Abstract
Protozoan parasites are remarkably sensitive to dinitroanilines such as oryzalin, which disrupt plant but not animal microtubules. To explore the basis of dinitroaniline action, we isolated 49 independent resistant Toxoplasma gondii lines after chemical mutagenesis. All 23 of the lines that we examined harbored single point mutations in α-tubulin. These point mutations were sufficient to confer resistance when transfected into wild-type parasites. Several mutations were in the M or N loops, which coordinate protofilament interactions in the microtubule, but most of the mutations were in the core of α-tubulin. Docking studies predict that oryzalin binds with an average affinity of 23 nM to a site located beneath the N loop of Toxoplasma α-tubulin. This binding site included residues that were mutated in several resistant lines. Moreover, parallel analysis of Bos taurus α-tubulin indicated that oryzalin did not interact with this site and had a significantly decreased, nonspecific affinity for vertebrate α-tubulin. We propose that the dinitroanilines act through a novel mechanism, by disrupting M-N loop contacts. These compounds also represent the first class of drugs that act on α-tubulin function.
INTRODUCTION
Microtubules are polymers constructed from α-β-tubulin heterodimers (Downing and Nogales, 1998a,b). These structures are rapidly assembled and disassembled to create essential components of eukaryotic cells, such as spindles and flagella. The dynamic nature of microtubules makes them susceptible to pharmacological agents. Microtubule-disrupting and microtubule-stabilizing drugs have provided great insight into tubulin and microtubule function; they also have tremendous practical use. Compounds that perturb microtubule dynamics are currently some of the most effective drugs to treat medical conditions, including cancer, gout, and helminth infection (Jordan et al., 1998). Dinitroanilines (oryzalin, ethafluralin, and trifluralin) disrupt the microtubules of plants, ranging from the single-celled alga Chlamydomonas reinhardtii to higher plants such as the monocot Eleusine indica (James et al., 1993; Anthony et al., 1998; Zeng and Baird, 1999). Dinitroanilines also disrupt the microtubules of protozoa, including both free-living species such as Tetrahymena and protozoan parasites such as Trypanosoma spp., Leishmania spp., Entamoeba spp., Plasmodium falciparum, Cryptosporidium parvum, and Toxoplasma gondii (Chan and Fong, 1990; Chan et al., 1991; Gu et al., 1995; Edlind et al., 1996; Stokkermans et al., 1996; Armson et al., 1999; Makioka et al., 2000a,b; Traub-Cseko et al., 2001). Remarkably, the activity of dinitroanilines is restricted to plants and protozoa; these compounds are ineffective against vertebrate or fungal microtubules (Chan and Fong, 1990; Hugdahl and Morejohn, 1993; Murthy et al., 1994; Edlind et al., 1996).
T. gondii is a member of the Apicomplexa, a phylum of parasites that includes several medically and agriculturally significant pathogens (Black and Boothroyd, 2000). Apicomplexans are obligate intracellular parasites; these protozoa only grow and replicate within host cells. Extracellular parasites released by host cell lysis must rapidly invade new host cells to stay alive. Despite these rigorous requirements for parasite survival, apicomplexans are some of the most widespread and damaging pathogens. Human infection by T. gondii can cause life-threatening illness in immunocompromised individuals and birth defects or miscarriage during fetal infection (Hill and Dubey, 2002). Other apicomplexans include Plasmodium and Cryptosporidium, parasites of considerable medical importance, and Theileria and Eimeria, animal pathogens with extensive impact on food production (Levene, 1988).
Parasites of the Apicomplexa are named for their distinctly polarized cell apex that contains a number of unique organelles that coordinate invasion of host cells. Apicomplexan parasites are surrounded by the pellicle, a composite structure formed by association of the plasma membrane with the inner membrane complex, an assemblage of flattened vesicles (Porchet and Torpier, 1977; Dubremetz and Torpier, 1978). There are two populations of microtubules in the invasive stages of apicomplexan parasites: subpellicular microtubules and spindle microtubules (Morrissette and Sibley, 2002a,b). Subpellicular microtubules are nondynamic; they maintain both apical polarity and the characteristic crescent shape of the parasite by interacting with the pellicle (Morrissette et al., 1997). Spindle microtubules form an intranuclear spindle to coordinate chromosome segregation. Both populations are critically important to parasite survival and replication. Although extracellular parasites are refractory to the effects of microtubule-disrupting drugs, during intracellular growth parasite microtubules are dynamic and are highly sensitive to disruption (Stokkermans et al., 1996).
In this work, we demonstrate that Toxoplasma resistance to oryzalin is associated with point mutations to α-tubulin. The point mutations are sufficient to confer oryzalin resistance when introduced into wild-type (sensitive) parasites. When mapped onto the structure of α-tubulin, most mutations cluster in the core of the protein. Using representative structures of Toxoplasma α-tubulin taken from a molecular dynamics trajectory, we find that oryzalin consistently docks to α-tubulin and binding is altered in several point mutants due to conformational changes. When a similar analysis is carried out on vertebrate α-tubulin, oryzalin has significantly lower affinity and does not bind to the same region.
MATERIALS AND METHODS
Mutagenesis and Selection of Oryzalin-resistant Lines
Toxoplasma tachyzoites (RH strain) were propagated in human foreskin fibroblast (HFF) cells in DMEM with 10% fetal bovine serum. Approximately 107 extracellular tachyzoites were mutagenized in 50 or 100 μg/ml N-nitroso-N-ethylurea (Sigma-Aldrich, St. Louis, MO) in serum-free minimal essential medium for 1 h at 37°C. The parasites were washed and inoculated into 20 T25 flasks containing HFF cells. All subpopulations were kept independent to prevent isolation of sibling lines. Oryzalin (Riedel-deHaen, Seelze, Germany) stock solutions were made up in dimethyl sulfoxide. Parasites were selected at either 0.5 or 2.5 μM oryzalin. Oryzalin-resistant parasites were single cell cloned in 96-well dishes of HFF cells (Roos et al., 1994).
Drug Assays
Toxoplasma has two discrete populations of microtubules: spindle microtubules and subpellicular microtubules. The characteristic elongated shape of Toxoplasma is maintained by the ∼6-μm-long subpellicular microtubules. Shortening these subpellicular microtubules with oryzalin converts parasites to a distinctive round shape. Parasites with shortened microtubules can continue to replicate until they lyse from a host cell. However, round parasites are incapable of invading new host cells and die (Morrissette and Sibley, 2002b). To assess oryzalin resistance in the individual lines, we scored parasite shape and subpellicular microtubule length in increasing concentrations of oryzalin. Parasites were grown overnight in HFF cells on coverslips in increasing concentrations of oryzalin. The coverslips were processed for immunofluorescence staining with a rabbit peptide antibody that specifically recognizes Toxoplasma β-tubulin (Morrissette and Sibley, 2002b). Parasites were scored as having 1) a wild-type or “elongated” phenotype in which the subpellicular microtubules are long and parasites have an elongated shape, or having 2) a “round” phenotype in which the parasites are noninvasive because the subpellicular microtubules are shortened making parasites round (Morrissette and Sibley, 2002b). Resistance to oryzalin was scored as the highest concentration of drug in which intracellular replicating parasites maintained an elongated shape and long subpellicular microtubules. Because long subpellicular microtubules impart parasites with the capacity to reinvade host cells, these microtubules are an indicator of the ability to proliferate in oryzalin.
Amplification of the α-Tubulin Genes from the Oryzalin-resistant Lines
Genomic DNA was isolated from resistant Toxoplasma lines. The α-tubulin gene was amplified from genomic DNA by using PFU turbo (Stratagene, La Jolla, CA) in an MJ research thermocycler with 30 cycles of annealing at 45°C for 30 s followed by 5 min of extension at 68°C. The primers GAGTCTCGTAGAGAAC AAGC (5′ untranslated region [UTR]) and CGTTTATACCTTCACCTTTTC (3′ UTR) amplified a 2.3-kb fragment of α-tubulin. Amplified sequences were purified (polymerase chain reaction purification kit; QIAGEN, Valencia, CA), quantified on a 1% agarose gel and sequenced.
Sequencing the α-Tubulin Genes from the Oryzalin-resistant Lines
The following primers were used to obtain sequence from α-tubulin by using the Big Dye II sequencing reaction (ABI, Foster City, CA). 5′ UTR: GAGTCTCGTAGAGAACAAGC; 3′ UTR: CGTTTATACCTTCACCTTTTC; 3′ exon 1: GCTACGCGGGAGATC; 5′ exon 2: GGTTACAGCGGAACTACG; 3′ exon 2: CGGTTCCCACAGTCATATC; 5′ exon 3: GCGTCAAATCTGCAAC; forward exon 3a: CCGTCCTTTCCACTCAC; reverse exon 3a: GTGAGTGGAAAGGACGG; forward exon 3b: GTACCGTGGTGATGTCGTC; reverse exon 3b: GACGACATCACCACGGTAC. Sequences were aligned and compared using the Sequencher program (Genecodes, Ann Arbor, MI).
Introduction of Novel Restriction Site
The QuikChange kit (Stratagene) was used to ablate a unique XbaI site in the second intron of α-tubulin and to introduce a novel BamH1 site by using the primers BamH1 RE site sense GTATCACCTCTTCCACCGGGATCCTATGACTGTGGGAACCG and BamH1 RE site antisense CGGTTCCCACAGTCATAGGATCCCGGTGGAAGAGGTGATAC. This alteration distinguishes the endogenous tubulin gene from the transgene to discriminate Toxoplasma transformants with allelic replacement from those with nonhomologous insertions.
Creation of α-Tubulin Point Mutations
The mutations Ser165Pro, Ser165Thr, Ser165Ala, Ile231Thr, and Thr239Ile were constructed using the QuikChange kit (Stratagene) to modify the sequence of wild-type genomic α-tubulin containing the BamH1 restriction site. The Ser165Pro construct was created with primers Ser165Pro sense GTTGACTACGGCAAGAAGCCGAAGCTGAACTTCTGC and Ser165Pro antisense GCAGAAGTTCAGCTTCGGCTTCTTGCCGTAGTCAAC. The Ser165Thr construct was created with primers Ser165Thr sense GTTGACTACGGCAAGAAGACGAAGCTGAACTTCTGCTCG and Ser165Thr antisense CGAGCAGAAGTTCAGCTTCGTCTTCTTGCCGTAGTCAAC. The Ser165Ala construct was created with primers Ser165Ala sense GTTGACTACGGCAAGAAGGCGAAGCTGAACTTCTGCTCG and Ser165Ala antisense CGAGCAGAAGTTCAGCTTCGCCTTCTTGCCGTAGTCAAC. The Ile231Thr construct was created with primers Ile231Thr sense GACTGATTGCCCAGGTCACCTCCTCCCTGACC and Ile231Thr antisense GGTCAGGGAGGAGGTGACCTGGGCAATCAGTC. The Thr239Ile construct was created with the primers Thr239Ile sense GCCCAGGTCATCTCCTCCCTGATCGCGTCTCTCCG and Thr239Ile antisense CGGAGAGACGCGATCAGGGAGGAGATGACCTGGGC. Double and triple mutant constructs were created using the same primers to sequentially introduce the individual point mutations. Because the Ile231Thr and the Thr239Ile primers overlap, the primers GACTGATTGCCCAGGTCACCTCCTCCCTGATC and GATCAGGGAGGAGGTGACCTGGGCAATCAGTC were used to introduce the Thr239Ile mutation into the Ile231Thr construct.
Transformation, Selection, and Analysis of Oryzalin-resistant Transformants
The linearized transgene constructs with the diagnostic restriction enzyme change and the point mutation were electroporated into RH strain tachyzoites. Approximately 107 tachyzoites were transfected with ∼8 μg of DNA by using electroporation parameters established previously (Soldati and Boothroyd, 1993). Parasites were selected in 0.5 μM oryzalin; resistant tachyzoites were single cell cloned (as described above). Individual clones were assayed for transformation by amplification of the α-tubulin gene by using the primers 5′ coding CAAAATGAGAGAGGTTATCAGC and 3′ coding TTAGTACTCGTCACCATAGCC. Amplified α-tubulin was digested with either XbaI or BamH1. Transformed parasites (allelic replacements and pseudodiploids) were assayed for their drug resistance by using increasing concentrations of oryzalin (see above).
Tubulin Structure Analysis
The amino acid sequences of Sus scrofa (pig) (gi:15988311) and T. gondii (gi:161937) α-tubulins were aligned using Clustal within the Vector NTI suite of programs (Higgins et al., 1996). The amino acid sequences of Toxoplasma α-tubulin (gi:161937) was fit to the structure of bovine tubulin (1JFF) by using the Swiss-Model Automated Comparative Protein Modeling Server (http://www.expasy.ch/swissmod/) in the first approach mode (Guex and Peitsch, 1997). The resulting structure was viewed with SwissPdb Viewer version 3.7 (http://www.expasy.ch/spdbv/).
Computational Techniques
To prepare α-tubulin for our docking studies, the missing residues (Gln35 to Lys60) of bovine α-tubulin were modeled based on the analogous bovine β-tubulin N loop structure (1JFF). This homology modeling was followed by energy minimization using the OPLSAA force-field in Tinker 4.1 (http://dasher.wustl.edu/ponder; Ponder and Richards, 1987). We created the T. gondii α-tubulin structure by mutating the completed bovine structure to the Toxoplasma α-tubulin sequence by using WHAT-IF (Vriend, 1990). The coordinates of the N-site GTP (1JFF) were obtained from the Protein Data Bank. The resulting complexes (Toxoplasma α-tubulin with GTP and bovine α-tubulin with GTP) were minimized, heated and equilibrated using NAMD, a package developed by the Theoretical and Computational Biophysics Group in the Beckman Institute for Advanced Sciences and Technology at the University of Illinois at Urbana-Champaign (Kale et al., 1999). Molecular dynamics simulations were performed using the CHARMM27 force-field with TIP3P solvent. Representative structures were taken every 500 ps from the 2.5-ns trajectory for use in the docking simulations. By using AutoDock3.0 (Morris et al., 1998), flexible docking of oryzalin onto each snapshot taken from the trajectory were performed. Thirty trial runs were carried out for each α-tubulin conformation followed by cluster analysis of the results.
RESULTS
Oryzalin-resistant Tachyzoites Have Diverse Phenotypes
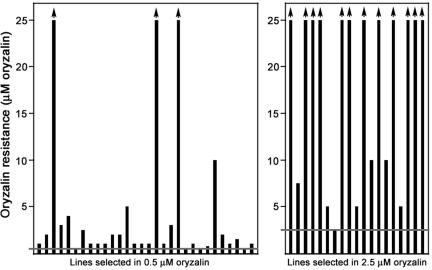
Although dinitroanilines are potent inhibitors of microtubules in plants and protozoa, their precise mode of action is unknown. To determine the molecular basis of dinitroaniline action, we generated oryzalin-resistant lines by using the chemical mutagen N-nitroso-N-ethylurea (Waldeland et al., 1983; Pfefferkorn, 1984; Dobrowolski and Sibley, 1996). Because Toxoplasma is haploid, genetic changes that confer resistance are readily detected, including mutations that are recessive in diploid organisms. We selected for resistance to either 0.5 or 2.5 μM oryzalin. In earlier work, we demonstrated that parasites treated with 0.5 μM oryzalin have shortened, nonfunctional subpellicular microtubules but intact spindle microtubules. In contrast, parasites treated with 2.5 μM oryzalin are missing all microtubules (Morrissette and Sibley, 2002b). The individual oryzalin-resistant Toxoplasma lines displayed distinct drug resistance profiles (Figure 1 and Table 1). Most of the 0.5 μM resistant lines were only moderately resistant to increased oryzalin concentrations. In contrast, the majority of the 2.5 μM resistant lines were resistant to >10-fold increases in oryzalin concentration.
Figure 1.
Left, levels of resistance to oryzalin displayed by the Toxoplasma lines selected in 0.5 μM oryzalin (horizontal gray line). Most lines selected for resistance to 0.5 μM oryzalin showed only slight resistance to increased oryzalin. Right, Toxoplasma lines selected in 2.5 μM oryzalin (horizontal gray line). Most lines selected for resistance to 2.5 μM oryzalin were highly resistant to increased oryzalin.
Table 1.
Properties of the oryzalin-resistant lines
| Amino acid substitution | Oryzalin selection | Maximum resistance, μM | Codon change | Location in protein |
|---|---|---|---|---|
| His8Tyr | 0.5 μM | 10.0 | CAC to TAC | β-sheet 1 |
| His28Gln | 0.5 μM | 1.0 | CAT to CAA | N loop |
| Phe52Ile | 0.5 μM | 2.5 | TTC to ATC | N loop |
| Phe52Leu | 2.5 μM | 2.5 | TTC to CTC | N loop |
| Leu136Phe | 2.5 μM | >25 | TTG to TTC | (β-sheet 4) |
| Asn139Lys | 0.5 μM | 1.0 | AAC to AAA | β-sheet 4 |
| Ser165Ala | 2.5 μM | 5.0 | TCG to GCG | β-sheet 5 |
| Ser165Pro | 0.5 μM | 1.0 | TCG to CCG | β-sheet 5 |
| Ser165Pro | 0.5 μM | 5.0 | TCG to CCG | β-sheet 5 |
| Ser165Thr | 0.5 μM | 2.0 | TCG to ACG | β-sheet 5 |
| Ser165Thr | 0.5 μM | >25 | TCG to ACG | β-sheet 5 |
| Ile231Thr | 0.5 μM | 1.0 | ATC to ACC | α-helix 7 |
| Ile235Val | 0.5 μM | 1.0 | ATC to GTC | α-helix 7 |
| Leu238Val | 0.5 μM | 1.0 | CTG to GTG | α-helix 7 |
| Thr239Ile | 2.5 μM | >25 | ACC to ATC | α-helix 7 |
| Arg243Cys | 2.5 μM | >25 | CGT to AGT | After α-helix 7 |
| Arg243Ser | 2.5 μM | >25 | CGT to TGT | After α-helix 7 |
| Val252Leu | 0.5 μM | >25 | GTG to TTG | (α-helix 8) |
| Ile275Thr | 0.5 μM | 0.5 | ATC to ACC | M loop |
| Ala295Val | 0.5 μM | 2.0 | GCT to GTT | Near M loop (α-helix 9) |
| Met301Thr | 0.5 μM | >25 | ATG to ACG | After α-helix 9 |
| Ile378Met | 0.5 μM | 3.0 | ATC to ATG | β-sheet 10 |
| Met391Ile | 0.5 μM | 1.0 | ATG to ATT | α-helix 11 |
| Tyr24His | C. reinhardtiia | Near N loop | ||
| Thr239Ile | E. indictab | (α-helix 7) | ||
| Met268Thr | E. indictab | Near M loop |
The Oryzalin-resistant Lines Harbor Different Single α-Tubulin Point Mutations
One potential mechanism for resistance to the dinitroanilines would be mutations to tubulin that alter drug binding or microtubule dynamics. The single α-tubulin gene from 23 of the 49 oryzalin-resistant lines was amplified and sequenced to identify base changes conferring amino acid substitutions to tubulin. Each line had a single point mutation to α-tubulin; collectively, there were 21 different amino acid substitutions at 16 positions in α-tubulin (Table 1). Ser165 was a particularly frequent target and was mutated in five of the 23 lines to threonine, proline, or alanine. Either isoleucine or leucine replaced Phe52 and either cysteine or serine replaced Arg243. The Thr239Ile point mutation was also previously identified in E. indica (goosegrass) as a dinitroaniline-resistant α-tubulin mutation (Anthony et al., 1998).
Tubulin Mutations Are Sufficient to Confer Oryzalin Resistance in Toxoplasma
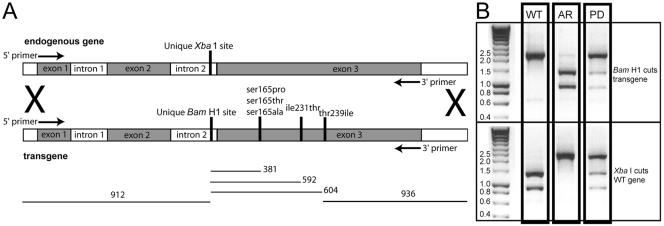
To demonstrate that the α-tubulin mutations were sufficient to confer oryzalin resistance in a wild-type background, they were introduced into the parental Toxoplasma strain. To distinguish wild-type α-tubulin from the α-tubulin transgene, intron 2 was altered to ablate a unique XbaI restriction site and to introduce a unique BamHI restriction site (Figure 2A). The BamHI restriction site is closely linked to the individual point mutations. Five single point mutations (Ser165Pro, Ser165Thr, Ser165Ala, Ile231Thr, and Thr239Ile) were individually introduced into this construct. To test whether the substitutions were additive in combination, the double and triple permutations of these substitutions were also made.
Figure 2.
(A) Diagram of the exon-intron structure of α-tubulin. Endogenous (wild-type) α-tubulin has a unique XbaI restriction site in intron 2. The transgene was altered to ablate the XbaI site and to introduce a unique BamHI site in intron 2. The point mutations Ser165Pro, Ser165Thr, Ser165Ala, Ile231Thr, and Thr239Ile were introduced in the transgene construct both individually and in double and triple combinations. The α-tubulin from transformants with oryzalin resistance was amplified with primers (arrows) that are internal to the 5′ and 3′ ends of the transgene and represent the 5′ end of exon 1 and 3′ end of exon 3. (B) Restriction enzyme analysis of the transformed oryzalin-resistant lines gives easily distinguishable patterns after amplification of the α-tubulin gene. The endogenous (WT) α-tubulin gene is cut by BamH1 and is not cut by XbaI. Conversely, the allelic replacement (AR, homologous integration) is cut by XbaI and is not cut by BamHI. Nonhomologous integration of the transgene creates a pseudodiploid (PD). In this case, both the BamHI and XbaI enzymes cut incompletely.
A linear construct of the α-tubulin gene containing the BamHI restriction site and the specific point mutation or mutations was transfected into wild-type (sensitive) Toxoplasma. After 24 h of recovery, the parasites were placed under selection in 0.5 μM oryzalin, the lowest concentration of drug that effectively selects for resistance. Resistant parasites were single cell cloned and expanded. The lines were assayed for allelic integration versus random insertion of the transgene by amplification of the α-tubulin gene from parasite lysate and restriction enzyme analysis of the product (Figure 2B). As a control, the wild-type α-tubulin gene with the restriction site change was transfected into Toxoplasma; resistant lines did not arise after selection in 0.5 μM oryzalin.
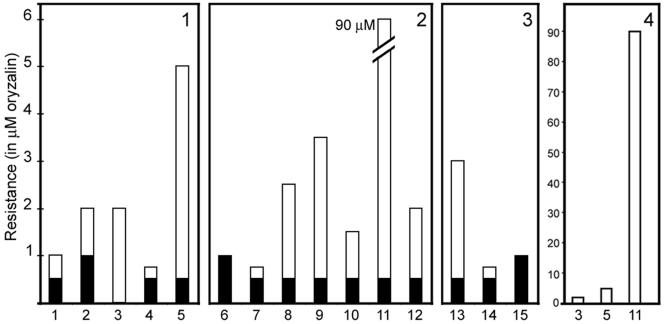
Transformation of the tubulin point mutation genes was sufficient to confer resistance to oryzalin in Toxoplasma (Figure 3). In the pseudodiploid (nonhomologous integration) state (Figure 3, black bars), point mutations conferred resistance to lower levels of oryzalin than did allelic replacements of the same constructs (Figure 3, white bars). Allelic replacement of wild-type tubulin with the α-tubulin point mutations conferred resistance to between 0.75 and 5.0 μM oryzalin. The single point mutation Thr239Ile conferred the highest level of resistance. In almost all cases, the double and triple point mutation combinations were less resistant than the single mutations, indicating that these point mutations were not additive in combination (Figure 3, panels 2 and 3). However, the double mutation Ser165Ala/Thr239Ile was highly resistant to oryzalin (>90 μM; Figure 3, panel 4), indicating that these substitutions are synergetic. Oryzalin is insoluble in aqueous solutions above ∼95 μM, making it impossible to determine the full degree of drug resistance of the Ser165Ala/Thr239Ile double mutation.
Figure 3.
Degree of oryzalin resistance was assessed for lines transformed with the α-tubulin constructs. The resistance of both homologous integrations (allelic replacements) and nonhomologous integrations (pseudodiploids) was measured for each construct. Allelic replacements (white bars) show greater oryzalin resistance than do pseudodiploids (black bars). Panel 1, single mutations (1) Ser165Pro, (2) Ser165Thr, (3) Ser165Ala, (4) Ile231Thr, and (5) Thr239Ile. Panel 2, double mutations (6) Ser165Pro/Ile231Thr, (7) Ser165Pro/Thr239Ile, (8) Ser165Thr/Ile231Thr, (9) Ser165Thr/Thr239Ile, (10) Ser165Ala/Ile231Thr, (11) Ser165Ala/Thr239Ile, and (12) Ile231Thr/Thr239Ile. Panel 3, triple mutations (13) Ser165Pro/Ile231Thr/Thr239Ile, (14) Ser165Thr/Ile231Thr/Thr239Ile, and (15) Ser165Ala/Ile231Thr/Thr239Ile. Allelic replacements of Ser165Pro/Ile231Thr and Ser165Ala/Ile231Thr/Thr239Ile were not obtained nor were nonhomologous integrations of Ser165Ala and Ser165Pro/Ile231Thr. Panel 4, synergism of the double point mutation Ser165Ala/Thr239Ile. (3) Ser165Ala, (5) Thr239Ile, and (11) Ser165Ala/Thr239Ile lines.
In some cases, two of the resistant lines from the mutagenesis screen contained the same α-tubulin point mutation but displayed distinct resistance profiles. The clearest example of this was seen with the Ser165Thr point mutation. Two lines harbored this substitution but showed dramatically different resistance profiles: one line was resistant to 2.0 μM oryzalin, whereas the other was resistant to >25 μM oryzalin (Table 1). When the Ser165Thr point mutation was introduced as an allelic replacement, the α-tubulin point mutation alone conferred resistance to 2.0 μM oryzalin (Figure 3). Therefore, the line with resistance to >25 μM oryzalin must have an additional mutation(s) superimposed upon the α-tubulin mutation to increase resistance to oryzalin. We hypothesize that lines with high levels of drug resistance (Figure 1) have additional mutations superimposed on the α-tubulin mutation to increase resistance to oryzalin.
Most Tubulin Mutations Localize to the Core of α-Tubulin
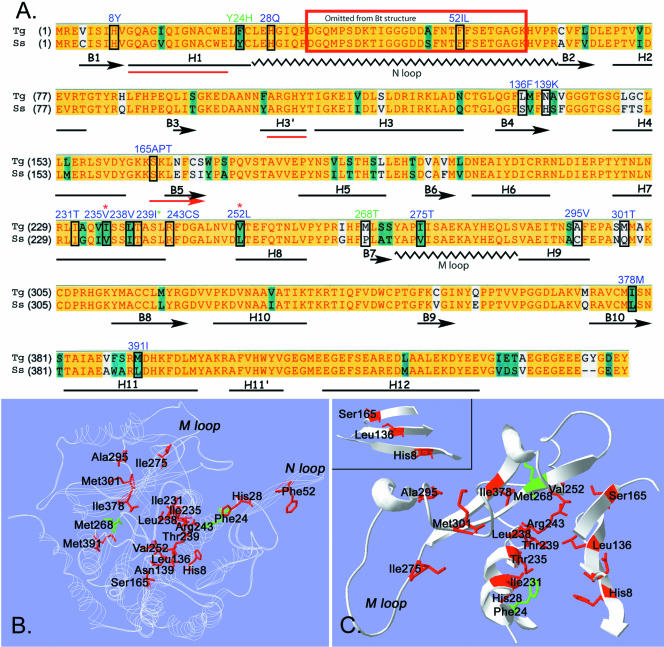
The 23 point mutations identified in this study were distributed throughout the linear sequence of Toxoplasma α-tubulin. To investigate how these amino acid mutations might affect tubulin function, we evaluated their conservation in other α-tubulins and their spatial distribution in a structural model of α-tubulin. Figure 4A is a Clustal alignment of S. scrofa α-tubulin (insensitive to the dinitroanilines) with Toxoplasma α-tubulin. Although some point mutations occur in residues that distinguish Toxoplasma (sensitive) from pig (insensitive) α-tubulin, many resistance mutations occur in conserved amino acids. The amino acid sequence of Toxoplasma α-tubulin was fit to the electron diffraction structure of bovine α-tubulin by using the Swiss-Model Automated Comparative Protein Modeling Server Web site in the first approach mode. The mutated amino acids in α-tubulin were identified within the modeled Toxoplasma α-tubulin structure. Two previously identified plant mutations Tyr24His and Met268Thr that confer dinitroaniline resistance were also included in our analysis (James et al., 1993; Yamamoto et al., 1998). The majority of the point mutations clustered within the core of α-tubulin (Figure 4B). Some of the mutated residues were distributed in striking patterns. For example, several of the mutated resides residues (Ile231, Ile235, and Thr239) localized along a single face of helix 7 in α-tubulin (Figure 4C). Arg243 and Leu238 were also in this immediate proximity. In addition, His8, Leu136, and Ser165 occupied essentially linear positions on three strands (β-strands 1, 4, and 5) of a β-sheet (Figure 4C, inset).
Figure 4.
(A) Clustal alignment of oryzalin-sensitive Toxoplasma (Tg) and oryzalin-insensitive pig (S. scrofa, Ss) α-tubulins. A yellow background represents amino acid identity, teal denotes residue similarity, and white indicates nonconserved residues. The distribution of α-helices, β-strands, and coils in bovine α-tubulin (black, below Clustal sequence) were adapted from Lowe et al. (2001) who solved the structure of bovine tubulin, but mapped it onto the amino acid sequence of pig tubulin. The three red lines (H1, H3′, and B5) indicate regions where the predicted secondary structure of Toxoplasma α-tubulin deviates from the bovine secondary structure. The boxed residues are mutated in the individual oryzalin-resistant lines. The blue-labeled residues were identified in Toxoplasma oryzalin-resistant lines in this study. Three dinitroaniline resistance mutations (two labeled in green and one with a green asterisk) were identified in previous work in Chlamydomonas and Eleusine. (B and C) The amino acid sequence of Toxoplasma α-tubulin was fit to the structure of bovine α-tubulin by using Swiss-Model automated comparative protein modeling. B shows a ribbon model of Toxoplasma α-tubulin (white). The residues that are mutated to confer oryzalin resistance are colored red. The plant mutations at residues 24 and 268 are colored green. The position of the N loop containing the Phe52 mutations is not necessarily accurate because this area was disordered in structural studies and was eliminated from the final model. C shows cut-away views of some of the α-tubulin residues that are mutated to confer oryzalin resistance. Many residues are in striking proximity and show specific orientations. For example, residues Ile231, Ile235, and Thr239 all occur on the same face of α-helix 7. Leu238, Arg243, and Val252 cluster in the same general area. The inset shows a β-sheet formed by β-strands 1, 4, and 5. The mutated residues His8, Leu136, and Ser165 are in a linear pattern across the β-sheet.
Some Mutations Localize to Domains That Coordinate Protofilament-Protofilament Contact
Structural studies have established the critically important role of two tubulin domains (the M loop and the N loop) in microtubule assembly. These loops coordinate lateral interactions between protofilaments to build a microtubule. Studies docking the tubulin dimer structure into high-resolution images of microtubules have established that M loops interact with N loops of laterally adjacent subunits (Li et al., 2002). Moreover, organisms with increased microtubule stability (such as arctic fish) have two amino acid substitutions in the α-tubulin M loop, Ala278Thr and Ser287Thr (Detrich et al., 2000). The oryzalin-resistant point mutation Ile275Thr was an M loop substitution, and three of the point mutations described here (His28Gln, Phe52Ile, and Phe52Leu) were in the N loop (Figure 4). These α-tubulin mutations may compensate for the destabilizing effects of the dinitroanilines by increasing the intrinsic stability of Toxoplasma microtubules.
Identification of the Oryzalin Binding Site
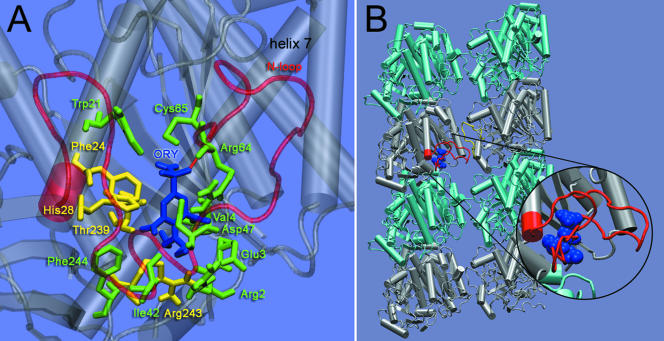
Although our analysis of the resistance mutations implies that oryzalin binds to α-tubulin, the spatial distribution of the mutations and the fact that many mutations were in the core of the protein suggested that the mutations did not directly define a binding site for the dinitroanilines. For this reason, we used computational techniques, with molecular dynamics simulations to generate multiple conformations for Toxoplasma α-tubulin, and flexible docking simulations to locate the binding site (see MATERIALS AND METHODS). The representative structures of Toxoplasma α-tubulin taken from this molecular dynamics trajectory, showed that oryzalin consistently docked to residues Arg2, Glu3, Val4, Trp21, Phe24, His28, Ile42, Asp47, Arg64, Cys65, Thr239, Arg243, and Phe244 (Figure 5A). The binding affinity at this site was 23 nM. This binding site placed oryzalin beneath the N loop (Figure 5). We also analyzed Toxoplasma α-tubulin containing point mutations that conferred resistance. For the mutants Thr239Ile, Ser165Ala, and Thr239Ile/Ser165Ala, oryzalin binding was eliminated at the site identified in the wild-type Toxoplasma α-tubulin.
Figure 5.
(A) Representative structure of oryzalin bound to α-tubulin as predicted by the docking simulations. The binding site consists of Arg2, Glu3, Val4, Trp21, Phe24, His28, Ile42, Asp47, Arg64, Cys65, Thr239, Arg243, and Phe244. Residues Phe24, His28, Thr239, and Arg243 (yellow) were mutated in resistant lines; all other binding site residues are colored green. The binding site lies beneath the N loop (translucent red). The SO2 group of oryzalin forms a hydrogen bond with the backbone amide (NH) of Arg64 (red). (B) Two protofilaments, each consisting of two α-β heterodimers, with bound oryzalin (blue). Oryzalin binds to α-tubulin (silver) beneath the N loop (red); this may interfere with lateral interactions between the N loop and the M loop (yellow) of the adjacent protofilament. The inset shows oryzalin bound beneath the N loop.
In parallel studies, we investigated the interaction of oryzalin with Bos taurus α-tubulin by docking oryzalin to a set of structures obtained from the molecular dynamics trajectories. The results indicated that oryzalin does not interact with equivalent residues in vertebrate α-tubulin. It has a significantly decreased overall affinity (>50-fold) for bovine tubulin and no true consensus binding site. The limited interaction of oryzalin with bovine tubulin does not imply a secondary binding site for oryzalin on bovine α-tubulin but indicates nonspecific, low-affinity interactions in single tubulin snapshots.
DISCUSSION
Analysis of oryzalin resistance in T. gondii implicates α-tubulin as the target for dinitroaniline action. We demonstrate here that all Toxoplasma lines with resistance to oryzalin have a point mutation in the single α-tubulin gene. These α-tubulin point mutations are sufficient to confer resistance to oryzalin in Toxoplasma. The preponderance of α-tubulin mutations identified in this study suggests that dinitroanilines bind to and act on α-tubulin. This is unusual, as all characterized compounds that perturb microtubule function bind to and act on β-tubulin (Nogales, 2000). Several of the resistance mutations are located in the M or N loops, regions of tubulin that mediate lateral adhesion of protofilaments (Downing and Nogales, 1998a; Nogales et al., 1998, 1999; Lowe et al., 2001). These mutations are predicted to counteract dinitroaniline action by hyperstabilizing microtubules. However, most mutations were found to be in the core of α-tubulin. We hypothesize that core mutations affect the conformation of the α-tubulin dinitroaniline binding site.
Docking simulations were used to identify the α-tubulin binding site for oryzalin. These simulations were based on a genetic algorithm where the results of individual searches are clustered based on their root mean square deviation. For each α-tubulin conformation, we found the consensus binding site (Figure 5A) was both the largest cluster and contained the binding conformations with the highest affinity. The binding affinity predicted for this site was 23 nM and is in agreement with experimentally determined values of 95-117 nM obtained with purified plant tubulin (Hugdahl and Morejohn, 1993; Murthy et al., 1994). The site identified by our docking simulations is located beneath the N loop and is composed of residues Arg2, Glu3, Val4, Trp21, Phe24, His28, Ile42, Asp47, Arg64, Cys65, Thr239, Arg243, and Phe244 (Figure 5A). Several resistance mutations (Phe24, His28, Thr239, and Arg243) map to this binding site. We propose that additional mutated residues in the core of α-tubulin also affect dinitroaniline binding at this site. For example, Thr239 is a binding site residue located on α-helix 7 along with mutated residues Ile231, Ile235, and Leu238 (Figures 4C and 5A). Presumably, mutation of any one of these other amino acids alters α-helix 7, the behavior of Thr239, and ultimately the properties of the binding site (conformation, flexibility, and dynamics). In a recent article, Blume et al. (2003) used analysis of surface electrostatic potential to suggest that dinitroanilines bind α-tubulin residues located at the dimer interface. They propose a binding site consisting of Cys4, His8, Leu136, Phe138, Thr239, Arg243, Asp251, Val252, and Asn253 based on a region of altered electrostatic potential between wild-type E. indica α-tubulin and the resistant α-tubulin Thr239Ile point mutation. Although their predicted binding site is in the same vicinity and shares three common residues (Val/Cys4, Thr239, and Arg243), the two predictions are distinct. Our binding site is not at the dimer-dimer interface, but rather directly behind the N-loop, suggesting a direct mechanism for affecting lateral contacts that lead to microtubule disruption (see below).
The selective action of the dinitroanilines on plant and protozoan tubulin suggests that only plant and protozoan α-tubulins bind these compounds. This is supported by our computational analysis docking oryzalin to a set of B. taurus α-tubulin structures obtained from molecular dynamics trajectories. Oryzalin has a decreased overall affinity for bovine tubulin and the trajectory snapshots do not contain a consistent oryzalin binding site. Plant and protozoan lineage-restricted residues do not directly account for the lack of a consensus binding site because binding site residues are not specific to plants and protozoa. We hypothesize that lineage restricted residues elsewhere in α-tubulin selectively create a binding pocket in plant and protozoan α-tubulin (Morrissette, Mitra, and Sept, unpublished data).
Beyond allowing us to identify the binding site on α-tubulin, our docking results suggest a mechanism for how dinitroanilines cause microtubule disassembly. When oryzalin was inserted into the α-tubulin binding site within the microtubule structure, it was situated beneath the N loop between protofilaments in the microtubule (Figure 5B). Studies fitting the tubulin dimer structure into high-resolution cryoelectron microscopy images of microtubules have established that protofilament-protofilament contact is mediated by N loops interacting with M loops of laterally adjacent subunits (Li et al., 2002). Oryzalin bound beneath the N loop may inhibit N loop interaction with the M loop of the adjacent protofilament (Figure 5B, enlargement) with the consequence of microtubule disruption. Indeed, the significance of M-N loop contact for microtubule stability can be appreciated by considering the action of taxol, a drug which stabilizes microtubules by reinforcing M-N loop interactions in β-tubulin (Snyder et al., 2001).
Dinitroaniline resistance was previously investigated in the single cell alga Chlamydomonas reinhardtii. Resistance in Chlamydomonas is rarely due to mutations to tubulin (James et al., 1988; James and Lefebvre, 1989; James et al., 1989, 1993; Lux and Dutcher, 1991; Schibler and Huang, 1991). Instead, some resistance alleles act as a multidrug resistance pump (James and Lefebvre, 1989), whereas others are regulators of tubulin polymerization. Although Chlamydomonas is haploid, it has two copies of both the α- and β-tubulin genes (Silflow and Rosenbaum, 1981). Presumably, tubulin-based resistance does not arise in Chlamydomonas because these mutations are masked by expression of the other wild-type tubulin gene. The β-tubulin mutations Lys350Met and Lys350Glu confer hypersensitivity to the microtubule-stabilizing drug taxol and resistance to dinitroanilines and other microtubule-disrupting drugs such as colchicine. These mutations function by hyperstabilizing microtubules and are not dinitroaniline specific (Schibler and Huang, 1991). The α-tubulin mutation Tyr24His was identified as a “step-up” mutation in a Chlamydomonas strain with low levels of dinitroaniline resistance and is the only specific dinitroaniline-resistance allele associated with tubulin (James et al., 1993).
The weed goosegrass rapidly acquires resistance to dinitroaniline herbicides (Anthony et al., 1998, 1999; Yamamoto et al., 1998; Anthony and Hussey, 1999; Zeng and Baird, 1999). These studies demonstrated that the α-tubulin mutations Thr239Ile and Met268Thr were associated with resistance to dinitroanilines and that the double point mutation (not observed in the original lines) produced higher oryzalin resistance than either of the single mutations alone. Multiple α- and β-tubulin genes are characteristic of higher plants and animals; this property complicated analysis of tubulin mutations in oryzalin-resistant goosegrass. To confer oryzalin resistance with α-tubulin transgenes, the α-tubulin point mutations had to be overexpressed by driving both α- and β-tubulin transgene expression with the powerful cauliflower mosaic virus 35S/maize alcohol dehydrogenase intron 1 hybrid promoter (Anthony et al., 1999). In contrast to goosegrass, Toxoplasma is haploid and has single α- and β-tubulin genes (Nagel and Boothroyd, 1988). It is therefore ideally suited for genetic analysis of recessive mutations that would be masked in a diploid organism, particularly one with redundant gene family members. These genetic qualities allowed the isolation of a large number of tubulin mutations that are partially recessive to wild-type tubulin.
It is well established that dinitroanilines selectively inhibit the microtubules of plants and protozoa and do not act on fungal or vertebrate tubulins (Chan and Fong, 1990; Chan et al., 1991; James et al., 1993; Gu et al., 1995; Edlind et al., 1996; Stokkermans et al., 1996; Anthony et al., 1998, 1999; Anthony and Hussey, 1999; Armson et al., 1999; Zeng and Baird, 1999; Makioka et al., 2000a,b; Traub-Cseko et al., 2001). Dinitroaniline disruption of plant microtubules has been exploited to develop these compounds as components of a variety of preemergence herbicides. The selective disruption of protozoan parasite microtubules by the dinitroanilines indicates that tubulin is a suitable chemotherapeutic target for control of parasite infection. Understanding the nature of the interaction between dinitroanilines and protozoan tubulin is key to the development of novel antiparasitic drugs with selective activity, appropriate pharmacokinetic properties, and low human toxicity. Moreover, studies on the mechanism of dinitroaniline disruption of microtubules will increase our understanding of the role of α-tubulin in microtubule polymerization.
Acknowledgments
We thank Eva Istvan and Jeremia Ory for discussions of the structural biology aspects of this work and Susan Dutcher for reading the manuscript. N.S.M. was partially supported by an Individual National Research Service Award fellowship F32 GM20484-01A1 and a National Institutes of Health training grant in Infectious Disease held by the Washington University School of Medicine (T32 AI-07017221). D.S. is supported by a grant from the Whitaker Foundation. L.D.S. is the recipient of a Scholar Award in Molecular Parasitology from the Burroughs Welcome Fund.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-07-0530. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-07-0530.
References
- Anthony, R.G., and Hussey, P.J. (1999). Dinitroaniline herbicide resistance and the microtubule cytoskeleton. Trends Plant Sci. 4, 112-116. [DOI] [PubMed] [Google Scholar]
- Anthony, R.G., Reichelt, S., and Hussey, P.J. (1999). Dinitroaniline herbicide-resistant transgenic tobacco plants generated by co-overexpression of a mutant alpha-tubulin and a beta-tubulin. Nat. Biotechnol. 17, 712-716. [DOI] [PubMed] [Google Scholar]
- Anthony, R.G., Waldin, T.R., Ray, J.A., Bright, S.W., and Hussey, P.J. (1998). Herbicide resistance caused by spontaneous mutation of the cytoskeletal protein tubulin. Nature 393, 260-263. [DOI] [PubMed] [Google Scholar]
- Armson, A., Kamau, S.W., Grimm, F., Reynoldson, J.A., Best, W.M., Mac-Donald, L.M., and Thompson, R.C. (1999). A comparison of the effects of a benzimidazole and the dinitroanilines against Leishmania infantum. Acta Trop. 73, 303-311. [DOI] [PubMed] [Google Scholar]
- Black, M.W., and Boothroyd, J.C. (2000). Lytic cycle of Toxoplasma gondii. Microbiol. Mol. Biol. Rev. 64, 607-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume, Y.B., Nyporko, A.Y., Yemets, A.I., and Baird, W.V. (2003). Structural modeling of the interaction of plant alpha-tubulin with dinitroaniline and phosphoroamidate herbicides. Cell Biol. Int. 27, 171-174. [DOI] [PubMed] [Google Scholar]
- Chan, M.M., and Fong, D. (1990). Inhibition of leishmanias but not host macrophages by the antitubulin herbicide trifluralin. Science 249, 924-926. [DOI] [PubMed] [Google Scholar]
- Chan, M.M., Triemer, R.E., and Fong, D. (1991). Effect of the anti-microtubule drug oryzalin on growth and differentiation of the parasitic protozoan Leishmania mexicana. Differentiation 46, 15-21. [DOI] [PubMed] [Google Scholar]
- Detrich, H.W., 3rd, Parker, S.K., Williams, R.C., Jr., Nogales, E., and Downing, K.H. (2000). Cold adaptation of microtubule assembly and dynamics. Structural interpretation of primary sequence changes present in the alpha- and beta-tubulins of Antarctic fishes. J. Biol. Chem. 275, 37038-37047. [DOI] [PubMed] [Google Scholar]
- Dobrowolski, J.M., and Sibley, L.D. (1996). Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell 84, 933-939. [DOI] [PubMed] [Google Scholar]
- Downing, K.H., and Nogales, E. (1998a). Tubulin and microtubule structure. Curr. Opin. Cell Biol. 10, 16-22. [DOI] [PubMed] [Google Scholar]
- Downing, K.H., and Nogales, E. (1998b). Tubulin structure: insights into microtubule properties and functions. Curr. Opin. Struct. Biol. 8, 785-791. [DOI] [PubMed] [Google Scholar]
- Dubremetz, J.F., and Torpier, G. (1978). Freeze fracture study of the pellicle of an eimerian sporozoite (Protozoa, Coccidia). J. Ultrastruct. Res. 62, 94-109. [DOI] [PubMed] [Google Scholar]
- Edlind, T., Li, J., and Katiyar, S. (1996). Expression of Cryptosporidium parvum beta-tubulin sequences in yeast: potential model for drug development. J. Eukaryot. Microbiol. 43, 86S. [DOI] [PubMed] [Google Scholar]
- Gu, L., Gaertig, J., Stargell, L.A., and Gorovsky, M.A. (1995). Gene-specific signal transduction between microtubules and tubulin genes in Tetrahymena thermophila. Mol. Cell. Biol. 15, 5173-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N., and Peitsch, M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714-2723. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., Thompson, J.D., and Gibson, T.J. (1996). Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266, 383-402. [DOI] [PubMed] [Google Scholar]
- Hill, D., and Dubey, J.P. (2002). Toxoplasma gondii: transmission, diagnosis and prevention. Clin. Microbiol. Infect. 8, 634-640. [DOI] [PubMed] [Google Scholar]
- Hugdahl, J.D., and Morejohn, L.C. (1993). Rapid and reversible high-affinity binding of the dinitroaniline herbicide oryzalin to tubulin from Zea mays L. Plant Physiol. 102, 725-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, S.W., and Lefebvre, P.A. (1989). Isolation and characterization of dominant, pleiotropic drug-resistance mutants in Chlamydomonas reinhardtii. Curr. Genet. 15, 443-452. [DOI] [PubMed] [Google Scholar]
- James, S.W., Ranum, L.P., Silflow, C.D., and Lefebvre, P.A. (1988). Mutants resistant to anti-microtubule herbicides map to a locus on the uni linkage group in Chlamydomonas reinhardtii. Genetics 118, 141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, S.W., Silflow, C.D., Stroom, P., and Lefebvre, P.A. (1993). A mutation in the alpha 1-tubulin gene of Chlamydomonas reinhardtii confers resistance to anti-microtubule herbicides. J. Cell Sci. 106, 209-218. [DOI] [PubMed] [Google Scholar]
- James, S.W., Silflow, C.D., Thompson, M.D., Ranum, L.P., and Lefebvre, P.A. (1989). Extragenic suppression and synthetic lethality among Chlamydomonas reinhardtii mutants resistant to anti-microtubule drugs. Genetics 122, 567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, A., Hadfield, J.A., Lawrence, N.J., and McGown, A.T. (1998). Tubulin as a target for anticancer drugs: agents which interact with the mitotic spindle. Med. Res. Rev. 18, 259-296. [DOI] [PubMed] [Google Scholar]
- Kale, L., Skeel, R., Bhandarkar, M., Brunner, R., Gursoy, A., Krawetz, N., Phillips, J., Shinozaki, A., Varadarajan, K., and Schulten, K. (1999). NAMD 2, Greater scalability for parallel molecular dynamics. J. Comput. Phys. 151, 283-312. [Google Scholar]
- Levene, N.D. (1988). The Protozoan Phylum Apicomplexa, Boca Raton, FL: CRC Press.
- Li, H., DeRosier, D., Nicholson, W., Nogales, E., and Downing, K. (2002). Microtubule structure at 8 A resolution. Structure 10, 1317-1328. [DOI] [PubMed] [Google Scholar]
- Lowe, J., Li, H., Downing, K.H., and Nogales, E. (2001). Refined structure of alpha beta-tubulin at 3.5A resolution. J. Mol. Biol. 313, 1045-1057. [DOI] [PubMed] [Google Scholar]
- Lux, F.G., 3rd, and Dutcher, S.K. (1991). Genetic interactions at the FLA10 locus: suppressors and synthetic phenotypes that affect the cell cycle and flagellar function in Chlamydomonas reinhardtii. Genetics 128, 549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makioka, A., Kumagai, M., Ohtomo, H., Kobayashi, S., and Takeuchi, T. (2000a). Effect of dinitroaniline herbicides on the growth of Entamoeba histolytica. J. Parasitol. 86, 607-610. [DOI] [PubMed] [Google Scholar]
- Makioka, A., Kumagai, M., Ohtomo, H., Kobayashi, S., and Takeuchi, T. (2000b). Effect of the antitubulin drug oryzalin on the encystation of Entamoeba invadens. Parasitol. Res. 86, 625-629. [DOI] [PubMed] [Google Scholar]
- Morris, G.M., Goodsell, D.S., Halliday, R.S., Huey, R., Hart, W.E., Belew, R.K., and Olson, A.J. (1998). Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19, 1639-1632. [Google Scholar]
- Morrissette, N.S., Murray, J.M., and Roos, D.S. (1997). Subpellicular microtubules associate with an intramembranous particle lattice in the protozoan parasite Toxoplasma gondii. J. Cell Sci. 110, 35-42. [DOI] [PubMed] [Google Scholar]
- Morrissette, N.S., and Sibley, L.D. (2002a). Cytoskeleton of apicomplexan parasites. Microbiol. Mol. Biol. Rev. 66, 21-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissette, N.S., and Sibley, L.D. (2002b). Disruption of microtubules uncouples budding and nuclear division in Toxoplasma gondii. J. Cell Sci. 115, 1017-1025. [DOI] [PubMed] [Google Scholar]
- Murthy, J.V., Kim, H.H., Hanesworth, V.R., Hugdahl, J.D., and Morejohn, L.C. (1994). Competitive inhibition of high-affinity oryzalin binding to plant tubulin by the phosphoric amide herbicide amiprophos-methyl. Plant Physiol. 105, 309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel, S.D., and Boothroyd, J.C. (1988). The alpha- and beta-tubulins of Toxoplasma gondii are encoded by single copy genes containing multiple introns. Mol. Biochem. Parasitol. 29, 261-273. [DOI] [PubMed] [Google Scholar]
- Nogales, E. (2000). Structural insights into microtubule function. Annu. Rev. Biochem. 69, 277-302. [DOI] [PubMed] [Google Scholar]
- Nogales, E., Whittaker, M., Milligan, R.A., and Downing, K.H. (1999). High-resolution model of the microtubule. Cell 96, 79-88. [DOI] [PubMed] [Google Scholar]
- Nogales, E., Wolf, S.G., and Downing, K.H. (1998). Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391, 199-203. [DOI] [PubMed] [Google Scholar]
- Pfefferkorn, E.R. (1984). Characterization of a mutant of Toxoplasma gondii resistant to aphidicolin. J. Protozool. 31, 306-310. [DOI] [PubMed] [Google Scholar]
- Ponder, J.W., and Richards, F.M. (1987). An efficient Newton-like method for molecular mechanics energy minimization of large molecules. J. Comput. Chem. 8, 1016-1026. [Google Scholar]
- Porchet, E., and Torpier, G. (1977). [Freeze fracture study of Toxoplasma and Sarcocystis infective stages (author's transl)]. Z. Parasitenkd. 54, 101-124. [DOI] [PubMed] [Google Scholar]
- Roos, D.S., Donald, R.G., Morrissette, N.S., and Moulton, A.L. (1994). Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 45, 27-63. [DOI] [PubMed] [Google Scholar]
- Schibler, M.J., and Huang, B. (1991). The colR4 and colR15 beta-tubulin mutations in Chlamydomonas reinhardtii confer altered sensitivities to microtubule inhibitors and herbicides by enhancing microtubule stability. J. Cell Biol. 113, 605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow, C.D., and Rosenbaum, J.L. (1981). Multiple alpha- and beta-tubulin genes in Chlamydomonas and regulation of tubulin mRNA levels after deflagellation. Cell 24, 81-88. [DOI] [PubMed] [Google Scholar]
- Snyder, J.P., Nettles, J.H., Cornett, B., Downing, K.H., and Nogales, E. (2001). The binding conformation of Taxol in beta-tubulin: a model based on electron crystallographic density. Proc. Natl. Acad. Sci. USA 98, 5312-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati, D., and Boothroyd, J.C. (1993). Transient transfection and expression in the obligate intracellular parasite Toxoplasma gondii. Science 260, 349-352. [DOI] [PubMed] [Google Scholar]
- Stokkermans, T.J., Schwartzman, J.D., Keenan, K., Morrissette, N.S., Tilney, L.G., and Roos, D.S. (1996). Inhibition of Toxoplasma gondii replication by dinitroaniline herbicides. Exp. Parasitol. 84, 355-370. [DOI] [PubMed] [Google Scholar]
- Traub-Cseko, Y.M., Ramalho-Ortigao, J.M., Dantas, A.P., de Castro, S.L., Barbosa, H.S., and Downing, K.H. (2001). Dinitroaniline herbicides against protozoan parasites: the case of Trypanosoma cruzi. Trends Parasitol. 17, 136-141. [DOI] [PubMed] [Google Scholar]
- Vriend, G. (1990). WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8, 52-56. [DOI] [PubMed] [Google Scholar]
- Waldeland, H., Pfefferkorn, E.R., and Frenkel, J.K. (1983). Temperature-sensitive mutants of Toxoplasma gondii: pathogenicity and persistence in mice. J. Parasitol. 69, 171-175. [PubMed] [Google Scholar]
- Yamamoto, E., Zeng, L., and Baird, W.V. (1998). Alpha-tubulin missense mutations correlate with antimicrotubule drug resistance in Eleusine indica. Plant Cell 10, 297-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L., and Baird, W.V. (1999). Inheritance of resistance to anti-microtubule dinitroaniline herbicides in an “intermediate” resistant biotype of Eleusine indica (Poaceae). Am. J. Bot. 86, 940. [PubMed] [Google Scholar]