Abstract
Phosphatidylinositol 4-kinaseβ (PI4Kβ) plays an essential role in maintaining the structural integrity of the Golgi complex. In a search for PI4Kβ-interacting proteins, we found that PI4Kβ specifically interacts with the GTP-bound form of the small GTPase rab11. The PI4Kβ-rab11 interaction is of functional significance because inhibition of rab11 binding to PI4Kβ abolished the localization of rab11 to the Golgi complex and significantly inhibited transport of vesicular stomatitis virus G protein from the Golgi complex to the plasma membrane. We propose that a novel function of PI4Kβ is to act as a docking protein for rab11 in the Golgi complex, which is important for biosynthetic membrane transport from the Golgi complex to the plasma membrane.
INTRODUCTION
Phosphoinositides are essential phospholipids that act as key regulators of many cellular processes (Payrastre et al., 2001). Although they are minor components of the biological membrane, phosphoinositides have been found to play an important role in the regulation of cell proliferation, apoptosis, cytoskeletal organization, and vesicular trafficking (Simonsen et al., 2001). Recently, several phosphatidylinositol 4-kinases (PI4-kinases) kinases have been cloned and characterized (Balla, 1998; Gehrmann and Heilmeyer, 1998). Based on their differential sensitivity for the lipid kinase inhibitor wortmannin, PI4-kinases have been grouped into two types (II and III). Two mammalian types III, wortmannin-sensitive PI4-kinases have been described, a 92-kDa protein designated as PI4Kβ, which is homologous to the Saccharomyces cerevisiae protein Pik1p (Balla et al., 1997; Meyers and Cantley, 1997), and PI4K230, a 230-kDa isoform homologous to the S. cerevisiae protein Stt4p (Gehrmann et al., 1999). Both Pik1p and Stt4p are PI4-kinases that are essential for yeast growth. Functional studies have shown that they cannot substitute for each other, suggesting that they have distinct cellular functions (Audhya et al., 2000). Stt4p is involved in maintaining vacuole- and cell wall integrity, and actin-related functions, whereas Pik1p and PI4Kβ control the structural integrity of the Golgi complex and regulate transport of newly synthesized proteins from the Golgi complex to the plasma membrane (Godi et al., 1999a; Audhya et al., 2000).
PI4Kβ is present in the cytoplasm where it is concentrated in the Golgi complex (Wong et al., 1997; Godi et al., 1999a). The recruitment of PI4Kβ to the Golgi complex is critically dependent on the small GTP-binding protein ARF1, which potently enhances phosphatidylinositol 4-phosphate [PI(4)P] synthesis and ultimately phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] production (Godi et al., 1999a). The enzyme activity of the yeast orthologue of PI4Kβ, Pik1p, is important in the secretory process because reduced PI(4)P levels inhibit secretion of newly synthesized proteins (Hama et al., 1999), and kinase dead Pik1p inhibits biosynthetic protein transport from the Golgi complex to the cell surface of S. cerevisiae (Walch-Solimena and Novick, 1999). In mammalian cells, however, kinase activity of PI4Kβ was reported to inhibit the transport of influenza hemagglutinin from the trans-Golgi network (TGN) to the apical cell surface of polarized Madin-Darby canine kidney cells (Bruns et al., 2002). In addition, kinase activity of PI4Kβ is modulated by frequenin, a myristoylated small Ca2+-binding protein that controls constitutive and regulated secretion (McFerran et al., 1998; Weisz et al., 2000).
To investigate the function of PI4Kβ, we searched for novel binding partners. Here, we report that rab11 in its GTP-bound state functionally interacts with PI4Kβ to regulate vesicular transport from the Golgi complex to the plasma membrane. In contrast to many other rab-interacting proteins, PI4Kβ is not recruited to the Golgi complex by rab11. Instead, PI4Kβ seems to act as a docking factor for rab11 on the Golgi complex. This novel link between rab11 and PI4Kβ provides new insights in the function of small G proteins and inositol lipid kinases in membrane transport.
MATERIALS AND METHODS
Materials
The rabbit polyclonal antibodies against PI4Kβ and rab11 were generous gifts of Rachel Meyers (Harvard Medical School, Boston, MA) (Meyers and Cantley, 1997) and Bruno Goud (Institut Curie, Paris, France), respectively. The rabbit antibody against the Myc-epitope (9E10) was from Upstate Biotechnology (Lake Placid, NY). Mouse monoclonal antibodies against the Myc-epitope (9E10) and green fluorescent protein (GFP) were from Roche Applied Science (Basel, Swiss), against the FLAG-epitope M2 were from Sigma-Aldrich (St. Louis, MO), and against GM130 and rab11 were from BD Biosciences (Franklin Lakes, NJ). The monoclonal antibody against the X31 influenza hemagglutinin NH epitope was described previously (Bottger et al., 1996). Secondary antibodies conjugated to the different CyDyes or to horseradish peroxidase were all from Jackson ImmunoResearch Laboratories (West Grove, PA).
DNA Constructs
The human PI4Kβ cDNA was obtained as described previously (de Graaf et al., 2002). pcDNA3.1 zeo+-PI4Kβ N406A, V710G (PI4KβE5) was made by error prone polymerase chain reaction (PCR) (Bottger et al., 2000), and pcDNA3-NH-rab11, pcDNA3-NH-rab11Q70L, and pcDNA3-NH-rab11S25N were described previously (Roberts et al., 2001). pGEX-rab4, pGEX-rab11, pGEX-rab11Q70L, pGEX-rab11S25N, pGBT9-PI4Kβ, pGBT9-rab4, pGBT9-rab5, pGBT9-rab6, pGBT9-rab11 (A and B isoforms), pGBT9-ARF1, pcDNA3.1 zeo+-PI4Kβ (aa 1-400), pcDNA3.1 zeo+-PI4Kβ (aa 401-801), and pcDNA3.1 zeo+-PI4Kβ (aa 401-516) were generated by standard molecular biology methods. Except for the two-hybrid experiments, all other assays with rab11 were done with rab11A. pEGFP-VSV-G tsO45 was a generous gift of Mark McNiven (Cao et al., 2000). Constructs generated by PCR were confirmed by dideoxy sequencing (Eurogentec, Seraing, Belgium).
Yeast Two-Hybrid Assays
S. cerevisiae strain AH109 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2:GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, ura3::MEL1UAS-MEL1TATA-LacZ) was used for two-hybrid assays. Standard rich medium (YPD) and synthetic complete medium (SC), supplemented with 2% glucose were used for the growth of yeast cells and for selection and maintenance of the plasmids. pGBT9-PI4Kβ was transformed into AH109 by using the lithium acetate method. Subsequently, the human brain MATCH-MAKER cDNA library in pACT2 (BD Biosciences Clontech, Palo Alto, CA) was transformed and interacting clones were isolated on selective SC medium (yeast nitrogen base, without amino acids) (BD Biosciences) supplemented with 2% glucose and nutrients as appropriate for the maintenance of plasmids (SC-Trp-Leu-Ade).
β-Galosidase Assay
S. cerevisiae strain Y190 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2:GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, ura3::MEL1UAS-MEL1TATA-LacZ) was used for the β-galactosidase assay. Y190 cells were transformed with the indicated constructs and grown on selective plates. Obtained colonies were grown, collected, and washed with phosphate-buffered saline (PBS) by centrifugation, resuspended in Z-buffer (0.1 M Tris/Triton X-100 buffer, pH 7.0) and stored at -80°C to permeabilize the cells. On thawing at 4°C, Z-buffer with O-nitrophenyl β-d-galactopyranoside (4 mg/ml) was added to initiate the reaction, which was stopped by addition of 1.0 M Na2CO3. The conversion of O-nitrophenyl β-d-galactopyranoside into galactose and O-nitrophenyl was measured colorometrically at 420 nm and expressed in Miller units (OD420 × 1000/OD600 × t (minutes) × volume (milliliters) (Dang et al., 1996).
Cell Culture and Transfections
Baby hamster kidney cells (BHK-21) and COS-1 cells were grown in DMEM supplemented with 7.5% fetal calf serum (Invitrogen, Paisley, United Kingdom). Transient transfections in BHK-21 cells were performed at 25% confluence by using FuGENE 6 (Roche Applied Science) according to the procedure of the manufacturer. Transient transfections in COS-1 cells were performed at 40% confluence by the DEAE-dextran method.
Glutathione S-Transferase (GST) Pull-Down Experiments
GST fusion proteins were produced in the E. coli strain BL21-CodonPlus-RIL (Stratagene, La Jolla, CA), and purified as described previously (Whitehead et al., 1999). COS-1 cells, grown in 10-cm-diameter dishes were transfected, lysed in 1 ml of ice-cold pull-down buffer (20 mM HEPES, pH 7.2, 100 mM NaCl, 20 mM MgCl2, 2 mM EDTA, 1 mM dithiothreitol, 0.1% Tween 20, protease inhibitor cocktail Complete (Roche Applied Science), supplemented with 10 mg/ml bovine serum albumin. Lysates were passed six times through a 25-gauge needle, and centrifuged for 5 min at 13.000 × g. GST-fusion protein coupled to glutathione (GSH)-agarose beads was added and samples were incubated by end over end rotation for 2 h at 4°C. Bound proteins were either analyzed by Western blotting or in an in vitro lipid kinase assay. Pull-down assays of GST-rab4, GST-rab5, GST-rab11 with [35S]methionine-labeled Myc-tagged PI4Kβ, produced by in vitro transcription-translation, were exactly done as described previously (Nielsen et al., 2000).
Fluorescence Microscopy
Cells grown on coverslips were fixed for 20 min in 4% formaldehyde in PBS, and permeabilized by a 5-min incubation with 0.2% Triton X-100 in PBS. Formaldehyde was quenched for 10 min by using 50 mM glycine in PBS. Cells were incubated for 60 min with primary and secondary antibodies diluted in 0.2% gelatin in PBS, embedded in Mowiol/PPD and analyzed in a Leica TCS-SP inverted spectral emission confocal-scanning microscope (Leica GmbH, Heidelberg, Germany) by using 488- and 568-nm excitation and a 63× APO-water immersion objective. Specific wavelength settings were selected for optimal detection of emission from the individual probes with minimal spill-over emission originating from unrelated probes. Images are representative of at least three independent experiments.
Vesicular Stomatitis Virus-G Protein-Green Fluorescent Protein (VSV-G-GFP) Transport Assay
VSV-G-GFP transport assay was performed as described previously (Chen et al., 1998) with minor modifications. BHK-21 cells were grown in DMEMHEPES (DMEM buffered with 25 mM HEPES, pH 7.4) supplemented with 7.5% fetal calf serum, and transfected with pEGFP-VSV-G and vectors encoding Myc-tagged PI4Kβ constructs and/or NH-tagged rab11S25N. The cells were cultured at 40°C in a humidified atmosphere and received 100 μg/ml cycloheximide 20 h after transfection. The cells were then incubated at 20°C for 2 h, and either fixed or transferred to 32°C. After 2 h, cells were fixed and processed for fluorescence microscopy. For biotinylation of plasma membrane proteins the cells were washed twice with ice-cold PBS, and incubated with 100 μg/ml sulfo-NHS-SS-biotin (Pierce Chemical, Rockford, IL) for 20 min on ice. Excess sulfo-NHS-SS-biotin was removed by washing with PBS, and the cells were quenched by incubation for 10 min with 50 mM glycine in PBS. Subsequently, the cells were lysed in immunoprecipitation (IP) buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10% glycerol, 0.5% Triton X-100, 0.1% SDS, 1 mM EDTA, protease inhibitor cocktail Complete; Roche Applied Science), and biotinylated proteins were isolated using streptavidin-Sepharose beads. VSV-G-GFP was detected by Western blotting by using anti-GFP, and the individual bands were quantified using a lumino-imager.
Miscellaneous Biochemical Assays
Lipid kinase activities were measured as described previously (de Graaf et al., 2002). Protein samples were separated on 10% SDS-PAA gels, and analyzed by Western blotting as described previously (de Graaf et al., 2002). Detection was done with chemiluminescence as described by the manufacturer (Lumi-light Plus; Roche Applied Science), and visualized using a multi-imager lumino-imager (FluorS; Bio-Rad, Hercules, CA).
RESULTS
Rab11 Is a Novel Binding Partner of PI4Kβ
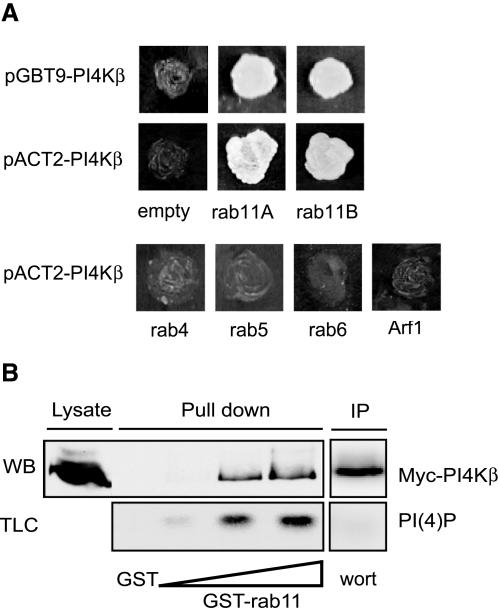
To find binding partners of PI4Kβ, we screened an adult human brain cDNA library with full-length PI4Kβ as bait by using the yeast two-hybrid system. From the 19 different clones that were able to grow under selective conditions (-Trp, -Leu, -Ade), we isolated two independent clones encoding rab11A and three independent clones encoding rab11B, the two isoforms of rab11 (Figure 1A). PI4Kβ in the library vector interacted equally well with either rab11A or rab11B in the bait vector, indicating that the interaction is caused by the bait and is independent of the two-hybrid plasmid. To define the specificity of the interaction, we analyzed several other small GTPases for their ability to yield reporter activation with PI4Kβ. We selected rab4 and rab5, which are both involved in endocytosis, and rab6, which has a role in retrograde intra-Golgi transport (Zerial and McBride, 2001). In addition we tested ARF1, which controls COPI and PI4Kβ recruitment to the Golgi complex and also genetically interacts with Pik1p (Godi et al., 1999a; Walch-Solimena and Novick, 1999). Besides a specific interaction of PI4Kβ with both rab11A and rab11B, no interaction was observed with rab4, rab5, rab6, or ARF1, demonstrating the specificity of PI4Kβ for rab11A and B (Figure 1A).
Figure 1.
Rab11 is a binding partner of PI4Kβ. (A) For yeast two-hybrid analysis, S. cerevisiae AH109 cells were transformed with the bait vector pGBT9 containing PI4Kβ and the prey vector pACT2 containing rab11A and rab11B, or with pACT2 alone as control (empty). Alternatively, S. cerevisiae cells were transformed with constructs that were exchanged from bait to prey vector: pGBT9 with rab11A, rab11B, rab4, rab5, rab6 or ARF1, and pGBT9 containing PI4Kβ or pACT2 alone as control (empty). Yeast cells were grown for 4 d on selective medium (-Trp, -Leu, -Ade), and images of the culture plates are shown. (B) COS-1 cells were grown in 50-cm2 dishes and transfected with 5 μg of cDNA encoding Myc-tagged PI4Kβ. After 2 d, cells were lysed in 1 ml of lysis buffer as described in MATERIALS AND METHODS, and 20 μl of this lysate was analyzed by Western blotting (Lysate). GST pull-down assays were performed as described in MATERIALS AND METHODS by using either GST (5 μg) or increasing amounts of GST-rab11 (2.5, 5, and 10 μg) (pull down). Proteins from the COS-1 lysate bound to GSH beads were split, either analyzed by Western blotting by using anti-Myc (WB), or subjected to an in vitro lipid kinase assay for the detection of PI(4)P (TLC). PI4Kβ, isolated from the COS-1 lysate by immunoprecipitation was subjected to an in vitro lipid kinase assay, and inhibition of lipid kinase activity was done using 1 μM wortmannin (wort) (IP).
We extended the results from the yeast two-hybrid assay by investigating the binding of PI4Kβ to rab11 in an in vitro GST pull-down assay. To this aim, we prepared detergent lysates of COS-1 cells transiently expressing Myc-tagged PI4Kβ and incubated these with increasing amounts of GST-rab11. PI4Kβ was retrieved in a dose-dependent manner on GST-rab11 beads, but not on control GST beads (Figure 1B). In addition, we characterized the lipid kinase activity by using in vitro lipid kinase measurements. The amount of PI(4)P formed was directly proportional to the amount of PI4Kβ captured on GST-rab11 beads, whereas control pull-downs with GST did not to contain kinase activity (Figure 1B). The observation that a type III, wortmannin-sensitive phosphoinositide kinase was retrieved on the beads containing rab11 confirmed the presence of PI4Kβ (Figure 1B).
PI4Kβ/rab11 Interaction Is Regulated by GTP but Not by Lipid Kinase Activity
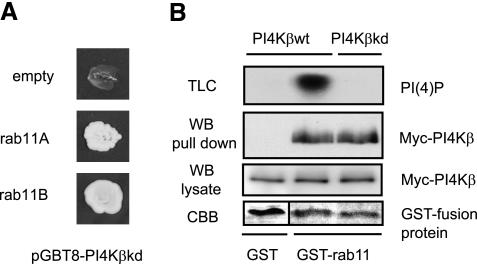
To investigate whether the enzyme activity of PI4Kβ was relevant for binding to rab11, we generated a kinase dead version of PI4Kβ (PI4Kβkd) by substituting the aspartic acid residue at position 656 for alanine (Godi et al., 1999a). Two-hybrid and pull down assays with the mutant showed that PI4Kβkd and PI4Kβ wild-type bound equally well to rab11 (Figure 2, A and B). The absence of PI(4)P on the thin layer chromatography confirmed that the mutant was devoid of kinase activity (Figure 2B).
Figure 2.
PI4Kβ/rab11 association is independent of lipid kinase activity. (A) For yeast two-hybrid assay, S. cerevisiae AH109 cells were transformed with the kinase dead mutant PI4KβD656A in the bait vector pGBT9 (pGBT9-PI4KβkDa), and rab11A or rab11B in pACT2, or with pACT2 alone as control (empty). Yeast cells were grown for 4 d on selective medium (-Trp, -Leu, -Ade), and images of culture plates are shown. (B) COS-1 cells were transfected with 5 μg of cDNA encoding Myc-tagged proteins PI4Kβ or PI4KβkDa, and detergent lysates (1 ml) were used in GST pull-down assays with 5 μg of GST or GST-rab11. Proteins bound to GSH beads were split, either analyzed by Western blotting by using anti-Myc (WB, pull down), or subjected to an in vitro lipid kinase assay (TLC). As controls, PI4Kβ and PI4KβkDa present in cell lysates were detected with anti-Myc (WB, lysate), and GST-fusion proteins were stained with Coomassie Brilliant Blue (CBB).
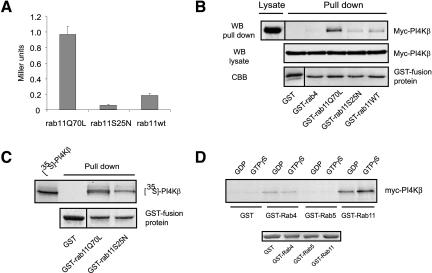
Because the function of rab GTP-binding proteins is regulated by cycles of guanine nucleotide hydrolysis, we next analyzed whether the interaction between PI4Kβ and rab11 was dependent on its guanine nucleotide state. To address this question, we generated point mutants of rab11 that are either deficient in GTP-hydrolysis (rab11Q70L), or poorly bind guanine nucleotide (rab11S25N) (Wilcke et al., 2000). First, we tested these mutants in a quantitative yeast two-hybrid assay. The yeast strain Y190 was transformed with pGBT9-rab11Q70L, -rab11S25N, or -rab11wt together with pAct2-PI4Kβ. After selection on appropriate plates, cells were lysed and β-galactosidase activity was measured. A 20 times higher β-galactosidase activity was found in cells expressing rab11Q70L and PI4Kβ, compared with cells containing rab11S25N (Figure 3A). As expected, rab11wt binds with an intermediate affinity. We then used these mutants fused to GST to assess their ability to bind Myc-PI4Kβ transiently expressed in COS-1 cells. Binding of PI4Kβ to rab11Q70L was superior compared with rab11S25N (Figure 2B). Again, wild-type rab11 bound PI4Kβ with intermediate affinity. Because we did not find binding to GST or GST-rab4, we conclude that PI4Kβ preferentially interacts with the GTP-bound form of rab11. Moreover, binding of GST-rab11Q70L to PI4Kβ in the pull down assays was direct because it could be recapitulated with 35S-labeled PI4Kβ produced in an in vitro transcription translation reaction (Figure 3C). In addition, we analyzed the role of GTP in the interaction of PI4Kβ/rab11 by using GST-rab11 in vitro loaded with either GDP or GTPγS. PI4Kβ preferably binds to the active GTP-bound form of rab11, which is in agreement with the results obtained with the rab11 mutants (Figure 3D). No interaction was seen with GST and rab5, and a very weak binding to rab4 was observed, which was independent of the nucleotide state of the small G protein. Finally, we also investigated whether PI4Kβ kinase activity is regulated by the interaction with rab11. To this aim, we performed in vitro lipid kinase assays in the presence of GST-rab11Q70L, GST-rab4, or GST and found the same amount of PI(4)P formed, no matter which fusion protein was present in the assay (our unpublished data). In conclusion, four independent assays demonstrate that PI4Kβ interacts directly with rab11 in a guanine nucleotide-dependent manner.
Figure 3.
PI4Kβ/rab11 association is regulated by GTP. (A) For quantitative yeast two-hybrid assay, S. cerevisiae Y190 cells were transformed with rab11Q70L, rab11S25N, or rab11wt in the bait vector pGBT9 and with PI4Kβ in pACT2. Yeast cells were grown on selective medium (-Trp, -Leu, -His), and β-galactosidase activity was determined as described in MATERIAL AND METHODS and expressed as Miller units. (B) COS-1 cells were transfected with 5 μg of cDNA encoding Myc-tagged PI4Kβ. After 2 d, cells were lysed and detergent lysates (1 ml), and 20 μl of this lysate was analyzed by Western blotting (Lysate). GST pull-down assays were performed using 5 μg of GST, GST-rab4, GST-rab11, GST-rab11Q70L, or GST-rab11S25N (pull down). Proteins bound to GSH beads were analyzed by Western blotting by using anti-Myc (WB, pull down). As controls, Myc-PI4Kβ present in cell lysates was detected with anti-Myc (WB, lysate), and the GST-fusion proteins were stained with Coomassie Brilliant Blue (CBB). (C) 35S-labeled PI4Kβ was produced by an in vitro transcription translation reaction, and subjected to a GST pull-down assay with 5 μg of GST, GST-rab11Q70L, or GST-rab11S25N. 35S-labeled PI4Kβ was detected by phosphorimaging. (D) 35S-labeled PI4Kβ was produced by an in vitro transcription translation reaction and subjected to a GST pull-down assay with 5 μg of GST, GST-rab4, GST-rab5, or GST-rab11. Preloading was done with either GDP or GTPγS as indicated. Bound 35S-labeled PI4Kβ was detected by phosphorimaging.
PI4Kβ Is Required for Recruitment of Active rab11 to the Golgi Complex
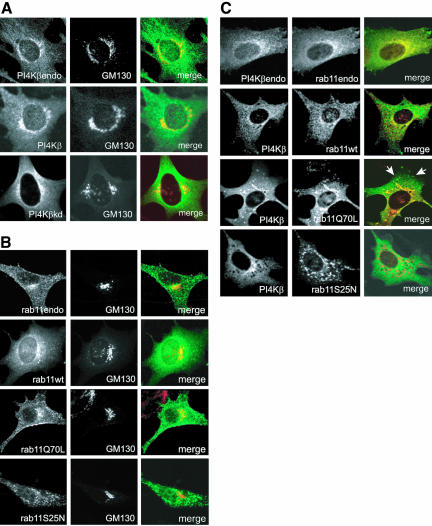
Because both PI4Kβ and rab11 behave as peripheral membrane proteins, we asked where the two proteins interact in the cell. We first determined the distribution of endogenous PI4Kβ in BHK-21 cells by using double-label confocal immunofluorescence microscopy. Whereas some PI4Kβ is present in the cytoplasm, most of it colocalized with both the trans-Golgi network protein TGN46 (our unpublished data) and the cis-medial Golgi matrix protein GM130 and in vesicular structures outside the Golgi (Figure 4A), confirming previous observations of others (Wong et al., 1997; Godi et al., 1999a). Moderately (approximately 5 times) overexpressed Myc-tagged PI4Kβ or PI4Kβkd showed the same distribution as endogenous PI4Kβ, documenting first, that overexpression does not cause mistargeting, and second, that kinase activity is not required for the localization of PI4Kβ to the Golgi complex (Figure 4A). We next determined the distribution of endogenous rab11, which was previously shown to localize to the Golgi complex and recycling endosomes (Ullrich et al., 1996; Chen et al., 1998). Both endogenous rab11 and exogenous Myc-tagged rab11 colocalized with GM130 (Figure 4B). Interestingly, transfected rab11Q70L localized predominantly to the Golgi complex, whereas rab11S25N distributed to punctuate cytoplasmic structures (Figure 4B). This localization seems to contrast with earlier observations of Chen and colleagues who reported a Golgi localization of GFP-rab11S25N (Chen et al., 1998). In our hands, however, GFP-rab11S25N constructs had a different intracellular localization than either untagged, or NH and Myc-tagged constructs, apparently due to the relatively large GFP portion of the fusion protein (PvBeH; our unpublished data). Thus, the localization experiments suggest that the guanine nucleotide state is an important determinant in the intracellular distribution of rab11.
Figure 4.
PI4Kβ and rab11Q70L colocalize in the Golgi complex. (A) BHK-21 cells were grown on coverslips and either transfected with the indicated constructs (Myc-PI4Kβ or Myc-PI4KβkDa) or left untreated. After fixation with 4% formaldehyde, endogenous PI4Kβ was labeled with rabbit anti-PI4Kβ (PI4Kβendo), whereas the Myc-tagged proteins were labeled with rabbit anti-Myc (left, green). The cis-medial Golgi marker GM130 was labeled with mouse monoclonal antibody (middle, red). After labeling with the appropriate Cy2- and Cy3-labeled secondary antibodies the cells were examined by confocal immunofluorescence microscopy. Merged pictures are shown in the right panel. (B) BHK-21 cells were grown on coverslips and either transfected with cDNAs encoding Myc-tagged rab11wt, rab11Q70L, or rab11S25N or left untreated. After fixation with 4% formaldehyde, endogenous rab11 was labeled with rabbit antibody (rab11endo), whereas the Myc-tagged rab11 mutants were labeled with rabbit anti-Myc (left, green). The cis-medial Golgi marker GM130 was labeled with mouse monoclonal antibodies (middle, red). After labeling with the appropriate Cy2- and Cy3-labeled secondary antibodies, the cells were examined by confocal immunofluorescence microscopy. Merged pictures are shown in the right panel. (C) BHK-21 cells were grown on coverslips and either cotransfected with cDNAs encoding Myc-tagged PI4Kβ and NH-tagged rab11wt, rab11Q70L, or rab11S25N, or they were left untreated. After fixation with 4% formaldehyde, endogenous PI4Kβ (PI4Kβendo) and Myc-PI4Kβ were labeled with rabbit polyclonal antibodies (left, green), whereas endogenous rab11 (rab11endo) and NH-tagged rab11 proteins were labeled with mouse monoclonal antibodies. After labeling with the appropriate Cy2- and Cy3-labeled secondary antibodies, the cells were examined by confocal immunofluorescence microscopy. Merged pictures are shown in the right panel. Arrows indicate colocalization of PI4Kβ and rab11Q70L in vesicular structures outside the Golgi complex.
Given the interaction between PI4Kβ and rab11 and their localization with respect to GM130, we expected that the complex would be associated with the Golgi complex. A clear colocalization of PI4Kβ and rab11 as well as coexpressed Myc-tagged PI4Kβ and NH-tagged rab11 was observed in the Golgi complex (Figure 4C). Rab11 was also distributed in punctate structures in the cytoplasm that, however, did not contain PI4Kβ. We next investigated the location of constitutively active rab11Q70L, which binds to PI4Kβ. Extensive colocalization of active rab11 with the lipid kinase was observed both in the Golgi complex and in vesicular structures in the cytoplasm (Figure 4C, arrows). In contrast, the dominant negative rab11S25N mutant distributed in punctuate structures in the cytoplasm that were devoid of PI4Kβ (Figure 4C).
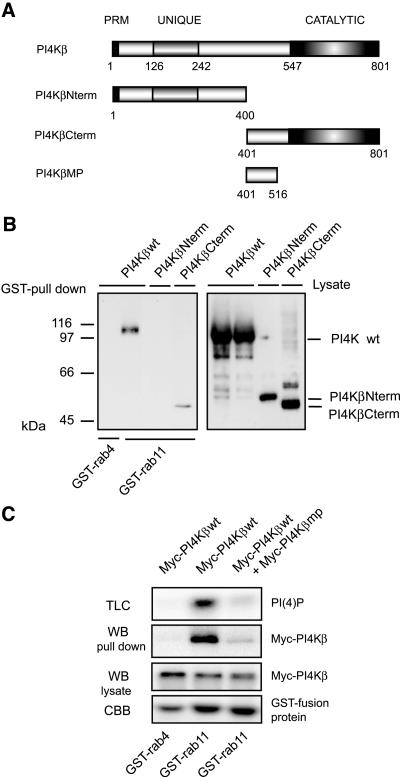
To investigate the function of the PI4Kβ/rab11 interaction, we first mapped the rab11-binding site on PI4Kβ by using the GST pull-down assay. Full-length Myc-tagged PI4Kβ or two truncation mutants representing the N- and C-terminal halves of the protein were expressed in COS-1 cells and detergent lysates of transfected cells were incubated with GST-rab11 or GST-rab4 as a control. Bound PI4Kβ was detected by Western blotting. The C-terminal half of PI4Kβ (PI4KβCterm, aa 401-801) retained the ability to bind GST-rab11, whereas the N-terminal half (PI4KβNterm, aa1-400) of the protein did not yield a signal above the negative control GST-rab4 (Figure 5A). We then evaluated whether the overexpression of the rab11-binding domain of PI4Kβ affected the interaction of full-length PI4Kβ with rab11. To preclude possible pleiotropic side effects related to the presence of the kinase domain (aa 547-801) (Meyers and Cantley, 1997) within the rab11-binding region, we performed the experiments with domain aa 401-516 (PI4Kβmp, middle piece), which lacks the kinase domain. In pull-down assays with GST-rab11 and lysates of COS-1 cells expressing full-length Myc-PI4Kβ, a sixfold overexpression of Myc-tagged PI4Kβmp induced a strong reduction in the amount of PI4Kβ retrieved on the GST-rab11 beads compared with incubations with control lysates prepared from cells expressing PI4Kβwt alone (Figure 5B, WB pull down). A similar reduction was observed in the levels of PI(4)P (Figure 5B, TLC). Together, these data show that rab11 interacts with the C-terminal part of PI4Kβ and that expression of the noncatalytic region in the C terminus of PI4Kβ (PI4Kβmp) inhibits the association between PI4Kβ and rab11.
Figure 5.
The rab11-binding site is located in the C terminus of PI4Kβ. (A) Schematic representation of PI4Kβ mutants. PRM, proline-rich motif; UNIQUE, lipid kinase unique domain; CATALYTIC, catalytic domain. (B) COS-1 cells were transfected with 5 μg cDNA encoding Myc-tagged PI4Kβwt (wild-type), PI4KβNterm (N-terminal domain, aa 1-400), or PI4KβCterm (C-terminal domain, aa 401-801). After 2 d, cells were lysed in 1 ml of lysis buffer, and GST pull-down assays were performed as described in the MATERIALS AND METHODS by using 5 μg of GST-rab4 or GST-rab11. Proteins bound to GSH beads were analyzed by Western blotting by using anti-Myc (GST pull down). Expression of the different constructs was analyzed by Western blotting by using anti-Myc (Lysate). (B) COS-1 cells were transfected with 5 μg of cDNA encoding Myc-tagged PI4Kβwt, or Myc-tagged PI4Kβwt in combination with Myc-tagged PI4Kβmp (middle piece, domain aa 401-516). GST pull-down assays were performed as in A. Proteins bound to GSH beads were split, either analyzed by Western blotting by using anti-Myc (WB, pull down) or subjected to an in vitro lipid kinase assay (TLC). As controls, PI4Kβ present in cell lysates was detected by Western blotting (WB, lysate), and the GST-fusion proteins were stained with Coomassie Brilliant Blue (CBB).
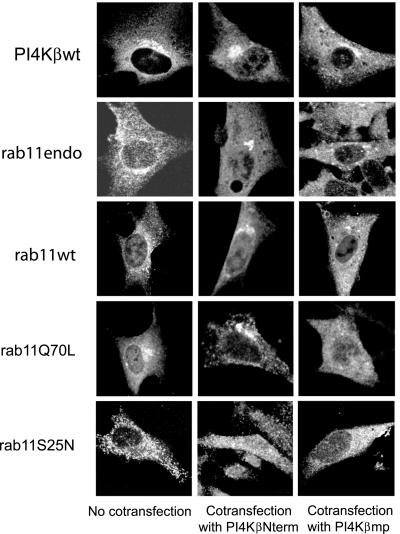
Given the presence of a PI4Kβ/rab11 complex in the Golgi, it became important to discriminate between the possibilities whether Golgi-associated PI4Kβ might recruit active rab11, or alternatively, whether cytoplasmic PI4Kβ is recruited by Golgi resident rab11. To address this question, we used the inhibitory PI4Kβmp construct that perturbs binding of the two proteins. If PI4Kβ is required for the localization of rab11 to the Golgi complex, expression of PI4Kβmp should not effect the localization of resident PI4Kβ, but interfere by competition with the localization of rab11. BHK-21 cells coexpressing FLAG-tagged PI4Kβ and Myc-tagged PI4Kβmp or control domain PI4KβNterm were labeled with antibodies against the epitope tags and analyzed by confocal fluorescence microscopy. Expression of PI4Kβmp did not affect the localization of PI4Kβwt (Figure 6). In contrast, in ∼35% of the cells coexpressing NH-rab11Q70L and Myc-PI4Kβmp, the rab11-binding domain of PI4Kβ completely removed rab11Q70L from the juxtanuclear region (Figure 6), while leaving the Golgi complex intact (our unpublished data). Similar results were obtained when endogenous rab11, or NH-tagged rab11wt were analyzed in cells expressing the rab11-binding domain of PI4Kβ (Figure 6). As expected, coexpression of control Myc-tagged PI4KβNterm did not affect the localization of either endogenous rab11, rab11wt, or rab11Q70L to the Golgi complex (Figure 6, middle). We did not find any effect of the two PI4Kβ constructs on the localization of the rab11S25N mutant, which poorly binds to PI4Kβ (Figure 3) and does not localize to the Golgi complex (Figure 6). Because the expression of PI4Kβmp prevented the association of rab11Q70L with the Golgi complex but did not interfere with the localization of PI4Kβ, we conclude that PI4Kβ is essential for the localization of active rab11 to the Golgi complex.
Figure 6.
PI4Kβ is required for localization of active rab11 in the Golgi complex. BHK-21 cells were grown on coverslips and either left untreated or transfected with the indicated constructs (FLAG-tagged PI4Kβwt, NH-tagged rab11wt, rab11Q70L, or rab11S25N) alone, or cotransfected with cDNAs encoding Myc-PI4KβNterm or Myc-PI4Kβmp. Cells were fixed in 4% formaldehyde and labeled with either mouse antibodies against rab11, against FLAG to detect PI4Kβ, or against NH to detect rab11 mutants, and rabbit anti-Myc was used to detect the PI4Kβ mutants. Cells were labeled with the appropriate Cy2- and Cy3-labeled secondary antibodies and examined by confocal fluorescence microscopy. Shown are the distributions of PI4Kβwt, endogenous rab11 (rab11endo) or rab11 mutants in cells that stained positive for Myc-tagged proteins.
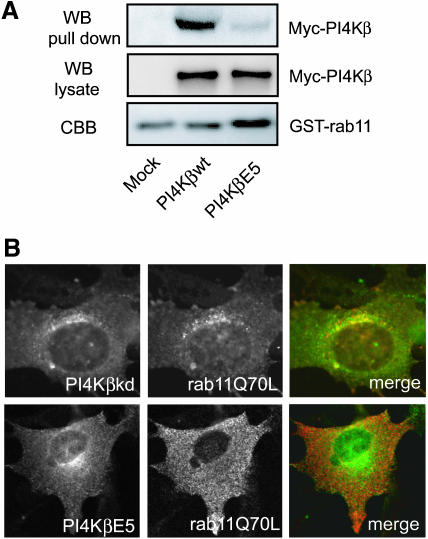
To further explore the role of PI4Kβ in the localization of rab11, we examined the effect of the kinase dead PI4Kβkd, which retained the ability to bind rab11 (Figure 2B), and of the double mutant PI4KβE5 on the distribution of rab11Q70L. PI4KβE5 contains a N406A mutation in the rab11-interacting domain, which perturbs binding to rab11, and a V710G mutation in the kinase domain, which eliminates kinase activity (our unpublished data). PI4KβΕ5 was obtained by random mutagenesis using error prone PCR followed by two-hybrid screening for nonbinding mutants. The absence of binding of PI4KβE5 to rab11 was confirmed using the GST pull-down assay (Figure 7A). Both mutants were similarly localized as PI4Kβwt, confirming that rab11 binding and enzyme activity are not required for the Golgi localization of PI4Kβ (Figure 7B). In contrast to the cells expressing PI4KβkDa, rab11Q70L is absent from the Golgi complex in cells expressing PI4KβE5. This observation confirms the notion that the binding of rab11 to PI4Kβ is critical for the presence of active rab11 in the Golgi complex (Figure 7B).
Figure 7.
PI4Kβ enzyme activity is not required for localization of rab11 in the Golgi complex. (A) COS-1 cells were either mock transfected or transfected with 5 μg of cDNA encoding Myc-tagged PI4Kβwt, or PI4KβΕ5. GST pull-down assays were performed as described in MATERIALS AND METHODS. Proteins bound to GSH beads were split and either analyzed by Western blotting by using anti-Myc (WB, pull down) or subjected to an in vitro lipid kinase assay (TLC). As controls, PI4Kβwt and PI4KβΕ5 present in cell lysates were detected by Western blotting (WB, lysate), and the GST-fusion proteins were stained with Coomassie Brilliant Blue (CBB). (B) BHK-21 cells were grown on coverslips and cotransfected with cDNAs encoding FLAG-tagged PI4KβkDa or PI4KβΕ5 (green, left), in combination with Myc-tagged rab11Q70L (red, middle). Cells were labeled with mouse anti-FLAG and rabbit anti-Myc, labeled with the appropriate Cy2- and Cy3-labeled secondary antibodies, and examined by confocal fluorescence microscopy. Merged pictures (right).
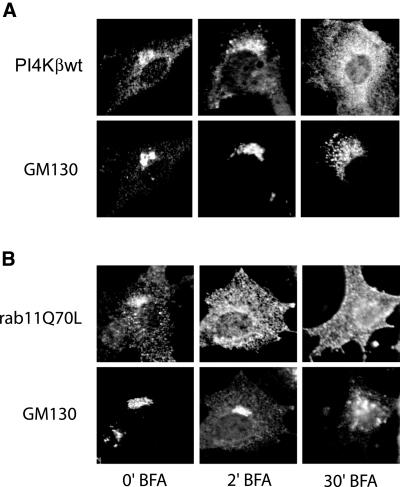
Finally, the role of PI4Kβ in the Golgi localization of rab11 was investigated using brefeldin A (BFA). This fungal metabolite inhibits a class of ARF1 exchange factors and prevents activation of ARF1 causing disassembly of the Golgi complex. A short BFA treatment (2 min) induced the release of rab11Q70L from the Golgi complex, whereas the distribution of PI4Kβ remained invariant and Golgi morphology was still intact as assessed by the distribution of GM130 (Figure 8). Longer treatment with BFA (4-5 min) resulted in the disassembly of the Golgi stacks and both PI4Kβ and rab11Q70L no longer colocalized with GM130. The order in which PI4Kβ and rab11 disappeared from the Golgi complex was identical in BHK-21 and Chinese hamster ovary cells (our unpublished data). From these results, we conclude that the PI4Kβ/rab11 association in the Golgi is sensitive to BFA and that the localization of PI4Kβ to the Golgi complex is independent of rab11.
Figure 8.
Effect of BFA on localization of PI4Kβ and rab11Q70L. BHK-21 cells were grown on coverslips and transfected with cDNA either encoding Myc-tagged PI4Kβ or Myc-tagged rab11Q70L. After 2 d, the cells were treated for the indicated time intervals with 2 μg/ml BFA. Cells were fixed in 4% formaldehyde and labeled with rabbit anti-Myc and mouse monoclonal anti-GM130, labeled with the appropriate Cy2- and Cy3-labeled secondary antibodies, and examined by confocal fluorescence microscopy.
Binding of rab11 to PI4Kβ Regulates Transport of VSV-GGFP
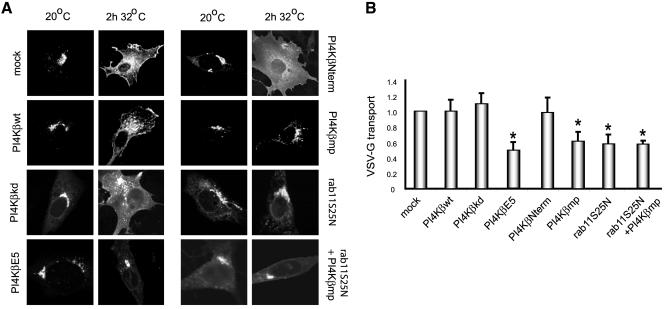
To analyze the role of the PI4Kβ-rab11 interaction in the transport of proteins from the Golgi complex to the plasma membrane, we used a thermosensitive mutant (tsO45) of VSV-G fused with GFP, which is retained in the endoplasmic reticulum (ER) at 40°C (Bergmann, 1989). VSV-G-GFP cDNA was cotransfected with the different Myc-tagged PI4Kβ constructs, and/or NH-tagged rab11S25N into BHK-21 cells. The localization of VSV-G-GFP in the cells expressing the different constructs was visualized by confocal fluorescence microscopy. On a rapid reduction of the temperature to 20°C, VSV-G-GFP moved out of the ER and accumulated in the Golgi complex in control cells (mock) as well as in all transfected cells expressing NH-tagged rab11S25N or any of the Myc-PI4Kβ constructs (Figure 9A), indicating that the PI4Kβ/rab11 association is not involved in ER-Golgi transport. After release of the block by a temperature shift to 32°C, VSV-G-GFP was transported to the cell surface with the same efficiency in mock transfectants and transfectants expressing wild-type PI4Kβ, kinase dead PI4Kβ, or control domain PI4KβNterm (Figure 9A). However, under conditions that endogenous rab11 is removed from the Golgi complex by expressing PI4KβE5 or PI4Kβmp (Figures 6 and 7), a strong inhibition in transport to the cell surface was observed. Similar results were obtained when rab11S25N, or a combination of PI4Kβmp and rab11S25N were coexpressed with VSV-G-GFP (Figure 9A). The inhibition of VSV-G transport by the expression of rab11S25N was in agreement with previous studies performed by Chen et al. (1998), who reported that inactive rab11 induces a decrease in transport kinetics rather than a delay. The similar morphological effect of the inhibiting constructs suggests that also in these cases the reduced transport to the plasma membrane is due to a reduced kinetics.
Figure 9.
PI4Kβ/rab11 complex coregulates VSV-G-GFP delivery to the plasma membrane. (A) BHK-21 cells were grown on coverslips and transfected with cDNAs encoding VSV-G-GFP alone (mock) or with VSV-G-GFP cDNA in combination with the Myc-tagged constructs PI4Kβwt, PI4KβkDa, PI4KβE5, PI4KβNterm, Myc-PI4Kβmp, and the combination of NH-tagged rab11S25N and Myc-tagged PI4Kβmp. Cells were grown for 20 h at 40°C, followed by 2 h at 20°C in the presence of 100 μg/ml cycloheximide, and finally incubated for 2 h at 32°C. Cells were fixed with 4% formaldehyde, labeled with rabbit anti-Myc or mouse anti-NH. After labeling with Cy3- and Cy5-conjugated secondary antibodies the cells were examined by confocal fluorescence microscopy. The distribution of VSV-G in the anti-Myc and anti-NH-positive cells is shown after the 20°C incubation period and after an incubation for 2 h at 32°C. (B) BHK-21 cells were grown in 75-cm2 dishes and cotransfected with cDNA encoding VSV-G-GFP and the indicated constructs. The cells were grown for 20 h at 40°C and subsequently incubated at the different temperatures as described in A. Plasma membrane proteins were biotinylated using the membrane impermeable agent sulfo-NHS-SS-biotin (100 μg/ml) and precipitated using streptavidin-Sepharose beads as described in MATERIALS AND METHODS. VSV-G-GFP bound to the beads and present in the cell lysate was detected by quantitative Western blotting by using a fluoroimager. VSV-G transport is expressed as the ratio of surface located VSV-G-GFP and VSV-G-GFP in the cell lysate and presented as a fraction ± SEM (n = 3, *p < 0.05) of the control value from mock-transfected cells.
In addition, cell surface delivery of VSV-G-GFP was assayed after biotinylation of plasma membrane proteins with membrane impermeable sulfo-NHS-SS-biotin, and isolation of biotinylated proteins on streptavidin beads. The amount of VSV-G-GFP captured on the beads was then measured by Western blotting with an antibody against GFP. As shown in Figure 9B, PI4KβE5, PI4Kβmp and rab11S25N significantly inhibited cell surface delivery by ∼45%, whereas PI4Kβ, control domain PI4KβNterm, and kinase dead PI4Kβ had no significant effect (Figure 9B). Coexpression of PI4Kβmp and rab11S25N did not additionally inhibit VSV-G transport, indicating that the two proteins indeed operate in the same pathway. The partial inhibition of VSV-G-GFP secretion may suggest the existence of an alternative transport pathway from the Golgi complex to the cell surface that is independent of PI4Kβ/rab11. More likely, because we found that the PI4Kβ/rab11 interaction is not abolished by coexpression of the rab11-binding domain of PI4Kβ or PI4KβE5, this may leave sufficient PI4Kβ/rab11 complexes intact to sustain the remaining transport activity that we observed. The nonsignificant effect of the kinase dead mutant of PI4Kβ on VSV-G transport stimulated us to investigate its role in other Golgi-mediated transport processes such as Golgi-to-endosome and Golgi-to-endoplasmic reticulum transport. The effect of overexpression of PI4Kβkd on these transport processes was investigated by analyzing the distribution of either the mannose-6-phospate/IGFII receptor (MPR300) and the KDEL-receptor (Bard et al., 2003; Umeda et al., 2003). No alteration in the distribution of both receptors was observed in the presence of the PI4Kβ kinase dead mutant, indicating that the lipid kinase activity is merely involved in the building of the Golgi complex than in transport processes from the Golgi. On the other hand, our results clearly show that the PI4Kβ/rab11 complex is involved in the VSV-G transport, indicating the PI4Kβ is functioning as a scaffold protein and involved in recruiting rab11 to the Golgi. Given the function of rab11 in early endosomes, we also investigated the role of the PI4Kβ/rab11 complex in endocytosis and recycling of transferrin-Rhodamine. However, no effects on the transferrin cycle were found in cells expressing wild-type and mutant PI4Kβ at expression levels of the constructs that yielded the secretion phenotype (our unpublished data).
DISCUSSION
In this study, we have identified rab11 as a novel binding partner of PI4Kβ. The interaction was specific for rab11 and did not occur with the other rab proteins that we tested. Binding of rab11 to PI4Kβ is regulated by GTP and occurs on the Golgi complex independently of the lipid kinase activity of PI4Kβ. The lipid kinase activity of PI4Kβ is in vitro not altered by rab11. The interaction of rab11 with PI4Kβ is essential for the localization of rab11 in the Golgi complex. Furthermore, the PI4Kβ/rab11 interaction was found to regulate the biosynthetic membrane transport from the Golgi complex to the cell surface.
Colocalization of PI4Kβ with active rab11 was most prominent in the Golgi complex, although the two proteins also codistributed to vesicular structures in the cytoplasm. The Golgi localization of active rab11 was abolished when the binding between PI4Kβ and active rab11 was perturbed. This indicates that PI4Kβ serves as a docking protein for active rab11 in the Golgi complex. Because PI4Kβ binds to the GTP-bound form of rab11, prior activation of rab11 is required for their interaction and sequestration of active rab11 in the Golgi complex. As for most rab proteins, the rab11-specific guanidine exchange factor is not known. Identification of factors involved in the in vivo activation of rab11 will therefore be required to fully understand recruitment of rab11 to the Golgi complex.
PI4Kβ was found to bind to both isoforms of rab11, rab11A and rab11B. So far, it is not known whether rab11A and rab11B have similar or different functions. The fact that PI4Kβ and most other known rab11-binding proteins interact with both isoforms suggests that rab11A and rab11B are functionally related proteins. The interaction of PI4Kβ with rab11 is unique with respect to two features: it is the only rab11-binding protein identified thus far that colocalizes with rab11 in the Golgi complex. Other rab11-binding proteins such as eferin, RIPs, and rabphilin11/rab11BP colocalize with rab11 in endosomal recycling systems (Hales et al., 2001; Prekeris et al., 2001). Also, PI4Kβ lacks the recently identified rab11-binding domain conserved in the C terminus of RIPs, which forms an amphipathic α-helix (Hales et al., 2001; Prekeris et al., 2001). These observations may reflect the functional difference in rab11 interactions because the rab11/PI4Kβ complex is involved in the biosynthetic protein transport from the Golgi complex, whereas the other rab11-interacting proteins are involved in the regulation of endocytic membrane recycling.
The functional interaction of rab11 with PI4Kβ in distal steps of the secretory pathway is consistent with earlier observations. Rab11 has previously been localized to the TGN and TGN-derived vesicles in neuroblastoma cells, and in addition, rab11S25N was shown to inhibit the transport of newly synthesized proteins from the trans-Golgi network to the plasma membrane (Urbe et al., 1993; Chen et al., 1998). On the other hand, a function of rab11 has also been reported in receptor recycling from early endosomes and transport from early endosomes to the TGN (Ullrich et al., 1996; Wilcke et al., 2000). These observations may provide new clues to the molecular principles defining the function of rab11 in the two distinct transport pathways. Conceivably, interactions of rab11 with distinct sets of binding proteins might account for the diversity in rab11 functions.
The interaction between PI4Kβ and rab11 provides a novel example of the interplay between rab-GTPases and enzymes involved in phosphatidylinositol metabolism. For instance, rab5 interacts with and recruits hVPS34 and p110β to early endosomes (Christoforidis et al., 1999b). These PI 3-kinases increase the local PI(3)P concentration that is required for docking of additional cytoplasmic proteins containing FYVE, PH, or PX domains on early endosomes (Christoforidis et al., 1999a; Kanai et al., 2001). Activation of rab5 produces a recruitment site for the assembly of oligomeric protein complexes involved in early endosome membrane dynamics. The observation that rab5 is an upstream regulator of PI-3-kinases, whereas we found that rab11 acts downstream of PI4Kβ, suggests that the interactions of rab-GTPases with PI 3- and PI 4-kinases may serve distinct purposes. A common denominator is that the activity of both kinases leads to the formation of a scaffold site for the assembly of distinct protein complexes containing a rab GTPase. The major difference, however, is that PI 3-kinase activity is required for rab5-mediated transport, whereas kinase activity of PI4Kβ is required for Golgi complex formation and not essential for transport. As such, the ARF1-dependent increase in PI(4)P and PI(4,5)P2 levels in the Golgi complex may recruit proteins involved in the assembly of the Golgi complex, such as spectrin (Godi et al., 1999b). On the other hand, complex formation of PI4Kβ with rab11 irrespective of its kinase activity is involved in the Golgi to plasma membrane transport, suggesting that PI4Kβ is functioning as a scaffold protein in the transport process. In conclusion, given the role of the PI4Kβ/rab11 complex in VSV-G transport, we anticipate that the characterization of proteins involved in complex formation of PI4Kβ and rab11 in the Golgi complex will provide novel and important clues that should help to unravel protein/lipid complexes regulating transport from the Golgi complex to the cell surface.
Acknowledgments
We thank Jannes Woudenberg, Irina Sorokina, Jarno Voortman, Willem Stoorvogel, Ellen van Dam, and Ben Distel for technical assistance and valuable discussions, and Jord Stam and Koert Burger for valuable discussions and critical reading of the manuscript. We also thank Rachel Meyers, Bruno Goud, Victor Hsu, Graham Warren, and Mark McNiven for generously providing reagents. This work was supported by the Algemene Levens Watenschappen (ALW) branch of the Netherlands Organization for Scientific Research (to P.v.d.S.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-12-0862. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-12-0862.
References
- Audhya, A., Foti, M., and Emr, S.D. (2000). Distinct roles for the yeast phosphatidylinositol 4-kinases, stt4p and pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell 11, 2673-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla, T. (1998). Phosphatidylinositol 4-kinases. Biochim. Biophys. Acta 1436, 69-85. [DOI] [PubMed] [Google Scholar]
- Balla, T., Downing, G.J., Jaffe, H., Kim, S., Zolyomi, A., and Catt, K.J. (1997). Isolation and molecular cloning of wortmannin-sensitive bovine type III phosphatidylinositol 4-kinases. J. Biol. Chem. 272, 18358-18366. [DOI] [PubMed] [Google Scholar]
- Bard, F., Mazelin, L., Pechoux-Longin, C., Malhotra, V., and Jurdic, P. (2003). Src regulates Golgi structure and KDEL receptor-dependent retrograde transport to the endoplasmic reticulum. J. Biol. Chem. 278, 46601-46606. [DOI] [PubMed] [Google Scholar]
- Bergmann, J.E. (1989). Using temperature-sensitive mutants of VSV to study membrane protein biogenesis. Methods Cell Biol. 32, 85-110. [DOI] [PubMed] [Google Scholar]
- Bottger, G., Barnett, P., Klein, A.T., Kragt, A., Tabak, H.F., and Distel, B. (2000). Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner. Mol. Biol. Cell 11, 3963-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottger, G., Nagelkerken, B., and van der Sluijs, P. (1996). Rab4 and Rab7 define distinct nonoverlapping endosomal compartments. J. Biol. Chem. 271, 29191-29197. [DOI] [PubMed] [Google Scholar]
- Bruns, J.R., Ellis, M.A., Jeromin, A., and Weisz, O.A. (2002). Multiple roles for phosphatidylinositol 4-kinase in biosynthetic transport in polarized Madin-Darby canine kidney cells. J. Biol. Chem. 277, 2012-2018. [DOI] [PubMed] [Google Scholar]
- Cao, H., Thompson, H.M., Krueger, E.W., and McNiven, M.A. (2000). Disruption of Golgi structure and function in mammalian cells expressing a mutant dynamin. J. Cell Sci. 113, 1993-2002. [DOI] [PubMed] [Google Scholar]
- Chen, W., Feng, Y., Chen, D., and Wandinger-Ness, A. (1998). Rab11 is required for trans-Golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol. Biol. Cell 9, 3241-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis, S., McBride, H.M., Burgoyne, R.D., and Zerial, M. (1999a). The Rab5 effector EEA1 is a core component of endosome docking. Nature 397, 621-625. [DOI] [PubMed] [Google Scholar]
- Christoforidis, S., Miaczynska, M., Ashman, K., Wilm, M., Zhao, L., Yip, S.C., Waterfield, M.D., Backer, J.M., and Zerial, M. (1999b). Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1, 249-252. [DOI] [PubMed] [Google Scholar]
- Dang, V.D., Bohn, C., Bolotin-Fukuhara, M., and Daignan-Fornier, B. (1996). The CCAAT box-binding factor stimulates ammonium assimilation in Saccharomyces cerevisiae, defining a new cross-pathway regulation between nitrogen and carbon metabolisms. J. Bacteriol. 178, 1842-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf, P., Klapisz, E.E., Schulz, T.K., Cremers, A.F., Verkleij, A.J., and van Bergen en Henegouwen, P.M. (2002). Nuclear localization of phosphatidylinositol 4-kinase beta. J. Cell Sci. 115, 1769-1775. [DOI] [PubMed] [Google Scholar]
- Gehrmann, T., and Heilmeyer, Jr., L.M.G. (1998). Phosphatidylinositol 4-kinases. Eur. J. Biochem. 253, 357-370. [DOI] [PubMed] [Google Scholar]
- Gehrmann, T., Heilmeyer, L,M.G., Jr., Suer, S., Herberg, F.W., Balla, A., Vereb, G., Mayr, G.W., and Heilmeyer, L.M., Jr. (1999). Functional expression and characterisation of a new human phosphatidylinositol 4-kinase PI4K230. Biochim. Biophys. Acta 1437, 341-356. [DOI] [PubMed] [Google Scholar]
- Godi, A., Pertile, P., Meyers, R., Marra, P., Di Tullio, G., Iurisci, C., Luini, A., Corda, D., and De Matteis, M.A. (1999a). ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1, 280-287. [DOI] [PubMed] [Google Scholar]
- Godi, A., Santone, I., Pertile, P., Marra, P., Di Tullio, G., Luini, A., Corda, D., and De Matteis, M.A. (1999b). ADP-ribosylation factor regulates spectrin skeleton assembly on the Golgi complex by stimulating phosphatidylinositol 4,5-bisphosphate synthesis. Biochem. Soc. Trans. 27, 638-642. [DOI] [PubMed] [Google Scholar]
- Hales, C.M., Griner, R., Hobdy-Henderson, K.C., Dorn, M.C., Hardy, D., Kumar, R., Navarre, J., Chan, E.K., Lapierre, L.A., and Goldenring, J.R. (2001). Identification and characterization of a family of Rab11 interacting proteins. J. Biol. Chem. 276, 39067-39075. [DOI] [PubMed] [Google Scholar]
- Hama, H., Schnieders, E.A., Thorner, J., Takemoto, J.Y., and DeWald, D.B. (1999). Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274, 34294-34300. [DOI] [PubMed] [Google Scholar]
- Kanai, F., Liu, H., Field, S.J., Akbary, H., Matsuo, T., Brown, G.E., Cantley, L.C., and Yaffe, M.B. (2001). The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 3, 675-678. [DOI] [PubMed] [Google Scholar]
- McFerran, B.W., Graham, M.E., and Burgoyne, R.D. (1998). Neuronal Ca2+ sensor 1, the mammalian homologue of frequenin, is expressed in chromaffin and PC12 cells and regulates neurosecretion from dense-core granules. J. Biol. Chem. 273, 22768-22772. [DOI] [PubMed] [Google Scholar]
- Meyers, R., and Cantley, L.C. (1997). Cloning and characterization of a wortmannin-sensitive human phosphatidylinositol 4-kinase. J. Biol. Chem. 272, 4384-4390. [DOI] [PubMed] [Google Scholar]
- Nielsen, E., Christoforidis, S., Uttenweiler-Joseph, S., Miaczynska, M., Dewitte, F., Wilm, M., Hoflack, B., and Zerial, M. (2000). Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 151, 601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payrastre, B., Missy, K., Giuriato, S., Bodin, S., Plantavid, M., and Gratacap, M. (2001). Phosphoinositides: key players in cell signalling, in time and space. Cell Signal. 13, 377-387. [DOI] [PubMed] [Google Scholar]
- Prekeris, R., Davies, J.M., and Scheller, R.H. (2001). Identification of a novel Rab11/25 binding domain present in eferin and Rip proteins. J. Biol. Chem. 276, 38966-38970. [DOI] [PubMed] [Google Scholar]
- Roberts, M., Barry, S., Woods, A., van der Sluijs, P., and Norman, J. (2001). PDGF-regulated rab4-dependent recycling of alphavbeta3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr. Biol. 11, 1392-1402. [DOI] [PubMed] [Google Scholar]
- Simonsen, A., Wurmser, A.E., Emr, S.D., and Stenmark, H. (2001). The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 13, 485-492. [DOI] [PubMed] [Google Scholar]
- Ullrich, O., Reinsch, S., Urbe, S., Zerial, M., and Parton, R.G. (1996). Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135, 913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda, A., Fujita, H., Kuronita, T., Hirosako, K., Himeno, M., and Tanaka, Y. (2003). Distribution and trafficking of MPR300 is normal in cells with cholesterol accumulated in late endocytic compartments: evidence for early endosome-to-TGN trafficking of MPR300. J. Lipid Res. 44, 1821-1832. [DOI] [PubMed] [Google Scholar]
- Urbe, S., Huber, L.A., Zerial, M., Tooze, S.A., and Parton, R.G. (1993). Rab11, a small GTPase associated with both constitutive and regulated secretory pathways in PC12 cells. FEBS Lett. 334, 175-182. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena, C., and Novick, P. (1999). The yeast phosphatidylinositol-4-OH kinase Pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1, 523-525. [DOI] [PubMed] [Google Scholar]
- Weisz, O.A., Gibson, G.A., Leung, S.M., Roder, J., and Jeromin, A. (2000). Overexpression of frequenin, a modulator of phosphatidylinositol 4-kinase, inhibits biosynthetic delivery of an apical protein in polarized Madin-Darby canine kidney cells. J. Biol. Chem. 275, 24341-24347. [DOI] [PubMed] [Google Scholar]
- Whitehead, B., Tessari, M., Carotenuto, A., van Bergen en Henegouwen, P.M., and Vuister, G.W. (1999). The EH1 domain of Eps15 is structurally classified as a member of the S100 subclass of EF-hand-containing proteins. Biochemistry 38, 11271-11277. [DOI] [PubMed] [Google Scholar]
- Wilcke, M., Johannes, L., Galli, T., Mayau, V., Goud, B., and Salamero, J. (2000). Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-Golgi network. J. Cell Biol. 151, 1207-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, K., Meyers, R., and Cantley, L.C. (1997). Subcellular Locations of Phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 272, 13236-13241. [DOI] [PubMed] [Google Scholar]
- Zerial, M., and McBride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2, 107-117. [DOI] [PubMed] [Google Scholar]