A tomato MADS-box gene, SlMADS1, impacts fruit ripening as a negative regulator.
Abstract
MADS-box genes encode a highly conserved gene family of transcriptional factors that regulate numerous developmental processes in plants. In this study, a tomato (Solanum lycopersicum) MADS-box gene, SlMADS1, was cloned and its tissue-specific expression profile was analyzed. The real-time polymerase chain reaction results showed that SlMADS1 was highly expressed in sepals and fruits; its expression level was increased with the development of sepals, while the transcript of SlMADS1 decreased significantly in accordance with fruit ripening. To further explore the function of SlMADS1, an RNA interference (RNAi) expression vector targeting SlMADS1 was constructed and transformed into tomato plants. Shorter ripening time of fruit was observed in SlMADS1-silenced tomatoes. The accumulation of carotenoid and the expression of PHYTOENE SYNTHETASE1 were enhanced in RNAi fruits. Besides, ethylene biosynthetic genes, including 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE1A, 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE6, 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE1, and 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE3, and the ethylene-responsive genes E4 and E8, which were involved in fruit ripening, were also up-regulated in silenced plants. SlMADS1 RNAi fruits showed approximately 2- to 4-fold increases in ethylene production compared with the wild type. Furthermore, SlMADS1-silenced seedlings displayed shorter hypocotyls and were more sensitive to 1-aminocyclopropane-1-carboxylate than the wild type. Additionally, a yeast two-hybrid assay revealed a clear interaction between SlMADS1 and SlMADS-RIN. These results suggest that SlMADS1 plays an important role in fruit ripening as a repressive modulator.
The ripening of fleshy fruit is a developmental biochemical process including numerous metabolic changes, such as changes in color, flavor, aroma, and nutrition. These changes not only make fruit assist in seed dispersal but also provide essential nutrition for human and animal diets (Ampomah-Dwamena et al., 2002; Giovannoni, 2004; Goff and Klee, 2006). In climacteric fruits such as tomato (Solanum lycopersicum), banana (Musa spp.), apple (Malus domestica), and pear (Pyrus communis), ethylene plays an important role in triggering the onset of ripening and is an essential factor for the ripening process (Abeles et al., 1973; Hiwasa et al., 2003). There are two key biosynthetic enzymes in ethylene biosynthesis, 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE (ACS) and 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE (ACO; Yang and Hoffman, 1984; Kende, 1993; Zarembinski and Theologis, 1994; Oetiker et al., 1997). It has been revealed that ethylene production and fruit ripening are strongly inhibited in SlACS2 RNA interference (RNAi) transgenic tomato fruits (Alexander and Grierson, 2002), and the expression level of SlACS2 is notably induced by exogenous ethylene (Olson et al., 1991; Lincoln et al., 1993; Barry et al., 1996, 2000). Furthermore, the expression of both SlACO1 and SlACO3 is significantly increased at the onset of tomato fruit ripening (Barry et al., 1996). Previous studies also indicate that RNAi inhibition of SlACO1 delays the ripening of climacteric fruits (Hamilton et al., 1990; Blume and Grierson, 1997; Giovannoni, 2001). These findings suggest that the normal function of ethylene biosynthesis is required for the ripening process.
Besides the functional ethylene synthesis, the abilities of ethylene perception and response are also necessary for ripening. E4 and E8 are two classical genes that are induced by ethylene (Lincoln et al., 1987). The expression of E4 in fruit is rapidly induced following exogenous ethylene induction (Lincoln and Fischer, 1988a). Meanwhile, the transcripts of E4 in fruit are suppressed through ethylene biosynthesis inhibition (Tigchelaar et al., 1978; Lincoln and Fischer, 1988b). In tomato, E8 is regulated by ethylene and is activated at the onset of fruit ripening (Peñarrubia et al., 1992; Kneissl and Deikman, 1996). The promoter of E8 has been characterized and is widely used to drive the expression of exogenous genes in transgenic tomato fruits (Sandhu et al., 2000; Krasnyanski et al., 2001; Kesanakurti et al., 2012).
Tomato is generally considered to be a model plant for studying fruit ripening. To date, a wide range of studies have been performed to uncover the mechanism of fruit ripening of tomato, and a lot of ripening-deficient mutants, such as ripening inhibitor (rin), never ripe (Nr), nonripening (nor), and color nonripening (cnr), have been found and investigated in tomato (Tigchelaar et al., 1973; Mizrahi et al., 1982; Wilkinson et al., 1995; Vrebalov et al., 2002). The rin mutant displays enlarged sepals and inhibited fruit ripening. This mutant phenotype has been attributed to the function of two MADS-box transcriptional factors, SlMADS-RIN and SlMADS-MC. SlMADS-RIN regulates fruit ripening and SlMADS-MC is involved in sepal development (Vrebalov et al., 2002). Besides SlMADS-RIN and SlMADS-MC, other MADS-box proteins also have been investigated in tomato. A prior study indicates that at least 36 MADS-box proteins have been found playing different and important biological roles in tomato, such as the determination of inflorescence and fruit ripening (Hileman et al., 2006). Among them, TOMATO AGAMOUS1 (TAG1), TOMATO AGAMOUS-LIKE1 (TAGL1), TOMATO MADS BOX4 (TM4 [TDR4, FUL1]), and TM6 have been investigated and identified to be associated with the development of fruits (Giovannoni, 2007). RNAi suppression of the TAG1 gene in tomato leads to misshapen fruits and homeotic conversion of stamens into petalloid organs (Pnueli et al., 1994; Pan et al., 2010), while TAGL1 plays an important role in regulating fruit ripening. The antisense suppression of TAGL1 results in ripening inhibition and pericarp thickness reduction. Furthermore, overexpression of TAGL1 leads to ripening-like sepals and enhanced lycopene fruits (Itkin et al., 2009; Vrebalov et al., 2009; Giménez et al., 2010). TM4 is a homolog of the Arabidopsis (Arabidopsis thaliana) FRUITFULL (FUL) gene and has also been reported to be related to fruit ripening (Busi et al., 2003). The expression of TM4 is repressed in the rin, cnr, and nor mutants (Seymour et al., 2002; Fujisawa et al., 2012). Additionally, TM6 transcripts mainly accumulate in carpel primordial and young fruits in tomato and have been considered to be involved in fruit ripening (Pnueli et al., 1994; Busi et al., 2003). Interestingly, these reported genes of the MADS-box family all function as positive regulators of ripening. In general, some inhibitors regulate these positive regulatory factors or are directly involved in the regulation of fruit ripening in other ways, out of consideration of the balance of the activities of these positive ripening regulators (Chung et al., 2010). It is reported that SlAP2a plays a role in fruit ripening as a negative regulator (Chung et al., 2010). Recently, SlERF6 was reported to influence carotenoid biosynthesis and additional ripening phenotypes as an inhibitor (Lee et al., 2012). However, to date, no inhibitor of fruit ripening in the MADS-box family has been reported in tomato.
Here, we cloned a MADS-box gene, SlMADS1 (GenBank accession no. AY294329), which has been reported as an inhibitor in vitro (Gaffe et al., 2011). The SlMADS1 protein belongs to the SEPALLATA (SEP) subfamily (Hileman et al., 2006). A prior report indicates that SlMADS1 transcripts mainly accumulate in fruits and that the accumulation decreases as fruits develop and ripen (Gaffe et al., 2011). However, SlMADS1 has not been functionally analyzed in tomato to date. In this study, RNAi repression of SlMADS1 was performed to investigate the exact role of SlMADS1 in tomato, and the results certify our supposition that SlMADS1 acts as an inhibitor in regulating fruit ripening.
RESULTS
SlMADS1 Transcripts Accumulate at High Levels in Sepals and Fruits
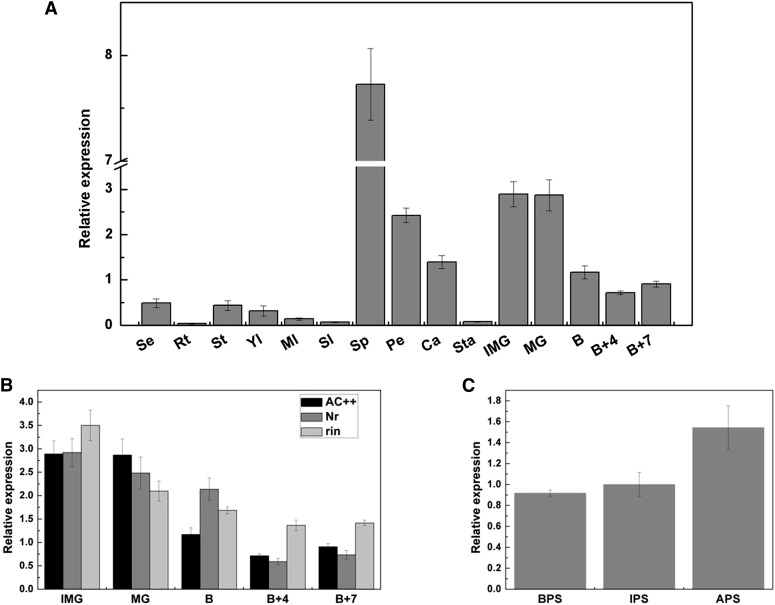
Based on the sequence in GenBank, full-length complementary DNA (cDNA) of SlMADS1 was cloned from tomato of cv Ailsa Craig. In order to explore its tissue-specific expression profile, real-time PCR was performed to analyze the accumulation of SlMADS1 transcripts in roots, stems, leaves, flowers, and a series of stages of fruits including normal and nonripening mutant fruits (Nr and rin). A low level of SlMADS1 was observed in seedlings, stems, and a series of stages of leaves (Fig. 1A). Almost no transcripts accumulated in roots (Fig. 1A). In tissues of flowers, a low level of SlMADS1 was detected in stamen, high levels were seen in carpel and petals, and the maximum level was displayed in sepals of flowers (Fig. 1A). Additionally, the SlMADS1 gene was highly expressed in immature green and mature green fruits, and a rapid declining trend was observed as fruit ripened (Fig. 1, A and B). A similar expression trend was observed in Nr and rin fruits, indicating that SlMADS1 expression is not impacted by the single-locus SlMADS-RIN and Nr (Fig. 1B). To further detect the expression of SlMADS1 in sepals, its transcripts were analyzed in different developmental stages of sepals. SlMADS1 mRNA was highly accumulated in flower sepals and increased with the development of sepals (Fig. 1C), which hinted that SlMADS1 may play a role during the development of sepals.
Figure 1.
Expression profile of SlMADS1 in tissues of cv Ailsa Craig and nonripening mutant fruits. A, Expression of SlMADS1 in cv Ailsa Craig as indicated: Se, seedlings; Rt, roots; St, stems; Yl, young leaves; Ml, mature leaves; Sl, senescent leaves; Sp, sepals of flower in anthesis; Pe, petals of flower in anthesis; Ca, carpels of flower in anthesis; Sta, stamens of flower in anthesis; IMG, immature green fruits; MG, mature green fruits; B, breaker fruits; B+4, 4 d after breaker fruits; B+7, 7 d after breaker fruits. B, Expression of SlMADS1 in cv Ailsa Craig (AC++), Nr, and rin fruits. C, Expression of SlMADS1 in sepals of cv Ailsa Craig. BPS, Sepals of flowers before pollination; IPS, sepals of flowers in pollination; APS, sepals of flowers after pollination.
Creation of SlMADS1-Silenced Lines
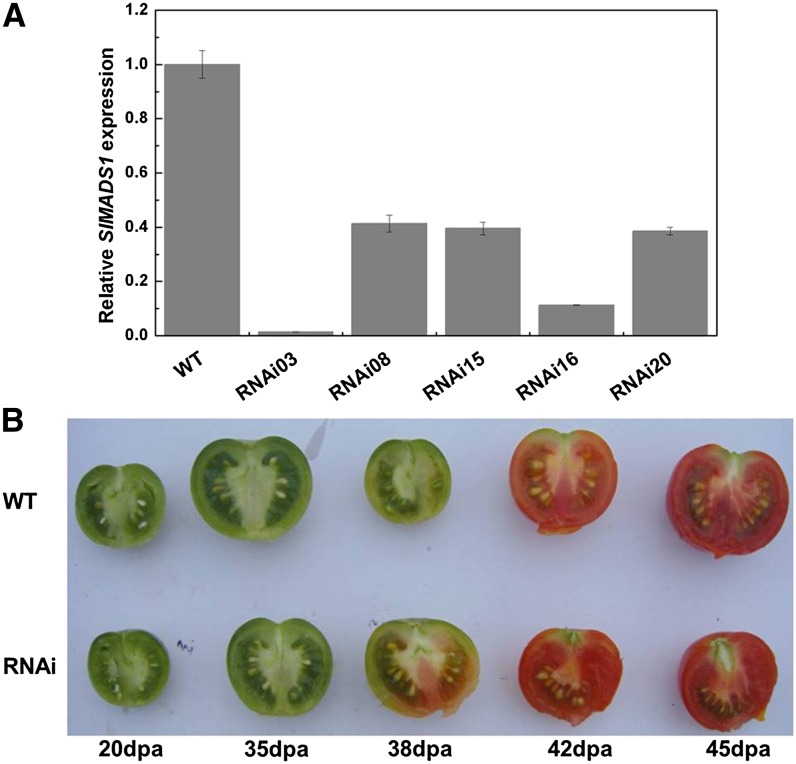
To gain further insight into the function of SlMADS1, an RNAi construct targeting the specific fragment of SlMADS1 was created and transformed into wild-type tomato plants via Agrobacterium tumefaciens-mediated T-DNA transfer. Five independent transgenic lines confirmed for transgene integration were selected for characterization. Real-time quantitative PCR (qPCR) results showed that SlMADS1 transcripts were significantly reduced in the transgenic lines compared with the wild type, and the most silenced SlMADS1 line, named RNAi-03, had a 99% reduction in breaker fruits and about 80% in seedlings (Fig. 2A and Supplemental Fig. S1). The expression of other members of the MADS-box family, including two SEP genes, SlMADS-RIN and SlMBP21, an AGAMOUS gene, TAGL1, and a FUL gene, TDR4, was also detected. TAGL1 and SlMADS-RIN were up-regulated, while the expression of TDR4 had no obvious change in SlMADS1-silenced fruits compared with the wild type (Supplemental Fig. S2, A, C, and D). In particular, SlMBP21, a homolog of SlMADS1 (Leseberg et al., 2008), was not impacted in SlMADS1-silenced lines (Supplemental Fig. S2B). These results indicated that the RNAi construct of SlMADS1 is specific and does not target to other MADS-box genes. Subsequently, three transgenic lines, RNAi-03, RNAi-16, and RNAi-20, were selected for further investigation.
Figure 2.
SlMADS1 repression phenotypes. A, Expression of SlMADS1 in RNAi lines and wild type (WT). RNAs were extracted for qPCR assay from breaker fruits of RNAi lines and the wild type. Three replications for each sample were performed. B, Genotypes are SlMADS1 RNAi lines (RNAi) and the wild type. The color of SlMADS1-silenced fruits changed earlier than in the wild type. [See online article for color version of this figure.]
SlMADS1 Impacts Fruit Ripening
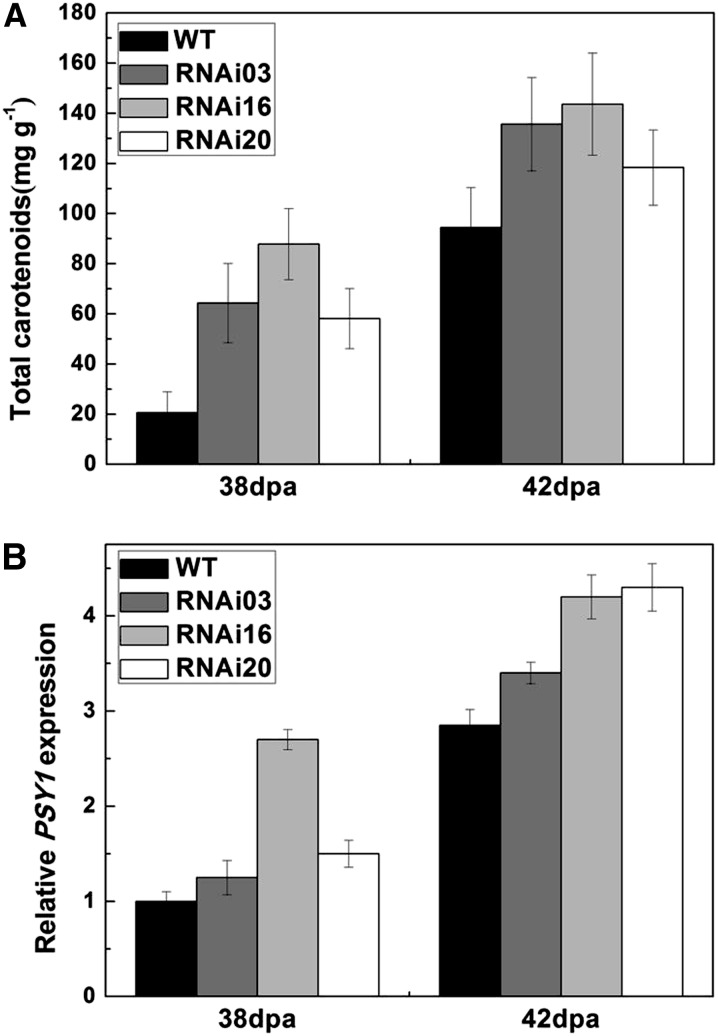
During the process of fruit development, we measured the time from anthesis to ripening and observed that the color of SlMADS1-silenced fruits changed earlier than wild-type fruits (Fig. 2B), and their ripening time was accelerated 3 to 6 d compared with the wild type (Table I). It has been shown that the dramatic change of pigmentation in ripening tomato fruits is caused by the accumulation of carotenoids (Fraser et al., 1994). In this study, the carotenoids in transgenic and wild-type fruits at 38 and 42 DPA were extracted and determined. As shown in Figure 3A, the accumulation of carotenoid in RNAi lines was much higher than in the wild type. Real-time PCR analysis results indicated that PHYTOENE SYNTHETASE1 (PSY1) was up-regulated in RNAi fruits both at 38 and 42 DPA (Fig. 3B).
Table I. Days from anthesis to breaker stage for control and SlMADS1-silenced lines.
| Tomato Line | Days |
|---|---|
| Wild type | 38.0 ± 0.50 |
| RNAi-03 | 31.8 ± 0.45 |
| RNAi-16 | 33.6 ± 0.48 |
| RNAi-20 | 34.6 ± 0.48 |
Figure 3.
Carotenoid accumulation and expression of PSY1 in SlMADS1-silenced and wild-type (WT) fruits. A, Analysis of carotenoid accumulation in 38- and 42-DPA fruits of transgenic SlMADS1 RNAi lines and the wild type. se is indicated for a minimum of three fruits per sample. B, Expression of PSY1 in 38- and 42-DPA fruits of transgenic SlMADS1 lines and the wild type.
Ethylene-Related and Ripening-Related Genes Are Significantly Up-Regulated in SlMADS1-Silenced Fruits
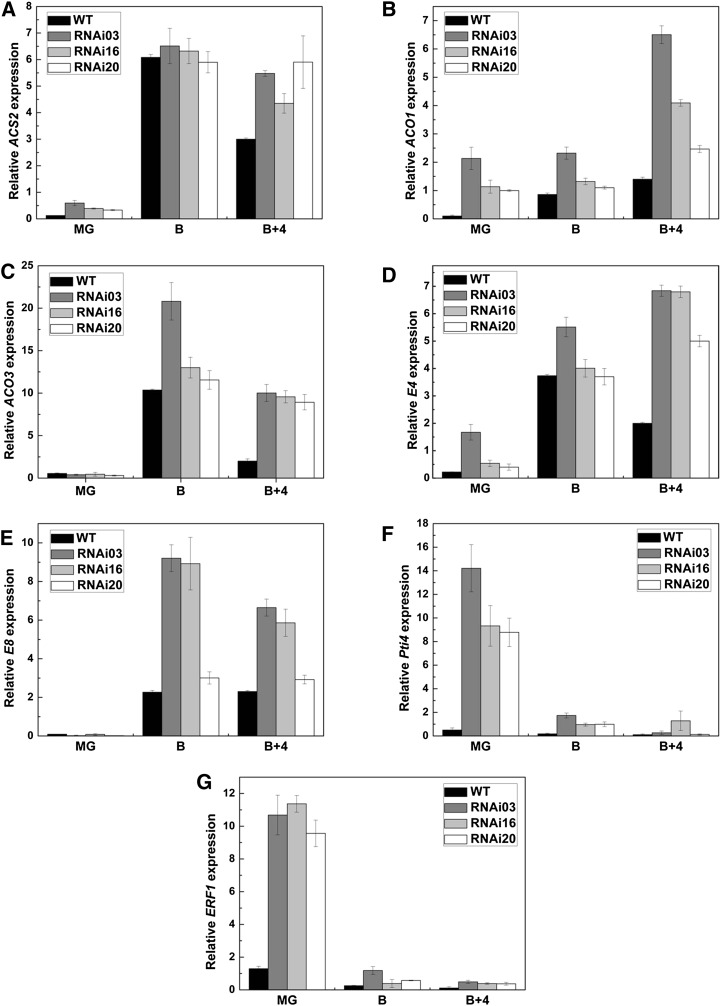
To further characterize the molecular regulation mechanism of SlMADS1 in fruit ripening, a set of ethylene-related and ripening-related genes in wild-type and transgenic tomato fruits were examined. Two ethylene biosynthetic genes, ACS2 and ACO3, were dramatically up-regulated in breaker + 4 d fruits of SlMADS1-silenced lines (Fig. 4, A and C), and the transcripts of another ethylene biosynthesis gene, ACO1, was also increased significantly in SlMADS1-silenced fruits at all stages (Fig. 4B). Furthermore, the expression of two ripening-related genes that responded specifically to ethylene, E4 and E8, was markedly increased in SlMADS1-silenced fruits at the breaker + 4 d stage (Fig. 4, D and E). These results indicated that SlMADS1 might inhibit fruit ripening by directly or indirectly impacting ethylene biosynthesis or ethylene response.
Figure 4.
Ripening- and ethylene-related gene expression in SlMADS1-silenced and wild-type (WT) fruits. RNAs were extracted for qPCR assay from mature green (MG), breaker (B), and breaker + 4-d (B+4) fruits of RNAi lines and the wild type. Three replications for each sample were used. A, Expression of ACS2 in RNAi lines and the wild type. B, Expression of ACO1 in RNAi lines and the wild type. C, Expression of ACO3 in RNAi lines and the wild type. D, Expression of E4 in RNAi lines and the wild type. E, Expression of E8 in RNAi lines and the wild type. F, Expression of Pti4 in RNAi lines and the wild type. G, Expression of ERF1 in RNAi lines and the wild type.
Additionally, two ethylene-responsive genes, ERF1 and Pti4, which have been reported to be factors associated with defense responses, were also analyzed. Dramatic increases were also detected in transgenic fruits at the mature green stage (Fig. 4, F and G), suggesting that SlMADS1 might play a role in the stress response.
More Ethylene Is Produced by SlMADS1-Silenced Lines
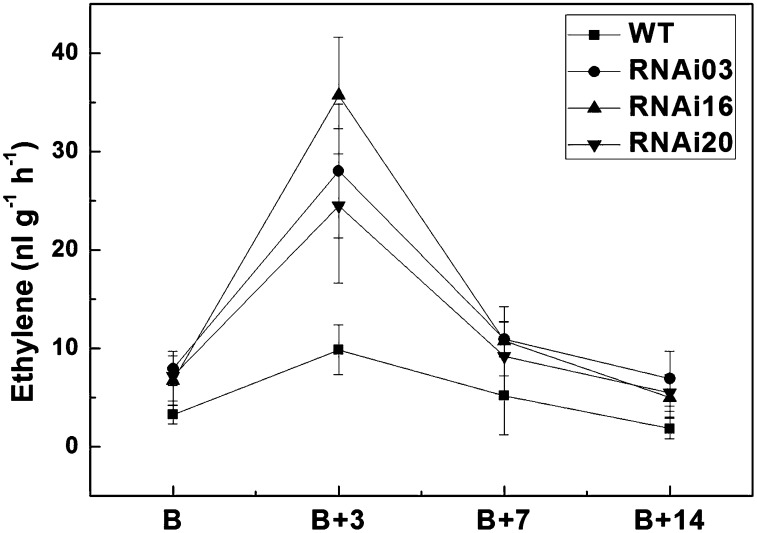
To further investigate the relationship between SlMADS1 and ethylene, we measured ethylene production during fruit development and ripening. SlMADS1 RNAi lines exhibited a rapid and massive increase in ethylene production at the breaker + 3 d, stage like the wild type, but SlMADS1 RNAi fruits produced approximately 2- to 4-fold more ethylene than the wild type during fruit ripening and remained at high levels even at breaker + 14 d (Fig. 5).
Figure 5.
Production of ethylene in control and SlMADS1-silenced lines. Fresh fruits of breaker (B), breaker + 3 d (B+3), breaker + 7 d (B+7), and breaker + 14 d (B+14) were sealed in air-tight vials, and 1 mL of gas was sampled from the headspace after 24 h. Values represent means of at least three individual fruits. Error bars represent se. WT, Wild type.
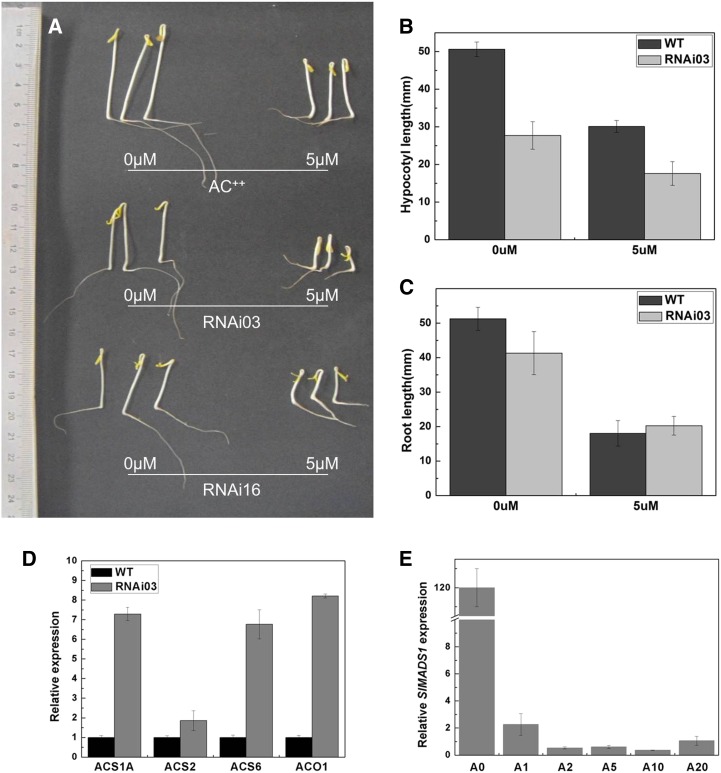
To ascertain if the high level of ethylene production in fruit tissues of SlMADS1 RNAi lines persisted in nonfruit tissues, an ethylene triple response assay was performed. Wild-type and SlMADS1-silenced seeds were germinated on Murashige and Skoog (MS) medium supplemented with or without the ethylene precursor 1-aminocyclopropane-1-carboxylate (ACC), which could be taken up by the roots and converted rapidly to ethylene. The elongation of hypocotyls and roots was detected 7 d after sowing. The results showed that the average length of hypocotyl elongation of RNAi lines was significantly shorter than that of the wild type both in the absence (0 µm) and presence (5.0 µm) of ACC (Fig. 6, A and B), while the root elongation of wild-type and RNAi lines was nearly identical in the above two conditions (Fig. 6, A and C).
Figure 6.
Ethylene triple response assay. A, Seedlings of wild-type Ailsa Craig (AC++) and RNAi lines (RNAi-03 and RNAi-16) treated with 0 and 5.0 µm ACC. B and C, Elongation of hypocotyl (B) and root (C) growth on different concentrations of ACC. Error bars represent ± se. D, Expression of ACS1A, ACS2, ACS6, and ACO1 in seedlings of RNAi lines and the wild type (WT). E, Expression of SlMADS1 in seedlings of the wild type treated with 0 (A0), 1.0 (A1), 2.0 (A2), 5.0 (A5), 10.0 (A10), and 20.0 (A20) µm ACC. [See online article for color version of this figure.]
To verify the triple response exhibited by silenced lines, the expression of SlMADS1 in RNAi and wild-type seedlings was detected. The result suggested that SlMADS1 expression was reduced at least 60% (Supplemental Fig. S1). The expression of ACS1A, ACS2, ACS6, and ACO1 was also detected by quantitative PCR, in order to further explore the triple response mechanism of SlMADS1-silenced seedlings. The results demonstrated that ACS1A, ACS6, and ACO1 were all up-regulated significantly in seedlings of RNAi lines in the absence of ACC (Fig. 6D), which suggested that silencing SlMADS1 could activate the expression of ethylene biosynthesis genes, while the transcripts of ACS2 were slightly increased in transgenic lines (Fig. 6D). The expression of SlMADS1 in cv Ailsa Craig seedlings decreased dramatically after the ACC treatment, and a slow declining trend was observed with the increased density of ACC (Fig. 6E), which suggested that SlMADS1 might be impacted by ACC or ethylene.
The Yeast Two-Hybrid Assay Demonstrates That SlMADS1 Interacts with SlMADS-RIN
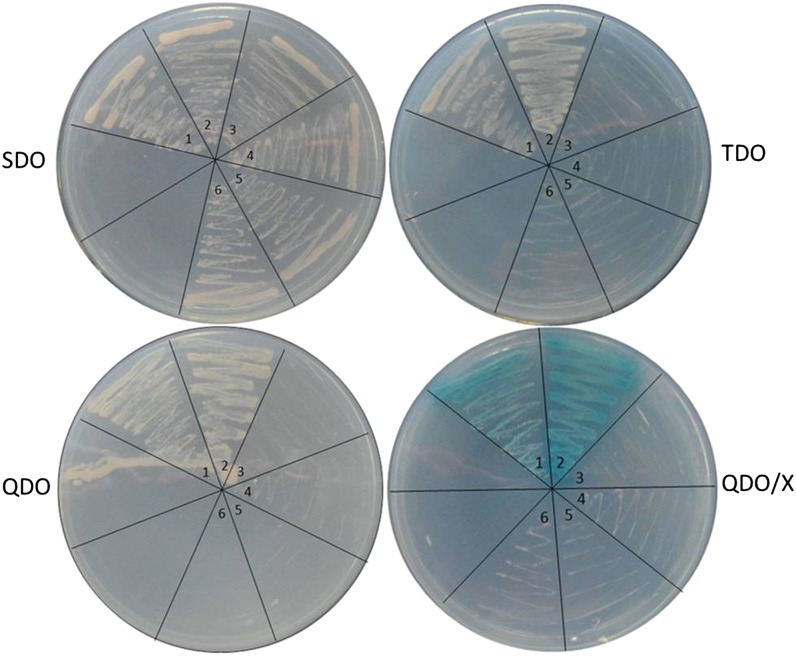
An essential regulator of tomato fruit ripening, SlMADS-RIN was preferentially selected for yeast two-hybrid assay. The open reading frame of SlMADS1 was amplified and cloned into pGBKT7 as the bait. Self-activation of pGBKT7-MADS1 was tested, and the result was negative (Fig. 7). The open reading frame of SlMADS-RIN was amplified and cloned into pGADT7 as the prey. An empty prey and bait vector was used as a negative control with each bait and prey construct, respectively. Figure 7 shows that the yeast grew on selective medium and turned blue on the 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-gal) indicator plate, suggesting that there exists an interaction between SlMADS1 and SlMADS-RIN in vivo.
Figure 7.
Yeast two-hybrid assay for SlMADS1 and SlMADS-RIN proteins. SDO, SD medium without Trp; TDO, SD medium without Trp, His, and adenine; QDO, SD medium without Trp, Leu, His, and adenine; QDO/X, SD medium without Trp, Leu, His, and adenine with X-α-Gal. Numbered wedges are as follows: 1, pGBKT7-MADS1 and pGADT7-RIN (interaction of SlMADS1 and SlMADS-RIN); 2, pGBKT7-53 and pGADT7-T (positive control); 3, pGBKT7-Lam and pGADT7-T (negative control); 4, pGBKT7-MADS1 (autoactivation assay); 5, pGBKT7 and pGADT7-RIN (empty bait vector); 6, pGBKT7-MADS1 and pGADT7 (empty prey vector). [See online article for color version of this figure.]
DISCUSSION
SlMADS1 Inhibits Ethylene Biosynthesis and Impacts Fruit Ripening as an Inhibitor
In higher plants, the ethylene biosynthesis pathway is well studied (Bleecker and Kende, 2000). Two modes of ethylene synthesis, system 1 and system 2, have been defined (McMurchie et al., 1972; Barry et al., 2000). System 1 contributes to providing basal ethylene in vegetative tissues and unripe fruits. System 2 produces a large amount of ethylene at the onset of fruit ripening (Yang and Oetiker, 1994; Nakatsuka et al., 1998). Two kinds of rate-limiting enzymes (ACS and ACO) in ethylene biosynthesis have been reported. ACS catalyzes the conversion of S-adenosyl-l-Met to ACC, and the conversion of ACC to ethylene is carried out by ACO (Kende, 1993). At least nine ACS genes (ACS1A, ACS1B, ACS2, ACS3, ACS4, ACS5, ACS6, ACS7, and ACS8) and five ACO genes (ACO1–ACO5) have been identified in tomato (Zarembinski and Theologis, 1994; Barry et al., 1996; Oetiker et al., 1997; Nakatsuka et al., 1998; Shiu et al., 1998; Sell and Hehl, 2005). It has been proposed that SlACS1A and SlACS6 are involved in system 1 and present in tomato fruits before the onset of ripening (Barry et al., 2000). Prior studies have reported that SlACS2 was an important factor to transit system 1 to system 2 (Nakatsuka et al., 1998; Barry et al., 2000). The fruit from RNAi repression of SlACS2 could not ripen normally (Oeller et al., 1991). Moreover, two ACO genes (SlACO1 and SlACO3) have been reported to contribute to triggering fruit ripening (Alexander and Grierson, 2002). The expression of SlACO3 is induced but transitory at the breaker stage, while SlACO1 expression is sustained during ripening (Barry et al., 1996; Nakatsuka et al., 1998).
In this study, we tested the expression of ACS2 in SlMADS1-silenced fruits and ACS1A and ACS6 in SlMADS1-silenced seedlings. The results showed that expression levels of all these ACS genes were noticeably higher in RNAi lines than in the wild type (Figs. 4A and 6D). Furthermore, the accumulation of the ACO transcripts (ACO1 and ACO3) in transgenic fruit was much higher than in the wild type (Figs. 4, B and C, and 6D). These results indicate that SlMADS1 might inhibit the expression of ethylene biosynthesis genes, then impact the ethylene biosynthesis in tomatoes, which was confirmed by ethylene determination of fruit and the triple response assay. SlMADS1 RNAi fruits produce more ethylene (Fig. 5). Also, the hypocotyl elongation of RNAi lines was shorter than in the wild type in the absence of ACC, and the RNAi seedlings were more sensitive to ACC than the wild type (Fig. 6, A and B), which indicated that more ethylene was probably produced in the RNAi transgenic plants than the wild type. These results suggest that SlMADS1 impacts ethylene biosynthesis both in vegetative organs and fruits.
E4 and E8 are well known as important ethylene-responsive genes during fruit ripening. E8 influences ethylene biosynthesis both in fruit and flower (Kneissl and Deikman, 1996). The expression of E4 is suppressed when high-level ethylene biosynthesis is inhibited by mutations that block fruit ripening (Tigchelaar et al., 1978). Our study showed that both of these genes were expressed highly in the transgenic fruits compared with the wild type (Fig. 4, D and E).
For SlMADS-RIN, TDR4 (TM4, FUL1), and TAGL1, three MADS-box proteins are necessary for the completion of fruit ripening (Vrebalov et al., 2002, 2009). Their expression levels were significantly up-regulated in SlMADS1-silenced fruits (Supplemental Fig. S2, A, C, and D). PSY1, a major regulator of metabolic flux toward downstream carotenoids, is induced by ethylene during fruit ripening (Fray and Grierson, 1993). In our study, the expression of PSY1 was notably increased in transgenic fruits (Fig. 3B). Furthermore, phenotype analysis demonstrated that SlMADS1-silenced fruits ripen in advance (Fig. 2B; Table I). These results suggest that suppressing the expression of SlMADS1 promotes the expression of ripening-related genes and accelerates the rate of ripening, indicating that SlMADS1 acts as an inhibitor in fruit ripening.
SlMADS1 Might Weaken the Activity of SlMADS-RIN
In recent years, more and more MADS-box genes have been identified and revealed to play positive roles in fruit ripening. Heterodimers, homodimers, or higher order complexes have been detected in MADS-domain proteins (Favaro et al., 2002; Shchennikova et al., 2004; de Folter et al., 2006). SlMADS-RIN is a classical and essential positive regulator of tomato fruit ripening among the MADS-box proteins and is associated with ethylene biosynthesis, ethylene perception, and ethylene response. As reported previously, ACS2 and ACS4 are bound by SlMADS-RIN (Ito et al., 2008; Martel et al., 2011; Fujisawa et al., 2012). ACO1 is influenced by SlMADS-RIN through the homeobox gene HB1, which interacts with the promoter of ACO1 (Lin et al., 2008; Martel et al., 2011). E8 is identified as a novel direct target of SlMADS-RIN, which can be rapidly induced following ethylene induction and during normal fruit ripening (Martel et al., 2011; Qin et al., 2012). In our study, ACO1, ACS2, and E8 are up-regulated markedly in SlMADS1-silenced lines, which suggests that these genes are negatively regulated by SlMADS1 (Fig. 4). Moreover, the yeast two-hybrid assay indicates that there is an interaction between SlMADS1 and SlMADS-RIN (Fig. 7). These results imply that SlMADS1 might bind to SlMADS-RIN and depress its activity, subsequently influence the expression of ethylene biosynthesis and response genes such as ACO1, ACS2, and E8, and then reduce the biosynthesis of ethylene and inhibit fruit ripening.
In summary, SlMADS1 plays an important role in fruit ripening as a repressive modulator by regulating ethylene biosynthesis directly or impacting ethylene biosynthesis and response indirectly by interacting with SlMADS-RIN. Although higher levels of a developmental regulatory cascade of this gene remain to be discovered, as a repressive regulator, SlMADS1 plays an important role in balancing the activities of positive ripening regulators and adds a new component to the emerging mechanisms regulating fleshy fruit ripening.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
In our experiments, we used plants of tomato (Solanum lycopersicum ‘Ailsa Craig’ AC++), a near-isogenic tomato line, as the wild type. The plants were planted in a greenhouse and watered daily. Transgenic cultures grew under standard greenhouse conditions (16-h-day/8-h-night cycle, 25°C/18°C day/night temperature, 80% humidity, and 250 µmol m−2 s−1 light intensity). Two generations of tomato plants were used in the experiments. Plants of the first generation (T0) came from tissue culture, and plants of the second generation (T1) were from seedlings. Flowers were tagged at anthesis. The ripening stages of tomato fruits were divided according to DPA and fruit color. In the wild type, immature green was defined as 20 DPA. Mature green was defined as 35 DPA and characterized as being green and shiny with no obvious color change. Breaker fruits were defined as fruits of 38 DPA with the color change from green to yellow. Other fruits of 4 d after breaker and 7 d after breaker were also used. All plant samples were immediately frozen with liquid nitrogen, mixed, and stored at –80°C until further use.
SlMADS1 Isolation
Total RNA of tomato was extracted using Trizol (Invitrogen) according to the manufacturer’s instructions. Then, 1 µg of total RNA was used to synthesize first-strand cDNA through reverse transcription-PCR using Moloney murine leukemia virus reverse transcriptase (Takara) with tailed oligo(dT)18 primer (5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTGTTTTTTTTTTTTTTTTTT-3′). One to 2 µL of cDNA was used to clone the full-length SlMADS1 gene with primers of SlMADS1-F (5′-ATGGGAAGAGGAAGAGTTG-3′) and dT-r (5′-GCTGTCAACGATACGCTACGTAACG-3′) through high-fidelity PCR (Prime START HS DNA polymerase; Takara). The amplified products were tailed by using the DNA A-Tailing kit (Takara) and linked with pMD18-T vector (Takara). Positive clones were picked out via Escherichia coli JM109 transformation and confirmed by sequencing (Invitrogen).
Construction of the SlMADS1 RNAi Vector and Plant Transformation
In order to down-regulate the expression of the SlMADS1 gene, an RNAi vector was constructed. A 515-bp specific DNA fragment of SlMADS1 was amplified with primers SlMADS1i-F (5′-CGGGGTACCAAGCTTGATTACTCCGTAGAAA-3′) and SlMADSi-R (5′-CCGCTCGAGTCTAGACAATGATACAAAAAATAC-3′), which had been tailed with HindIII/KpnI and XhoI/XbaI restriction sites at the 5′ end, respectively. Then, the amplified products were digested with HindIII/XbaI and KpnI/XhoI and linked into the pHANNIBAL plasmid at the HindIII/XbaI restriction site in the sense orientation and at the KpnI/XhoI restriction site in the antisense orientation. Finally, the double-stranded RNA expression unit, containing the cauliflower mosaic virus 35S promoter, SlMADS1 fragment in the antisense orientation, PDK intron, SlMADS1 fragment in the sense orientation, and OCS terminator, was purified and inserted into the plant binary vector pBIN19 with SacI and XbaI restriction sites.
The generated binary plasmids were translated into Agrobacterium tumefaciens LBA4404 strain, and A. tumefaciens-mediated transformation was performed following the protocols described by Chen et al. (2004). The transgenic plants were detected with primers NPTII-F (5′-GACAATCGGCTGCTCTGA-3′) and NPTII-R (5′-AACTCCAGCATGAGATCC-3′). The positive transgenic plants were selected and used for subsequent experiments.
Quantitative Real-Time PCR Analysis
Total RNAs of tissues of cv Ailsa Craig, Nr, rin, and transgenic lines were extracted using Trizol (Invitrogen) according to the manufacturer’s instructions. Quantitative real-time PCR was performed using the SYBR Premix Ex Taq II kit (Takara) in a 10-μL total sample volume (5.0 μL of 2× SYBR Premix Ex Taq, 1.0 μL of primers, 1.0 μL of cDNA, and 3 μL of distilled, deionized water). To remove the effect of genomic DNA and the template from the environment, no-template control and no-reverse transcription control experiments were performed. Additionally, three replications for each sample were used, and standard curves were run simultaneously. Tomato SlCAC (Expósito-Rodríguez et al., 2008) and SlEF1α (Expósito-Rodríguez et al., 2008) were used as internal standards. The primers SlMADS1(RT)-F and SlMADS1(RT)-R (Table II) were used to determine the expression levels of SlMADS1 in the wild type, Nr and rin, and transgenic lines. Furthermore, the expression levels of other MADS-box genes, including SlMADS-RIN (Vrebalov et al., 2002), SlMBP21 (Leseberg et al., 2008), TAGL1 (Busi et al., 2003; Vrebalov et al., 2009), and TDR4 (TM4, FUL1; Seymour et al., 2002; Bemer et al., 2012), as well as fruit ripening-related, carotenoid biosynthesis, and ethylene biosynthesis and response genes, such as E4 (Lincoln et al., 1987; Peñarrubia et al., 1992), E8 (Kneissl and Deikman, 1996), ACO1, ACO3, and ACS2 (Griffiths et al., 1999; Alexander and Grierson, 2002), PSY1 (Fray and Grierson, 1993), Pti4 (Chakravarthy et al., 2003), and ERF1 (Li et al., 2007), were determined simultaneously. Primers are shown in Table II and Supplemental Table S1.
Table II. Details of primers for qPCR amplification.
| Primer Name | Primer Sequence (5′–3′) | Product |
|---|---|---|
| bp | ||
| SlCAC | CCTCCGTTGTGATGTAACTGG | 173 |
| ATTGGTGGAAAGTAACATCATCG | ||
| SlEF1α | ACCTTTGCTGAATACCCTCCATTG | 150 |
| CACACTTCACTTCCCCTTCTTCTG | ||
| SlMADS1 | GTGTAGCTGGATTTCCACTTCG | 175 |
| GCCGCTGCATTCACCTCAT | ||
| E4 | AGGGTAACAACAGCAGTAGCA | 167 |
| CCCAACCTCCGTCTTCAC | ||
| E8 | GGCACCATTCAACATACCG | 242 |
| CTTTCACCGAAGAAGCACG | ||
| PSY1 | AGAGGTGGTGGAAAGCAA | 298 |
| TCTCGGGAGTCATTAGCAT | ||
| ACO1 | ACAAACAGACGGGACACGAA | 181 |
| CTCTTTGGCTTGAAACTTGA | ||
| ACO3 | CAAGCAAGTTTATCCGAAAT | 113 |
| CATTAGCTTCCATAGCCTTC | ||
| ACS2 | GAAAGAGTTGTTATGGCTGGTG | 107 |
| GCTGGGTAGTATGGTGAAGGT | ||
| ERF1 | TTTTAGTATCGGATGGACG | 102 |
| GGCGGAGAAACAGAAGTA | ||
| Pti4 | CTCTAAGCGTCGGATGGTC | 150 |
| AATGTCTTCCTTTCGGTGTTT |
Carotenoid Extraction
A 1.0-g sample of each line was cut from pericarp in a 5-mm-wide strip around the equator of 38- and 42-DPA fruits. Then, 10 mL of 60:40 (v/v) hexane:acetone was added, and total carotenoids of wild-type and RNAi line fruits were extracted. The extract was centrifuged at 4,000g for 5 min, and the absorbance of the supernatant was measured at 450 nm. Carotenoid content was calculated with the following equation: total carotenoid (mg mL−1) = 4×(optical density at 450 nm) × 10 mL/1 g (Fray and Grierson, 1993; Forth and Pyke, 2006). Three independent experiments were performed for each sample.
Ethylene Measurements
Fruits of beaker, beaker + 3 d, beaker + 7 d, and beaker + 14 d were harvested and placed in open 100-mL jars for 3 h to minimize the effect of wound ethylene caused by picking. Jars were then sealed and incubated at room temperate for 24 h, and 1 mL of headspace gas was injected into a Hewlett-Packard 5890 series gas chromatograph equipped with a flame ionization detector. Samples were compared with reagent-grade ethylene standards of known concentration and normalized for fruit weight (Chung et al., 2010).
Ethylene Triple Response Assay
The seeds of wild-type plants were sterilized and sown on MS medium supplemented with 0, 0.5, 1.0, 2.0, 5.0, 10.0, and 20.0 µm ACC and then cultured in the dark at 25°C. Meanwhile, T1 seeds of RNAi lines were sterilized and sown on MS medium supplemented with 0 and 5.0 µm ACC and then cultured in the same conditions as the wild type. Hypocotyl and root elongation were measured 7 d after sowing, and at least 20 seedlings were measured for each culture. To further explore the molecular mechanism of the triple response of transgenic lines, the expression of ACS1A, ACS2, ACS6, and ACO1 in the wild type and transgenic lines was measured by qPCR. The expression of SlMADS1 was also detected in wild-type seedlings treated with 0, 1.0, 2.0, 5.0, 10.0, and 20.0 µm ACC.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed using the MATCHMAKER GAL4 Two-Hybrid System III according to the manufacturer’s protocol (Clontech). The open reading frame of SlMADS1 was amplified by PCR with the primer pair SlMADS1(Y)-F (5′-CCGGAATTCATGGGAAGAGGAAGAGTTG-3′) and SlMADS(Y)-R (5′-CGCGGATCCTTAAAGCATCCATCCATGAATA-3′). The PCR products were digested using EcoRI and SalI and cloned into the EcoRI/SalI site of the pGBKT7 bait vector to obtain the vector pGBKT7-MADS1. Then, pGBKT7-MADS1 vector was translated into Y2HGold. The Y2HGold with bait was plated on synthetic dropout (SD) medium lacking Trp and SD medium lacking Trp, His, and adenine to test the self-activation of pGBKT7-MADS1. In parallel, the open reading frame of SlMADS-RIN was also amplified by primers SlRIN(Y)-F (5′-CCGGAATTCATGGGTAGAGGGAAAGTAGA-3′) and SlRIN(Y)-R (5′-CGCGGATCCTCATAGATGTTTATTCAT-3′). The product was cloned into the pGADT7 vector and translated into Y187. Subsequently, Y2HGold with bait and Y187 with prey were cultured together in 2× YPDA (yeast extract, peptone, and dextrose medium supplemented with adenine hemisulfate) medium for 24 h. After that, these cultures were cultured on SD medium lacking Trp and Leu to select for diploids containing prey and bait vectors. After 2 to 5 d, fresh diploid cells were plated on SD medium lacking Trp, Leu, His, and adenine with X-α-Gal to judge whether SlMADS1 can interact with SlMADS-RIN or not. Plates were incubated for 3 to 7 d at 30°C. An empty prey and bait vector was used as a negative control with each bait and prey construct, respectively. Meanwhile, positive controls were cultured. The assays were repeated at least three times with fresh transformants.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers SlMADS1 (AY294329), E4 (S44898), E8 (X13437), PSY1 (EF157835), ACO1 (NM_001247095), ACO3 (Z54199), ACS2 (AY326958), ERF1 (AY077626), and Pti4 (U89255).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SlMADS1 expression in seedlings of RNAi lines and wild type.
Supplemental Figure S2. Other MADS-box gene expression in SlMADS1-silenced and wild-type fruits.
Supplemental Table S1. Details of other MADS-box gene primers for qPCR amplification.
Glossary
- RNAi
RNA interference
- cDNA
complementary DNA
- ACC
1-aminocyclopropane-1-carboxylate
- qPCR
quantitative PCR
- MS
Murashige and Skoog
- SD
synthetic dextrose
- SD
synthetic dropout
References
- Abeles F, Morgan P, Saltveit M., Jr (1973) Ethylene in Plant Biology. Academic Press, New York [Google Scholar]
- Alexander L, Grierson D. (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot 53: 2039–2055 [DOI] [PubMed] [Google Scholar]
- Ampomah-Dwamena C, Morris BA, Sutherland P, Veit B, Yao JL. (2002) Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol 130: 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CS, Blume B, Bouzayen M, Cooper W, Hamilton AJ, Grierson D. (1996) Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J 9: 525–535 [DOI] [PubMed] [Google Scholar]
- Barry CS, Llop-Tous MI, Grierson D. (2000) The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol 123: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, Rossetto PdB, Angenent GC, de Maagd RA. (2012) The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24: 4437–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Kende H. (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Blume B, Grierson D. (1997) Expression of ACC oxidase promoter-GUS fusions in tomato and Nicotiana plumbaginifolia regulated by developmental and environmental stimuli. Plant J 12: 731–746 [DOI] [PubMed] [Google Scholar]
- Busi MV, Bustamante C, D’Angelo C, Hidalgo-Cuevas M, Boggio SB, Valle EM, Zabaleta E. (2003) MADS-box genes expressed during tomato seed and fruit development. Plant Mol Biol 52: 801–815 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Tuori RP, D’Ascenzo MD, Fobert PR, Després C, Martin GB. (2003) The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15: 3033–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Hackett R, Walker D, Taylor A, Lin Z, Grierson D. (2004) Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol 136: 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee JM, McQuinn R, Chung JD, Klein P, Giovannoni J. (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64: 936–947 [DOI] [PubMed] [Google Scholar]
- de Folter S, Shchennikova AV, Franken J, Busscher M, Baskar R, Grossniklaus U, Angenent GC, Immink RGH. (2006) A Bsister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J 47: 934–946 [DOI] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA. (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R, Immink RG, Ferioli V, Bernasconi B, Byzova M, Angenent GC, Kater M, Colombo L. (2002) Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Mol Genet Genomics 268: 152–159 [DOI] [PubMed] [Google Scholar]
- Forth D, Pyke KA. (2006) The suffulta mutation in tomato reveals a novel method of plastid replication during fruit ripening. J Exp Bot 57: 1971–1979 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM. (1994) Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol 105: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fray RG, Grierson D. (1993) Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol Biol 22: 589–602 [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Shima Y, Higuchi N, Nakano T, Koyama Y, Kasumi T, Ito Y. (2012) Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 235: 1107–1122 [DOI] [PubMed] [Google Scholar]
- Gaffe J, Lemercier C, Alcaraz JP, Kuntz M. (2011) Identification of three tomato flower and fruit MADS-box proteins with a putative histone deacetylase binding domain. Gene 471: 19–26 [DOI] [PubMed] [Google Scholar]
- Giménez E, Pineda B, Capel J, Antón MT, Atarés A, Pérez-Martín F, García-Sogo B, Angosto T, Moreno V, Lozano R. (2010) Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS ONE 5: e14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J. (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. (2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Goff SA, Klee HJ. (2006) Plant volatile compounds: sensory cues for health and nutritional value? Science 311: 815–819 [DOI] [PubMed] [Google Scholar]
- Griffiths A, Barry C, Alpuche-Solis AG, Grierson D. (1999) Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. J Exp Bot 50: 793–798 [Google Scholar]
- Hamilton A, Lycett G, Grierson D. (1990) Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature 346: 284–287 [Google Scholar]
- Hileman LC, Sundstrom JF, Litt A, Chen M, Shumba T, Irish VF. (2006) Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol Biol Evol 23: 2245–2258 [DOI] [PubMed] [Google Scholar]
- Hiwasa K, Kinugasa Y, Amano S, Hashimoto A, Nakano R, Inaba A, Kubo Y. (2003) Ethylene is required for both the initiation and progression of softening in pear (Pyrus communis L.) fruit. J Exp Bot 54: 771–779 [DOI] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. (2009) TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T. (2008) DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Kende H. (1993) Ethylene biosynthesis. Annu Rev Plant Biol 44: 283–307 [Google Scholar]
- Kesanakurti D, Kolattukudy PE, Kirti PB. (2012) Fruit-specific overexpression of wound-induced tap1 under E8 promoter in tomato confers resistance to fungal pathogens at ripening stage. Physiol Plant 146: 136–148 [DOI] [PubMed] [Google Scholar]
- Kneissl ML, Deikman J. (1996) The tomato E8 gene influences ethylene biosynthesis in fruit but not in flowers. Plant Physiol 112: 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnyanski SF, Sandhu J, Domier LL, Buetow DE, Korban SS. (2001) Effect of an enhanced CaMV 35S promoter and a fruit-specific promoter on uida gene expression in transgenic tomato plants. In Vitro Cell Dev Biol Plant 37: 427–433 [Google Scholar]
- Lee JM, Joung JG, McQuinn R, Chung MY, Fei Z, Tieman D, Klee H, Giovannoni J. (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Leseberg CH, Eissler CL, Wang X, Johns MA, Duvall MR, Mao L. (2008) Interaction study of MADS-domain proteins in tomato. J Exp Bot 59: 2253–2265 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhu B, Xu W, Zhu H, Chen A, Xie Y, Shao Y, Luo Y. (2007) LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep 26: 1999–2008 [DOI] [PubMed] [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D. (2008) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln JE, Campbell AD, Oetiker J, Rottmann WH, Oeller PW, Shen NF, Theologis A. (1993) LE-ACS4, a fruit ripening and wound-induced 1-aminocyclopropane-1-carboxylate synthase gene of tomato (Lycopersicon esculentum): expression in Escherichia coli, structural characterization, expression characteristics, and phylogenetic analysis. J Biol Chem 268: 19422–19430 [PubMed] [Google Scholar]
- Lincoln JE, Cordes S, Read E, Fischer RL. (1987) Regulation of gene expression by ethylene during Lycopersicon esculentum (tomato) fruit development. Proc Natl Acad Sci USA 84: 2793–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln JE, Fischer RL. (1988a) Diverse mechanisms for the regulation of ethylene-inducible gene expression. Mol Gen Genet 212: 71–75 [DOI] [PubMed] [Google Scholar]
- Lincoln JE, Fischer RL. (1988b) Regulation of gene expression by ethylene in wild-type and rin tomato (Lycopersicon esculentum) fruit. Plant Physiol 88: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ. (2011) The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol 157: 1568–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurchie EJ, McGlasson WB, Eaks IL. (1972) Treatment of fruit with propylene gives information about the biogenesis of ethylene. Nature 237: 235–236 [DOI] [PubMed] [Google Scholar]
- Mizrahi Y, Zohar R, Malis-Arad S. (1982) Effect of sodium chloride on fruit ripening of the nonripening tomato mutants nor and rin. Plant Physiol 69: 497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka A, Murachi S, Okunishi H, Shiomi S, Nakano R, Kubo Y, Inaba A. (1998) Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol 118: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeller PW, Lu MW, Taylor LP, Pike DA, Theologis A. (1991) Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254: 437–439 [DOI] [PubMed] [Google Scholar]
- Oetiker JH, Olson DC, Shiu OY, Yang SF. (1997) Differential induction of seven 1-aminocyclopropane-1-carboxylate synthase genes by elicitor in suspension cultures of tomato (Lycopersicon esculentum). Plant Mol Biol 34: 275–286 [DOI] [PubMed] [Google Scholar]
- Olson DC, White JA, Edelman L, Harkins RN, Kende H. (1991) Differential expression of two genes for 1-aminocyclopropane-1-carboxylate synthase in tomato fruits. Proc Natl Acad Sci USA 88: 5340–5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan IL, McQuinn R, Giovannoni JJ, Irish VF. (2010) Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J Exp Bot 61: 1795–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñarrubia L, Aguilar M, Margossian L, Fischer RL. (1992) An antisense gene stimulates ethylene hormone production during tomato fruit ripening. Plant Cell 4: 681–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Hareven D, Rounsley SD, Yanofsky MF, Lifschitz E. (1994) Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6: 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Wang Y, Cao B, Wang W, Tian S. (2012) Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J 70: 243–255 [DOI] [PubMed] [Google Scholar]
- Sandhu JS, Krasnyanski SF, Domier LL, Korban SS, Osadjan MD, Buetow DE. (2000) Oral immunization of mice with transgenic tomato fruit expressing respiratory syncytial virus-F protein induces a systemic immune response. Transgenic Res 9: 127–135 [DOI] [PubMed] [Google Scholar]
- Sell S, Hehl R. (2005) A fifth member of the tomato 1-aminocyclopropane-1-carboxylic acid (ACC) oxidase gene family harbours a leucine zipper and is anaerobically induced. DNA Seq 16: 80–82 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Manning K, Eriksson EM, Popovich AH, King GJ. (2002) Genetic identification and genomic organization of factors affecting fruit texture. J Exp Bot 53: 2065–2071 [DOI] [PubMed] [Google Scholar]
- Shchennikova AV, Shulga OA, Immink R, Skryabin KG, Angenent GC. (2004) Identification and characterization of four chrysanthemum MADS-box genes, belonging to the APETALA1/FRUITFULL and SEPALLATA3 subfamilies. Plant Physiol 134: 1632–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu OY, Oetiker JH, Yip WK, Yang SF. (1998) The promoter of LE-ACS7, an early flooding-induced 1-aminocyclopropane-1-carboxylate synthase gene of the tomato, is tagged by a Sol3 transposon. Proc Natl Acad Sci USA 95: 10334–10339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigchelaar E, McGlasson W, Buescher R. (1978) Genetic regulation of tomato fruit ripening. HortScience 13: 508–513 [Google Scholar]
- Tigchelaar E, Tomes M, Kerr E, Barman R. (1973) A new fruit ripening mutant, non-ripening (nor). Rep Tomato Genet Coop 23: 33 [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJM, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J, et al (2009) Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ. (1995) An ethylene-inducible component of signal transduction encoded by never-ripe. Science 270: 1807–1809 [DOI] [PubMed] [Google Scholar]
- Yang S, Oetiker J. (1994) The role of ethylene in fruit ripening. ISHS Acta Hortic 398: 167–178 [Google Scholar]
- Yang SF, Hoffman NE. (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 155–189 [Google Scholar]
- Zarembinski TI, Theologis A. (1994) Ethylene biosynthesis and action: a case of conservation. Plant Mol Biol 26: 1579–1597 [DOI] [PubMed] [Google Scholar]