | 4 | MSP | BOT |
| Li [26] | 2007 | China | 52.0 (36–89) | 27 | 40 | 0 | 40 | MSP | BOT |
| Makarla [27] | 2005 | USA | 51.5 (20–86) | 7 | 16 | 0 | 23 | MSP | BOT |
| 2 | 14 | NT | |||||||
| Ma [19] | 2005 | China | NA | 42 | 38 | 0 | 80 | MSP | AT |
| Ibanez [20] | 2004 | USA | 62.0 | 16 | 19 | 0 | 10 | MSP | BOT |
| 0 | 10 | NT | |||||||
| Rathi [28] | 2002 | USA | 56.0 | 20 | 29 | 6 | 33 | MSP | BOT |
| Yoon [21] | 2001 | USA | NA | 8 | 12 | 0 | 10 | BSP | NT |
BOT: benign ovarian tissues AT: adjacent tissues; NT: normal ovarian tissues of cancer-free patients or healthy people; NA: not available; MSP: methylation-specific polymerase chain reaction; MS-MLPA: methylation-specific multiplex ligation-dependent probe amplification; BSP: bisulfite sequencing PCR; M: RASSF1A methylated; U: RASSF1A unmethylated.
The results of the NOS showed that most studies had a hospital control except for Li [26] and had not matched the control according to the potential confounder except for Ibanez [20]. The score of the studies ranged from 5 to 8 with a median score of 7. Detail information of the NOS was shown in Table 2.
Table 2. The quality assessment of included studies.
| Newcastle-Ottawa Scale* | ||||||||||
| Author | 1 | 2 | 3 | 4 | 5A | 5B | 6 | 7 | 8 | Total |
| Ho [22] | Yes | Yes | No | Yes | Yes | No | Yes | Yes | Yes | 7 |
| Montavon [16] | Yes | Yes | No | Yes | Yes | No | Yes | Yes | Yes | 7 |
| Bhagat [23] | Yes | Yes | No | Yes | Yes | No | Yes | Yes | Yes | 7 |
| He [24] | Yes | Yes | No | Yes | Yes | No | Yes | Yes | Yes | 7 |
| Chen [17] | Yes | Yes | No | Yes | Yes | No | Yes | Yes | Yes | 7 |
| Shen [25] | Yes | Yes | No | No | Yes | No | Yes | Yes | Yes | 6 |
| Wu [18] | Yes | Yes | No | Yes | No | No | Yes | Yes | Yes | 5 |
| Li [26] | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | 8 |
| Makarla [27] | Yes | Yes | No | Yes | Yes | No | Yes | Yes | Yes | 7 |
| Ma [19] | Yes | No | No | Yes | Yes | No | Yes | Yes | Yes | 5 |
| Ibanez [20] | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | 8 |
| Rathi [28] | Yes | Yes | No | Yes | Yes | No | Yes | Yes | Yes | 7 |
| Yoon [21] | Yes | Yes | No | Yes | Yes | No | Yes | Yes | Yes | 7 |
1 indicates case definition and diagnostic appropriate; 2, consecutive patients or case having a good representation; 3, community controls; 4, control with no the history of study disease; 5A, according to the most important factor to select and analyze control; 5B, according to the second important factor to select and analyze control; 6, ascertainment of exposure by blinded interview or record; 7, same method of ascertainment used for cases and controls; and 8, non-response rate the same for cases and controls.
Meta-analysis
The x 2-based Cochran Q statistic test and I 2 statistics did not detect a significant heterogeneity among the included studies (I2 = 29.0%, Q = 16.9, P = 0.15). Then, we used a fixed-effects model to assess the association between RASSF1A promoter methylation and ovarian cancer. In the meta-analysis, RASSF1A promoter methylation frequency was significantly associated with ovarian cancer (Summary OR was 11.17, 95% CI = 7.51–16.61) (Fig. 2).
Figure 2. The estimates for RASSF1A methylation frequency associated with ovarian cancer in the meta-analysis.
Meta-regression and Subgroup Analysis
Although tests did not find a significant heterogeneity across the studies, we first conducted meta-regression to explore potential sources of heterogeneity. Based on previous studies we assumed that heterogeneity may come from the population subgroup, sample size and control sample type. We conducted a multiple regression model with three variables (i.e. population subgroup, publication year, and case sample size). In the end, no source of significant heterogeneity was found (Table 3). We performed a subgroup analysis to further evaluate the source of the heterogeneity according to populations, case sample size and control type.
Table 3. Meta-regression analysis.
| 95% CI | ||||
| Heterogeneity sources | Coefficient | Lower | Upper | P |
| Population | −1.10 | −2.91 | 0.71 | 0.23 |
| Publication year | −0.09 | −0.34 | 0.16 | 0.48 |
| Case sample size | −0.12 | −2.39 | 2.16 | 0.92 |
In the subgroup analysis, the OR between the RASSF1A promoter methylation and ovarian cancer was 14.76 (95% CI = 5.73–38.01) in Asians and 6.85 (95% CI = 3.46–13.58) in Caucasians under the fixed-effects model. Similarly, the OR for the case sample size subgroup was 8.93 (95% CI = 4.43–18.42) in the <50 case group under the fixed-effects model, and 11.19 (95% CI = 4.83–25.96) in the ≥50 case group under the random-effects model. In the subgroup analysis of control type, the OR was 8.43 (95% CI = 4.03–17.63) in the BAT group including benign ovarian tissues and adjacent tissues under the random-effects model and 16.50 (95% CI = 6.82–39.88) in the NT group under the fixed-effects model (Table 4).
Table 4. Subgroup analysis of the association between RASSF1A promoter methylation and ovarian cancer.
| Cancer | Normal | M-H pooled OR† | D+L pooled OR‡ | Heterogeneity | |||||
| Group | M+ | N | M+ | N | OR (95% CI) | OR (95% CI) | I2 (%) | P | τ2 |
| Total | 364 | 763 | 32 | 438 | 11.17 (7.51–16.61) | 9.26 (5.30–16.19) | 29.0 | 0.153 | 0.27 |
| Population subgroup | |||||||||
| Asians | 249 | 510 | 22 | 314 | 13.97 (8.54–22.85) | 14.76 (5.73–38.01) | 55.0 | 0.038 | 0.74 |
| Caucasians | 115 | 253 | 10 | 124 | 6.85 (3.46–13.58) | 5.97 (2.96–12.05) | 0.0 | 0.724 | <0.01 |
| Case sample size | |||||||||
| <50 | 90 | 216 | 8 | 142 | 8.93 (4.33–18.42) | 6.96 (3.27–14.81) | 0 | 0.473 | <0.01 |
| ≥50 | 243 | 547 | 24 | 296 | 12.26 (7.62–19.72) | 11.19 (4.83–25.96) | 51.3 | 0.055 | 0.57 |
| Control type | |||||||||
| BAT | 356 | 743 | 29 | 336 | 8.95 (5.91–13.55) | 8.43 (4.03–17.63) | 48.3 | 0.031 | 0.67 |
| NT | 164 | 331 | 3 | 102 | 16.50 (6.82–39.88) | 12.82 (5.08–32.37) | 0.0 | 0.566 | <0.01 |
BAT: benign ovarian tissues and adjacent tissues; NT: normal ovarian tissues of cancer-free patients or healthy people.
the fixed-effects model.
the random-effects model.
Sensitivity Analysis
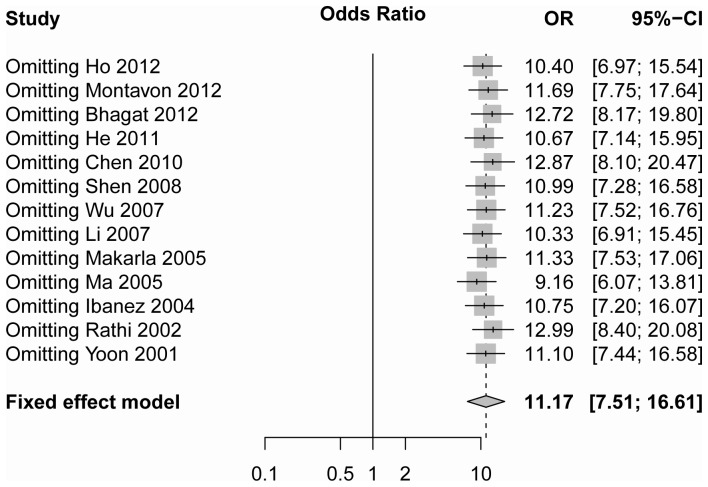
According to sensitivity analysis the odds ratio ranged from 9.16 (95% CI = 6.07–13.81) to 12.99 (95% CI = 8.40–20.08) by omitting a single study under the random-effect model (Fig. 3). No single study was found to affect the pooled OR as indicated by sensitivity analysis.
Figure 3. The sensitivity analysis by omitting a single study under the fixed-effects method.
Publication Bias
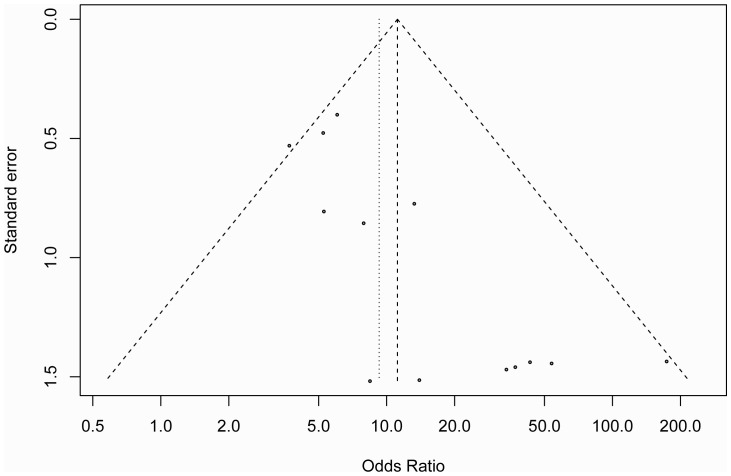
The study by Peters [29] showed that type I error rates for the Egger’s test [13] are higher than those for the alternative regression test when the summary estimates are lnORs. Therefore, Begg’s rank correlation, a funnel plot and the Peters test were employed to assess the publication bias of the literature. The shape of the Begg’s funnel plot in Figure 4 showed a possible asymmetry but the Peters test did not detect publication bias (P = 0.48) and the Begg’s rank correlation also did not detect publication bias (P = 0.14). The fail-safe number (Z = 48.23, Nfs0.05 = 851.98, Nfs0.01 = 415.53) also suggested that the study had a very small degree of publication bias.
Figure 4. The Begg’s funnel plot for assessment of publication bias in the meta-analysis (each study is represented by an point).
Discussion
RASSF1A modulates multiple apoptotic, tubulin dynamics and cell cycle checkpoint pathways. Some studies found that the overexpression of RASSF1A promotes apoptosis and cell cycle arrest in cancer cell lines [9], [30]. RASSF1A can be inactivated by gene mutation and promoter methylation and the latter accounted for the vast majority of cases [9]. RASSF1A promoter methylation is one of the most frequent epigenetic inactivation events detected in human cancer and leads to silencing of RASSF1A expression [31]. Loss of RASSF1A expression has been reported in lung, breast, bladder, gastric, cholangiocarcinoma and oesophageal sqaumous cell carcinoma primary tumors [32]–[36].
The present meta-analysis included thirteen articles with 763 cases and 438 controls. The RASSF1A methylation level of the cancer group was significantly higher than the control group. The pooled odds ratio under fixed-effect model was 11.17 (95% CI = 7.51–16.61) in the cancer cases versus the controls. The result demonstrated that RASSF1A promoter methylation was associated with ovarian cancer, which was consistent with previous studies [22], [23].
The summary OR was 14.76 (95% CI = 5.73–38.01) in Asians and 6.85 (95% CI = 3.46–13.58) in Caucasians. The association between RASSF1A promoter methylation and ovarian cancer in Asians was stronger than that in Caucasians. Similarly,Fraser’s study observed divergence of DNA methylation between African and European population, which may be due to methylation-associated SNPs (mSNPs) and complex epistasis or gene × environment interactions [37].
For sample size, the summary OR was 8.93 (95% CI = 4.43–18.42) in the <50 case group and 11.19 (95% CI = 4.83–25.96) in the ≥50 case group. In another subgroup the summary OR changed a little between <50 case group and ≥50 case group. The ORs for the different control subgroup were 8.43 (95% CI = 4.03–17.63) for the BAT group and 16.50 (95% CI = 6.82–39.88) for the NT group. This result showed that the difference in frequency of RASSF1A promoter methylation between the case group and the NT group was greater than that between the case group and the BAT group. The result indicated that benign ovarian tissues and adjacent tissues had a higher frequency of RASSF1A promoter methylation than normal ovarian tissue, which also suggested that RASSF1A promoter methylation may play an important role in the pathogenesis of ovarian carcinoma [23], [38].
There were some limitations in the study. The first limitation is potential confounder. Because there was a lack of information about variables in the control group, we only considered three variables (population, case sample size, and control type) in the subgroup analysis. Another possible confounding factors could exist in the meta-analysis. The second limitation is publication bias. Although no significant publication bias was found according to the Peter’s test, negative and unpublished studies may lead to some bias.
In conclusion, hypermethylation of RASSF1A promoter was found to be associated with ovarian cancer according to the meta-analysis, which suggested that the promoter methylation of RASSF1A is a potentially useful biomarker in the carcinogenic process of ovarian cancer. The present study requires confirmation through well-designed prospective studies.
Supporting Information
PRISMA Checklist.
(DOCX)
Acknowledgments
The authors thank Helen Neumann for editing the article.
Funding Statement
The authors have no support or funding to report.
References
- 1. Ozdemir F, Altinisik J, Karateke A, Coksuer H, Buyru N (2012) Methylation of tumor suppressor genes in ovarian cancer. Exp Ther Med 4: 1092–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee JY, Jeong W, Lim W, Lim CH, Bae SM, et al. (2013) Hypermethylation andpost-transcriptional regulation of DNA methyltransferases in the ovarian carcinomas of the laying hen. PLoS One 8: e61658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hall M, Gourley C, McNeish I, Ledermann J, Gore M, et al. (2013) Targeted anti-vascular therapies for ovarian cancer: current evidence. Br J Cancer 108: 250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barnholtz-Sloan JS, Schwartz AG, Qureshi F, Jacques S, Malone J, et al. (2003) Ovarian cancer: changes in patterns at diagnosis and relative survival over the last three decades. Am J Obstet Gynecol 189: 1120–1127. [DOI] [PubMed] [Google Scholar]
- 5. Hesson LB, Cooper WN, Latif F (2007) The role of RASSF1A methylation in cancer. Dis Markers 23: 73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barton CA, Hacker NF, Clark SJ, O’Brien PM (2008) DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol Oncol 109: 129–139. [DOI] [PubMed] [Google Scholar]
- 7. Bondurant AE, Huang Z, Whitaker RS, Simel LR, Berchuck A, et al. (2011) Quantitative detection of RASSF1A DNA promoter methylation in tumors and serum of patients with serous epithelial ovarian cancer. Gynecol Oncol 123: 581–587. [DOI] [PubMed] [Google Scholar]
- 8. Teodoridis JM, Hall J, Marsh S, Kannall HD, Smyth C, et al. (2005) CpG islandmethylation of DNA damage response genes in advanced ovarian cancer. Cancer Res 65: 8961–8967. [DOI] [PubMed] [Google Scholar]
- 9. Donninger H, Vos MD, Clark GJ (2007) The RASSF1A tumor suppressor. J Cell Sci 120: 3163–3172. [DOI] [PubMed] [Google Scholar]
- 10. Imura M, Yamashita S, Cai LY, Furuta J, Wakabayashi M, et al. (2006) Methylation and expression analysis of 15 genes and three normally-methylated genes in 13 Ovarian cancer cell lines. Cancer Lett 241: 213–220. [DOI] [PubMed] [Google Scholar]
- 11. Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 12. Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50: 1088–1101. [PubMed] [Google Scholar]
- 13. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robert R (1979) The file drawer problem and tolerance for null results. Psychological bulletin 86: 638–641. [Google Scholar]
- 15. Jiang WH, Chen BW, Zheng JG, Li WQ, Li J (2011) Application of meta Package of R in the Meta-Analysis. The Journal of Evidence-Based Medicine 11: 305–309. [Google Scholar]
- 16. Montavon C, Gloss BS, Warton K, Barton CA, Statham AL, et al. (2012) Prognostic and diagnostic significance of DNA methylation patterns in high grade serous ovarian cancer. Gynecol Oncol 124: 582–588. [DOI] [PubMed] [Google Scholar]
- 17. Chen DL, Chen SX, Feng J, Zhang CL, Liu YZ, et al. (2010) Detection and clinical significance of RASSF1A gene methylation in epithelial ovarian tumors. Chinese Journal of Cancer 20: 712–714. [Google Scholar]
- 18. Wu Q, Lothe RA, Ahlquist T, Silins I, Trope CG, et al. (2007) DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol Cancer 6: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma L, Zhang JH, Liu FR, Zhang X (2005) Hypermethylation of promoter region of RASSF1A gene in ovarian malignant epithelial tumors. Zhonghua Zhong Liu Za Zhi 27: 657–659. [PubMed] [Google Scholar]
- 20. Ibanez de Caceres I, Battagli C, Esteller M, Herman JG, Dulaimi E, et al. (2004) Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res 64: 6476–6481. [DOI] [PubMed] [Google Scholar]
- 21. Yoon JH, Dammann R, Pfeifer GP (2001) Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int J Cancer 94: 212–217. [DOI] [PubMed] [Google Scholar]
- 22. Ho CM, Huang CJ, Huang CY, Wu YY, Chang SF, et al. (2012) Promoter methylation status of HIN-1 associated with outcomes of ovarian clear cell adenocarcinoma. Mol Cancer 11: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhagat R, Chadaga S, Premalata CS, Ramesh G, Ramesh C, et al. (2012) Aberrant promoter methylation of the RASSF1A and APC genes in epithelial ovarian carcinoma development. Cellular Oncology 35: 473–479. [DOI] [PubMed] [Google Scholar]
- 24.He Y, Jin MS, Du DL, Wang CZ (2011) The study of RASSF 1A gene methylationin ovarian cancer and peritoneal washing. Journal of Bengbu Medical College 36: 35–37+41.
- 25. Shen WJ, Dai DQ, Guo KJ, Li XM (2008) RASSF1A and BRCA1 and p16 gene aberrant methylation detection and its clinical significance in epithelial ovarian cancer. Chinese Journal of Cancer Prevention and Treatment 15: 530–533. [Google Scholar]
- 26.Li QR, Liu PS, Zhang Y (2007) The detection of RASSF1A gene methylation in ovarian cancer tissue and serum and its clinical significance Journal of Shandong University (Medical Sciences): 1046–1049.
- 27. Makarla PB, Saboorian MH, Ashfaq R, Toyooka KO, Toyooka S, et al. (2005) Promoter hypermethylation profile of ovarian epithelial neoplasms. Clin Cancer Res 11: 5365–5369. [DOI] [PubMed] [Google Scholar]
- 28. Rathi A, Virmani AK, Schorge JO, Elias KJ, Maruyama R, et al. (2002) Methylation profiles of sporadic ovarian tumors and nonmalignant ovaries from high-risk women. Clin Cancer Res 8: 3324–3331. [PubMed] [Google Scholar]
- 29. Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L (2006) Comparison of two methods to detect publication bias in meta-analysis. JAMA 295: 676–680. [DOI] [PubMed] [Google Scholar]
- 30. Agathanggelou A, Cooper WN, Latif F (2005) Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res 65: 3497–3508. [DOI] [PubMed] [Google Scholar]
- 31. Pfeifer GP, Dammann R (2005) Methylation of the tumor suppressor gene RASSF1A in human tumors. Biochemistry (Mosc) 70: 576–583. [DOI] [PubMed] [Google Scholar]
- 32. Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, et al. (2001) Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst 93: 691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Byun DS, Lee MG, Chae KS, Ryu BG, Chi SG (2001) Frequent epigenetic inactivation of RASSF1A by aberrant promoter hypermethylation in human gastric adenocarcinoma. Cancer Res 61: 7034–7038. [PubMed] [Google Scholar]
- 34. Kuroki T, Trapasso F, Yendamuri S, Matsuyama A, Alder H, et al. (2003) Allele loss and promoter hypermethylation of VHL, RAR-beta, RASSF1A, and FHIT tumor suppressor genes on chromosome 3 p in esophageal squamous cell carcinoma. Cancer Res 63: 3724–3728. [PubMed] [Google Scholar]
- 35. Lee MG, Kim HY, Byun DS, Lee SJ, Lee CH, et al. (2001) Frequent epigenetic inactivation of RASSF1A in human bladder carcinoma. Cancer Res 61: 6688–6692. [PubMed] [Google Scholar]
- 36. Wong N, Li L, Tsang K, Lai PB, To KF, et al. (2002) Frequent loss of chromosome 3p and hypermethylation of RASSF1A in cholangiocarcinoma. J Hepatol 37: 633–639. [DOI] [PubMed] [Google Scholar]
- 37. Fraser HB, Lam LL, Neumann SM, Kobor MS (2012) Population-specificity of human DNA methylation. Genome Biol 13: R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi YL, Kang SY, Shin YK, Choi JS, Kim SH, et al. (2006) Aberrant hypermethylation of RASSF1A promoter in ovarian borderline tumors and carcinomas. Virchows Arch 448: 331–336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA Checklist.
(DOCX)