Abstract
Despite the apparent overall structural stability of the nuclear pore complex during interphase, at least two nucleoporins have been shown to move dynamically on and off the pore. It is not yet certain what contribution nucleoporin mobility makes to the process of nuclear transport or how such mobility is regulated. Previously, we showed that Nup98 dynamically interacts with the NPC as well as bodies within the nucleus in a transcription-dependent manner. We have extended our studies of dynamics to include Nup153, another mobile nucleoporin implicated in RNA export. In both cases, we found that although only one domain is essential for NPC localization, other regions of the protein significantly affect the stability of association with the pore. Interestingly, like Nup98, the exchange of Nup153 on and off the pore is inhibited when transcription by Pol I and Pol II is blocked. We have mapped the regions required to link Nup98 and Nup153 mobility to transcription and found that the requirements differ depending on which polymerases are inhibited. Our data support a model whereby transcription of RNA is coupled to nucleoporin mobility, perhaps ultimately linking transport of RNAs to a cycle of remodeling at the nuclear pore basket.
INTRODUCTION
The vertebrate nuclear pore complex is stably anchored in the nuclear envelope through attachments to the nuclear lamina (Daigle et al., 2001); however, during the M-phase of each cell cycle the nuclear pore is disassembled into soluble component subcomplexes (Vasu and Forbes, 2001; Burke and Ellenberg, 2002; Suntharalingam and Wente, 2003). Most of the ∼30 nuclear pore proteins (nucleoporins or Nups) appear to be essential structural elements of the pore and are only released when the NPC undergoes ordered breakdown at mitosis (Belgareh et al., 2001; Daigle et al., 2001; Beaudouin et al., 2002). However, a few nucleoporins have been shown to dynamically associate with the nuclear pore during interphase when the structure remains intact. Nup153 and Nup98 move on and off the pore, and Gle1, Rae1/Gle2, Nup98, and Nup50/NPAP60 shuttle between the nucleus and cytoplasm (Pritchard et al., 1999; Daigle et al., 2001; Griffis et al., 2002; Lindsay et al., 2002; Kendirgi et al., 2003). Nup50/NPAP60 has been proposed to function as a cofactor that aids in directing import complexes into the nucleus and speeds their dissociation in a manner similar to Nup2 in yeast (Dilworth et al., 2001; Gilchrist et al., 2002; Lindsay et al., 2002). However, it is not yet certain how the mobility of the other proteins contribute to their functions in nuclear trafficking.
Nup153 is a large, 1476-amino acid protein that is comprised of multiple domains. The N-terminal domain contains sequences necessary for binding to the nuclear envelope (amino acids [aa] 1-144), the nuclear pore via the Nup160 complex [aa 39-339], Tpr [aa 228-439], and RNA [aa 250-400]; Sukegawa and Blobel, 1993; Enarson et al., 1998; Dimaano et al., 2001; Hase and Cordes, 2003). The central domain of Nup153 contains four C2-C2 zinc fingers, constituting a novel Ran-GDP-binding domain, and the C-terminal domain contains numerous FG and FxFG repeats providing both receptor docking sites (Shah and Forbes, 1998; Nakielny et al., 1999) as well as an interface for binding Lamin B (Smythe et al., 2000). Nup153 resides on the basket of the nuclear pore, and in a recent EM study it was proposed that Nup153 might extend the length of the fibers that form the basket (Fahrenkrog et al., 2002).
Nup98 resides on both faces of the nuclear pore and, like Nup153, is composed of multiple domains (Griffis et al., 2002). The N-terminal half of the protein consists of two domains containing FG and GLFG repeats separated by a domain that serves as the binding site for Rae1/Gle2 (Pritchard et al., 1999). Nup98 can be expressed in two forms, Nup98 and a Nup98/Nup96 polyprotein. The Nup98 C-terminal domain encodes an autoproteolytic activity that either releases an 8-kDa peptide from the C-terminus of the Nup98 form or cleaves apart the Nup98 and Nup96 proteins (Fontoura et al., 1999; Hodel et al., 2002).
Previously we showed that the intranuclear mobility of Nup98 is dependent on ongoing transcription, raising the possibility that Nup98 could be interacting directly with the transcription machinery or with transcripts and RNP complexes as they are produced (Griffis et al., 2002). Antibody injection experiments implicated both Nup98 and Nup153 in the export of multiple classes of RNA from the nucleus of Xenopus oocytes (Powers et al., 1997; Ullman et al., 1999). Nups98 and 153 have been shown to interact with the same nucleoporin subcomplex, the Nup107-Nup160-Nup133-Nup96 complex (Vasu et al., 2001). Nup98 binds to Nup96, the C-terminal half of the Nup98/Nup96 polyprotein, but the direct binding partner of Nup153 within this complex is not known (Fontoura et al., 1999; Hodel et al., 2002). Both proteins are also known to bind to multiple soluble RNA transport factors (Pritchard et al., 1999; Zolotukhin and Felber, 1999; Bachi et al., 2000; Kuersten et al., 2002). However, there is as yet no known Nup98 binding partner that would clearly link this nucleoporin to the transcription machinery.
To begin to elucidate the link between nucleoporin mobility and transcription, we have defined the regions within Nup98 that are important for directing the protein to the nuclear pore and the regions that link protein mobility to ongoing transcription. Additionally, we have analyzed the dynamics of Nup153 localization and found that, like Nup98, multiple regions within Nup153 influence the relative stability of its association with the nuclear pore. Furthermore, we observed that the exchange of Nup153 at the nuclear pore is dependent on ongoing transcription. Interestingly, the link between mobility and Pol I vs. Pol II transcription could be traced to distinct domains within Nup98 and Nup153.
MATERIALS AND METHODS
DNA Constructs
Full-length GFP-Nup98, GFP-Nup98 C-terminus and GFP-GLFG+C-term have all been previously described (Griffis et al., 2002, 2003). The Nup98 GLFG(N) + C-term construct (amino acids 221-369 + 505-920) was created by cutting the GFP-GLFG construct using the HindIII sites in the vector and following amino acid 369, the fragment was then ligated to the GFP-Nup98 C-term construct cut with HindIII. The Nup98 GLFG(C) + C-term construct (aa 362-920) was created by PCR amplifying the Nup98 sequence with restriction sites at both ends. After digestion, the insert was ligated to pEGFP-C1 (Clontech Laboratories, Palo Alto, CA) cut with EcoRI and SmaI. The N-term + GLFG-GFP construct was created by inserting a SmaI site at amino acid 505 of full-length Nup98; the fragment was then cut from the backbone using ClaI and SmaI and then ligated into pEGFP-N3 cut with AccI and SmaI.
The full-length human Nup153 gene (Dimaano et al., 2001) was cut out from pBluescript (Stratagene, La Jolla, CA) and placed into pEGFP-C2 using XhoI and SacII. Nup153 deletion constructs, GFP-Nup153-NZ (aa 1-892) GFPNup153-N (aa 1-610), and GFP-Nup153 PTD (aa 1-339), were made in this vector by placing stops in the desired places within the coding sequence of the cDNA. In the case of GFP-Nup153-PTD, two stops were inserted after amino acid 339; the remaining 3′ end of the gene was removed by digesting the construct with EcoRV, which cuts after amino acid 346 and SmaI, which cuts in the multicloning site of the vector, 3′ of the insert. After digesting, the construct was gel purified and religated. The RNA-binding domain of Nup153 (aa 250-400) was PCR amplified and inserted into vector pEGFP-C1. All constructs generated by PCR, and mutagenesis was confirmed by sequencing.
Cell Culture and Immunofluorescence
HeLa and tsBN2 cells were maintained as previously described (Griffis et al., 2002). BHK cells were maintained in high-glucose DMEM supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 μg/ml penicillin, and 100 U/ml streptomycin. Transfections were performed using Fugene 6 (Roche, Indianapolis, IN) according to manufacturer's instructions. For live cell imaging, cells were plated into Lab-Tek II chambered coverglasses (Nunc, Rochester, NY), and before imaging, the medium was replaced with an imaging medium composed of DMEM without phenol-red, supplemented with 25 mM HEPES, 20% FBS, and penicillin/streptomycin. Actinomycin D (Actinomycin C1, Roche) and 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB; Calbiochem, La Jolla, CA) were used at 5 and 75 μg/ml, respectively, for 4 h before imaging. Immunofluorescence was performed as previously described (Griffis et al., 2002). Antibodies used in this study were anti-hNup98, anti-xNup153, and goat anti-rabbit Alexa-545 (Molecular Probes, Eugene, OR) and were used at 1:3000, 1:200, and 1:1000, respectively. To optimize the nuclear rim signal, cells transiently expressing the various GFP-Nup98 constructs were simultaneously fixed and permeabilized on ice for 10 min using 2% paraformaldehyde and 0.2% Triton X-100.
Photobleaching and Data Analysis
Photobleaching experiments were performed as previously described (Griffis et al., 2002). In all cases, cells expressing low amounts of the GFP-fusion proteins were selected for analysis. For fluorescence recovery after photobleaching (FRAP) experiments where the nuclear rim was analyzed, the pinhole of the confocal was set to one airy unit to give a smaller optical slice. To minimize nonspecific photobleaching during long time course FRAP experiments, a 2-s delay was added between each scan. To select nuclear bodies that lay outside of nucleoli, cells were viewed by phase to identify nucleoli before photobleaching. When fitting the recovery data for full-length GFP-Nup153 and GFP-Nup153-NZ, the following equation was used to fit the data points:
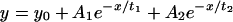 |
For GFP Nup153-N and other datasets that fit with a single exponential equation, we used the equation: y = y0 + A1e-x/t. These equations, along with the values determined for their constants, were used for extrapolating the recovery of each construct to determine the theoretical maximal recovery.
RESULTS
Multiple Domains Contribute to the Mobility of Nup153
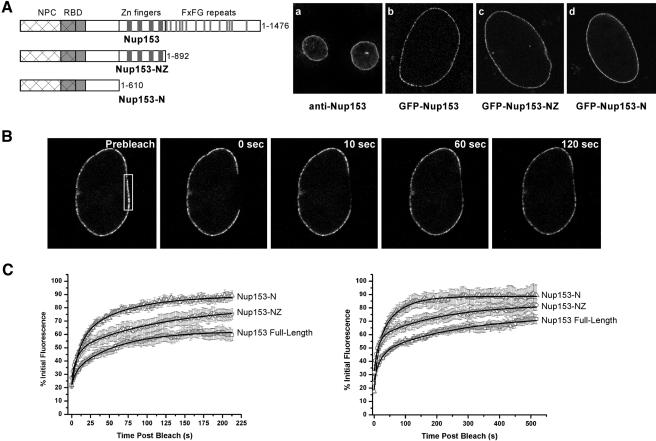
Nup153 has been shown to be a highly dynamic protein that can rapidly associate with and dissociate from the nuclear pore complex (Daigle et al., 2001). Although the N-terminus mediates direct interaction with the NPC, possible contributions of other domains of Nup153 to the dynamics of this interaction have not been assessed. To address this question, we began by establishing the steady state localization in live HeLa cells of three GFP-tagged Nup153 constructs (Figure 1A): full-length Nup153 (aa 1-1476), a Nup153 truncation that contains the N-terminal and zinc-finger domains (Nup153-NZ, aa 1-892), and a second truncation that retains only the N-terminus (Nup153-N, aa 1-610). In agreement with the previous findings of Burke and colleagues, each construct displayed a steady-state localization at the nuclear pore identical to the endogenous protein (Enarson et al., 1998; Figure 1A).
Figure 1.
Truncating Nup153 does not affect NPC targeting but accelerates FRAP. (A) The full-length and truncated versions of Nup153 diagrammed were all expressed as GFP-fusion proteins and visualized in live cells (narrow vertical stripes indicate individual FxFG repeats, FG repeats are not included; thick vertical stripes represent C2-C2 zinc-fingers). The pore targeting of each construct (b-d) was compared with the pore localization seen when anti-Nup153 antibodies were used to stain XL-177 cells (a). (B) In a cell expressing full-length GFP-Nup153, the section of the nuclear envelope indicated by the boxed region was photobleached, and the recovery was monitored every 1.5 s for 120 s after bleaching. (C) The recovery of the Nup153 constructs was monitored and averaged for several independent experiments with recovery times of either 212 s (left graft) or 518 s (right graph) after photobleaching (n ≥ 7 for each construct).
We then addressed the dynamics of these different forms of Nup153 by assessing FRAP. As previously described, the fluorescence of full-length GFP-Nup153 at the NPC recovered rapidly after photobleaching of a section of the nuclear envelope (Figure 1B). In our experiments, GFP-Nup153 recovered to 71.5 ± 3.5% over a time course of 518 s and had recovered half of its fluorescence (T1/2 of recovery) by ∼41 s (n = 10). These data are not identical to previously reported values; however, differences could perhaps result from the number of copies of GFP at the N-terminus of Nup153 (two copies in a previous study by Daigle et al. [2001] and one copy in this study). The recovery of GFP-Nup153 is significantly faster than what we see when GFP-Nup98 is photobleached at the nuclear pore.
We next performed FRAP assays with each of the GFP-Nup153 constructs over both a short time course, to provide optimal resolution of initial rates of recovery (140 measurements >212 s), and a long time course, to better illustrate the later, slow phase of recovery (140 measurements >518 s). Interestingly, we found that Nup153-N was the most mobile of the Nup153 constructs. It had the fastest initial rate of recovery and reached its maximal recovery (89.5 ± 7.7%) after the shortest amount of time (T1/2 ≈ 18 s, n = 6). The Nup153-NZ construct recovered more slowly than the N construct (T1/2 ≈ 25 s, n = 9), but still to a greater degree (82.0 ± 4.1%), and faster than the full-length protein.
The recovery of Nup153 at the pore appeared to occur in two phases, a result that suggests the existence of two or more populations of this nucleoporin at the nuclear pore. Both full-length Nup153 and Nup153-NZ displayed a fast initial phase of recovery, likely representing a highly mobile population. This is followed by a second, slow phase with a gradual return of fluorescence over the course of several minutes, corresponding to a pool of Nup153 that is more stably associated with the NPC. Indeed, the recovery curves for full-length Nup153 and Nup153-NZ are best fit by double exponential functions, indicating two distinct populations (see MATERIALS AND METHODS). In contrast, only a single rapid phase of recovery was observed for Nup153-N, and this was best fit by a single exponential function, indicative of only one population. These findings suggest that the Zn finger domain makes a significant contribution toward stabilizing a fraction of Nup153 at the NPC. Furthermore, the lack of full recovery to prebleach levels seen with Nup153 suggests the possibility of an additional, immobile subpopulation of protein that is substantially reduced in constructs lacking the repeat domain.
Previously we showed that the central GLFG repeat domain of Nup98 acts synergistically with the C-terminal NPC-binding sequences in targeting to the pore; in the absence of the repeat domain the protein is undetectable at the NPC in live cells (Griffis et al., 2003). Removal of the FxFG repeat domain from Nup153 does not alter the steady state localization of the protein; however, loss of this domain does accelerate the rate of exchange at the NPC. Further removal of the zinc finger domain to produce Nup153-N not only increased the rate of recovery at the pore, but also altered the kinetic parameters of recovery. Thus, although the relative contribution may be less dramatic than what is seen for Nup98, domains of Nup153 with no intrinsic targeting activity also influence the association of Nup153 with the NPC.
Two Regions within the Nup98 GLFG Domain Independently Enhance Nuclear Pore Targeting
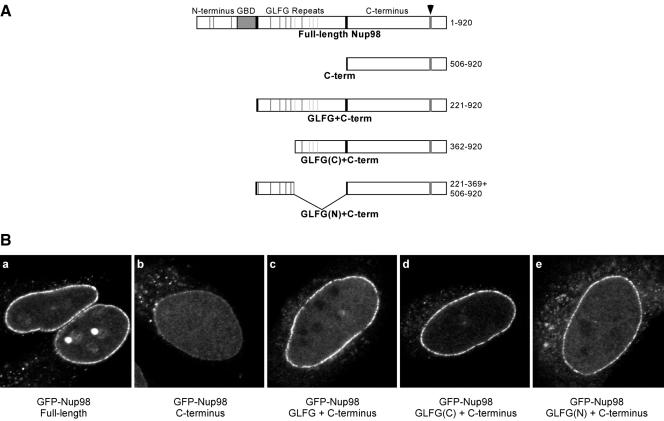
We have observed synergistic contributions of nucleoporin repeat domains on the pore targeting of two dynamic nucleoporins, Nup98 and Nup153. One possible explanation for these effects is a direct interaction between the nucleoporin repeat domain and another protein of the NPC that results in increased stability of pore binding. A second possibility is that the repeat domain could bind a transport factor with multiple NPC interaction domains, which, by mediating binding to a second nucleoporin, would stabilize Nup98 or Nup153 at the pore. One candidate molecule that could dock repeat domains to the pore in such a manner is the RNA export factor TAP/NXF1. TAP/NXF1 shows a weak steady state localization at the nuclear envelope and possesses two distinct sites for binding to nucleoporin repeats. To test whether the TAP/NXF1 binding was required for synergistic NPC targeting of dynamic nucleoporins, we turned to Nup98 where we have mapped TAP/NXF1 binding to the N-terminal half of the central GLFG repeat domain (Blevins et al., 2003). We localized two new Nup98 constructs (Figure 2A): one containing the N-terminal, TAP/NXF1-binding, portion of the GLFG repeat domain (aa 221-369) fused to the C-terminus (aa 506-920), and the second containing the C-terminal portion of the GLFG repeat domain (aa 362-505) through to the end of the protein. Interestingly, when expressed in HeLa cells, both constructs target to the pore as efficiently as the fusion that contains the full GLFG repeat domain (Figure 2B, d and e). Therefore, we conclude that NPC targeting of Nup98 cannot be enhanced solely via an interaction with TAP/NXF1.
Figure 2.
The GLFG domain of Nup98 confers nonspecific enhancement of NPC targeting. (A) The constructs diagrammed were made as GFP-fusion proteins to monitor pore targeting (GBD = Gle2 binding domain; the arrowhead indicates the site of autoproteolytic cleavage, and narrow stripes indicate individual GLFG repeats; FG repeats are not included). (B) The localization of the indicated constructs in HeLa cells that were simultaneously fixed and permeabilized.
Nup153 Mobility Is Sensitive to Transcription Inhibitors
Previous work has suggested that Nup153 is intimately linked to the process of RNA export. As with Nup98, injection of anti-Nup153 antibodies blocked the export of multiple classes of RNA (Ullman et al., 1999) and overexpression of domains of Nup153 blocked mRNA export in cultured cells (Bastos et al., 1996); Nup153 also binds a variety of RNA export factors (Nakielny et al., 1999; Bachi et al., 2000; Hofmann et al., 2001; Kuersten et al., 2002). However, unlike Nup98, Nup153 is a bona fide RNA-binding protein (Dimaano et al., 2001; Ball et al., 2004).
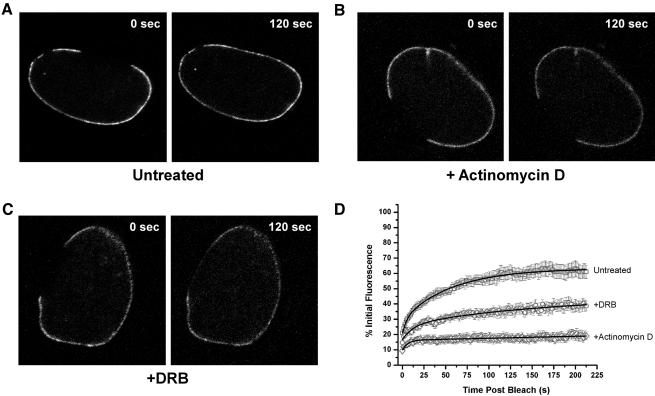
Given these many links to RNA trafficking as well as the dynamic interaction of Nup153 with the NPC, we asked whether the exchange of Nup153 on and off the pore might be dependent on transcription. The mobility of full-length GFP-Nup153 was assessed by FRAP assays in either untreated HeLa cells, or in HeLa cells treated with DRB or actinomycin D to inhibit RNA polymerase II or RNA polymerases I and II, respectively. Figure 3A shows an untreated HeLa cell expressing GFP-Nup153 immediately after photobleaching and 120 s later when the GFP-Nup153 has recovered most of its initial fluorescence at the nuclear rim. In contrast, when cells were treated with actinomycin D (Figure 3B), the recovery of Nup153 fluorescence at nuclear pores was severely inhibited. To ensure that the inhibition of recovery that we saw was not due to the induction of heterochromatin, we also tested cells that were treated with a lower concentration of actinomycin D (0.5 μg/ml) and found identical results (our unpublished results). Treatment with DRB produced an intermediate effect (Figure 3C); GFP-Nup153 recovered more slowly than in untreated cells, but to a greater extent than in actinomycin D-treated cells (61.0 ± 4.2% in untreated, 38.8 ± 3.3% in DRB-treated, and 19.1 ± 2.5% in actinomycin D-treated cells). The recovery of fluorescence after 212 s in cells with or without inhibitor treatment was quantitated for several experiments (untreated, n = 11; DRB treated, n = 9; actinomycin D treated, n = 8) and these data are presented in Figure 3D.
Figure 3.
Nup153 mobility is sensitive to transcription inhibitors. (A) A HeLa cell expressing GFP-Nup153 was photobleached, and images are shown immediately and 120 s after photobleaching. The same protocol was followed for cells treated with actinomycin D (B) and DRB (C). (D) Several independent experiments were performed on cells with or without transcription inhibitors. The data were quantitated and averaged and are presented graphically (n ≥ 5 for each condition).
Recovery of fluorescence in a photobleached area must necessarily involve both the dissociation of bleached protein as well as the association of unbleached protein. The simplest interpretation of the effect of transcription inhibitors might be that in the absence of export cargo Nup153 dissociates from the pore but cannot rebind. To test this hypothesis, we performed inverse FRAP or iFRAP analyses (Dundr et al., 2002) in which we bleached a large portion of the nuclear envelope and then measured loss of fluorescence from the unbleached area as bleached protein exchanges for the unbleached protein. In untreated cells, GFP-Nup153 dissociated from the pore and equilibrated between bleached and unbleached areas. However, we found that GFP-Nup153 did not dissociate from the unbleached area in actinomycin D-treated cells (our unpublished results). Thus the absence of new transcripts prevents the release of Nup153 from the pore; inhibition of transcription may also block binding to the pore, but we cannot independently measure this half of the equilibrium. iFRAP experiments also revealed that the truncated proteins, as predicted, were able to dissociate from the pore at a faster rate than the full-length protein in untreated cells.
Nup153 plays a role in nuclear protein trafficking; it is a binding site for several transport factors including Importin β/Karyopherin β, Crm1/Exportin1, and Transportin/Karyopherin β2. Nup153 has also been shown to bind directly to Ran (Saitoh et al., 1996; Nakielny et al., 1999). Therefore, it was possible that the release of Nup153 from the pore was also linked to the Ran cycle or Ran-dependent nuclear transport. To evaluate this, FRAP studies were performed on the full-length GFP-Nup153, GFP-Nup153-NΖ, and GFP-Nup153-N constructs expressed in tsBN2 cells at 34 and 40°C. However, no difference was observed in the recovery at either temperature (our unpublished results). Therefore, the exchange of Nup153 at the nuclear pore is linked to transcription but not to the Ran gradient or Ran-dependent nuclear transport.
Nup98 in Different Compartments of the Nucleus Is Selectively Sensitive to Transcription Inhibitors
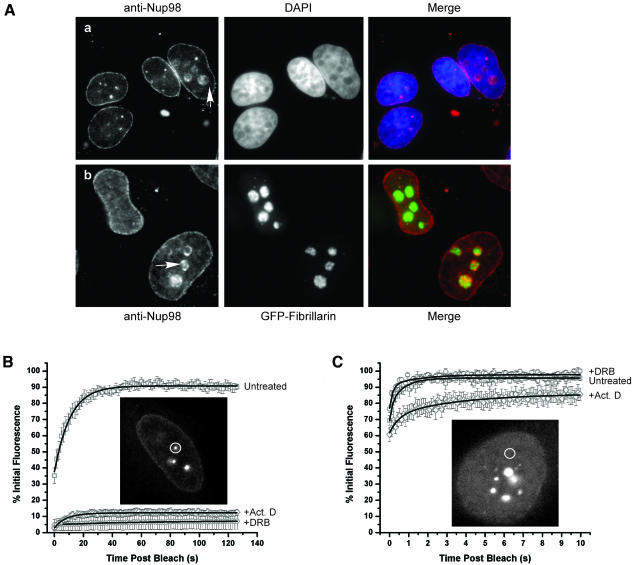
In addition to the fraction of Nup98 found at the nuclear pore, Nup98 is also found throughout the nucleoplasm and is concentrated in nuclear GLFG bodies. When observing either endogenous human Nup98 or a cell line that stably expresses GFP-Nup98, we have occasionally seen what appears to be free Nup98 protein or intact GLFG bodies localized within nucleoli. Figure 4A illustrates the localization of endogenous Nup98 in the nucleoli of a group of cells costained with either DAPI or GFP-Fibrillarin. Sample nucleoli containing Nup98 are indicated with arrows. When viewed in 3D stacks of images, Nup98 in nucleoli frequently appears to be surrounding the edges of the compartment in a cage-like manner (our unpublished results). Nucleolar Nup98 does not colocalize precisely with markers for canonical nucleolar subcompartments such as Fibrillarin (which marks the dense fibrillar component) or UBF (which marks the fibrillar centers), although there is partial overlap with a marker of the granular component, B23 (Figure 4A; our unpublished results).
Figure 4.
Nup98 in different compartments is selectively sensitive to transcription inhibitors. (A) Endogenous Nup98 was detected in HeLa cells and colocalized in nucleoli with either DAPI (a) or GFP-Fibrillarin (b). (B) Nup98 bodies that are excluded from nucleoli were photobleached in untreated cells as well as cells treated with either actinomycin D or DRB. The results were quantitated and averaged and are presented in B. Inset: a cell before photobleaching the body indicated with the circle. (C) Nucleoplasmic Nup98 was photobleached in cells treated as in B using a modified protocol that allows for more rapid data acquisition (n ≥ 7 for each condition). Inset: an area of Nup98 staining in the nucleoplasm before photobleaching.
Previously, we have shown that Nup98 is unable to exchange in and out of nuclear bodies in cells treated with either actinomycin D or DRB (Griffis et al., 2002). However, the inhibition seen with DRB was not as complete as that seen with actinomycin D. In cells treated with DRB, some GLFG bodies did not recover at all after photobleaching, whereas others recovered slowly; when the dataset is averaged, this produces an aggregate effect of slow, partial recovery. In contrast, treatment with actinomycin D completely inhibits mobility of Nup98 in every GLFG body tested. Actinomycin D blocks transcription by both RNA Polymerases I and II, whereas DRB blocks only RNA Polymerase II transcription. We therefore speculated that the GLFG bodies where Nup98 mobility was unaffected by DRB might be those localized within nucleoli, a nuclear domain where transcription is carried out by Pol I and thus not perturbed by DRB treatment. We repeated the FRAP assays in DRB-treated cells and chose only GLFG bodies that were clearly excluded from nucleoli when cells were viewed by phase microscopy. Strikingly, we now found that the inhibition by DRB treatment was as pronounced as the inhibition produced by actinomycin D (Figure 4B). Conversely, when the nucleolar bodies were photobleached in DRB treated cells, we found that they still recovered fluorescence, albeit somewhat slower than nucleoplasmic bodies in untreated cells (our unpublished results).
Movement of Nup98 both in and out of GLFG bodies and on and off the nuclear pore is strongly dependent on ongoing transcription (Figure 4A and Griffis et al., 2002). We now asked whether movement of the free nucleoplasmic pool of GFP-Nup98 is likewise coupled to ongoing transcription. To test this, the mobility of Nup98 was assayed in cells with or without transcription inhibitors. We found that only actinomycin D affected the mobility of nucleoplasmic Nup98; GFP-Nup98 in actinomycin D-treated cells bleached to a greater extent and recovered more slowly and to a lesser degree than in untreated or DRB-treated cells (Figure 4C).
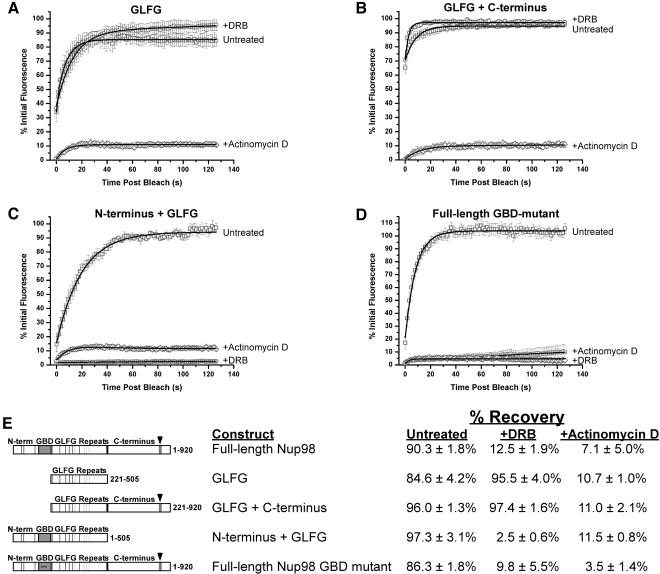
Different Domains of Nup98 Are Required for Sensitivity to Actinomycin D and DRB
Understanding the process by which mobility of a nucleoporin within the nucleus is linked to ongoing transcription could reveal new information about the coupling of RNA transcription, processing, and export. As a first step toward elucidating this mechanistic network, we set out to map determinants within Nup98 that are required to confer sensitivity to inhibitors of transcription. Our previous work indicated that the GLFG domain confers on Nup98 a characteristic mobility that is slower than the rate of diffusion within the nucleus. Additionally, this domain is both necessary and sufficient for targeting to the GLFG bodies (Griffis et al., 2002). We asked therefore, whether the GLFG domain was also sufficient to make the exchange of Nup98 in and out of nuclear bodies sensitive to actinomycin D. Figure 5A presents quantitation of the mobility of GFP-GLFG in cells with and without transcription inhibitors. When transcription is blocked by actinomycin D, the mobility of the GLFG domain is clearly diminished to the same extent as the mobility of full-length Nup98 (Figure 5A). In contrast, when cells were treated with the Pol II specific inhibitor, DRB, we did not see any effect on the mobility of the GLFG domain construct. To assess the basis for this differential effect, we made a series of constructs, each of which contains the GLFG domain along with additional sequences from Nup98 (Figure 5E). Cells were transfected with these constructs, and fluorescence recovery after photobleaching was quantitated in the presence or absence of transcription inhibitors. We found that the addition of the C-terminal domain did not alter the mobility under any condition (Figure 5B), although as expected a substantial amount of Nup98 was now at the NPC. In contrast, when the N-terminal domain was fused to the GLFG domain, the resulting protein was not found at the nuclear pore. Moreover, exchange in and out of GLFG bodies was now sensitive to both actinomycin D and DRB (Figure 5C).
Figure 5.
Nup98 requires different domains for sensitivity to different transcription inhibitors. (A-D) GLFG bodies containing the indicated GFP-tagged constructs were photobleached as above in the presence or absence of transcription inhibitors. (n ≥ 5 for every condition). (E) The mobility of the constructs illustrated were assayed under multiple conditions, and the amount of recovery 120 s after photobleaching is presented in the table.
The N-terminal region of Nup98 contains the binding site for the mRNA export factor Rae1/Gle2 (Gle2-binding domain; GBD). Rae1/Gle2 has been proposed to bind to mRNA, and this protein could potentially provide the link between the mobility of Nup98 and Pol II transcription. To test whether Gle2 binding was the essential contribution of the N-terminal domain, the GBD alone (amino acids 156-221) was added to the construct used in Figure 5B; this form of Nup98 remained mobile in the presence of DRB (unpublished data). To further test whether Gle2 binding was necessary, although not sufficient for the link between mobility and Pol II transcription, point mutations that were previously shown to abrogate binding between Nup98 and Gle2 (L200P, L203P, and R204P; Pritchard et al., 1999) were generated in the context of full-length GFP-Nup98. These mutations abolished Gle2 binding as assayed by both coimmunoprecipitation of Nup98 and Gle2 from cell extracts, and in vitro binding assays (our unpublished results). Despite loss of detectable Gle2 binding, the mobility of this mutant Nup98 remained fully sensitive to inhibition by DRB (Figure 5D). Thus the N-terminal domain, but unexpectedly, not Gle2 binding, confers Pol II specific inhibition of Nup98 mobility. It appears that the central GLFG domain is directly coupled to some process affected by actinomycin D, but the addition of the N-terminal repeats is required for Nup98 to interact with an additional protein or set of proteins that link the mobility of Nup98 to Pol II transcription
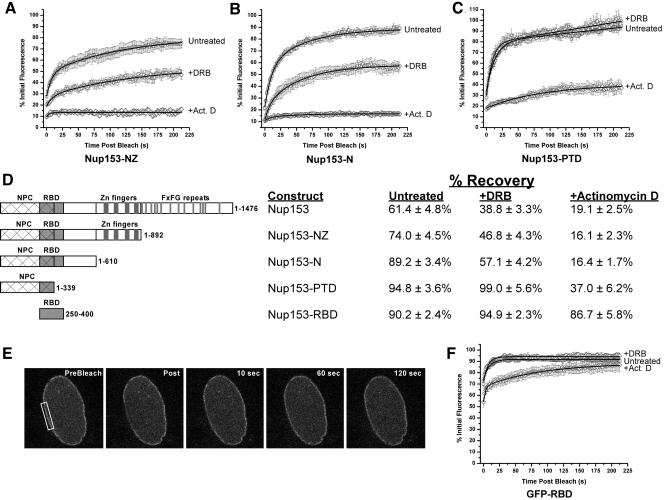
Distinct Domains Couple Nup153 Mobility to Pol I and Pol II Transcription
As we had done for Nup98, we wanted to determine whether it was possible to dissect sequences within Nup153 that are required for the interplay between Nup153 mobility and transcription. We began by testing the Nup153 deletion constructs in FRAP assays in the presence of either actinomycin D or DRB. The recovery of Nup153-NΖ showed the same degree of sensitivity to both inhibitors, as did the full-length protein (Figure 6A). The dispensability of the repeat domain was unexpected given our finding that Nup98 requires the GLFG repeat domain to link its mobility to transcription. We further found that the zinc-fingers of Nup153 are not required for sensitivity to transcription inhibitors. The mobility of the Nup153-N construct remained sensitive to both actinomycin D and DRB (Figure 6B). This domain contains pore targeting sequences but also contains an RNA-binding domain and sequences required for binding to the nucleoporin Tpr. To test whether the requirements for actinomycin D and DRB sensitivity can be distinguished in the context of Nup153 mobility, we made a further deletion that retained only the minimal Nup153-pore targeting domain (Nup153-PTD, aa 1-339). This protein has a steady-state localization at the nuclear pore, however, its expression level was lower than that of any other construct and expression was seen only in BHK cells (we were unable to detect expression in HeLa cells, unpublished data). When we tested this construct in a FRAP assay we found that GFP-Nup153-PTD recovered faster (T1/2 ≈ 13 s) and with a smaller immobile fraction (94.83 ± 3.58% recovery, n = 10) than other Nup153 constructs. Most striking, this protein was not sensitive to DRB treatment (Figure 6C). Thus, sequences between amino acids 340 and 610 are required for linking the mobility of Nup153 with Pol II transcription.
Figure 6.
A region between amino acids 340 and 610 is required for Nup153 DRB sensitivity. (A-C) The Nup153-NΖ, -N, and -PTD constructs were all evaluated by FRAP assays in untreated cells as well as cells treated with transcription inhibitors. The data were quantitated and are presented for each construct (n ≥ 6 for each). (D) This table shows the recovery of each Nup153 construct used in this study at 212 s after photobleaching for each indicated condition. (E) The RNA-binding domain of Nup153 was fused to GFP and expressed in HeLa cells. The panels show the recovery of The GFP-RBD after the section of the nuclear envelope indicated by the boxed region was photobleached. The recovery of GFP-RBD at the nuclear pore in the presence or absence of transcription inhibitors is shown in the graph (F; n ≥ 7 for each condition).
We were unable to determine if the RNA-binding function was required for Nup153 mobility, because complete removal of this domain abrogates pore targeting and we have not yet succeeded in generating point mutations that specifically eliminate association with RNA. However, we asked whether the mobility of the RNA-binding domain is itself sensitive to transcription inhibitors. To do this, we produced a GFP-tagged Nup153 RNA-binding domain (GFP-RBD, aa 250-400). This protein localizes primarily to the nucleoplasm but with a small pool at the NPC (Figure 6E). We then assessed the mobility of the GFP-RBD protein using FRAP assays. When FRAP of the small pool at the nuclear pore was quantitated, we found that only 30% of the initial fluorescence was lost during bleaching, and the bleached area recovered exceptionally fast, with a T1/2 of recovery of ∼5 s (Figure 6F, n = 8). We believe that we were only able to bleach a small amount of the rim fluorescence because of the high mobility of the protein. As with GFP alone, recovery is too rapid to allow measurement of the full extent of bleaching. Additionally, GFP-RBD was not sensitive to DRB, and in cells treated with actinomycin D, recovered to approximately the same extent as in untreated cells but recovery was somewhat slowed (T1/2 ≈ 14 s, n = 8 vs. untreated T1/2 ≈ 5 s, n = 8). Therefore we conclude that the intact RNA-binding domain of Nup153 is necessary but not sufficient to link Nup153 mobility to Pol II transcription.
DISCUSSION
In this study we have investigated the localization and dynamics of two mobile components of the nuclear pore complex, Nup98 and Nup153. In addition to their characterized pore targeting sequences, we found that other domains within both Nup98 and Nup153 significantly influence the stability of their association with the nuclear pore. Like Nup98, dynamic exchange of Nup153 at the pore is dependent on ongoing Pol I and Pol II transcription. We were able to identify the domains within these proteins that make their mobility sensitive to RNA polymerase inhibitors and found that transcription of RNA is essential for the release of two different nucleoporins from the NPC, perhaps through an effect on NPC remodeling during RNA export.
Dynamics of Nup153 at the NPC
The fact that Nup153 dynamically exchanges at the nuclear pore is paradoxical when one considers previous data indicating that Nup153 plays a structural role at the pore. Nuclei reconstituted from a Xenopus egg extract depleted of Nup153 contained pores that lacked several other components of the nuclear basket, including Nup98 and Tpr (Walther et al., 2001). Additional evidence pointing toward a structural role for Nup153 was obtained in recent RNAi knockdown experiments in which Nup153 was required for proper localization of Tpr to the nuclear pore (Hase and Cordes, 2003). In photobleaching assays, we found that GFP-Tpr is far more stably associated with the NPC than is Nup153 (our unpublished results). Thus, either Nup153 plays a transient role in targeting Tpr or a fraction of Nup153 must be stably anchored at the NPC.
The existence of multiple populations of Nup153 at the nuclear pore may reconcile these seemingly paradoxical roles of structural scaffold and mobile nucleoporin. Analysis of FRAP data supports the existence of at least two pools of mobile Nup153. The recovery curves for full-length Nup153 and Nup153-NZ are best fit with double exponential functions, which account for both the rapid initial recovery and the subsequent slower recovery. The fact that we consistently see an immobile fraction after photobleaching any NPC-associated Nup153 construct could be an indication that there is, in fact, a third population of Nup153 at the pore that does not freely exchange. Approximately 30% of the full-length Nup153 fluorescence did not recover during the time course of our longest FRAP experiments. If we extrapolated using the equations that best fit the existing recovery data, the eventual maximal recovery would only be 76%. Therefore, ∼25% of Nup153 appears to be stably bound to the nuclear pore. This fraction of Nup153 is similar to the amount which Hase and Cordes estimated would have to be stably associated with the NPC to account for Tpr binding (Hase and Cordes, 2003).
Multiple Domains Contribute to the Stability of Nup98 and Nup153 at the NPC
Nup153-NZ and Nup153-N constructs, which both lack the FxFG domain, both localized entirely to the nuclear pore; however, they exchanged on and off the pore much faster than the full-length protein. Additionally, because the Nup153-N mutant recovers without the second slow phase seen with Nup153 and Nup153-NZ, sequences between amino acids 610 and 892 of Nup153 must influence the establishment of the slowly exchanging population. It seems most likely that the FxFG and zinc-finger regions, although not primarily responsible for targeting, do serve to stabilize Nup153 at the pore. Multiple binding interactions have been reported for these domains that could serve to stabilize Nup153 at the NPC. The loss of such secondary interactions would therefore increase the rate and change the kinetic parameters of Nup153 exchange.
In vivo, the C-terminal domain of Nup98 by itself does not efficiently associate with the pore; addition of the GLFG domain greatly enhances pore targeting (Griffis et al., 2003). This points out another apparent contradiction in that the stability of both Nup98 and Nup153 at the nuclear pore is significantly enhanced by the addition of domains that do not target to nuclear pores when expressed independently. We can envision three possible models to explain how such auxiliary domains could enhance targeting: 1) the domain could have a regulatory function and under appropriate circumstances promote a conformation of the pore targeting domain with increased affinity for the NPC; 2) the domain could bind a factor, perhaps a receptor/cargo complex, that bridges an interaction with the NPC; and 3) these domains could themselves have low affinity but high avidity interactions at the NPC, which only occur when they are brought into close apposition to the pore via the primary targeting domain.
We originally hypothesized that a mobile nucleoporin could be directed to the NPC by binding of a transport factor/cargo complex to the repeat domain. This could occur through either of the first two mechanisms mentioned above. We found, however, that either half of the Nup98 repeat domain enhanced pore targeting equally well. Because the mRNA export factor TAP/NXF1 binds only to the N-terminal half, we were thus able to rule out TAP/NXF1 binding as a requirement for the synergistic targeting effect. It is possible that each half of the GLFG domain could bind to a distinct transport receptor, with either interaction being sufficient to enhance pore targeting; however, we have never seen a reduction of Nup98 at the NPC when either protein export or the Ran cycle are blocked. Consequently, it seems unlikely that transport factor/cargo complexes serve either as an activating signal or as a physical bridge for pore targeting. Given the existing data, it seems most likely that auxiliary domains interact directly with one or more proteins components of the NPC. Indeed, it could be that GLFG or FxFG repeats interact with the pore by integrating into the proposed hydrophobic central transport channel (Ribbeck and Gorlich, 2002). In Nup98 the relative contribution of this stabilizing interaction would be far more substantial than for Nup153. Such an interaction could be nonspecific for individual repeats. Indeed, if this model for general association of repeat domains at the NPC is correct, the Nup98 FG repeats upstream of the Gle2 binding domain should similarly enhance targeting, and experiments to test this model are underway.
Nucleolar Nup98
We consistently observe nucleolar localization of endogenous Nup98 in a fraction of cells. Others have previously noted some association of Nup98 with nucleoli either when Nup98 is overexpressed or when Rev is coexpressed (Fontoura et al., 1999; Zolotukhin and Felber, 1999). Within nucleoli, Nup98 did not colocalize with any of the well-characterized nucleolar subcompartments. A similar lack of association of the SRP particle with characterized compartments led Politz et al. (2002) to propose that there may be additional compartments within the nucleolus that have not been previously described. Nup98 also may associate with such an as yet uncharacterized subdomain of the nucleolus.
A unique aspect of Nup98 localization is its assembly into intranuclear structures termed GLFG bodies (Griffis et al., 2002). We occasionally observed GLFG bodies within nucleoli, either with antibody staining of endogenous protein or in a cell line stably expressing GFP-Nup98. It is not certain whether these bodies formed de novo within nucleoli or migrated to this compartment from the nucleoplasm. Nonetheless, it is striking that although GFP-Nup98 mobility in nucleoplasmic bodies was severely reduced by DRB, within the nucleolus GFP-Nup98 mobility was unaffected by this inhibitor of Pol II transcription, an activity not associated with the nucleolus. In contrast, when actinomycin D was used to block both Pol I and Pol II, the mobility of Nup98 within nucleoli was strongly inhibited. These results substantiate a connection between Pol I transcription and Nup98 mobility and underscore the significance of transcriptional inhibitor effects by demonstrating that an inhibitor has no influence on Nup98 mobility where the target polymerase is not active.
Mapping Domains That Link Nucleoporin Mobility and Transcription
As we previously reported for Nup98, the association and dissociation of Nup153 from the pore was highly dependent on ongoing transcription. Inhibition of both Pol I and Pol II transcription, and therefore the bulk of RNA transport, prevented dynamic exchange of Nup153 at the NPC. Distinct from the effect on Nup98 within GLFG bodies, blocking only Pol II transcription with DRB had an intermediate effect on Nup153 at the nuclear pore and eliminated only a fraction of the recovery.
To better understand how the mobility of Nup98 and Nup153 are linked to transcription, we mapped the domains required to confer sensitivity to transcription inhibitors. Such mapping of Nup98 dynamics at the nuclear pore was not possible since multiple domains are required for efficient pore targeting; therefore, we monitored the mobility of Nup98 in nuclear GLFG bodies where the GLFG domain was sufficient to target Nup98. Unexpectedly, exchange of this domain in and out of nuclear bodies was affected by actinomycin D but not by DRB. When only Pol II is inhibited by DRB, the N-terminal repeat region of Nup98 must be present in addition to the central GLFG domain for mobility to be affected. This requirement for the N-terminal repeats to link mobility to Pol II transcription is reminiscent of a previous study in which this domain contributed to the ability of Nup98 fusion proteins to transform fibroblasts and to activate luciferase transcription (Kasper et al., 1999). Therefore, the N-terminus may contribute a specific interaction that directly ties Nup98 to the process of Pol II transcription.
Curiously, whereas the GLFG domain of Nup98 is necessary and sufficient to confer actinomycin D sensitivity, the exchange of Nup153 at the nuclear pore remained sensitive to transcription inhibitors in the absence of its FxFG repeat domain. The only domain of Nup153 required for actinomycin D sensitivity was the N-terminal pore targeting sequences, residues 1-339. In an interesting parallel to Nup98, this minimal region was not sufficient when only Pol II transcription was inhibited. DRB sensitivity required the additional contribution of amino acids 340-610. This region of Nup153 contains the last 60 amino acids of the characterized RNA-binding domain as well as a portion of the reported Tpr-binding site. However, we found that residues 1-339 of Nup153 retain homoribopolymer-binding activity, although a possibly more subtle defect in binding cellular RNA cargoes has not been assessed (our unpublished results).
We also determined that residues 1-339 bind to Tpr in vitro, further narrowing the Tpr-binding site to residues 228-339 (our unpublished results). Therefore, DRB sensitivity is not conferred solely through binding of Nup153 to either RNA or Tpr. In both Nup98 and Nup153, the link between transcription and mobility requires targeting sequences plus, in the case of Pol II specific transcription, an additional domain of each nucleoporin that do not share any obvious features in common. Given the diverse nature of these domains, it is not certain whether the mobilities of Nup98 and Nup153 are coupled to transcription through common or distinct mechanisms.
It is not yet fully clear why components of the nuclear pore complex, such as Nup98 and Nup153, would need to dynamically associate with the NPC. Several previous reports have indicated that alterations of the nuclear pore may influence its translocation properties. Feldherr and Akin showed that injection of either SV40-transformed cytoplasm or protein kinase C into untransformed cells can increase the apparent size cutoff of the NPC (Feldherr et al., 1992; Feldherr and Akin, 1995). Additionally, electron microscopy studies have revealed distortions in the structure of the nuclear basket and central transporter when large RNP particles move through the pore (Kiseleva et al., 1996, 1998). We have shown that only treatment with drugs that block production of RNA cargoes decreases the mobility of Nup98 and Nup153 at the nuclear pore; treatments that impair protein translation or protein export have no effect. These results suggest the possibility that nucleoporins might be actively removed from the pore as part of RNP export, either through association with the RNP particle or in order to allow the RNP to pass through the pore. Therefore, it is possible that the remodeling that is seen at the nuclear basket is caused by partial dissociation of Nup153 and Nup98. When RNA cargoes are eliminated by blocking transcription of Pol I and Pol II messages, the nuclear pore may no longer require remodeling, and thus exchange of Nup153 and Nup98 would not take place. Removing only Pol II messages with DRB treatment could produce the less severe reduction in mobility that we observe for Nup153 by only eliminating a portion of the total cargo moving through the pore.
In summary, we have characterized the binding and dynamic exchange of two mobile components of the nuclear pore basket, Nups 98 and 153. We find that their steady state localizations and exchange kinetics differ significantly. Thus, although their mutual dependence on transcription suggests that they function in a common process, they appear to participate in distinct ways. Future identification of the binding partners for each of the domains shown here to link mobility and transcription should provide an indication of whether there is a common pathway. Importantly, characterization of binding partners should also begin to unravel the mechanism linking transcription and nuclear pore dynamics.
Acknowledgments
We thank Melanie Blevins and Erica Phillips for excellent technical assistance; Drs. Criss Hartzell and Keith Berland for helpful discussion; and Dr. Win Sale for critical reading of the manuscript. We are grateful to Dr. Volker Cordes for providing us with GST-Tpr constructs, Dr. Larry Gerace for providing us with GFP-Tpr and anti-Tpr serum, Dr. Mary Dasso for providing us with the anti-UBF antibody, and Dr. Sui Huang for providing us with the GFP-B23. This work was supported by Grant GM-59975 from the National Institutes of Health (to M.A.P.) and by NIH Grant GM-61275 (to K.S.U.). E.R.G. is a predoctoral trainee of the NIH (T32 GM08367-13), and C.D. was supported by a Ruth L. Kirchstein individual NRSA from the NIH.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-10-0743. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-10-0743.
References
- Bachi, A. et al. (2000). The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6, 136-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball, J.R., Dimaano, C., and Ullman, K.S. (2004). The RNA binding domain within the nucleoporin Nup153 associates preferentially with single-stranded RNA. RNA 10, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos, R., Lin, A., Enarson, M., and Burke, B. (1996). Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134, 1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin, J., Gerlich, D., Daigle, N., Eils, R., and Ellenberg, J. (2002). Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108, 83-96. [DOI] [PubMed] [Google Scholar]
- Belgareh, N. et al. (2001). An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154, 1147-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins, M.B., Smith, A.M., Phillips, E.M., and Powers, M.A. (2003). Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J. Biol. Chem. 278, 20979-20988. [DOI] [PubMed] [Google Scholar]
- Burke, B., and Ellenberg, J. (2002). Remodelling the walls of the nucleus. Nat. Rev. Mol. Cell. Biol. 3, 487-497. [DOI] [PubMed] [Google Scholar]
- Daigle, N., Beaudouin, J., Hartnell, L., Imreh, G., Hallberg, E., Lippincott-Schwartz, J., and Ellenberg, J. (2001). Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154, 71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth, D.J., Suprapto, A., Padovan, J.C., Chait, B.T., Wozniak, R.W., Rout, M.P., and Aitchison, J.D. (2001). Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153, 1465-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano, C., Ball, J.R., Prunuske, A.J., and Ullman, K.S. (2001). RNA association defines a functionally conserved domain in the nuclear pore protein Nup153. J. Biol. Chem. 276, 45349-45357. [DOI] [PubMed] [Google Scholar]
- Dundr, M., Hoffmann-Rohrer, U., Hu, Q., Grummt, I., Rothblum, L.I., Phair, R.D., and Misteli, T. (2002). A kinetic framework for a mammalian RNA polymerase in vivo. Science 298, 1623-1626. [DOI] [PubMed] [Google Scholar]
- Enarson, P., Enarson, M., Bastos, R., and Burke, B. (1998). Amino-terminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma 107, 228-236. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog, B., Maco, B., Fager, A.M., Koser, J., Sauder, U., Ullman, K.S., and Aebi, U. (2002). Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J. Struct. Biol. 140, 254-267. [DOI] [PubMed] [Google Scholar]
- Feldherr, C., and Akin, D. (1995). Stimulation of nuclear import by simian virus 40-transformed cell extracts is dependent on protein kinase activity. Mol. Cell Biol. 15, 7043-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldherr, C.M., Lanford, R.E., and Akin, D. (1992). Signal-mediated nuclear transport in simian virus 40-transformed cells is regulated by large tumor antigen. Proc. Natl. Acad. Sci. USA 89, 11002-11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura, B.M., Blobel, G., and Matunis, M.J. (1999). A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 144, 1097-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist, D., Mykytka, B., and Rexach, M. (2002). Accelerating the rate of disassembly of karyopherin. cargo complexes. J. Biol. Chem. 277, 18161-18172. [DOI] [PubMed] [Google Scholar]
- Griffis, E.R., Altan, N., Lippincott-Schwartz, J., and Powers, M.A. (2002). Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell 13, 1282-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis, E.R., Xu, S., and Powers, M.A. (2003). Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol. Biol. Cell 14, 600-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase, M.E., and Cordes, V.C. (2003). Direct interaction with nup153 mediates binding of tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell 14, 1923-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel, A., Hodel, M., Griffis, E., Hennig, K., Ratner, G., Xu, S., and Powers, M. (2002). The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin nup98. Mol. Cell 10, 347-358. [DOI] [PubMed] [Google Scholar]
- Hofmann, W. et al. (2001). Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J. Cell Biol. 152, 895-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper, L.H., Brindle, P.K., Schnabel, C.A., Pritchard, C.E., Cleary, M.L., and van Deursen, J.M. (1999). CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell. Biol. 19, 764-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendirgi, F., Barry, D.M., Griffis, E.R., Powers, M.A., and Wente, S.R. (2003). An essential role for hGle1 nucleocytoplasmic shuttling in mRNA export. J. Cell Biol. 160, 1029-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva, E., Goldberg, M., Daneholt, B., and Allen, T.D. (1996). RNP export is mediated by structural reorganization of the nuclear basket. J. Mol. Biol. 260, 304-311. [DOI] [PubMed] [Google Scholar]
- Kiseleva, E., Goldberg, M.W., Allen, T.D., and Akey, C.W. (1998). Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. J. Cell Sci. 111, 223-236. [DOI] [PubMed] [Google Scholar]
- Kuersten, S., Arts, G.J., Walther, T.C., Englmeier, L., and Mattaj, I.W. (2002). Steady-state nuclear localization of exportin-t involves RanGTP binding and two distinct nuclear pore complex interaction domains. Mol. Cell. Biol. 22, 5708-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, M.E., Plafker, K., Smith, A.E., Clurman, B.E., and Macara, I.G. (2002). Npap60/Nup50 is a tri-stable switch that stimulates importin-alpha:beta-mediated nuclear protein import. Cell 110, 349-360. [DOI] [PubMed] [Google Scholar]
- Nakielny, S., Shaikh, S., Burke, B., and Dreyfuss, G. (1999). Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 18, 1982-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J.C., Lewandowski, L.B., and Pederson, T. (2002). Signal recognition particle RNA localization within the nucleolus differs from the classical sites of ribosome synthesis. J. Cell Biol. 159, 411-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, M.A., Forbes, D.J., Dahlberg, J.E., and Lund, E. (1997). The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J. Cell Biol. 136, 241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, C.E., Fornerod, M., Kasper, L.H., and van Deursen, J.M. (1999). RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J. Cell Biol. 145, 237-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck, K., and Gorlich, D. (2002). The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21, 2664-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, H., Cooke, C.A., Burgess, W.H., Earnshaw, W.C., and Dasso, M. (1996). Direct and indirect association of the small GTPase Ran with nuclear pore proteins and soluble transport factors: studies in Xenopus laevis egg extracts. Mol. Biol. Cell 7, 1319-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, S., and Forbes, D.J. (1998). Separate nuclear import pathways converge on the nucleoporin nup153 and can be dissected with dominant-negative inhibitors [In Process Citation]. Curr. Biol. 8, 1376-1386. [DOI] [PubMed] [Google Scholar]
- Smythe, C., Jenkins, H.E., and Hutchison, C.J. (2000). Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 19, 3918-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukegawa, J., and Blobel, G. (1993). A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell 72, 29-38. [DOI] [PubMed] [Google Scholar]
- Suntharalingam, M., and Wente, S.R. (2003). Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell 4, 775-789. [DOI] [PubMed] [Google Scholar]
- Ullman, K.S., Shah, S., Powers, M.A., and Forbes, D.J. (1999). The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell 10, 649-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu, S., Shah, S., Orjalo, A., Park, M., Fischer, W.H., and Forbes, D.J. (2001). Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155, 339-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu, S.K., and Forbes, D.J. (2001). Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 13, 363-375. [DOI] [PubMed] [Google Scholar]
- Walther, T.C., Fornerod, M., Pickersgill, H., Goldberg, M., Allen, T.D., and Mattaj, I.W. (2001). The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20, 5703-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin, A.S., and Felber, B.K. (1999). Nucleoporins nup98 and nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J. Virol. 73, 120-127. [DOI] [PMC free article] [PubMed] [Google Scholar]