Strigolactones stimulate internode elongation by regulating cell division using a gibberellin-independent pathway.
Abstract
Strigolactone (SL) mutants in diverse species show reduced stature in addition to their extensive branching. Here, we show that this dwarfism in pea (Pisum sativum) is not attributable to the strong branching of the mutants. The continuous supply of the synthetic SL GR24 via the root system using hydroponics can restore internode length of the SL-deficient rms1 mutant but not of the SL-response rms4 mutant, indicating that SLs stimulate internode elongation via RMS4. Cytological analysis of internode epidermal cells indicates that SLs control cell number but not cell length, suggesting that SL may affect stem elongation by stimulating cell division. Consequently, SLs can repress (in axillary buds) or promote (in the stem) cell division in a tissue-dependent manner. Because gibberellins (GAs) increase internode length by affecting both cell division and cell length, we tested if SLs stimulate internode elongation by affecting GA metabolism or signaling. Genetic analyses using SL-deficient and GA-deficient or DELLA-deficient double mutants, together with molecular and physiological approaches, suggest that SLs act independently from GAs to stimulate internode elongation.
In plants, a single hormone can regulate multiple and diverse aspects of plant growth and development (Vogler and Kuhlemeier, 2003; Davies, 2010). Consequently, most developmental processes such as secondary growth, lateral root development, or hypocotyl elongation are influenced by multiple hormones that interact and integrate environmental factors to optimize plant growth and reproduction (Davies, 2010). Important questions in plant biology relate to how these different plant hormones interact to regulate particular processes and the relative roles of each plant hormone according to environmental conditions, developmental stages, and tissues (Nemhauser et al., 2006; Kuppusamy et al., 2009).
Plant height is a major target in plant breeding for improving seed yield, biomass, and standing ability. Most plant hormones have been shown to affect this trait, with auxin (Cleland, 2010), GA (Ingram et al., 1986; Hedden, 2003), and brassinosteroid (BR; Clouse and Sasse, 1998; Jager et al., 2007) promoting internode elongation and with abscisic acid, ethylene (Ross and Reid, 1986), and jasmonic acid (Heinrich et al., 2013) having an inhibitory effect (Davies, 2010; Ross et al., 2011). GA is one of the key determinants of plant height, as suggested by the different strategies designed to reduce plant height in several crops. Genetically manipulating GA biosynthesis (e.g. rice [Oryza sativa] gene Semi dwarf1) or GA signaling pathways (e.g. wheat [Triticum aestivum] Reduced height) to generate dwarf and semidwarf high-yield varieties in rice and wheat was particularly successful during the “Green Revolution” (Peng et al., 1999; Sasaki et al., 2002; Hedden, 2003; Salamini, 2003). In pea (Pisum sativum), the stem length gene LE, encoding a GA 3-oxidase (PsGA3ox1; Lester et al., 1997; Martin et al., 1997), controls internode length and is involved in plant height variation. The le-1 allele is found in most dwarf pea cultivars and generates one of the seven traits studied by Mendel (Ellis et al., 2011; Reid and Ross, 2011). Genetic studies have convincingly demonstrated that BR is also a major endogenous regulator of internode elongation (Clouse and Sasse, 1998; Ross et al., 2011). Based on several lines of evidence, including the rapid growth response during gravitropism and phototropism, auxin is also a major factor controlling internode elongation (Cleland, 2010).
Different signals may affect separate components of a given process or have distinct modes of action. In contrast to hypocotyl elongation, in which only cell elongation is concerned (Gendreau et al., 1997), internode elongation is the result of both cell division and cell elongation, and hormones have different effects on these two mechanisms. Whereas GA affects both cell division and cell elongation, auxin and BR mostly control cell elongation. Molecular mechanisms involved in interactions among hormone signaling pathways are being deciphered, and different classes of cross regulation of a given process by multiple hormones can be described (Kuppusamy et al., 2009). One of the most common schemes of cross regulation is the modulation of the metabolism of one hormone by another. For instance, an important mechanism by which auxin stimulates internode elongation is the increase of bioactive GA levels (Ross et al., 2000, 2003). Additionally, elevated levels of jasmonic acid, a stress-related hormone, can inhibit internode elongation by repressing transcript levels of GA biosynthesis genes (Heinrich et al., 2013). Genetic studies combined with physiology (hormone-level quantifications, hormone response) are very powerful to identify cross talk or cross regulation (Kuppusamy et al., 2009) of a given process by multiple hormones. For the control of internode elongation, pea, with its simple architecture, has been intensively used for such analyses (Ross et al., 2011). In pea stems, auxin increases transcript levels of the LE gene (PsGA3ox1) and down-regulates transcript levels of the SLENDER (SLN) GA catabolism gene (encoding a GA deactivation 2-oxidase [PsGA2ox1]; Lester et al., 1999), leading to increased levels of GA1, the main bioactive GA in pea (Lester et al., 1999; O’Neill and Ross, 2002; Ross et al., 2011). This ability of auxin to promote or maintain levels of bioactive GA in pea stems is also observed in other tissues (roots, pod) and in other species, such as Arabidopsis (Arabidopsis thaliana; Frigerio et al., 2006), barley (Hordeum vulgare; Wolbang et al., 2004), and tobacco (Nicotiana tabacum; Wolbang and Ross, 2001), with some changes in the specific GA metabolism gene that is regulated (Reid et al., 2011).
The growth repressor DELLA proteins are major players in GA signaling and are rapidly degraded in response to GA treatment (Silverstone et al., 2001; Jiang and Fu, 2007). In Arabidopsis, it was shown that genes encoding the GA biosynthetic enzymes GA3ox1 and GA20ox2 are direct DELLA targets, indicating the involvement of DELLA proteins in the feedback regulation of GA biosynthesis by GA treatment (Zentella et al., 2007). Whereas five DELLA genes are present in Arabidopsis (GIBBERELLIN INSENSITIVE, REPRESSOR OF ga1-3 [RGA], RGA-LIKE1 [RGL1], RGL2, and RGL3; Olszewski et al., 2002), the pea genome contains only two DELLA genes, LA and CRY (Weston et al., 2008). In pea, it was shown that auxin regulation of GA metabolism genes was DELLA independent, as indole-3-acetic acid (IAA) regulation of GA genes was also observed in the slender la cry-s line, a DELLA-deficient background (O’Neill et al., 2010; Ross et al., 2011).
In 2008, the role of strigolactones (SLs) in the control of shoot branching was demonstrated (Gomez-Roldan et al., 2008; Umehara et al., 2008). SL-deficient and SL-response mutants identified in pea (ramosus [rms]), rice (dwarf [d]), Arabidopsis (more axillary growth [max]) and petunia (Petunia hybrida; decreased apical dominance [dad]) display increased shoot branching (Ongaro and Leyser, 2008; Beveridge et al., 2009; McSteen, 2009). SLs are carotenoid-derived compounds that were already known for their roles in symbiotic and parasitic interactions in the rhizosphere (Akiyama et al., 2005; López-Ráez et al., 2009). Mutant-based approaches have identified several genes of the SL biosynthesis and signaling pathways (Beveridge and Kyozuka, 2010). The chloroplastic initial steps of the SL biosynthesis pathway involve D27 (Lin et al., 2009), a β-carotene isomerase, modifying all-trans-β-carotene into 9-cis-β-carotene (Alder et al., 2012) and two carotenoid cleavage dioxygenases, CCD7 (RMS5/D17/MAX3/DAD3; Booker et al., 2004; Johnson et al., 2006; Zou et al., 2006; Drummond et al., 2009) and CCD8 (RMS1/D10/MAX4/DAD1; Sorefan et al., 2003; Snowden et al., 2005; Arite et al., 2007), to produce the SL intermediate carlactone (Alder et al., 2012). Another putative SL biosynthesis enzyme has been identified, the cytochrome P450 MAX1 (Booker et al., 2005), acting very likely after carlactone formation; however, its biochemical function is not known yet.
Similar to the GA signaling pathway, the SL signaling pathway involves an F-box protein (RMS4/D3/MAX2; Stirnberg et al., 2002, 2007; Ishikawa et al., 2005; Johnson et al., 2006) and an α/β-fold hydrolase (D14/DAD2; Arite et al., 2009; Gao et al., 2009; Liu et al., 2009; Waters et al., 2012). In contrast to the GA receptor GIBBERELLIN-INSENSITIVE DWARF1, which has lost the enzymatic activity of the members of the α/β-fold hydrolase superfamily, DAD2 retains hydrolytic activity, leading to products without SL-like activity (Hamiaux et al., 2012). Direct binding of the synthetic SL, GR24, with the petunia DAD2 or the rice D14 protein (Hamiaux et al., 2012; Kagiyama et al., 2013) pinpoints the DAD2/D14 α/β-fold hydrolase as the best candidate for the SL receptor. In Arabidopsis and pea, the SL response involves the TCP (for TB1, CYCLOIDEA, and PCF domain) transcription factor BRANCHED1 (BRC1/PsBRC1; Aguilar-Martínez et al., 2007; Braun et al., 2012), homologous to the maize (Zea mays) TEOSINTE BRANCHED1 (TB1; Hubbard et al., 2002; Doebley et al., 2006). Branching of pea or Arabidopsis brc1 mutant plants is not repressed by treatment with the synthetic SL, GR24 (Brewer et al., 2009; Braun et al., 2012); therefore, these mutants are considered as SL-response mutants.
Since the discovery that SLs represent a novel class of plant hormones, several functions of SLs other than the control of branching have been identified through the examination of SL-deficient and SL-response mutants and by examining the effects of treatments with the synthetic SL GR24. Developmental processes such as lateral root formation, adventitious root formation, root hair elongation, and interfascicular cambium development have been shown to be regulated by SL (Agusti et al., 2011; Kapulnik et al., 2011; Ruyter-Spira et al., 2011; Rasmussen et al., 2012). The use of SL and auxin signaling mutants suggests that SLs act predominantly downstream of auxin to stimulate secondary growth (Agusti et al., 2011), whereas for lateral root formation, SLs would modulate auxin levels and/or signaling and ethylene synthesis for root hair elongation (Koltai, 2011). Whether SLs act directly or in coordination with other hormones to regulate these processes will need further investigation. Consequently, it appears that, as for other plant hormones, SLs are involved in many aspects of plant development.
Another phenotype of the SL-deficient and SL-response mutants observed in various species (pea, Arabidopsis, rice, petunia, tomato [Solanum lycopersicum], and Lotus japonicus) is their shorter stature in comparison with the wild type (Beveridge et al., 1996; Napoli, 1996; Stirnberg et al., 2002; Ishikawa et al., 2005; Zou et al., 2006; Kohlen et al., 2012; Liu et al., 2013). Previously, it was shown in rice that this reduced plant height was in part a consequence of the increased branching, as manual removal of tillers in the rice htd1/d17 SL-deficient mutant led to an increase in overall plant height (Zou et al., 2006). However, the overall plant height was not completely restored to that of comparative wild-type plants, and this incomplete restoration could have been due to the diversion of energy from the main stem to the tillers prior to their removal or to their SL deficiency. Umehara et al. (2008) have shown that SLs supplied in the hydroponics medium can substantially restore the height of SL-deficient rice mutants. Here, we used the pea model to investigate the role of SL in the regulation of internode elongation. We set out to investigate the origin of the dwarfism of SL-deficient and SL-response mutants in pea, to confirm the role of SL on internode elongation, and to analyze whether SL controls cell elongation and/or cell division. A possible interaction with GA was tested by evaluating two types of putative SL action: the effect of SLs on levels of the main bioactive GA in pea, GA1, and their possible influence on GA signaling, in particular on DELLA proteins. Overall, our results indicate that SL and GA act independently to stimulate internode elongation in pea.
RESULTS
Reduced Internode Length of Pea SL Mutants Is Not Caused by Their Increased Shoot Branching
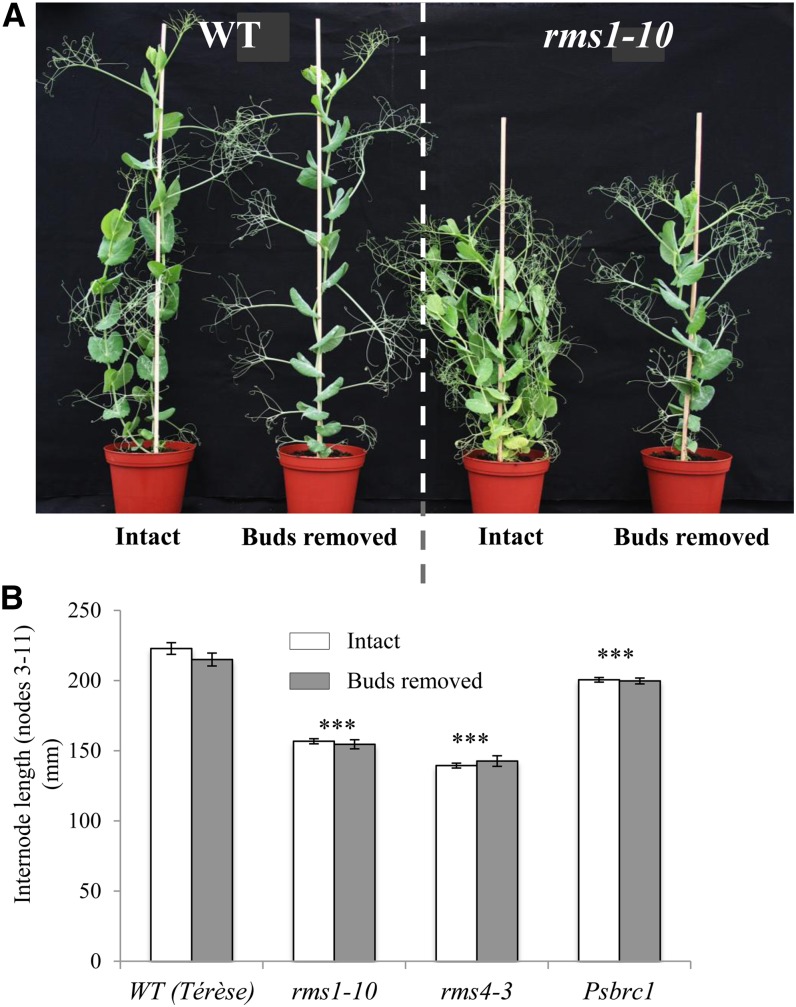
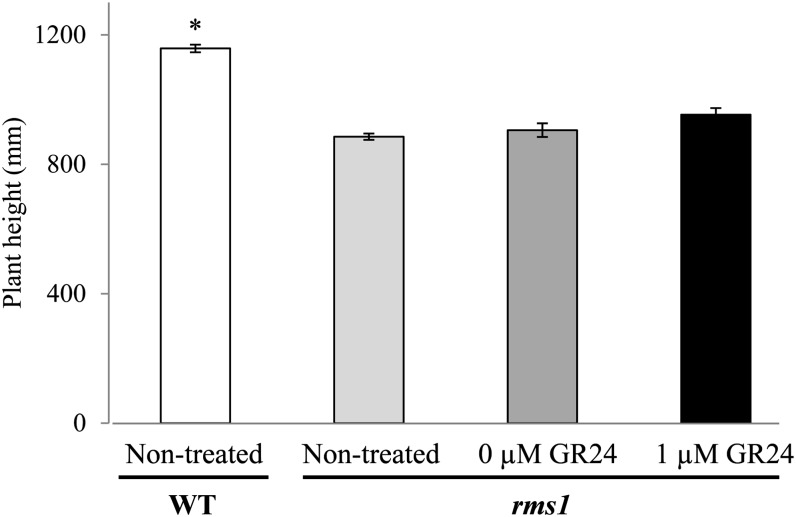
To investigate whether the reduced height of the pea SL-related mutants is due to their high branching, we determined the impact of manual bud removal on internode lengths of wild-type, rms1, rms4, and Psbrc1 plants. As reported previously, on a tall background, the rms1 SL-deficient and rms4 SL-insensitive mutants exhibited not only a high-branching phenotype but also a relative dwarf phenotype (Beveridge et al., 1996, 1997b). Internode length of the rms1-10 SL-deficient mutant and the rms4-3 SL-response mutant was reduced to 70% and 63%, respectively, of the dwarf wild-type line cv Térèse (Fig. 1). In contrast, Psbrc1, which is also unable to respond to SL, showed a significant reduction in internode length but was far less affected than rms1 and rms4 (90% of the wild type; Braun et al., 2012; Fig. 1B). For all genotypes, bud removal had no significant impact on internode length (Fig. 1). Similarly, further examination of the effect of shoot branching on stem elongation revealed that bud outgrowth inhibited by exogenous treatment of the synthetic SL GR24 directly to all axillary buds of SL-deficient rms1-1 mutant plants (Dun et al., 2013) did not lead to a significant increase in plant height (Fig. 2). Our results suggest that SLs may affect internode elongation independently of their function in bud outgrowth inhibition and that the reduced stature of the SL-related mutants is not simply a consequence of their increased branching phenotype. Manual removal of axillary buds along the stem, however, did induce a significant increase in stem diameter and leaf size in all genotypes tested, indicating that this response is SL independent and is possibly due to reallocation of resources (Supplemental Fig. S1).
Figure 1.
Reduced height of the rms SL mutants is not caused by their strong shoot branching. A, Phenotypes of wild-type (WT; cv Térèse) and rms1-10 plants, intact or with axillary buds removed. B, Internode lengths between node 3 and node 11 were measured when plants were 30 d old. Axillary buds were manually removed every 2 or 3 d. Data are means ± se (n = 7–8). Asterisks denote significant differences from the wild type (***P < 0.001, Student’s t test). [See online article for color version of this figure.]
Figure 2.
Inhibiting bud growth by directly treating buds with GR24 does not restore reduced rms1 plant height. Axillary buds of wild-type (WT) or rms1-1 plants (cv Parvus) were either left untreated (nontreated) or treated with 0 or 1 μm GR24 when the leaf was first open and retreated the following day. Bud lengths (Dun et al., 2013) and plant height were measured when the plants were 35 d old. Data are means ± se (n = 14–16). The asterisk denotes a significant difference from the rms1 0 μm GR24-treated plant (*P < 0.05, Student’s t test).
SL Promotes Internode Elongation in rms1 But Not in rms4 When Supplied to the Roots
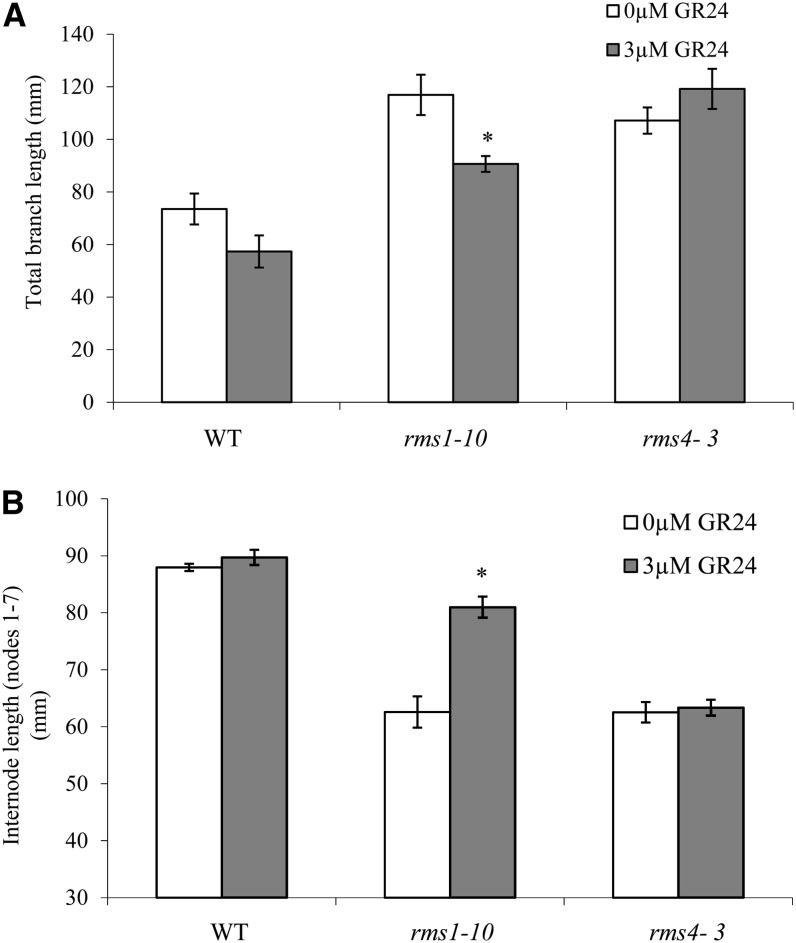
To confirm that SL promotes internode elongation, GR24 (3 µm) was continuously supplied to the roots of wild-type, rms1, and rms4 plants via hydroponics. After 16 d of GR24 treatment, total branch length was significantly reduced for rms1 but not for the wild type or rms4 (Fig. 3A). Generally, the strong basal branching at nodes 1 and 2 of dwarf pea lines (including the wild-type cv Térèse) is not inhibited by GR24 treatment using hydroponics (Boyer et al., 2012; Supplemental Fig. S1). The wild-type line displays branching only at these nodes, whereas rms1 and rms4 produce branches at basal and upper nodes. Upper branches in this dwarf background are inhibited by GR24 treatment in the SL-deficient rms1 mutant but not in the rms4 SL-response mutant. GR24 treatment caused a significant increase in internode length for the rms1 SL-deficient mutant but not for the rms4 SL-response mutant (Fig. 3B). No significant effect of GR24 on internode length was observed for the wild type (Fig. 3B).
Figure 3.
SL supplied via hydroponics increases the internode length of rms1 SL-deficient mutant plants but not of rms4 SL-response mutant plants. Six-day-old wild-type (WT), rms1-10, and rms4-3 plants (cv Térèse) were supplied with 0 or 3 μm GR24 via hydroponics for 16 d, after which the sum of all lateral buds and branches at nodes 1 to 5 (A) and the internode length between nodes 1 and 7 (B) were measured. Data are means ± se (n = 7–8). Asterisks denote significant effects of GR24-treated versus control-treated plants (*P < 0.05, Student’s t test).
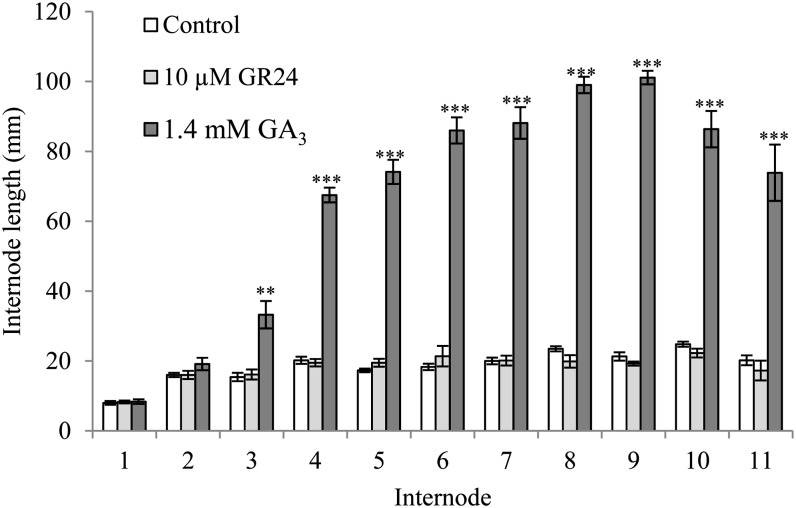
To determine if SL can function similarly to GAs in promoting main stem internode elongation in pea, we compared the response of rms1 SL-deficient mutant plants with treatment of the shoot tip with or without GR24 or GA3. GA3 applied to the shoot tip caused significant increases in length of all internodes except the first two nodes that were already elongated at the time of treatment. In contrast, GR24 at a dose more than 10 times greater than what is effective at axillary bud inhibition (10 µm) had no significant effect whatever the node and even on internodes that were newly forming in the shoot tip at the time of treatment (Fig. 4). These data suggest that, unlike GA, GR24 may not function in the shoot tip to regulate internode elongation.
Figure 4.
SL treatment to the shoot tip does not affect the growth of the main stem. The main shoot apex of 7-d-old rms1-11 plants (cv Térèse) with 2.5 expanded leaves was treated on two consecutive days with 0 or 10 μm GR24 or 1.4 mm GA3. Internode lengths were measured 23 d after treatment. Data are means ± se (n = 7–10). Asterisks denote significant differences from the control (**P < 0.01, ***P < 0.001, Student’s t test).
Mutations Affecting SL (rms1) and GA (le) Levels Have Additive Effects on Branching and Internode Elongation
The LE gene (PsGA3ox1) encodes a GA 3-oxidase that is able to convert GA20 to the bioactive GA1 (Lester et al., 1997; Martin et al., 1997). The enzyme encoded by the le-1 allele shows reduced activity, resulting in a 10- to 20-fold reduction of GA1 levels in elongating internodes of dwarf le-1 mutant plants compared with tall LE plants (Ross et al., 1992). We have previously shown that the rms1 mutation reduces plant height in different genetic backgrounds (Figs. 1 and 2; Beveridge et al., 1997b). Notably, a similar reduction is also observed in the rms1-10 line, obtained from the dwarf wild-type line cv Térèse, which contains the le-1 mutation affecting GA1 levels.
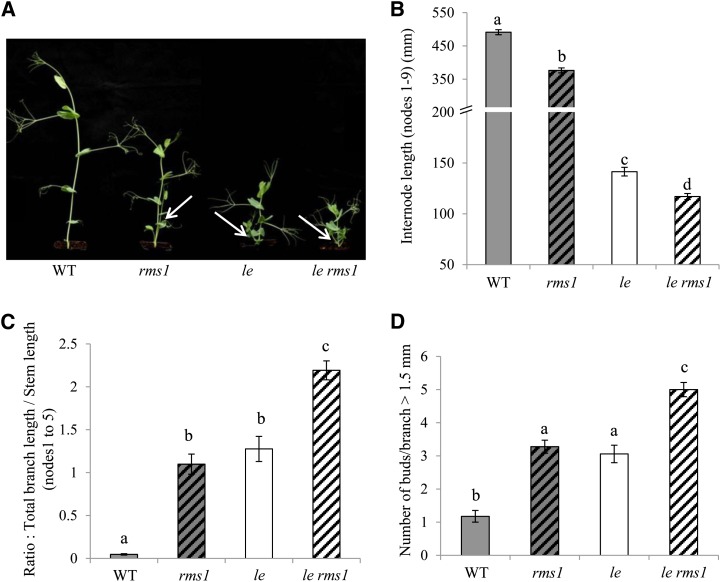
To compare the effects of SL and GA on internode elongation and to investigate if GA or SL impact on each other’s response, we used a series of four lines deficient in GA1 and/or SLs (le or LE and rms1 or RMS1) in the cv Térèse background (Fig. 5A). Branching and internode lengths of 15-d-old plants were measured. A deficiency in GA1 levels caused by the le-1 allele gave rise to a 71% decrease in internode length in an RMS1 background and 69% in an rms1 background. The rms1 mutation led to a significant reduction in internode length: 23% in an LE background and 17% in an le-1 background (Fig. 5, A and B). Clearly, the effect of SLs on internode elongation is not as strong as the effect of GA. However, SL does significantly stimulate internode elongation irrespective of the level of GA, and vice versa. Importantly, le-1 rms1 double mutant plants are shorter in stature than single le-1 and rms1 mutants, suggesting that phenotypes are additive and that SL and GA control internode elongation independently (Fig. 5, A and B).
Figure 5.
The effect of the rms1-10 and le mutations on both internode length and branching is additive. A, Fifteen-day-old wild-type (WT), rms1, le, and le rms1 plants. White arrows indicate the basal branching. B, Internode length between nodes 1 and 9. Data are means ± se (n = 18). Values with different lowercase letters are significantly different from one another (ANOVA, P < 0.01). C, Ratio of total branch length to total stem length (n = 18). Values with different lowercase letters are significantly different from one another (ANOVA, P < 0.001). D, Number of buds or branches from nodes 1 to 5 with a length greater than 1.5 mm (n = 18). Values with different lowercase letters are significantly different from one another (ANOVA, P < 0.001). [See online article for color version of this figure.]
The effect of GA on branching is suggested by the common use of the le-1 allele in pea crops to reduce internode length and increase basal branching. In rice, reducing endogenous GA levels by overexpression of GA2ox resulted in reduced stem elongation and increased tillering (Lo et al., 2008). We often quantify branching by measuring branch length; such a measure is strongly dependent on internode length. As such, here, we quantified branching of the four lines by the ratio of total branch length to main stem length (from nodes 1 to 5). Using this ratio as a measure of branching, the double le-1 rms1 mutant was more branched than single le-1 and rms1 mutants (Fig. 5C). We also estimated branching by the number of buds or branches with a size greater than 1.5 mm (Fig. 5D). Using this estimation, the double le-1 rms1 mutant also displayed more branch/bud outgrowth than single le-1 and rms1 mutants. These additive results in double mutant plants indicate that le-1 and rms1 mutations independently induce branching and, hence, that SL and GA control branching independently.
SLs Stimulate Stem Growth by Increasing Cell Number But Not Cell Length
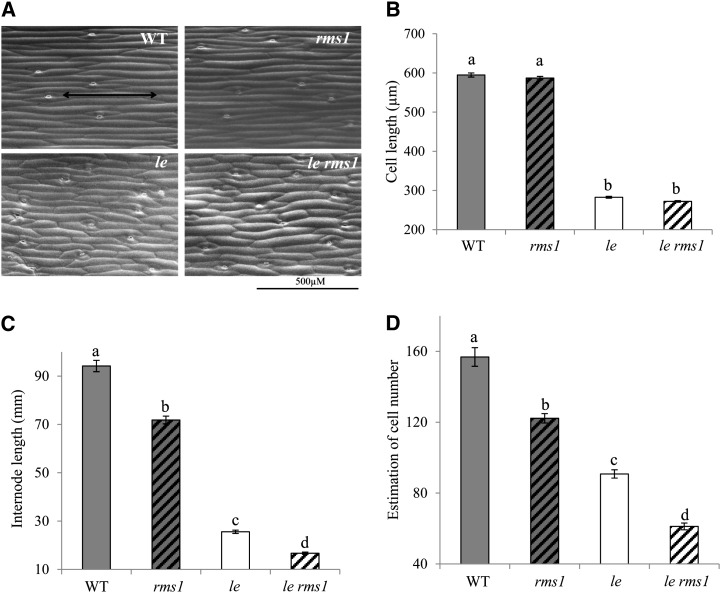
To investigate whether SLs increase cell number and/or cell length to control the height of the plant, and to compare the effect of SLs with GA, we quantified epidermal cell size at a given elongated internode (internode 4) of 25-d-old wild-type, le-1, rms1, and le-1 rms1 plants using scanning electronic microscopy (Fig. 6A). The number of cells was estimated for each plant by the ratio of internode length to cell length. Irrespective of the LE genotype, rms1 significantly affected cell number but not cell length (Fig. 6, B and C). Consistent with what is known for GA, cell length and number were both strongly decreased in GA-deficient le-1 plants in comparison with LE plants (Fig. 6, A, B, and D; Reid, 1983). These results indicate that SLs affect cell division rather than cell elongation to stimulate internode elongation. Both le-1 and rms1 mutations caused a significant increase in cell width (Supplemental Fig. S2).
Figure 6.
SLs regulate internode elongation by affecting cell number. A, Epidermal cells of internode 4 of the wild type (WT; top left), rms1 (top right), le (bottom left), and le rms1 (bottom right) in scanning electronic microscopy. B, Epidermal cell length (µm) in internode 4 of the wild type, rms1, le, and le rms1. Data are means ± se (n = 7–8 plants with about 100 cells per plant). Values with different lowercase letters are significantly different from one another (ANOVA, P < 0.05). C, Internode 4 length (mm) of the wild type, rms1, le, and le rms1. Values with different lowercase letters are significantly different from one another (ANOVA, P < 0.01). D, Estimation of cell number in internode 4 of the wild type, rms1, le, and le rms. Values with different lowercase letters are significantly different from one another (ANOVA, P < 0.001).
GA1 Content Is Not Correlated with the Dwarf Phenotype of the rms Mutants
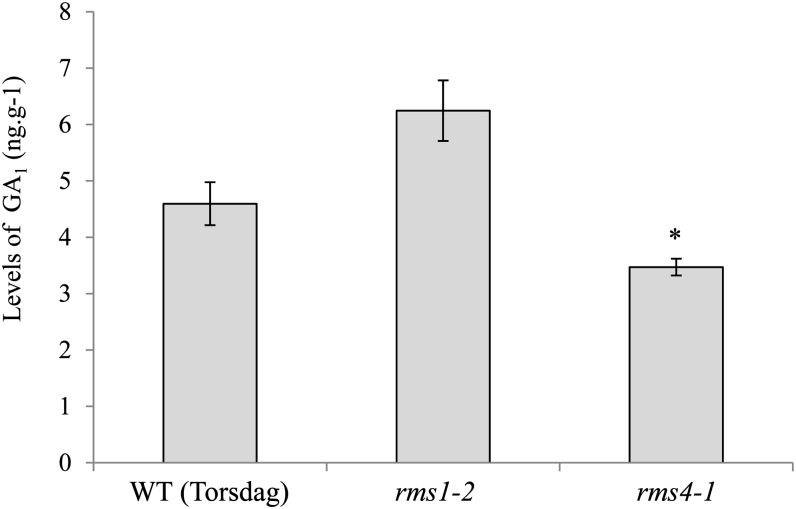
To investigate in more detail a possible interaction between SL and GA in the control of internode elongation, we determined whether SL affects the levels of GA1, the main bioactive GA in pea. GA1 levels were quantified from elongating internodes of the tall line cv Torsdag (LE) and the corresponding SL mutant lines rms1 and rms4. There was a small, although statistically significant (P < 0.05), reduction in GA1 content in the SL-response mutant rms4 compared with the wild type (Fig. 7). This reduction is unlikely to be physiologically significant, given the log/linear relationship between GA content and elongation in this species (Ross et al., 2011). GA1 levels in rms1 internodes were not significantly different from those of the wild type. To investigate further whether SLs are involved in the regulation of GA, transcript levels of genes involved in GA biosynthesis (PsGA20ox1 and PsGA3ox1 [LE]) and catabolism (PsGA2ox1 [SLN] and PsGA2ox2) were analyzed in expanding stem tissue 24 h after GR24 treatment (3 µm) via the root system. Transcript levels of the RMS5/PsCCD7 SL biosynthesis gene, known to be feedback down-regulated by SL in epicotyl, were analyzed as a positive control for GR24 treatment in the epicotyl (Supplemental Fig. S3). As expected, RMS5 was highly expressed in the rms1 and rms4 mutant epicotyls in comparison with the wild type, and the GR24 treatment caused a strong down-regulation of RMS5 transcript level in the rms1 SL-deficient mutant, while there was no effect of GR24 on RMS5 expression in the epicotyl of the rms4 SL-response mutant (Supplemental Fig. S3). Transcript levels of the four genes controlling GA levels were not significantly affected by SL treatment in expanding stem tissue of any genotype (Supplemental Figs. S4 and S5). These results support the hypothesis that the SL and GA pathways do not interact in the control of internode elongation. However, significant increases in expression of the GA biosynthesis gene PsGA3ox1 and the GA deactivation genes PsGA2ox1 and PsGA2ox2 were observed in the SL mutants relative to the wild type (Supplemental Figs. S4B and S5). Accordingly, a slight decrease in PsGA3ox1 transcript levels (although not significant) was observed after GR24 treatment in rms1 but not in rms4. Elevated transcript levels of the two GA deactivation genes in the rms4 SL-response mutant might explain the reduced levels of GA1 in this mutant (Fig. 7; Supplemental Fig. S5).
Figure 7.
GA1 levels in expanding stem tissue of wild-type (WT; Torsdag), rms1-2, and rms4-1 15-d-old plants. Data are means ± se (n = 3). The asterisk denotes a significant difference from wild-type plants (*P < 0.05, Student’s t test).
Hence, the dwarf phenotype of both rms SL mutants cannot be explained by low levels of GA1 in expanding tissue. This strengthens the previous genetic data (Fig. 5) that the action of SL on internode elongation is independent of GA biosynthesis.
SLs Regulate Internode Elongation in a DELLA-Deficient Background
To determine if the reduction of internode length due to SL deficiency requires functional DELLA proteins, we compared the internode length of SL-deficient and nondeficient lines in a background with no functional DELLA proteins (la cry-s; O’Neill et al., 2010). The double mutant la cry-s shows a slender phenotype with very long internodes, particularly the basal internodes. If SL and DELLA proteins act in the same pathway to control internode elongation, the dwarf phenotype caused by the rms mutations should not be observed in a genetic background where DELLA proteins are nonfunctional.
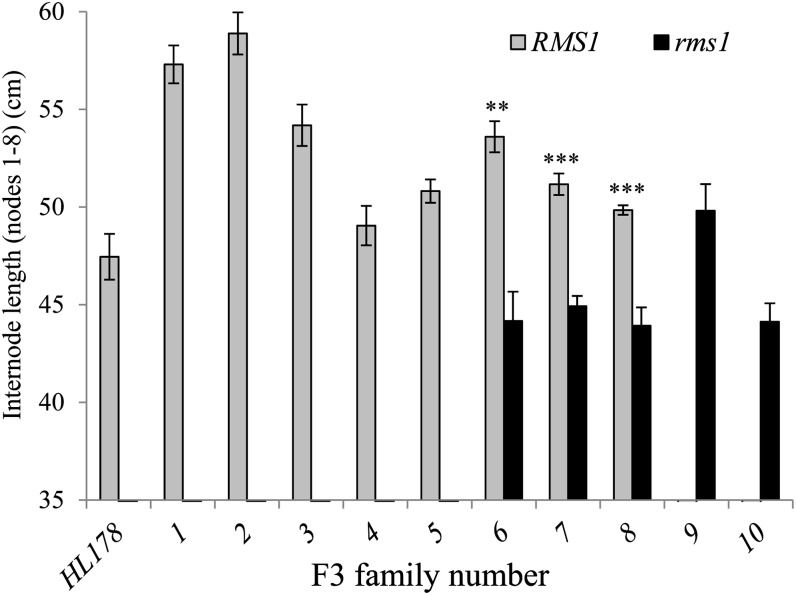
A strong variation in internode length between RMS1 F3 families and rms1 F3 families was observed, despite all these families being fixed for la cry-s and le-1 (as both lines, cv Térèse and HL178, contain the le-1 allele). RMS1 families had globally longer internodes (mean ± se = 53.1 ± 1.59 mm) than the rms1 families (mean ± se = 45.4 ± 1.12 mm; Student’s t test, P < 0.01; Fig. 8). In each of the three F3 families segregating for RMS1/rms1, a strong and significant effect of the rms1 mutation was observed on internode length (Student’s t test, P < 0.01 for family 6 and P < 0.001 for families 7 and 8; Fig. 8). These results indicate that functional DELLA proteins are not needed for SL to regulate internode elongation.
Figure 8.
SLs control internode elongation in a della background. Internode length between nodes 1 and 8 were measured in the della line HL178 (la cry-s) and in F3 della families fixed for la cry-s and fixed for RMS1 (1–5) or rms1 (9 and 10) or segregating for RMS1/rms1 (6–8). For families 6 to 8, the gray bars correspond to a mixture of RMS1/RMS1 and RMS1/rms1 plants (n = 11–28 except for rms1 F3 6–8, for which n = 6–8). Asterisks denote significant differences from corresponding rms1 plants (**P < 0.01, ***P < 0.001, Student’s t test).
DELLA Proteins Are Not Degraded by SL in the Root Tip of Arabidopsis
Because pea is not amenable to transformation, we used the Arabidopsis transgenic line carrying the RGA promoter::GFP-RGA fusion (Silverstone et al., 2001) to test if SL application affects the levels of the DELLA protein. RGA is one of the five DELLA proteins present in Arabidopsis. Mutations at the RGA locus suppress the dwarf phenotype of the GA-deficient ga1-3 mutant (Silverstone et al., 1997). The GFP-RGA fusion protein, localized to the nucleus in transgenic Arabidopsis seedlings, is rapidly degraded by GA treatment (Silverstone et al., 2001). The Arabidopsis max SL mutants have also been described to have reduced stature (Stirnberg et al., 2002; Booker et al., 2004), so we surmised that any effect of SL on DELLA proteins would also be conserved between pea and Arabidopsis.
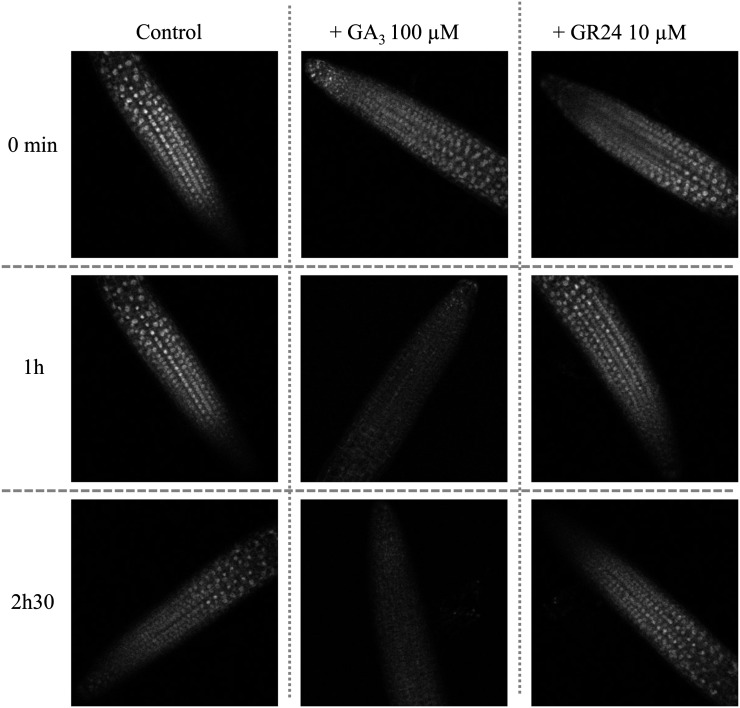
The pRGA::GFP-RGA line was used to follow the levels and the localization of the GFP-RGA fusion protein after GR24 treatment in root tips of Arabidopsis seedlings. Root tips of transgenic plants expressing pRGA::GFP-RGA were treated with water or GA3 (100 µm) as negative and positive controls, respectively, or with GR24 (10 µm) and observed 1 and 2.5 h after treatment using confocal microscopy (Fig. 9). GFP signal in the nuclei decreased slightly after 2.5 h in water but was maintained in the nuclei, while the GA3 treatment induced a strong decrease of GFP fluorescence in the nuclei. No degradation of GFP-RGA in the nuclei was observed 1 or 2.5 h after the GR24 treatment (Fig. 9) or after 4 h (data not shown). These results are in accordance with the results shown above and altogether suggest that SLs do not interact with the GA signaling pathway.
Figure 9.
GA3 but not GR24 treatment affects the expression of the GFP-RGA protein in root apex of Arabidopsis. Roots of transgenic plants (rga/ga1-3 background) expressing the pRGA::GFP-RGA fusion were observed using confocal laser microscopy. Shown are three-dimensional projections of the fluorescence images of root tips treated with control solution (left column), 100 µm GA3 (middle column), or 10 µm GR24 (right column) at different times as indicated.
Dose Response of GA Application on the Wild Type, rms1, and rms4
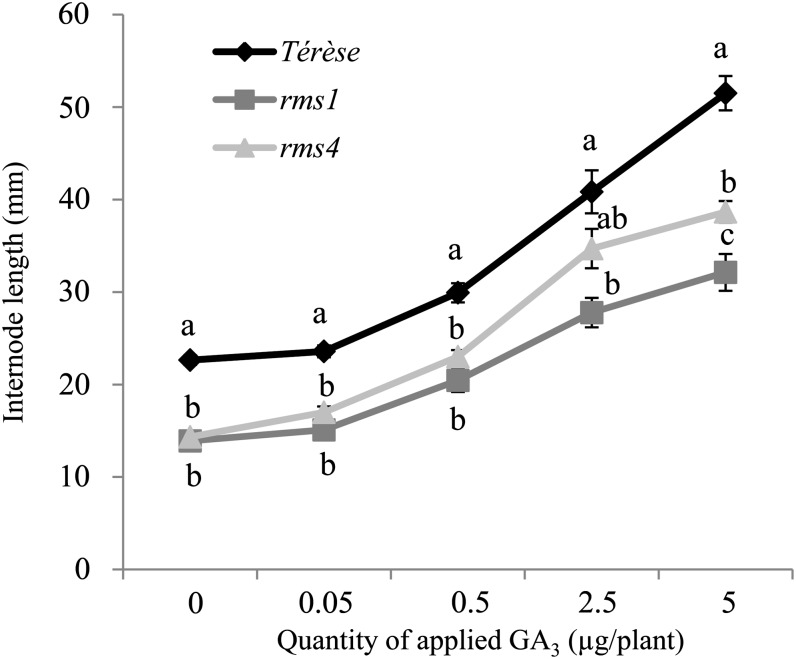
To test if the SL-related mutants are affected in GA response, a dose-response experiment was performed where GA3 was applied to expanding stipules at node 3 of 8-d-old plants and the length of internode 4 was measured 8 d after application. Both mutants responded to the application of GA3, and 2.5 µg of GA3 restored the internode length of the mutants to the wild-type control. However, GA application did not remove the difference in internode elongation between the mutants and the wild type (Fig. 10).
Figure 10.
SL-related mutants respond to GA3. Length of internode 4 of wild-type (diamonds), rms1 (squares), and rms4 (triangles) plants after treatment with various quantities of GA3 (µg plant−1). GA3 was applied on stipules at node 3 when the plants were 8 d old. Measurements were made 8 d after treatment. Data are means ± se (n = 8–12). Values with different lowercase letters are significantly different from one another (ANOVA, P < 0.05).
DISCUSSION
SLs Stimulate Stem Growth Independently from Its Effect on Branching
Pea grafting experiments have demonstrated that the reduced plant height observed in rms1 SL-deficient plants is partly restored by wild-type rootstocks (Beveridge et al., 1997b). This could be a consequence of the majority of branching in the rms1 shoot being inhibited by the wild-type rootstock or due to a supply of SL from the wild-type rootstock to the SL-deficient shoot. More recently, Umehara et al. (2008) demonstrated that the decreased plant height of d10 SL-deficient mutant rice plants was restored to that of the wild type when grown with GR24 in the hydroponic medium, indicating a possible role for SL in the main stem, but again this was associated with reduced branching.
Here, we show that depleted SL levels are more directly responsible for the reduced height of the pea rms1 SL-deficient mutant. Indeed, inhibiting the outgrowth of the buds along the rms1 mutant stem by applying the synthetic SL, GR24, specifically to the axillary buds led to only a minimal and insignificant increase in plant height, as was also observed when all buds along the stem were manually removed (Figs. 1 and 2). By contrast, the removal of axillary buds in rice led to a significant but partial restoration of reduced plant height in the htd1/d17/Osccd7 SL-deficient mutant but had no significant effect on plant height in the wild type (Zou et al., 2006). This difference between pea and rice could be attributed to the huge difference of tiller numbers between the rice htd1 mutant and the wild type (36 and one tillers, respectively); these tillers may act as a particularly strong sink; hence, their removal may substantially increase resources available to the main shoot for growth. In maize, a SL-independent action of the TB1 transcription factor to inhibit branching has been demonstrated, and, in contrast to other species, including rice, the branching of the Zmccd8 mutant is particularly mild relative to Tb1. It is proposed that the dwarf phenotype of Zmccd8 is not the result of competition for resources between the main stem and tillers, as the Tb1 mutant displaying high branching is not affected in plant height (Guan et al., 2012). Similarly, in pea, the Psbrc1 mutant is taller than rms mutants, although this comparison is less informative given that Psbrc1 in pea has generally reduced branching compared with rms mutants (Braun et al., 2012).
As shown previously in rice (Umehara et al., 2008), continuously supplying GR24 to pea roots via hydroponics led to a significant increase of internode elongation in rms1 SL-deficient plants but not in rms4 SL-response mutant plants (Fig. 3B). This supports a role of SL in controlling plant height via an RMS4-dependent mechanism. This mechanism may be independent of the competition for resources between growing branches and the main stem, since branch suppression by bud removal or direct application of SL to buds of pea has only minor effects on stem length. While we did not achieve a complete restoration of internode elongation in rms1, this may be related to the dose, duration, or starting point of GR24 treatment. Therefore, we suggest that, in addition to regulating shoot branching (Gomez-Roldan et al., 2008; Umehara et al., 2008), secondary growth (Agusti et al., 2011), and various aspects of root development (Koltai, 2011; Brewer et al., 2013), SLs also contribute to the regulation of internode elongation in the main shoot.
This function does not seem to occur via PsBRC1, a gene mostly expressed in axillary buds, as the pea Psbrc1 mutant is not as reduced in plant height as the SL pea mutants (Fig. 1; Braun et al., 2012). The shade-avoidance response of the brc1 mutant in Arabidopsis further demonstrates the lack of BRC1 influence over stem elongation. In response to simulated shade (light conditions enriched in far-red light), wild-type Arabidopsis plants exhibit strongly reduced branching, but the Arabidopsis brc1 mutant was partially insensitive, showing little reduction of branching (Gonzalez-Grandio et al., 2013). In contrast, when considering other shade-avoidance responses, including stem elongation, brc1 mutants responded similarly to the wild type, suggesting that BRC1 is involved in the control of branching but not of stem elongation in response to light enriched in far-red light (Gonzalez-Grandio et al., 2013). Whether SL biosynthesis and/or MAX2 are also regulated by these light conditions will need further investigation (Finlayson et al., 2010). Altogether, these observations add further support to the notion that SL can influence primary stem growth independently of its effect on axillary bud outgrowth. Nevertheless, it is still unclear why Psbrc1 showed reduced plant height that was not changed when buds were manually removed together with a reduced stem width in both intact plants and plants with buds removed. Novel functions for BRC1 are being discovered for this transcription factor, such as the regulation of flowering transition in axillary buds of Arabidopsis (Niwa et al., 2013), and further investigations are needed to have a good understanding of the mutant phenotype.
Tissue Specificity of SL Action on Cell Division
Among the three major plant hormones promoting stem elongation (IAA, GA, and BR), only GA has been shown to have an important effect on cell division, in addition to affecting cell elongation (Fig. 5; Achard et al., 2009; Lee et al., 2012). In pea, the length of epidermal cells in the internode is similar between rms1 SL-deficient mutant plants and wild-type plants. Similarly, in rice, no difference in cell size was observed in the culm of the wild type and the htd1/Osccd7 SL-deficient mutant (Zou et al., 2005). This suggests that SL does not act on cell elongation but rather stimulates cell division to regulate internode elongation. This effect of SLs on cell division to increase plant height is in accordance with their effect on stimulating cambium-like cell divisions, as observed in the inflorescence stem of Arabidopsis and in the stem of other plants, including Eucalyptus species (Agusti et al., 2011). By contrast, it was shown that SLs repress mesocotyl elongation in dark-grown rice seedlings (Hu et al., 2010). This was also achieved by regulating cell division but not cell size in this tissue located between the coleoptile node and the cotyledon (Hu et al., 2010). Consequently, SLs appear to regulate cell division but with varying effects according to tissue. Whereas SLs would repress cell division in axillary meristems via the transcription factor PsBRC1 and repress cell division in the mesocotyl of dark-grown rice plants (Hu et al., 2010), SLs would promote cell division in the stem to stimulate longitudinal growth and secondary growth of the stem. Whether the stimulatory effect of SLs on internode elongation and their effect on secondary growth are independent will need further investigation, in particular by identifying the transcription factor(s) involved in these responses. Candidates might be found among the TCP family of transcription factors, comprising 24 members in Arabidopsis including BRC1. Several members of this plant-specific protein family have been shown to activate or repress the transcription of cell cycle genes, very likely in coordination with other proteins (Berckmans and De Veylder, 2009; Martín-Trillo and Cubas, 2010).
Applying GR24 to the shoot tip of SL-deficient rms1 plants had no effect on main stem internode elongation (Fig. 4), suggesting that SL function in stem elongation is unlikely to be performed at the shoot tip. Nevertheless, in dicotyledons, stem tissues result from cell divisions occurring in a region called the rib meristem/rib zone, located at the base of the shoot apical meristem and consequently not directly at the surface of the shoot apex (Kerstetter and Hake, 1997). The strong difference between the GR24 and GA1 effect when applied to the shoot tip is intriguing. It is possible that GR24 treatment could not reach the rib zone, where cells divide and elongate rapidly. One important characteristic of SL molecules is their low stability, which is essential for their role in the rhizosphere to signal the presence of a host root (Parniske, 2008). Moreover, SL reception seems to involve the hydrolysis of SLs into inactive compounds by the D14/DAD2 α/β-fold hydrolase (Hamiaux et al., 2012). Therefore, the continuous supply of SL may be essential (Smith and Waters, 2012) to observe an effect on internode elongation, which is a prolonged process, whereas axillary bud outgrowth is more a “switch on/off” process. This would explain why we observed an effect of GR24 on shoot elongation using hydroponics with continuous feeding and not by a single GR24 application to the shoot tip. In contrast, axillary bud outgrowth can be inhibited by a single GR24 application directly onto the bud.
An indirect growth effect due to the smaller root system of Zmccd8 has been suggested in maize to explain the dwarf phenotype of the mutant, which displays fewer nodal roots and shorter primary root than the wild type (Guan et al., 2012). In Arabidopsis, SLs have been shown to control different aspects of root growth (main and lateral roots, root hair; Koltai, 2011; Ruyter-Spira et al., 2011), and it is possible that root supply of GR24 influences root growth and, indirectly, shoot growth. Nevertheless, in pea, and in normal nutritive conditions, the root system is not much affected in the SL-deficient and SL-response rms mutants (Beveridge et al., 1997a; Foo et al., 2013; data not shown). By contrast, in another legume, L. japonicus, while transgenic LjCCD7-silenced SL-deficient plants have strongly reduced internode lengths, they have roots with higher biomass and a longer primary root than wild-type plants (Liu et al., 2013). Consequently, an indirect effect of the root system on internode length is unlikely to play a major role in the SL-related mutants based purely on effects on root growth. Of course, it remains possible that SL signaling in the roots regulates the level of another mobile signal that affects elongation.
SL and GA Stimulate Stem Growth Independently
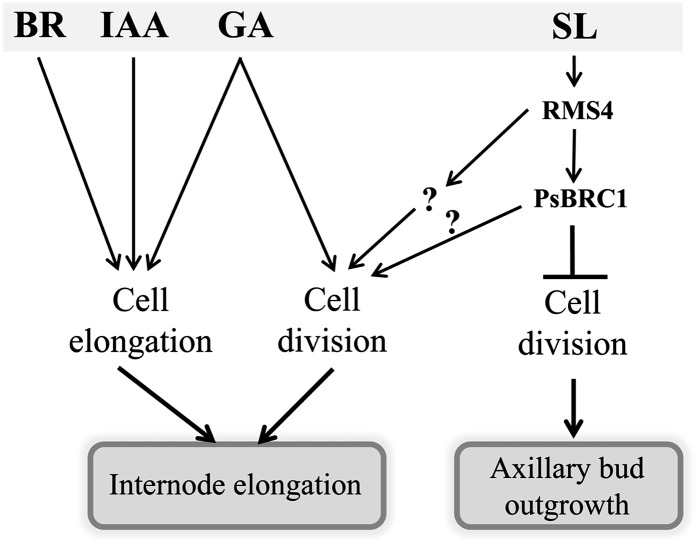
It is tempting to speculate that SLs in the roots or stem affect the levels of other signals that control stem elongation by affecting cell division, such as GA. Here, different approaches (genetic, physiological, molecular) were used to test whether SLs affect GA levels or signaling to control internode elongation. GA1 levels were not correlated with dwarfism, and no effect of GR24 application on the transcript level of GA metabolism genes was found. We showed that SL-deficient and SL-response mutants were able to respond to GA treatment and that functional DELLA proteins were not necessary for the SL regulation of internode elongation. Combined, our data strongly indicate that SLs do not act via GA to stimulate internode elongation in pea. Some results remain puzzling and will need further study, such as the reduced GA1 levels observed in the SL-response rms4 mutant. Nevertheless, it does not change our conclusion that SLs, independently of GA, stimulate cell division for the control of internode elongation in an RMS4-dependent pathway and possibly in a PsBRC1-independent pathway (Fig. 11). Another candidate for a SL-mediated signal in roots might be xylem-translocated cytokinins (CKs), which are considered to be anticorrelated in content with shoot SL signaling (Dun et al., 2009). CKs are known to play an important role in cell division, and xylem-CK content is known to be suppressed by reduced SL signaling. Unlike rms mutants, Psbrc1 mutants do not contain depleted xylem sap CK levels, consistent with their taller stature. CKs are well known to affect cell proliferation in the shoot apical meristem, and reducing CK levels by overexpression of a catabolic CYTOKININ OXIDASE gene gives plants with shoot apical meristem containing fewer cells and slower leaf formation. Nevertheless, the role of CKs in the control of internode elongation still needs further investigation, as opposite results have been obtained in different species. Overexpression of a cytokinin oxidase to reduce endogenous levels of CK resulted in severe dwarf plants in tobacco due to both slower development and shorter internodes (Werner et al., 2001), but longer internodes were observed in transgenic poplar (Populus tremula × tremuloides) trees (Nieminen et al., 2008).
Figure 11.
Model integrating the role of auxin (IAA), BR, GA, and SL on internode elongation in pea. Arrows indicate activation, and bars indicate repression. IAA and BR promote internode elongation by activating cell elongation; GA promotes internode elongation by activating both cell elongation and cell division; SL promotes internode elongation by cell division only independently from GA, via RMS4 and a transcription factor that, very likely, is not PsBRC1. In axillary buds, SL inhibits cell division via RMS4 and PsBRC1.
CONCLUSION
SLs were proposed to be central modulators of shoot architecture by modulating and coordinating shoot branching and secondary growth (Agusti and Greb, 2013). This study emphasizes the role of SLs in repressing or inducing the activities of different types of meristems to adjust shoot architecture to environmental conditions. We have shown by exogenous SL applications that control of branching and internode elongation can be independent. Whether, in natural conditions, these multiple actions of SLs are coordinated or not needs further investigation (Agusti and Greb, 2013).
MATERIALS AND METHODS
Plant Material
The dwarf branching pea (Pisum sativum) mutants rms1-10 (M3T-884), rms1-11 (M3T-988), and rms4-3 (M3T-946) were obtained in the dwarf (le) cv Térèse and are described by Rameau et al. (1997). The rms1-1 mutant (WL5237) obtained in the tall line Parvus and the rms4-1 mutant (K164) obtained in the tall line cv Torsdag are described by Arumingtyas et al. (1992). The rms1-2T mutant line was obtained by backcrossing the rms1-2 allele (derived from the cv Weitor background; Beveridge et al., 1997b) into the wild-type line Torsdag three times. The Psbrc1 used in this study corresponds to BC3 (Psbrc1 × Térèse). The mutant Psbrc1, identified by a TILLING (targeting-induced local lesions in genomes) approach from cv Caméor, is described by Braun et al. (2012). In the four LE/le, RMS1/rms1 lines, le RMS1 corresponds to cv Térèse and le rms1 corresponds to the rms1-10 branching mutant. The LE RMS1 line was obtained by backcrossing nine times the LE allele from Torsdag in cv Térèse. The LE rms1 line was obtained by crossing LE RMS1 with the branching rms1-10 mutant.
For Figure 8, the line HL178 la cry-s double mutant was crossed with the rms1-10 line, and several slender F2 individuals (la cry-s) were selected phenotypically. Different F3 families, fixed for la cry-s, were followed that were either fixed for RMS1 or rms1 or segregating for RMS1. The branching phenotype of rms1 plants was generally easy to identify; nevertheless, plants were genotyped for RMS1 in the segregating F3 families. Because the rms1 mutant and the double la cry-s mutant are not available in the same genetic background, comparisons of internode length were done for rms1 F3 individuals (rms1 rms1) and nonbranching F3 individuals (rms1 RMS1 and RMS1 RMS1) within the same segregating families (families 6–8 in Fig. 8). Other fixed rms1 (families 9 and 10 in Fig. 8) and RMS1 F3 (families 1–5 in Fig. 8) families were also analyzed for comparison.
The Arabidopsis (Arabidopsis thaliana) transgenic line expressing the GFP-RGA protein under the control of the RGA promoter is described by Silverstone et al. (2001).
Growing Conditions and Phenotype Measurements
Pea plants were grown in 16-h-photoperiod glasshouse conditions as described (Braun et al., 2012). For Figures 2 and 4, pea plants were grown in an 18-h photoperiod as described (Dun et al., 2013). Nodes were numbered acropetally from the first scale leaf as node 1. Internodes were numbered acropetally, with internode 1 extending between node 1 and node 2. The indicated stage (leaves expanded) corresponds to the number of nodes with fully expanded leaves. Bud and branch lengths were measured with digital calipers. For longer culture, one or two plants were cultivated per 2-L pot. To study the effect of branching on internode elongation, axillary buds were removed every day.
Hormonal Treatments
For Figure 2, solutions applied directly to the axillary buds contained 50% ethanol, 2% polyethylene glycol 1450, 0.1% acetone, and 0 or 1 µm (3 ng) GR24.
For shoot apex treatments (Fig. 4), 2 µL of solution containing 0 or 10 µm (6 ng) GR24 or 1.4 mm GA3 (0.1 µg; Sigma) in 0.2% Tween 20 with 1% acetone and 5% ethanol was applied with a pipette to the unexpanded stipules of the shoot tip.
For hydroponics treatments, culture was as described by Braun et al. (2012), except that 33-L polyvinyl chloride opaque pots were used, one for each genotype. Acetone or GR24 (dissolved in acetone) was added to the hydroponic culture solution to give a final concentration of 0 or 3 μm GR24 and 0.01% acetone. The hydroponic culture solution was continuously aerated by an aquarium pump and was replaced weekly. One day after the beginning of the treatment, expanding stem tissue (internode 5) and epicotyls were harvested for gene expression analyses.
For GA treatment, 2 µL of solution containing GA3 (Sigma) at the concentrations specified dissolved in 95% ethanol was applied to each of the two stipules at node 3 of the main stem on 8-d-old plants.
Gene Expression Analyses
Elongating internodes were harvested, total RNA was isolated and quantified, complementary DNA was synthesized, and real-time PCR gene expression analyses were performed as described (Braun et al., 2012). The analysis was done in two independent experiments. Primer sequences were as follows for GA synthesis genes and GA deactivation genes: PsGA20ox1, 5′-CATTCCATTAGGCCAAATTTCAAT-3′ and 5′-CTGCCCTATGTAAACAACTCTTGTATCT-3′; PsGA3ox1, 5′-TTCGAGAACTCTGGCCTCAAG-3′ and 5′-ATGTTCCTGCTAACTTTTTCATGGTT-3′; PsGA2ox1, 5′-CACAACCAATCAAGAACACAATTTC-3′ and 5′-CCCTTCTGCATCAAATCAAG-3′; PsGA2ox2, 5′-CCCTCCTGACCCCAGTGAAT-3′ and 5′-CTCACACTCACAAATCTTCCATTTG-3′. Transcript levels for the different genes were expressed relative to the expression of the EF1α gene (Johnson et al., 2006).
Segregation Studies
The cleaved-amplified polymorphic sequence marker for genotyping RMS1 was based on amplification of the PCR fragment using primers RMS1-118F (5′-TTGGTTGGACTTCACTTTGAGG-3′) and RMS1-984R (5′-CACAACAATCAGCAATGACAGC-3′) and digestion with the Cfr13I enzyme (Fermentas).
Hormone Quantification
Harvested material (expanding internodes of 15-d-old plants) was extracted in cold 80% methanol and prepared for analysis by ultra-performance liquid chromatography-mass spectrometry as described previously (Tivendale et al., 2012). The internal standard was [2H2]GA1, provided by Prof. Lewis Mander (Australian National University). The system consisted of a Waters Acquity H-series ultra-performance liquid chromatograph coupled to a Waters Xevo triple quadrupole mass spectrometer. A Waters Acquity UPLC BEH C18 column (2.1 × 100 mm × 1.7 µm particles) was used, with mobile phases A = 1% acetic acid (v/v) in water and B = acetonitrile. The program ran from 100% A to 85% A/15% B over 2 min, then to 77.5% A/22.5% B at 7 min. GA1 eluted at approximately 6 min. The flow rate was 0.35 mL min−1, and the column temperature was 35°C. Mass spectrometry was conducted in the electrospray and multiple reaction monitoring modes, monitoring negative ions. The ion source temperature was 130°C, the desolvation gas was nitrogen at 950 L h−1, the cone gas was nitrogen at 50 L h–1, the desolvation temperature was 450°C, the capillary voltage was 2.7 kV, and the cone voltage was 46 V. The transitions monitored, with collision energies in parentheses, were mass-to-charge ratio (m/z) 347.3 to 229.1 (28 V) and m/z 347.3 to 273.1 (22 V) for endogenous GA1 and m/z 3 49.3 to 231.1 (28 V) and m/z 349.3 to 275.1 (22 V) for deuterated GA1. The dwell time was 120 ms per channel.
Cell Measurements Using Scanning Electronic Microscopy
Freshly sampled tissues were cooled to −33°C by a Peltier cooling stage (Deben) and observed with a Hirox SH-1500 bench-top scanning electronic microscope. Internode 4 of 16-d-old plants was used, and its length was measured (eight plants per genotype). The epidermis cells of this internode were directly observed with a scanning electron microscope (MEB Hirox SH-1500). About 100 cells per plant were analyzed with ImageJ software (http://rsbweb.nih.gov/ij/), representing a total of 770 to 1,000 cells per genotype. The cell number in each internode 4 was estimated by the ratio of internode 4 length to mean cell length.
Confocal Laser Microscopy
Detection of GFP fluorescence was performed with a Zeiss LSM 710 confocal microscope. The excitation wavelength was 488 and 561 nm, and emission was collected at 565 to 720 nm for root tip imaging. For the experiments, root tips from 4-d-old transgenic plants expressing pRGA::GFP-RGA were mounted on standard microscope slides in the presence of water (5‰ acetone), 100 µm GA3 (5‰ acetone), or 10 µm GR24 (5‰ acetone).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ403160 (RMS5), X91658 and U58830 (PsGA20ox1), AF001219 and U85045 (PsGA3ox1), AF100955 (PsGA2ox1), and AF100954 (PsGA2ox2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Stem and stipule diameters of intact plants and plants with buds removed.
Supplemental Figure S2. Cell width of epidermal internode.
Supplemental Figure S3. Transcript levels of the SL biosynthesis gene RMS5 after GR24 supply.
Supplemental Figure S4. Transcript levels of GA biosynthesis genes after GR24 supply.
Supplemental Figure S5. Transcript levels of GA catabolism genes after GR24 supply.
Acknowledgments
We thank Sandrine Bonhomme (IJPB, Versailles) and Evelyne Costes (UMR AGAP, INRA Montpellier) for comments on the manuscript, François-Didier Boyer (ICSN, Gif-sur-Yvette) for kindly supplying GR24, and Noel Davies (Central Science Laboratory, University of Tasmania) for assistance with GA analyses.
Glossary
- BR
brassinosteroid
- IAA
indole-3-acetic acid
- SL
strigolactone
- CK
cytokinin
- m/z
mass-to-charge ratio
References
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GT, Genschik P. (2009) Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol 19: 1188–1193 [DOI] [PubMed] [Google Scholar]
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusti J, Greb T. (2013) Going with the wind: adaptive dynamics of plant secondary meristems. Mech Dev 130: 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusti J, Herold S, Schwarz M, Sanchez P, Ljung K, Dun EA, Brewer PB, Beveridge CA, Sieberer T, Sehr EM, et al. (2011) Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc Natl Acad Sci USA 108: 20242–20247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351 [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50: 1416–1424 [DOI] [PubMed] [Google Scholar]
- Arumingtyas EL, Floyd RS, Gregory MJ, Murfet IC. (1992) Branching in Pisum: inheritance and allelism tests with 17 ramosus mutants. Pisum Genet 24: 17–31 [Google Scholar]
- Berckmans B, De Veylder L. (2009) Transcriptional control of the cell cycle. Curr Opin Plant Biol 12: 599–605 [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Dun EA, Rameau C. (2009) Pea has its tendrils in branching discoveries spanning a century from auxin to strigolactones. Plant Physiol 151: 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Kyozuka J. (2010) New genes in the strigolactone-related shoot branching pathway. Curr Opin Plant Biol 13: 34–39 [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Murfet IC, Kerhoas L, Sotta B, Miginiac E, Rameau C. (1997a) The shoot controls zeatin riboside export from pea roots: evidence from the branching mutant rms4. Plant J 11: 339–345 [Google Scholar]
- Beveridge CA, Ross JJ, Murfet IC. (1996) Branching in pea: action of genes Rms3 and Rms4. Plant Physiol 110: 859–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Symons GM, Murfet IC, Ross JJ, Rameau C. (1997b) The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft transmissible signal(s). Plant Physiol 115: 1251–1258 [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O. (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell 8: 443–449 [DOI] [PubMed] [Google Scholar]
- Boyer FD, de Saint Germain A, Pillot JP, Pouvreau JB, Chen VX, Ramos S, Stévenin A, Simier P, Delavault P, Beau JM, et al (2012) Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: molecule design for shoot branching. Plant Physiol 159: 1524–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot JP, Boutet-Mercey S, Dalmais M, Antoniadi I, Li X, Maia-Grondard A, Le Signor C, Bouteiller N, et al. (2012) The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol 158: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. (2009) Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150: 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Koltai H, Beveridge CA. (2013) Diverse roles of strigolactones in plant development. Mol Plant 6: 18–28 [DOI] [PubMed] [Google Scholar]
- Cleland RE (2010) Auxin and Cell Elongation. Springer, Dordrecht, The Netherlands [Google Scholar]
- Clouse SD, Sasse JM. (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Davies PJ, editor (2010) Plant Hormones: Biosynthesis, Signal Transduction, Action! Springer, Dordrecht, The Netherlands [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. (2006) The molecular genetics of crop domestication. Cell 127: 1309–1321 [DOI] [PubMed] [Google Scholar]
- Drummond RS, Martínez-Sánchez NM, Janssen BJ, Templeton KR, Simons JL, Quinn BD, Karunairetnam S, Snowden KC. (2009) Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiol 151: 1867–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA. (2013) Dynamics of strigolactone function and shoot branching responses in Pisum sativum. Mol Plant 6: 128–140 [DOI] [PubMed] [Google Scholar]
- Dun EA, Hanan J, Beveridge CA. (2009) Computational modeling and molecular physiology experiments reveal new insights into shoot branching in pea. Plant Cell 21: 3459–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis TH, Hofer JM, Timmerman-Vaughan GM, Coyne CJ, Hellens RP. (2011) Mendel, 150 years on. Trends Plant Sci 16: 590–596 [DOI] [PubMed] [Google Scholar]
- Finlayson SA, Krishnareddy SR, Kebrom TH, Casal JJ. (2010) Phytochrome regulation of branching in Arabidopsis. Plant Physiol 152: 1914–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Yoneyama K, Hugill CJ, Quittenden LJ, Reid JB. (2013) Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol Plant 6: 76–87 [DOI] [PubMed] [Google Scholar]
- Frigerio M, Alabadí D, Pérez-Gómez J, García-Cárcel L, Phillips AL, Hedden P, Blázquez MA. (2006) Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol 142: 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Qian Q, Liu X, Yan M, Feng Q, Dong G, Liu J, Han B. (2009) Dwarf 88, a novel putative esterase gene affecting architecture of rice plant. Plant Mol Biol 71: 265–276 [DOI] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H. (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Grandio E, Poza-Carrion C, Sorzano CO, Cubas P. (2013) BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis Plant Cell 25: 834–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JC, Koch KE, Suzuki M, Wu S, Latshaw S, Petruff T, Goulet C, Klee HJ, McCarty DR. (2012) Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol 160: 1303–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC. (2012) DAD2 is an α/βhydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22: 2032–2036 [DOI] [PubMed] [Google Scholar]
- Hedden P. (2003) The genes of the Green Revolution. Trends Genet 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Heinrich M, Hettenhausen C, Lange T, Wünsche H, Fang J, Baldwin IT, Wu J. (2013) High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of Nicotiana attenuata stems. Plant J 73: 591–606 [DOI] [PubMed] [Google Scholar]
- Hu Z, Yan H, Yang J, Yamaguchi S, Maekawa M, Takamure I, Tsutsumi N, Kyozuka J, Nakazono M. (2010) Strigolactones negatively regulate mesocotyl elongation in rice during germination and growth in darkness. Plant Cell Physiol 51: 1136–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard L, McSteen P, Doebley J, Hake S. (2002) Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162: 1927–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram TJ, Reid D, MacMillan J. (1986) The quantitative relationship between gibberellin A1 and internode growth in Pisum sativum L. Planta 168: 414–420 [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46: 79–86 [DOI] [PubMed] [Google Scholar]
- Jager CE, Symons GM, Nomura T, Yamada Y, Smith JJ, Yamaguchi S, Kamiya Y, Weller JL, Yokota T, Reid JB. (2007) Characterization of two brassinosteroid C-6 oxidase genes in pea. Plant Physiol 143: 1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Fu X. (2007) GA action: turning on de-DELLA repressing signaling. Curr Opin Plant Biol 10: 461–465 [DOI] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C. (2006) Branching genes are conserved across species: genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol 142: 1014–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama M, Hirano Y, Mori T, Kim SY, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T. (2013) Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18: 147–160 [DOI] [PubMed] [Google Scholar]
- Kapulnik Y, Delaux PM, Resnick N, Mayzlish-Gati E, Wininger S, Bhattacharya C, Séjalon-Delmas N, Combier JP, Bécard G, Belausov E, et al. (2011) Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233: 209–216 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Hake S. (1997) Shoot meristem formation in vegetative development. Plant Cell 9: 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Lammers M, Pollina T, Tóth P, Haider I, Pozo MJ, de Maagd RA, Ruyter-Spira C, Bouwmeester HJ, et al. (2012) The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol 196: 535–547 [DOI] [PubMed] [Google Scholar]
- Koltai H. (2011) Strigolactones are regulators of root development. New Phytol 190: 545–549 [DOI] [PubMed] [Google Scholar]
- Kuppusamy KT, Walcher CL, Nemhauser JL. (2009) Cross-regulatory mechanisms in hormone signaling. Plant Mol Biol 69: 375–381 [DOI] [PubMed] [Google Scholar]
- Lee LY, Hou X, Fang L, Fan S, Kumar PP, Yu H. (2012) STUNTED mediates the control of cell proliferation by GA in Arabidopsis. Development 139: 1568–1576 [DOI] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB. (1997) Mendel’s stem length gene (Le) encodes a gibberellin 3β-hydroxylase. Plant Cell 9: 1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB. (1999) Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J 19: 65–73 [DOI] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z, Yan C, Jiang B, Su Z, Li J, et al. (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Novero M, Charnikhova T, Ferrandino A, Schubert A, Ruyter-Spira C, Bonfante P, Lovisolo C, Bouwmeester HJ, Cardinale F. (2013) Carotenoid cleavage dioxygenase 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus. J Exp Bot 64: 1967–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wu C, Fu Y, Hu G, Si H, Zhu L, Luan W, He Z, Sun Z. (2009) Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta 230: 649–658 [DOI] [PubMed] [Google Scholar]
- Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JA, Chen LJ, Yu SM. (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez JA, Matusova R, Cardoso C, Jamil M, Charnikhova T, Kohlen W, Ruyter-Spira C, Verstappen F, Bouwmeester H. (2009) Strigolactones: ecological significance and use as a target for parasitic plant control. Pest Manag Sci 65: 471–477 [DOI] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P. (1997) Mendel’s dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc Natl Acad Sci USA 94: 8907–8911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P. (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- McSteen P. (2009) Hormonal regulation of branching in grasses. Plant Physiol 149: 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C. (1996) Highly branched phenotype of the petunia dad1-1 mutant is reversed by grafting. Plant Physiol 111: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nieminen K, Immanen J, Laxell M, Kauppinen L, Tarkowski P, Dolezal K, Tähtiharju S, Elo A, Decourteix M, Ljung K, et al. (2008) Cytokinin signaling regulates cambial development in poplar. Proc Natl Acad Sci USA 105: 20032–20037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Daimon Y, Kurotani K, Higo A, Pruneda-Paz JL, Breton G, Mitsuda N, Kay SA, Ohme-Takagi M, Endo M, et al. (2013) BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis Plant Cell 25: 1228–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F. (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell (Suppl) 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill DP, Davidson SE, Clarke VC, Yamauchi Y, Yamaguchi S, Kamiya Y, Reid JB, Ross JJ. (2010) Regulation of the gibberellin pathway by auxin and DELLA proteins. Planta 232: 1141–1149 [DOI] [PubMed] [Google Scholar]
- O’Neill DP, Ross JJ. (2002) Auxin regulation of the gibberellin pathway in pea. Plant Physiol 130: 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro V, Leyser O. (2008) Hormonal control of shoot branching. J Exp Bot 59: 67–74 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al. (1999) ‘Green Revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Rameau C, Bodelin C, Cadier D, Grandjean O, Miard F. (1997) New ramosus mutants at loci Rms1, Rms3 and Rms4 resulting from the mutation breeding program at Versailles. Pisum Genet 29: 7–12 [Google Scholar]
- Rasmussen A, Mason MG, De Cuyper C, Brewer PB, Herold S, Agusti J, Geelen D, Greb T, Goormachtig S, Beeckman T, et al (2012) Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol 158: 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB. (1983) Internode length in Pisum: do the internode length genes effect growth in dark-grown plants? Plant Physiol 72: 759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB, Davidson SE, Ross JJ. (2011) Auxin acts independently of DELLA proteins in regulating gibberellin levels. Plant Signal Behav 6: 406–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB, Ross JJ. (2011) Mendel’s genes: toward a full molecular characterization. Genetics 189: 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, O’Neill DP, Rathbone DA. (2003) Auxin-gibberellin interactions in pea: integrating the old with the new. J Plant Growth Regul 22: 99–108 [Google Scholar]
- Ross JJ, O’Neill DP, Smith JJ, Kerckhoffs LH, Elliott RC. (2000) Evidence that auxin promotes gibberellin A1 biosynthesis in pea. Plant J 21: 547–552 [DOI] [PubMed] [Google Scholar]
- Ross JJ, Reid JB. (1986) Internode length in Pisum: the involvement of ethylene with the gibberellin insensitive erectoides phenotype. Physiol Plant 67: 673–679 [Google Scholar]
- Ross JJ, Reid JB, Dungey HS. (1992) Ontogenetic variation in levels of gibberellin A1 in Pisum: implications for the control of stern elongation. Planta 186: 166–171 [DOI] [PubMed] [Google Scholar]
- Ross JJ, Weston DE, Davidson SE, Reid JB. (2011) Plant hormone interactions: how complex are they? Physiol Plant 141: 299–309 [DOI] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, et al. (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol 155: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamini F. (2003) Hormones and the green revolution. Science 302: 71–72 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al (2002) Green Revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP. (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PY, Martínez EC, Sun TP. (1997) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Waters MT. (2012) Strigolactones: destruction-dependent perception? Curr Biol 22: R924–R927 [DOI] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. (2005) The DECREASED APICAL DOMINANCE1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17: 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, et al (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17: 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Ottoline Leyser HM. (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J 50: 80–94 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HM. (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Tivendale ND, Davidson SE, Davies NW, Smith JA, Dalmais M, Bendahmane AI, Quittenden LJ, Sutton L, Bala RK, Le Signor C, et al (2012) Biosynthesis of the halogenated auxin, 4-chloroindole-3-acetic acid. Plant Physiol 159: 1055–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Vogler H, Kuhlemeier C. (2003) Simple hormones but complex signalling. Curr Opin Plant Biol 6: 51–56 [DOI] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM. (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmülling T. (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98: 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston DE, Elliott RC, Lester DR, Rameau C, Reid JB, Murfet IC, Ross JJ. (2008) The pea DELLA proteins LA and CRY are important regulators of gibberellin synthesis and root growth. Plant Physiol 147: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang CM, Chandler PM, Smith JJ, Ross JJ. (2004) Auxin from the developing inflorescence is required for the biosynthesis of active gibberellins in barley stems. Plant Physiol 134: 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang CM, Ross JJ. (2001) Auxin promotes gibberellin biosynthesis in decapitated tobacco plants. Planta 214: 153–157 [DOI] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al (2007) Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Chen Z, Zhang S, Zhang W, Jiang G, Zhao X, Zhai W, Pan X, Zhu L. (2005) Characterizations and fine mapping of a mutant gene for high tillering and dwarf in rice (Oryza sativa L.). Planta 222: 604–612 [DOI] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L. (2006) The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J 48: 687–698 [DOI] [PubMed] [Google Scholar]