ABA regulates bud outgrowth responses to the ratio of red to far-red light, extending the known hormonal pathways associated with the regulation of branching and shade avoidance.
Abstract
Low red light/far-red light ratio (R:FR) serves as an indicator of impending competition and has been demonstrated to suppress branch development. The regulation of Arabidopsis (Arabidopsis thaliana) rosette bud outgrowth by the R:FR and the associated mechanisms were investigated at several levels. Growth under low R:FR suppressed outgrowth of the third from topmost bud (bud n-2) but not that of the topmost bud. Subsequently increasing the R:FR near the time of anthesis promoted bud n-2 outgrowth and reduced topmost bud growth. Buds from specific rosette positions, exhibiting divergent fates to increased R:FR, were harvested 3 h after modifying the R:FR and were used to conduct ATH1 microarray-based transcriptome profiling. Differentially expressed genes showed enrichment of light signaling and hormone-related Gene Ontology terms and promoter motifs, most notably those associated with abscisic acid (ABA). Genes associated with ABA biosynthesis, including the key biosynthetic gene NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (NCED3), and with ABA signaling were expressed at higher levels in the responsive bud n-2, and increasing the R:FR decreased their expression only in bud n-2. ABA abundance in responsive buds decreased within 12 h of increasing the R:FR, while indole-3-acetic acid levels did not change. A role for ABA in repressing bud outgrowth from lower positions under low R:FR was demonstrated using the nced3-2 and aba2-1 ABA biosynthesis mutants, which showed enhanced branching and a defective bud n-2 outgrowth response to low R:FR. The results provide evidence that ABA regulates bud outgrowth responses to the R:FR and thus extend the known hormonal pathways associated with the regulation of branching and shade avoidance.
The shoot-branching habit generates much of the unique form of the plant and provides the scaffold upon which the shoot system is elaborated. In many species, branching is a plastic trait that is modified by intrinsic and environmental signals to produce a form suitable for the ecological/evolutionary context in which the plant grows. Appropriate branching maximizes the utilization of resources and reproductive success to increase fitness (Juenger and Bergelson, 2000; Lortie and Aarssen, 2000; Bonser and Aarssen, 2003; Weinig et al., 2003). Branching is also an important agricultural trait that has been selected for during the domestication and improvement of many crops (Doebley et al., 1997; Wacker et al., 2002; Doust et al., 2004, 2007; Li et al., 2006; Bachlava et al., 2010). Branching contributes to yield and quality and end use parameters that are of interest to breeders, producers, and ultimately consumers of agricultural products (Zarrough et al., 1983; Peng et al., 1994; García del Moral and García del Moral, 1995; Zhao et al., 2006; Boe and Beck, 2008).
Shoot branches arise through a multistep process beginning with the formation of an axillary meristem in the leaf axil (Bennett and Leyser, 2006). The axillary meristem then generates an axillary bud through expansion of the meristem and the production of a few leaves and/or leaf primordia. The axillary bud may subsequently grow out to form a branch or it may remain dormant or semidormant for an indefinite period of time. In Arabidopsis (Arabidopsis thaliana), the formation of axillary buds and the outgrowth of buds to form branches occur in a basipetal wave coincident with the initiation of flowering under long days (Hempel and Feldman, 1994). While buds at upper positions of the rosette grow out to form branches in many ecotypes, buds at lower positions have less outgrowth potential and often remain arrested.
Branching plasticity appears to manifest at the level of bud outgrowth (Kebrom et al., 2006; Leyser, 2009; Finlayson et al., 2010). Auxin has long been known to contribute to branching, although the mechanistic basis of its function remains somewhat ambiguous (Waldie et al., 2010; Domagalska and Leyser, 2011). Auxin promotes apical dominance through a pathway whereby auxin derived from the main shoot apex inhibits the outgrowth of lower axillary buds, a form of correlative inhibition. The effect of auxin on the bud is indirect, as apically sourced auxin does not enter the bud. Contrasting theories contend that auxin effects on bud outgrowth are mediated by a second messenger(s) or that they result from competition between the main shoot and the axillary bud for auxin export in the polar auxin transport stream, although the two theories are not necessarily exclusive (Bennett et al., 2006; Brewer et al., 2009; Prusinkiewicz et al., 2009). Cytokinins have been considered as candidates for an auxin second messenger, as they have the potential to promote bud outgrowth and cytokinin abundance in or near the bud correlates with bud fate (Emery et al., 1998; Tanaka et al., 2006). Other data provide evidence that a strigolactone-derived hormone associated with the pea (Pisum sativum) RAMOSUS pathway and the Arabidopsis MORE AXILLARY GROWTH pathway could be an auxin second messenger (Brewer et al., 2009). However, there is additional evidence to suggest that this hormone(s), which acts to repress branching, may exert its effects by modulating the capacity of the polar auxin transport stream (Bennett et al., 2006; Prusinkiewicz et al., 2009).
Branching is modulated by several environmental factors, including light. Both photosynthetic photon flux density (PPFD) and the red light/far-red light ratio (R:FR) exert a strong influence on branch development, and interactions between the two parameters are known to occur (Kasperbauer, 1971; Deregibus et al., 1983; Casal et al., 1985, 1986; Davis and Simmons, 1994; Robin et al., 1994; Wan and Sosebee, 1998; Donohue and Schmitt, 1999; Kebrom et al., 2006; Finlayson et al., 2010; Su et al., 2011). Many plants monitor the R:FR as a measure of proximal competition, and they use this information to modify growth and development accordingly (Ballaré, 1999). In a noncompetitive environment where the R:FR is high, responsive plants maintain a moderate balance between main shoot elongation and branch growth. The vegetation of nearby competitors reflects far-red light, resulting in a reduction in the R:FR, and promotes the shade-avoidance syndrome (SAS) in responsive plants like Arabidopsis (Smith, 1995; Franklin and Whitelam, 2005; Casal, 2012). One output of the SAS is repressed branch development, while elongation of the main shoot is promoted. The effects of the R:FR are transduced largely by phytochrome B (phyB) into changes in the expression of a variety of genes associated with hormone biosynthesis and signaling and bud development (Kebrom et al., 2006; Finlayson et al., 2010; Kebrom et al., 2010; Su et al., 2011). Ultimately, the SAS may reduce branch numbers, but in Arabidopsis its effects are most apparent in terms of the relationship between the outgrowth of branches at different positions (Finlayson et al., 2010; Su et al., 2011), an effect attributed to correlative inhibition.
In spite of the considerable advances that have accumulated over many years of study, gaps still exist in our understanding of branching, and especially of the regulation of branching by light. In this study, a transcriptome profiling approach was employed to examine the response of specific axillary buds to R:FR signals that regulate bud outgrowth. It was anticipated that this approach might generate new hypotheses concerning the regulation of bud outgrowth that could subsequently be tested.
RESULTS
The Fate of Buds at Specific Positions Can Be Selectively Controlled by the R:FR
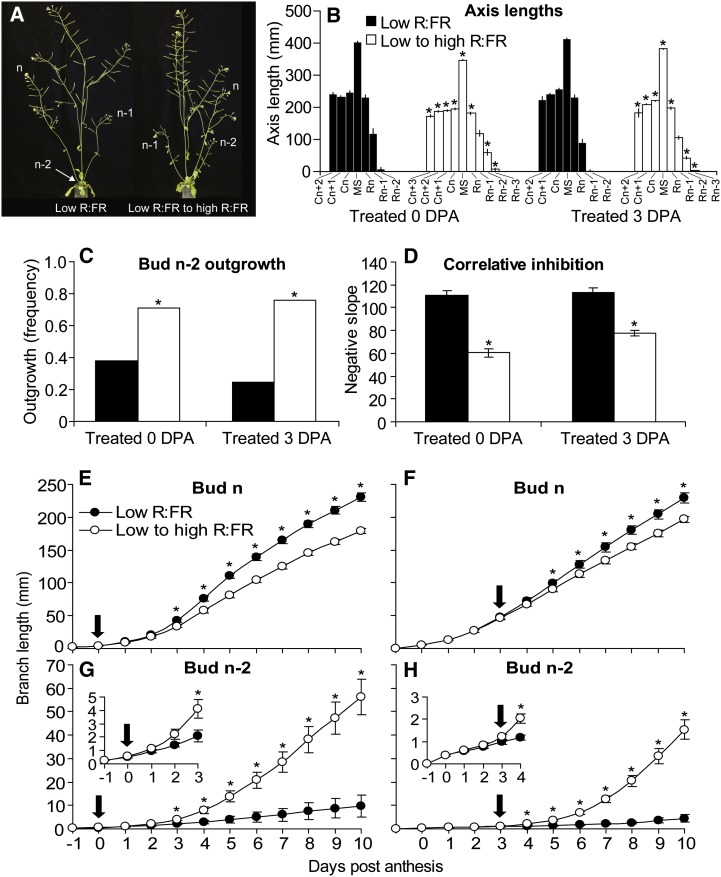
Previous studies demonstrated that the R:FR can have profound effects on the outgrowth of axillary buds in both sorghum (Sorghum bicolor) and Arabidopsis (Kebrom et al., 2006; Finlayson et al., 2010; Su et al., 2011). To investigate this phenomenon in greater detail, the effect of the R:FR on the fate of Arabidopsis buds at different positions in the rosette and the bases for their responses were explored. Preliminary experiments indicated that low the R:FR applied after axillary buds had formed did not rapidly alter bud development. Therefore, an alternative strategy of increasing the R:FR provided to plants previously grown under low R:FR was pursued. This treatment simulates a scenario where gaps in a canopy are created by disturbance (wind, hail, herbivores, etc.) after shade-avoidance phenotypes have been established. Plants were grown under low R:FR from 1 d after sowing and then exposed to high R:FR on 0 DPA to investigate the effect on the topmost bud (bud n). A companion experiment to target the response of the third bud from the top (bud n-2) to the R:FR employed a similar approach, increasing the R:FR at 3 DPA to permit bud n-2 to developmentally progress to the point where it was competent to respond rapidly to the change in the R:FR. Corresponding controls were maintained under low R:FR. Plants grown in this manner showed severe shade-avoidance phenotypes (Fig. 1A). The average number of rosette leaves was 5.2 for each treatment group. There was no effect of altering the R:FR on rosette leaf numbers, since they were already formed at the time of treatment. Increasing the R:FR at either 0 or 3 DPA uniformly suppressed the elongation of the main shoot and cauline branches but differentially affected rosette branch elongation, depending on position (Fig. 1B). The frequency of bud n-2 outgrowth was elevated in plants given high R:FR at both times (Fig. 1C). The lengths of the rosette branches at the top three positions were used to calculate a correlative inhibition index, as described previously (Finlayson et al., 2010; Su et al., 2011). This index integrates the timing of the initiation of bud outgrowth and the elongation rate of branches from the top three sequential rosette positions, providing a quantitative estimate of branching vigor. Increasing the R:FR reduced the correlative inhibition and thus increased branching strength (Fig. 1D).
Figure 1.
A, Phenotypes of wild-type Col-0 grown under low R:FR and then either maintained under low R:FR or provided with high R:FR at 0 DPA, assessed at 10 DPA. B to D, Axis lengths (B), frequency of bud n-2 outgrowth (C), and correlative inhibition index (D) of wild-type Col-0 grown as above, assessed at 10 DPA. Cn, Cauline branch; MS, main shoot; Rn, rosette branch. E to H, Elongation versus time of bud n (E and F) and bud n-2 (G and H) primary rosette buds of wild-type Col-0 grown under low R:FR and then either maintained under low R:FR or provided with high R:FR at 0 DPA (E and G) or 3 DPA (F and H). Anthesis occurred on day 0, and arrows indicate the time of R:FR increase. Asterisks indicate significant differences between R:FR treatments at α = 0.05. Data are means ± se of three to five independent experiments; total n = 45 to 75. [See online article for color version of this figure.]
Bud n was slightly larger at the start of outgrowth compared with bud n-2 (Supplemental Fig. S1, A and B). In both cases, small leaves and flowers were apparent. Daily bud length measurements revealed contrasting responses of bud n and bud n-2 to increased R:FR (Fig. 1, E–H). Bud n began to grow near anthesis regardless of the R:FR, and its elongation was gradually reduced by increasing the R:FR (Fig. 1, E and F). Conversely, bud n-2 was almost completely arrested under low R:FR. Bud n-2 elongation responded within 24 h of increasing the R:FR at 3 DPA (Fig. 1H) or within 72 h when the R:FR was increased at 0 DPA (Fig. 1G).
In summary, high R:FR could be used to rapidly and robustly promote the outgrowth of bud n-2 of plants previously grown under low R:FR, especially when the treatment was applied at 3 DPA.
Analysis of Axillary Bud Genes Showing Differential Expression Responses to the R:FR
The preceding experimental protocol was used as a basis to explore the rapid changes in the transcriptome of unelongated bud n and bud n-2 in response to changes in the R:FR using Affymetrix ATH1 genome arrays. Arrays were hybridized with probe generated from buds harvested 3 h after increasing the R:FR on 0 DPA (bud n) and 3 DPA (bud n-2). Buds at these stages were poised to begin elongation (Fig. 1, E and H). The analysis design and general outputs are presented in Supplemental Figure S1. Statistical analysis produced four major groups of differentially regulated genes. Group 1 was specific for R:FR effects (386 features), group 2 was specific for bud position effects (6,162 features), group 3 was specific for the combined effects of R:FR and bud position (1,208 features), and group 4 was specific for any interaction effects (2,048 features). Together, these groups contained a total of 9,804 nonredundant, differentially expressed features (Supplemental Fig. S1). A pairwise Student’s t test for the effect of R:FR was then applied to this set, and genes that showed significant effects of the R:FR in bud n-2 were retained to investigate the processes involved in the regulation of bud outgrowth by the R:FR. Genes were finally clustered into two groups: those showing lower expression in response to elevated R:FR in bud n-2 and those showing higher expression (Supplemental Fig. S1; Supplemental Table S1).
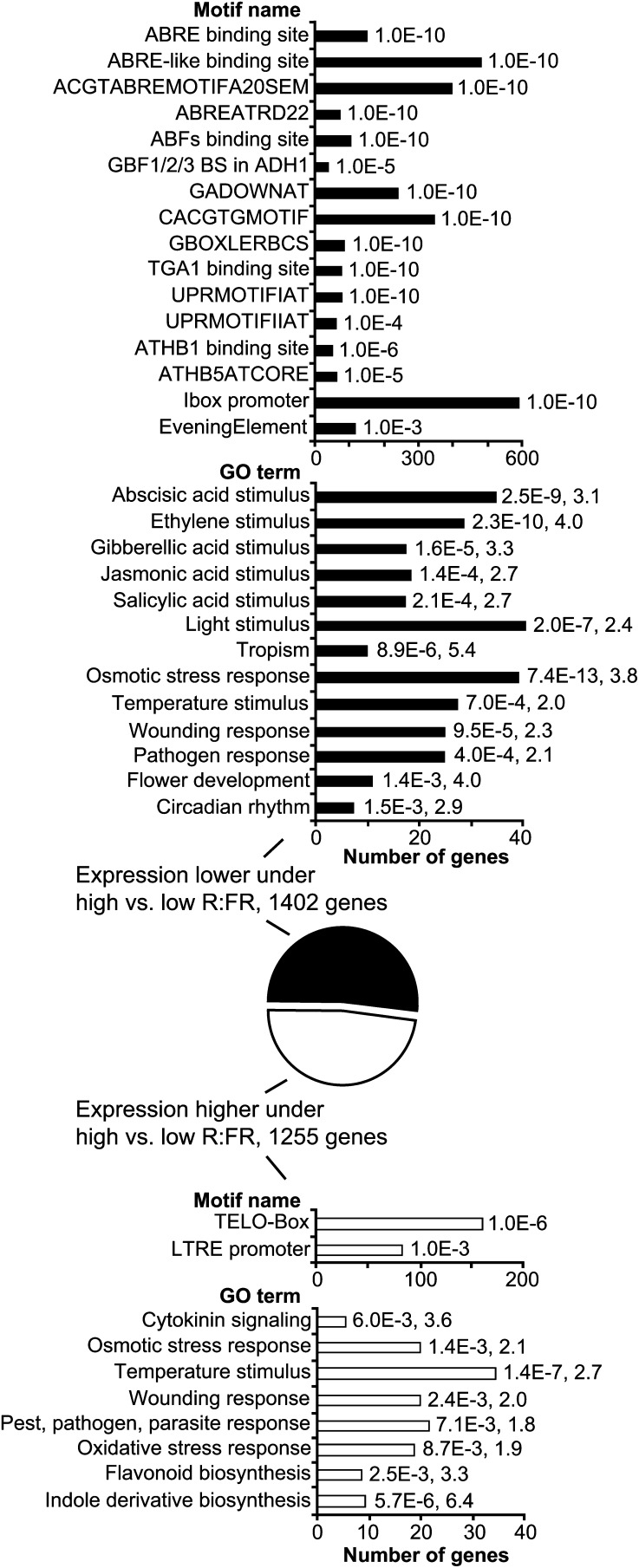
A total of 1,402 genes showed lower expression in response to elevated R:FR in bud n-2 (Fig. 2). Cluster analysis discovered overrepresented Gene Ontology (GO) terms and promoter motifs in this set. Many of the promoter motifs identified were based on variations of the G-box, although a few non-G-box motifs were also found. G-box motifs included those associated with both light and abscisic acid (ABA) signaling. GO terms associated with a variety of hormones, including ABA, were overrepresented, as were likely outputs of ABA signaling, such as osmotic stress response. Light signaling GO terms were also overrepresented. The converse comparison of genes that showed increased expression in response to elevated R:FR in bud n-2 produced a total of 1,255 genes (Fig. 2). Only two promoter motifs were identified in this set. GO terms associated with osmotic stress, temperature stimulus, and wounding overlapped with those from the previous set. Cytokinin signaling was the only hormone GO term discovered in the set of genes with increased expression following exposure to high R:FR.
Figure 2.
Summary of motif and GO analyses of clusters containing genes with lower or higher expression in bud n-2 under high versus low R:FR. Numbers associated with each bar represent P values and enrichment values (where applicable).
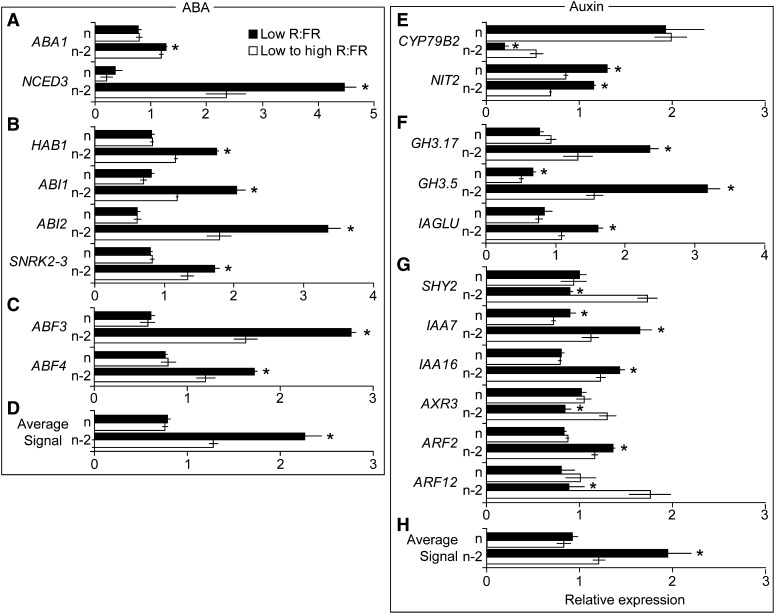
Genes involved in ABA biosynthesis, signal transduction, and transcriptional regulation showed expression patterns that varied by bud position and the R:FR. ABA biosynthetic gene expression was higher in bud n-2 versus bud n, with the key NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (NCED3) gene showing the greatest differential (Fig. 3A). Increasing the R:FR resulted in a sharp decline in NCED3 mRNA abundance in bud n-2, suggesting that ABA levels might also decline. This possibility was supported by similar expression patterns apparent in several ABA signal transduction genes (Fig. 3B) and ABA-responsive transcription factor genes (Fig. 3C). Based on averaged ABA-responsive gene expression (Fig. 3D), ABA signaling was elevated in bud n-2 versus bud n and decreased in bud n-2 exposed to high R:FR. The expression responses of a selection of the genes used to derive the average ABA signal response are presented in Supplemental Figure S2. The majority of genes identified as positively regulated by ABA showed decreased expression in bud n-2 provided with high R:FR, and expression was generally lower in bud n compared with bud n-2. A very small proportion of these genes showed significant expression differences in response to the R:FR in bud n, indicating that the effects of the R:FR on ABA responses were specific for bud n-2.
Figure 3.
Selected gene expression profiles in bud n and bud n-2 of wild-type Col-0 grown under low R:FR and then either maintained under low R:FR or provided with high R:FR for 3 h at 0 DPA (bud n) or 3 DPA (bud n-2). A to D, ABA biosynthesis (A), core signal transduction (B), signal transduction transcription factor (C), and average signal of ABA-responsive (D) genes. E to H, IAA biosynthesis (E), conjugation (F), signal transduction transcription factor (G), and average signal of IAA-responsive (H) genes. Data are means ± se with n = 3. Asterisks indicate significant differences between light treatments, within bud position, at α < 0.05.
The expression of two genes involved in indole-3-acetic acid (IAA) biosynthesis showed opposing responses to the R:FR in bud n-2 (Fig. 3E), while the expression of IAA-conjugating genes was suppressed by high R:FR (Fig. 3F). The expression responses of the individual IAA-responsive transcription factors to the R:FR varied considerably (Fig. 3G) and, therefore, did not provide support for auxin signaling status, but the average expression of auxin-responsive genes (Fig. 3H) was reduced in bud n-2 under high R:FR, indicating that auxin signaling might be repressed. Trends in IAA-related gene expression between bud positions were not as clear as those for ABA, but the average expression of auxin-responsive genes was generally elevated in bud n-2 compared with bud n. The expression patterns of a selection of the genes used to derive the average IAA signal response are presented in Supplemental Figure S3. Most of the genes identified as positively (or negatively) regulated by IAA showed corresponding decreased (or increased) expression in bud n-2 provided with high R:FR.
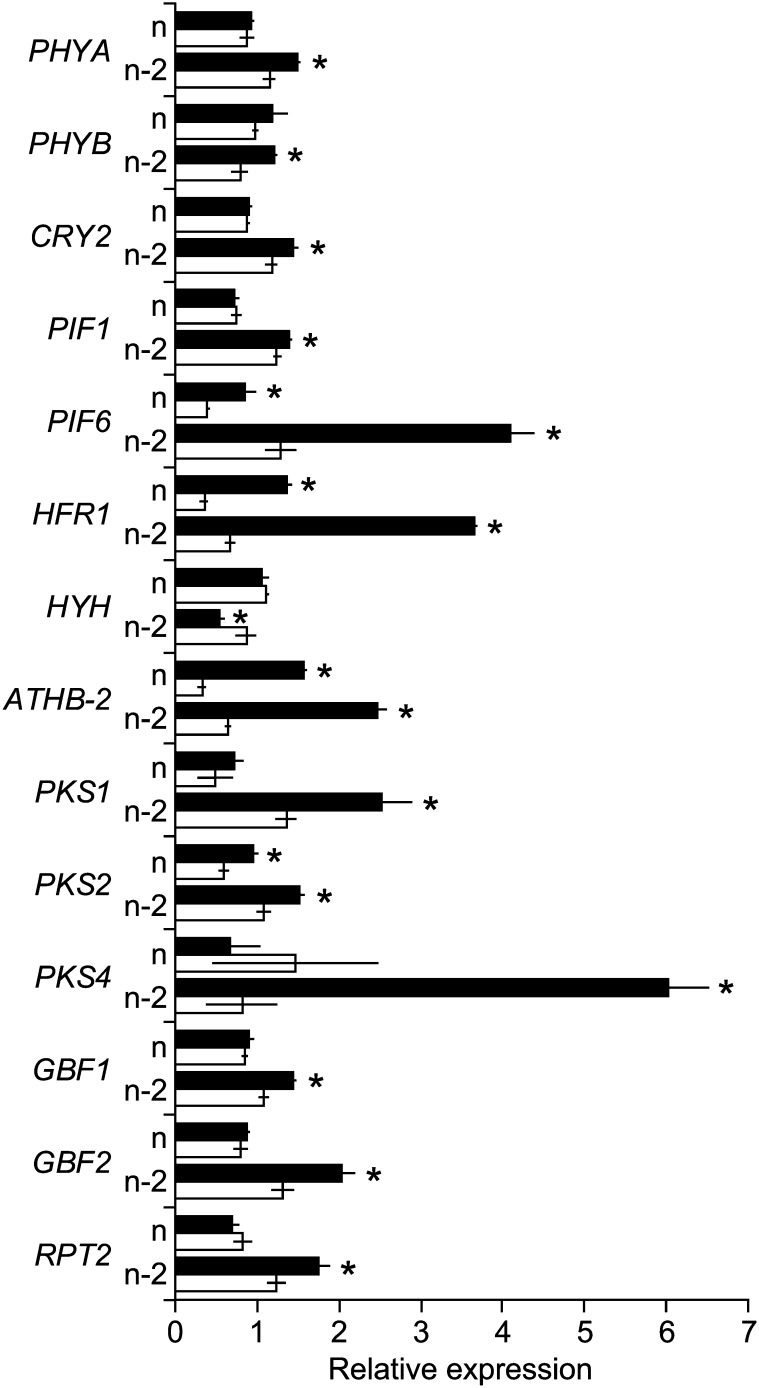
The expression of several light signaling genes responded to the R:FR in bud n-2 (Fig. 4). High R:FR reduced the expression of ARABIDOPSIS THALIANA HOMEOBOX PROTEIN2 (Carabelli et al., 1993) and LONG HYPOCOTYL IN FAR-RED (Sessa et al., 2005), which respectively promote (Steindler et al., 1999) and inhibit (Lorrain et al., 2009) shade-avoidance responses. High R:FR enhanced the expression of HY5-HOMOLOG (HYH) and reduced the expression of PHYTOCHROME KINASE SUBSTRATE4, and these changes are important for the termination of shade signaling by sunflecks with high R:FR (Sellaro et al., 2011). Low R:FR enhanced the expression of several G-box binding factor genes, including G-BOX BINDING FACTOR1 (GBF1), and GBF1 interacts physically with HYH, antagonizing its function (Singh et al., 2012). Low R:FR also enhanced the expression of PHYTOCHROME-INTERACTING FACTOR1 (PIF1) and PIF6 (Leivar and Quail, 2011) and of the photoreceptor genes PHYA, PHYB (Devlin et al., 2003), and CRYPTOCHROME 2. In all these cases, the level of expression and/or the response to the R:FR was stronger in bud n-2 than in bud n.
Figure 4.
Expression profiles of light signaling-related genes in bud n and bud n-2 of wild-type Col-0 grown under low R:FR and then either maintained under low R:FR or provided with high R:FR for 3 h at 0 DPA (bud n) or 3 DPA (bud n-2). Data are means ± se with n = 3. Asterisks indicate significant differences between light treatments, within bud position, at α < 0.05.
Characterization of ABA and IAA Abundances in Axillary Buds in Response to the R:FR
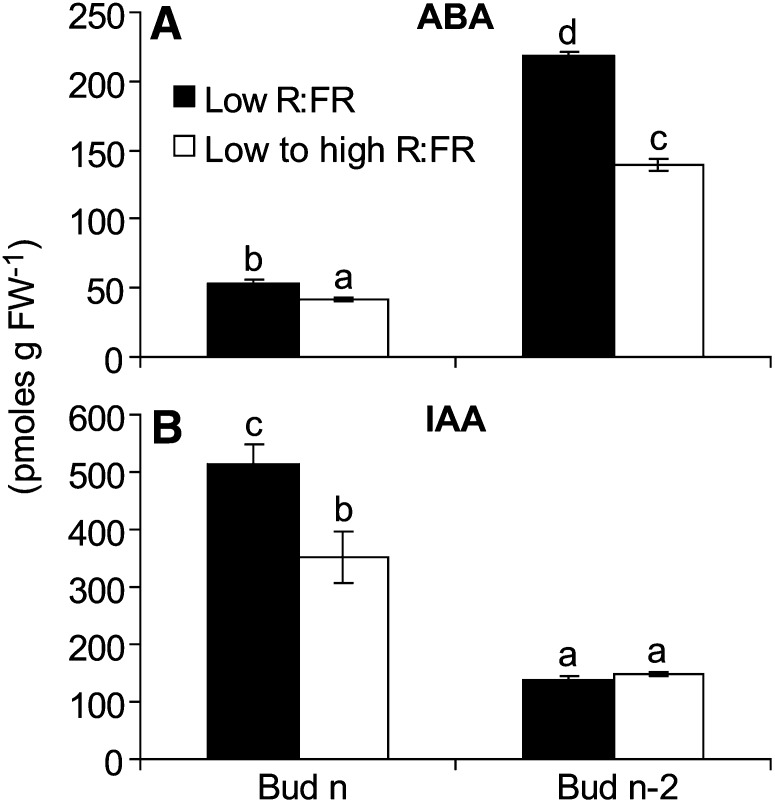
The microarray analysis provided evidence that a variety of hormones were associated with bud outgrowth responses to position and light signals. Since the transcriptome data implicating ABA were very strong, its abundance was assessed in buds from plants given treatments identical to those used for the microarray experiments. The abundance of IAA was also measured in these buds as a benchmark, since several previous studies have shown that IAA levels either increase (Hillman et al., 1977; Gocal et al., 1991; Galoch et al., 1998; Balla et al., 2002) or decrease (Emery et al., 1998; Mader et al., 2003) in buds during the transition to outgrowth. The level of ABA in bud n-2 declined within 12 h following the increase in the R:FR (Fig. 5A). The ABA level in bud n was much lower than in bud n-2 and also decreased with high R:FR, but the magnitude of the bud n response was small. IAA abundance decreased in bud n in response to increasing the R:FR (Fig. 5B). IAA levels were lower in bud n-2 than in bud n but were not altered by the R:FR.
Figure 5.
Abundances of ABA (A) and IAA (B) in bud n and bud n-2 of wild-type Col-0 grown under low R:FR and then either maintained under low R:FR or provided with high R:FR for 12 h at 0 DPA (bud n) or 3 DPA (bud n-2). Bars with different letters are significantly different at α < 0.05. Data are means ± se with n = 3 to 4. FW, Fresh weight.
Genetic Test of the Roles of the ABA Biosynthetic Genes NCED3 and ABA DEFICIENT2 (ABA2) in Regulating Branching Responses to the R:FR
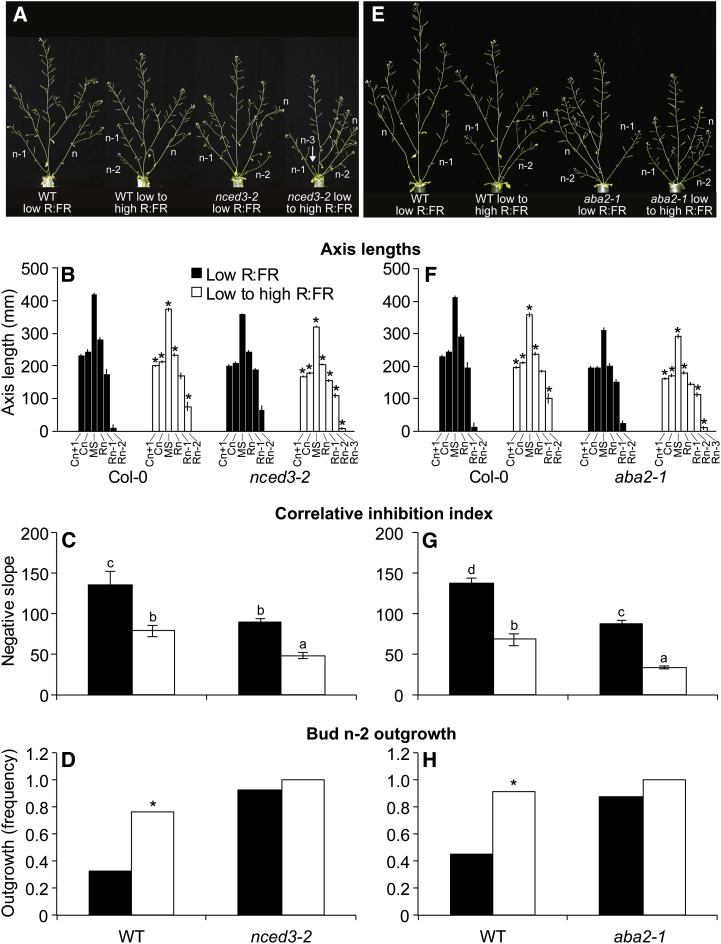
In conjunction with the microarray analysis, the survey of ABA abundance provided additional evidence that ABA participates in bud outgrowth responses to the R:FR. The role of ABA, therefore, was tested by measuring key architectural parameters of the ABA biosynthesis mutants nced3-2 and aba2-1 grown using the same conditions employed for transcriptome analysis of bud n-2 (low R:FR from 1 d after sowing, then continued low R:FR, or provided with high R:FR at 3 DPA). The growth of both mutants under these conditions was less robust than that of the wild type, with aba2-1 especially showing substantial pleiotropy, including a diminutive rosette with very small leaves. The plant height response of these lines to increased R:FR was similar to that of the wild type, demonstrating that neither gene is required for R:FR responsiveness in general (Fig. 6, A, B, E, and F). While increasing the R:FR reduced the correlative inhibition index of all three genotypes, the values of this parameter were lower in nced3-2 and aba2-1 compared with the wild type under both light regimens (Fig. 6, C and G). The frequency of wild-type bud n-2 outgrowth increased with increased R:FR (Fig. 6, D and H). Although the frequency of bud n-2 outgrowth was elevated in nced3-2 and aba2-1 compared with the wild type under low R:FR, increasing the R:FR did not significantly increase the value of this parameter (Fig. 6, D and H). Rosette leaf numbers were similar (ranging from 4.9 to 5.4), as were cauline leaf numbers (ranging from 1.9 to 2.0), and total leaf numbers varied by less than 0.5 leaves. Thus, ABA restricts bud outgrowth from bud n-2 and lower in the rosette. Buds from lower positions of nced3-2 (positions n-2 and n-3) contained only about half as much ABA as the wild type (Supplemental Fig. S4A) but had similar levels of IAA (Supplemental Fig. S4B). Bud n and bud n-2 of aba2-1 had less than 25% of the ABA as the wild type (Supplemental Fig. S5). These results show that the inhibition of lower position bud elongation induced by low R:FR is attenuated in nced3-2 and aba2-1 compared with the wild type, an effect that can be attributed to the deficiency of ABA.
Figure 6.
A, B, E, and F, Phenotypes (A and E) and axis lengths (B and F) of wild-type Col-0 and nced3-2 (A and B) and aba2-1 (E and F) grown under low R:FR and then either maintained under low R:FR or provided with high R:FR at 3 DPA, measured at 10 DPA. Cn, Cauline branch; MS, main shoot; Rn, rosette branch; WT, wild type. C, D, G, and H, Correlative inhibition index (C and G) and frequency of bud n-2 outgrowth (D and H) of wild-type Col-0 and nced3-2 (C and D) and aba2-1 (G and H) grown as above, assessed at 10 DPA. Bars with different letters are significantly different at α < 0.05. Asterisks indicate significant differences between R:FR treatments at α < 0.05. Data are means ± se with n = 30 (nced3-2) or 24 (aba2-1). [See online article for color version of this figure.]
DISCUSSION
ABA Is a Regulator of Branching and Bud Responses to the R:FR
Drawing from the results of the gene expression analysis that demonstrated that enrichment of ABA regulated promoter motifs and GO terms, the hypothesis that ABA was a key component of the repression of bud outgrowth by low R:FR was generated and tested. ABA levels were high in bud n-2 under dormancy-promoting low R:FR and were found to rapidly decrease when bud n-2 was promoted to outgrow by high R:FR. A role for ABA in the process was confirmed by the phenotypes of the nced3-2 and aba2-1 ABA biosynthesis mutants, which showed enhanced branching and defective bud outgrowth responses to increased R:FR. ABA specifically modulates branching responses to the R:FR by restricting the outgrowth of buds from lower positions in the rosette. The correlative inhibition index integrates the timing of bud outgrowth and branch elongation rates; while the correlative inhibition index of the ABA biosynthesis mutants was lower than that of the wild type under both low and high R:FR, the index decreased with exposure to high R:FR in all three genotypes, suggesting that NCED3 and ABA2 are not essential for this response. Therefore, the R:FR modulation of the outgrowth of lower buds and correlative inhibition appear to be controlled, at least in part, by independent mechanisms.
Even at high R:FR, wild-type ABA levels were lower in the topmost rosette buds, which always form branches, than in the lower buds. Also, even at high R:FR, ABA-deficient plants showed enhanced bud n-2 outgrowth, reduced topmost branch elongation, and a reduced correlative inhibition index compared with the wild type. Thus, ABA is important in regulating branch development in response to the R:FR, but it may also contribute to position-specific bud outgrowth and branch elongation patterns.
The possibility that ABA may regulate branching has been considered many times previously. ABA levels in dormant axillary buds of a variety of species have been observed to decrease in response to decapitation of the main shoot (Knox and Wareing, 1984; Gocal et al., 1991; Mader et al., 2003) and in response to fruit removal, a treatment that also promoted bud outgrowth (Tamas et al., 1979). Additionally, early work investigating the regulation of branching by the R:FR in Xanthium strumarium and tomato (Solanum lycopersicum) suggested that low R:FR promoted ABA accumulation while high R:FR reduced ABA levels and also permitted bud outgrowth (Tucker and Mansfield, 1972; Tucker, 1977). Although elevated ABA levels have often been shown to be associated with bud dormancy, there is little evidence demonstrating a direct role of endogenous ABA in regulating bud outgrowth in intact plants. However, several studies using exogenous ABA (Arney and Mitchell, 1969; Chatfield et al., 2000; Cline and Oh, 2006), the carotenoid (and ABA) biosynthesis inhibitor fluridone (Le Bris et al., 1999), or excised nodes of transgenic ABA-insensitive plants (Arend et al., 2009) have implied a role for ABA in restricting bud outgrowth in pea, Arabidopsis, Ipomoea spp., tomato, rose (Rosa spp.), and poplar (Populus spp.). A recent study also identified increased ABA-related gene expression in bud-containing tissues of Arabidopsis exposed to low R:FR, which was associated with a general reduction in branch numbers (González-Grandío et al., 2013). However, ABA levels were not assessed, and a functional test using a sextuple ABA receptor mutant did not indicate a role for ABA in the regulation of branching. Our results here support these earlier studies but extend them with genomic, biochemical, and genetic evidence that ABA acts as a regulator of branching and branching responses to the R:FR.
How does ABA integrate into the known branching regulatory apparatus? Several possibilities exist: ABA may act downstream of auxin, possibly as a second messenger; ABA may act downstream of strigolactones, possibly as a second messenger; ABA may act independently of IAA and strigolactones. A recent study found that ABA levels correlate positively with strigolactone levels, and it was postulated that ABA may regulate strigolactone production (López-Ráez et al., 2010). While this association should not be ignored, it is noteworthy that our study shows that various aspects of ABA signaling and output were dramatically altered in buds induced to outgrow; therefore, while ABA may alter strigolactone levels, it is likely to have significant impacts on bud physiology independent of strigolactones. Grossmann and Hansen (2001) proposed that IAA effects on branching are evoked by IAA-induced ethylene, which in turn induces ABA and suppresses bud outgrowth. While this is an intriguing hypothesis, and it might be satisfying to develop a model that integrates all known branching components into a single pathway, the fact that this has not occurred in spite of intensive research over many decades may indicate that linearity is not a feature of this process. It is possible that bud outgrowth is regulated incrementally by multiple pathways and through interactions between pathways.
This study demonstrates that ABA regulates branching responses to the R:FR but does not clearly define where ABA is acting. Given that ABA levels in bud n-2 decreased rapidly in response to increased R:FR and that ABA signaling in this bud also decreased, it is reasonable to hypothesize that ABA influences bud outgrowth by acting locally in the bud. When bud n-2 was provided with high R:FR, neither ABA abundance nor the expression of most of the genes associated with ABA declined to levels equivalent to those observed in bud n under either light treatment. This may suggest that ABA and related gene expression must reach a minimum threshold to permit bud outgrowth. The effect of ABA on the process could also be influenced by other outgrowth-regulating pathways, as described above. While the data are consistent with a bud-localized role for ABA, it is also possible that ABA exerts its effects in a systemic manner. Further experimentation is required to clarify these potential modes of action.
The most direct effectors of bud outgrowth appear to be transcription factors, like maize (Zea mays) TEOSINTE BRANCHED1 and GRASSY TILLERS1 and Arabidopsis BRANCHED1 (BRC1), that function in a bud-autonomous manner as repressors (Hubbard et al., 2002; Aguilar-Martínez et al., 2007; Finlayson, 2007; Whipple et al., 2011). These transcription factors may act as integrators of other branching signals, such as those transduced by various hormones (Aguilar-Martínez et al., 2007; Finlayson, 2007). Arabidopsis BRC1 was previously shown to be necessary for branching responses to the R:FR and phyB deficiency (Finlayson et al., 2010). These findings were confirmed in a subsequent study that suggested that BRC1 is necessary for the maintenance of ABA-related responses in buds (González-Grandío et al., 2013). Defining the relationship between ABA signaling and branching integrator function is an obvious direction for future research.
Shade Avoidance and ABA
An association between shade avoidance and ABA physiology has been documented in a variety of systems in addition to those related to buds described above, but a functional link has not previously been established. ABA levels in sunflower (Helianthus annuus) and tomato leaves were elevated under low R:FR (Kurepin et al., 2007; Cagnola et al., 2012), while they decreased in mesocotyls of maize given an end-of-day far-red light treatment, although the signal was not transduced through phyB (Dubois et al., 2010). phyB loss of function also resulted in increased ABA abundance in mature Arabidopsis leaves but reduced ABA sensitivity (González et al., 2012). This phenomenon was linked to reduced water deficit stress tolerance in phyB-deficient plants but was not associated with shade avoidance. Conversely, ABA was not found to play a role in Rumex palustris petiole elongation responses to the R:FR (Pierik et al., 2011), and previous transcriptome profiling studies exploring shade avoidance in Arabidopsis rosette leaves and entire young seedlings have not concluded that ABA-related gene expression was altered (Sessa et al., 2005; Carabelli et al., 2007; Tao et al., 2008; Kozuka et al., 2010; Leivar et al., 2012), suggesting that the tissues employed may respond in a different manner that does not involve ABA. Therefore, while it seems clear that ABA levels and signal output may be responsive to the R:FR, it is likely that the roles of specific hormones in shade avoidance are organ/tissue specific.
Light Signaling Pathways in Unelongated Buds Are Responsive to the R:FR
Different organs show specific patterns of transcriptome responses to the R:FR. In tomato, the stem shows stronger responses of flavonoid, cell wall carbohydrate, and photosynthesis (dark reactions) genes than the leaves (Cagnola et al., 2012). In our experiments with Arabidopsis, bud n-2 showed stronger responses of ABA synthesis and signaling genes than bud n. It is noteworthy that several light signaling genes also showed larger responses to the R:FR in bud n-2 than in bud n. These genes included those encoding photoreceptors (PHYA, PHYB, CRYPTOCHROME 2) and downstream players (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN2, HYH, PHYTOCHROME KINASE SUBSTRATE4, GBF1). The latter suggests that similar light signals (high or low R:FR) reaching both buds would find a bud-specific status of the light perception signaling cascade, which would in turn cause the bud-specific hormonal responses. It is tempting to speculate that a PIF or GBF1 could bind the canonical GBOX motif (GBOXLERBCS) present at −77 to −85 in the NCED3 promoter in a bud context-specific manner. Alternatively, the lower intrinsic expression levels of both light signaling and ABA-related genes in bud n under low R:FR might limit a further decrease when the R:FR increases.
CONCLUSION
The integrated study of bud responses to the R:FR and position cues demonstrates that buds from different positions in the rosette of Arabidopsis retain individual physiological and molecular characteristics; thus, they are not equivalent. This uniqueness should be considered when conducting experiments on branch development. Direct comparisons between, and inferences about, the development of buds/branches from dissimilar positions may not be wholly justified.
Increasing the R:FR from low to high resulted in profound changes in the bud transcriptome, with a substantial number of genes showing interaction effects of bud position and R:FR. Since bud n and bud n-2 exhibit contrasting responses to the R:FR in terms of outgrowth, it is likely that some of the genes showing interaction effects contribute to the divergence in growth.
The definition of ABA as a regulator of bud outgrowth adds further complexity to the branching milieu and raises the question of how ABA integrates with other known branching regulators. ABA abundances were rapidly responsive to growth-promoting high R:FR in bud n-2, while IAA levels were not. Therefore, it is tempting to speculate that ABA is acting independently of auxin in this system. However, it is also possible that auxin in the polar auxin transport stream indirectly influences the accumulation of ABA in the buds prior to changes in bud IAA levels. A variety of ABA biosynthesis genes were expressed in the buds, and the expression of some was responsive to the R:FR. These data suggest that the changes in ABA levels in buds were due to local biosynthesis/metabolism; however, it is also possible that ABA is transported into, or out of, the bud, perhaps via the action of recently discovered ABA transporters (Kang et al., 2010; Kuromori et al., 2010).
Bud outgrowth was very sensitive to the R:FR in plants exhibiting severe shade-avoidance phenotypes resulting from long-duration growth under low R:FR. Increasing the R:FR permitted the outgrowth of previously arrested buds. In the natural environment, this response may allow the plant to take advantage of serendipitous openings in the canopy by promoting late branching and increasing the amount of seed produced.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Columbia ecotype (Col-0) of Arabidopsis (Arabidopsis thaliana) was used throughout. Wild-type Col-60000 seed was obtained from the Arabidopsis Biological Resource Center (Ohio State University). aba2-1 and nced3-2 have been described previously (Léon-Kloosterziel et al., 1996; Urano et al., 2009).
To investigate the effect of exposing plants grown under low R:FR to high R:FR at either 0 or 3 DPA, seeds were stratified for 3 d at 4°C and sown in 50-mL plastic conical tubes (cut to the 25-mL mark) filled with Metro-Mix 200 potting mixture. Plants were grown under 18-h-light/6-h-dark photoperiods with 24°C/18°C day/night temperatures in a growth chamber providing 180 μmol m−2 s−1 PPFD and were fertilized weekly with 1 mL of 1× Hoagland solution. The low R:FR treatment (R:FR = 0.08) was initiated 1 d after sowing and was continued until 10 DPA or was discontinued on the day of anthesis or 3 DPA by turning off far-red diodes to increase the R:FR to 3.5 without altering the PPFD. Both low- and high-R:FR conditions were maintained in the same growth chamber using a barrier in the middle to prevent light from one side of the chamber reaching the other. Light was provided using a mixture of fluorescent (F48T12/CW/VHO; Philips Lighting) and compact fluorescent (CF30EL/TWIST; Osram Sylvania Products) lamps with an overhead array of 735-nm light-emitting diodes (L735-01AU; Epitex) mounted in a clear acrylic sheet to provide supplemental far-red light. Light was measured with a Li-1800 spectroradiometer (Licor Biosciences). The R:FR was calculated as the quantum flux density from 655 to 665 nm divided by the quantum flux density from 725 to 735 nm. The spectra of the light sources are provided in Supplemental Figure S6.
Architectural and Branch Elongation Analyses
Architectural characteristics and branch elongation were measured at 10 DPA as described by Finlayson et al. (2010) except that the correlative inhibition index was calculated for each record individually.
Transcriptome Analysis
Unelongated axillary buds less than 2.5 mm at the topmost (bud n) and third from topmost (bud n-2) rosette leaf positions of plants grown under both continuous low R:FR and low to high R:FR treatments were harvested at 3 h after altering the R:FR. RNA was extracted using Trizol. Transcriptome analysis was performed by the Nottingham Arabidopsis Seed Center using Affymetrix ATH1 genome chips. For each R:FR treatment/bud position combination, three biological replicates, each composed of RNA extracted from a pool of approximately 15 buds, were used to conduct the microarray analysis. The microarray data are available through the Gene Expression Omnibus (GSE42415) and NASCarrays (NASCARRAYS-561).
Microarray data analysis was performed using GeneSpring GX software version 11.0 and other tools. CEL files corresponded to three biological replicates for each of the four bud position (bud n and bud n-2) and light treatment (low R:FR and high R:FR) combinations: bud n low R:FR, bud n low to high R:FR, bud n-2 low R:FR, and bud n-2 low to high R:FR. The MAS5 summarization algorithm was used to normalize the data based on the median values, and the data were filtered to retain only features that were flagged “present” in all three replicates of at least one R:FR treatment/bud position combination, thus excluding features that were not reliably detected. This resulting set was subjected to a two-way factorial ANOVA to identify differentially expressed features (P < 0.05), and false positives were subsequently controlled using q to apply the Benjamini-Hochberg method to estimate the false discovery rate, with q < 0.05. The statistics (ANOVA and false discovery rate) produced three major categories of features showing significant expression responses: main effect of the R:FR (2,483), main effect of bud position (9,011), and interaction effect of the R:FR and bud position (2,048). The statistical grouping resulted in features occurring in overlapping categories. Venn diagrams were used to sort the features into groups that were specific for R:FR effects (386), bud position effects (6,162), combined effects of the R:FR and bud position (1,208), and interaction effects (2,048). These genes were further filtered by a paired Student’s t test (P < 0.05) for significant R:FR effects in bud n-2. The resulting output was clustered by the direction of the bud n-2 expression response to the R:FR. Supplemental Figure S1 provides an overview of the data analysis strategy and general outputs. Gene clusters are provided in Supplemental Table S1.
Hormone-responsive genes for the quantification of ABA and IAA signal outputs were identified from the stringent set described by Goda et al. (2008). Expression values for genes exhibiting repressed expression in response to the hormone were inverted by using the negative of the log value to calculate the fold change before averaging.
Promoter Motif and GO Analysis
Promoter motif analysis was performed using the Athena Web site (www.bioinformatics2.wsu.edu/cgi-bin/Athena/cgi/home.pl), and GO analysis was performed using the GO enrichment tool on the AtCOECiS Web site (http://bioinformatics.psb.ugent.be/ATCOECIS/; Vandepoele et al., 2009).
Analysis of Hormone Abundance
Phytohormones in unelongated buds were quantified using isotope dilution selected ion monitoring gas chromatography-mass spectrometry. Bud n was harvested following light treatment on the day of anthesis into a 1.7-mL microfuge tube chilled in liquid N2. Bud n-2 was harvested in the same manner, but following treatment at 3 DPA. Bud mass was estimated after transfer to new frostless chilled tubes. Approximately 30 buds were pooled per sample replicate. A mixture of stable isotope-labeled [2H6]ABA (0.5 ng) and [13C6]IAA (1 ng) was added to each sample replicate. The buds were extracted twice with 500 μL of methanol warmed to 55°C and then once with 500 μL of 80% ethanol warmed to 55°C, centrifuging and pooling the cleared supernatants after each extraction. The pooled extracts were dried, and the residue was resuspended in 800 μL of chloroform and partitioned against 1 mL of water adjusted to pH 9.0 with NH4OH. The aqueous fraction was recovered, adjusted to pH 5.0 with acetic acid, and partitioned against 1 mL of ethyl acetate. The organic fraction was dried and then methylated with ethereal diazomethane. Samples were analyzed on an Agilent 7890A/5975C XL gas chromatograph-mass spectrometer equipped with a 0.25-mm × 30-m DB-5MS column (0.25-m film) using pulsed splitless injection (7693A). Helium was used as the carrier gas at 0.75 mL min−1. The inlet was maintained at 250°C, and the oven was ramped from 45°C (2.25-min initial hold) to 250°C at 40°C min−1, held at 250°C for 3 min, and then ramped to 290°C at 40°C min−1. The ion source temperature was maintained at 230°C, and the quadrupole was heated to 150°C. The ion source was operated in electron-impact mode, and both scan and selected ion data were acquired. Two sets of ions were monitored for each hormone, and the larger fragment was used for quantification (ABA, 162, 166, 190, and 194 mass-to-charge ratio; IAA, 130, 136, 189, and 195 mass-to-charge ratio).
Statistics
Statistics associated with the microarray analysis are described above. For other data, comparisons between means were made using ANOVA followed by Tukey’s test or using a two-tailed Student’s t test with α < 0.05. Comparisons between frequencies were made using Fisher’s exact probability test with α < 0.05.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Bud phenotypes and microarray analysis overview.
Supplemental Figure S2. Expression of ABA-responsive genes.
Supplemental Figure S3. Expression of IAA-responsive genes.
Supplemental Figure S4. ABA and IAA abundance in buds of WT Col-0 and nced3-2.
Supplemental Figure S5. ABA abundance in buds of WT Col-0 and aba2-1.
Supplemental Figure S6. Spectra of light sources used in the experiments.
Supplemental Table S1. List of genes showing expression changes in response to the R:FR in bud n-2.
Glossary
- PPFD
photosynthetic photon flux density
- R:FR
red light:far-red light ratio
- SAS
shade-avoidance syndrome
- bud n
topmost bud
- bud n-2
third bud from the top
- GO
Gene Ontology
- ABA
abscisic acid
- IAA
indole-3-acetic acid
- Col-0
Columbia ecotype
References
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arend M, Schnitzler J-P, Ehlting B, Hänsch R, Lange T, Rennenberg H, Himmelbach A, Grill E, Fromm J. (2009) Expression of the Arabidopsis mutant ABI1 gene alters abscisic acid sensitivity, stomatal development, and growth morphology in gray poplars. Plant Physiol 151: 2110–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arney SE, Mitchell DL. (1969) The effect of abscisic acid on stem elongation and correlative inhibition. New Phytol 68: 1001–1015 [Google Scholar]
- Bachlava E, Tang S, Pizarro G, Schuppert GF, Brunick RK, Draeger D, Leon A, Hahn V, Knapp SJ. (2010) Pleiotropy of the branching locus (B) masks linked and unlinked quantitative trait loci affecting seed traits in sunflower. Theor Appl Genet 120: 829–842 [DOI] [PubMed] [Google Scholar]
- Balla J, Blažková J, Reinöhl V, Procházka S. (2002) Involvement of auxin and cytokinins in initiation of growth of isolated pea buds. Plant Growth Regul 38: 149–156 [Google Scholar]
- Ballaré CL. (1999) Keeping up with the neighbours: phytochrome sensing and other signalling mechanisms. Trends Plant Sci 4: 97–102 [DOI] [PubMed] [Google Scholar]
- Bennett T, Leyser O. (2006) Something on the side: axillary meristems and plant development. Plant Mol Biol 60: 843–854 [DOI] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16: 553–563 [DOI] [PubMed] [Google Scholar]
- Boe A, Beck DL. (2008) Yield components of biomass in switchgrass. Crop Sci 48: 1306–1311 [Google Scholar]
- Bonser SP, Aarssen LW. (2003) Allometry and development in herbaceous plants: functional responses of meristem allocation to light and nutrient availability. Am J Bot 90: 404–412 [DOI] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. (2009) Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150: 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnola JI, Ploschuk E, Benech-Arnold T, Finlayson SA, Casal JJ. (2012) Stem transcriptome reveals mechanisms to reduce the energetic cost of shade-avoidance responses in tomato. Plant Physiol 160: 1110–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Possenti M, Sessa G, Ciolfi A, Sassi M, Morelli G, Ruberti I. (2007) Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev 21: 1863–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Sessa G, Baima S, Morelli G, Ruberti I. (1993) The Arabidopsis Athb-2 and -4 genes are strongly induced by far-red-rich light. Plant J 4: 469–479 [DOI] [PubMed] [Google Scholar]
- Casal JJ. (2012) Shade avoidance. The Arabidopsis Book 10: e0157, /10.1199/tab.0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Deregibus VA, Sánchez RA. (1985) Variations in tiller dynamics and morphology in Lolium multiflorum Lam. vegetative and reproductive plants as affected by differences in red/far-red irradiation. Ann Bot (Lond) 56: 553–559 [Google Scholar]
- Casal JJ, Sánchez RA, Deregibus VA. (1986) The effect of plant density on tillering: the involvement of R/FR ratio and the proportion of radiation intercepted per plant. Environ Exp Bot 26: 365–371 [Google Scholar]
- Chatfield SP, Stirnberg P, Forde BG, Leyser O. (2000) The hormonal regulation of axillary bud growth in Arabidopsis. Plant J 24: 159–169 [DOI] [PubMed] [Google Scholar]
- Cline MG, Oh C. (2006) A reappraisal of the role of abscisic acid and its interaction with auxin in apical dominance. Ann Bot (Lond) 98: 891–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MH, Simmons SR. (1994) Tillering response of barley to shifts in light quality caused by neighboring plants. Crop Sci 34: 1604–1610 [Google Scholar]
- Deregibus VA, Sánchez RA, Casal JJ. (1983) Effects of light quality on tiller production in Lolium spp. Plant Physiol 72: 900–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA. (2003) A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol 133: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. (2011) Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol 12: 211–221 [DOI] [PubMed] [Google Scholar]
- Donohue K, Schmitt J. (1999) The genetic architecture of plasticity to density in Impatiens capensis. Evololution 53: 1377–1386 [DOI] [PubMed] [Google Scholar]
- Doust A. (2007) Architectural evolution and its implications for domestication in grasses. Ann Bot (Lond) 100: 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust AN, Devos KM, Gadberry MD, Gale MD, Kellogg EA. (2004) Genetic control of branching in foxtail millet. Proc Natl Acad Sci USA 101: 9045–9050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois PG, Olsefski GT, Flint-Garcia S, Setter TL, Hoekenga OA, Brutnell TP. (2010) Physiological and genetic characterization of end-of-day far-red light response in maize seedlings. Plant Physiol 154: 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery RJN, Longnecker NE, Atkins CA. (1998) Branch development in Lupinus angustifolius L. II. Relationship with endogenous ABA, IAA and cytokinins in axillary and main stem buds. J Exp Bot 49: 555–562 [Google Scholar]
- Finlayson SA. (2007) Arabidopsis Teosinte Branched1-like1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol 48: 667–677 [DOI] [PubMed] [Google Scholar]
- Finlayson SA, Krishnareddy SR, Kebrom TH, Casal JJ. (2010) Phytochrome regulation of branching in Arabidopsis. Plant Physiol 152: 1914–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC. (2005) Phytochromes and shade-avoidance responses in plants. Ann Bot (Lond) 96: 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galoch E, Zielihska M, Burkacka-Laukajtys E. (1998) The effect of decapitation on the levels of IAA and ABA in the lateral buds of Betula pendula Roth. Acta Physiol Plant 20: 399–403 [Google Scholar]
- García del Moral MB, García del Moral LF. (1995) Tiller production and survival in relation to grain yield in winter and spring barley. Field Crops Res 44: 85–93 [Google Scholar]
- Gocal GFW, Pharis RP, Yeung EC, Pearce D. (1991) Changes after decapitation in concentrations of indole-3-acetic acid and abscisic acid in the larger axillary bud of Phaseolus vulgaris L cv Tender Green. Plant Physiol 95: 344–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- González CV, Ibarra SE, Piccoli PN, Botto JF, Boccalandro HE. (2012) Phytochrome B increases drought tolerance by enhancing ABA sensitivity in Arabidopsis thaliana. Plant Cell Environ 35: 1958–1968 [DOI] [PubMed] [Google Scholar]
- González-Grandío E, Poza-Carrión C, Sorzano COS, Cubas P. (2013) BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25: 834–850, /10.1105/tpc.112.108480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann K, Hansen H. (2001) Ethylene-triggered abscisic acid: a principle in plant growth regulation? Physiol Plant 113: 9–14 [Google Scholar]
- Hempel FD, Feldman LJ. (1994) Bi-directional inflorescence development in Arabidopsis thaliana: acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192: 276–286 [Google Scholar]
- Hillman JR, Math VB, Medlow GC. (1977) Apical dominance and the levels of indole acetic acid in Phaseolus lateral buds. Planta 134: 191–193 [DOI] [PubMed] [Google Scholar]
- Hubbard L, McSteen P, Doebley J, Hake S. (2002) Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162: 1927–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger T, Bergelson J. (2000) The evolution of compensation to herbivory in scarlet gilia, Ipomopsis aggregata: herbivore-imposed natural selection and the quantitative genetics of tolerance. Evolution 54: 764–777 [DOI] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y. (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA 107: 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperbauer MJ. (1971) Spectral distribution of light in a tobacco canopy and effects of end-of-day light quality on growth and development. Plant Physiol 47: 775–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom TH, Brutnell TP, Finlayson SA. (2010) Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ 33: 48–58 [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Burson BL, Finlayson SA. (2006) Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol 140: 1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JP, Wareing PF. (1984) Apical dominance in Phaseolus vulgaris L.: the possible roles of abscisic acid and indole-3-acetic acid. J Exp Bot 35: 239–244 [Google Scholar]
- Kozuka T, Kobayashi J, Horiguchi G, Demura T, Sakakibara H, Tsukaya H, Nagatani A. (2010) Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol 153: 1608–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepin LV, Emery RJN, Pharis RP, Reid DM. (2007) Uncoupling light quality from light irradiance effects in Helianthus annuus shoots: putative roles for plant hormones in leaf and internode growth. J Exp Bot 58: 2145–2157 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K. (2010) ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci USA 107: 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bris M, Michaux-Ferriere N, Jacob Y, Poupet A, Barthe P, Guigonis J-M, Le Page-Degivry M-T. (1999) Regulation of bud dormancy by manipulation of ABA in isolated buds of Rosa hybrida cultured in vitro. Aust J Plant Physiol 26: 273–281 [Google Scholar]
- Leivar P, Quail PH. (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH. (2012) Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JAD, Koornneef M. (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2009) The control of shoot branching: an example of plant information processing. Plant Cell Environ 32: 694–703 [DOI] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T. (2006) Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol 170: 185–193 [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Kohlen W, Charnikhova T, Mulder P, Undas AK, Sergeant MJ, Verstappen F, Bugg TDH, Thompson AJ, Ruyter-Spira C, et al. (2010) Does abscisic acid affect strigolactone biosynthesis? New Phytol 187: 343–354 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Trevisan M, Pradervand S, Fankhauser C. (2009) Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J 60: 449–461 [DOI] [PubMed] [Google Scholar]
- Lortie CJ, Aarssen LW. (2000) Fitness consequences of branching in Verbascum thapsus (Scrophulariaceae). Am J Bot 87: 1793–1796 [PubMed] [Google Scholar]
- Mader JC, Emery RJN, Turnbull CGN. (2003) Spatial and temporal changes in multiple hormone groups during lateral bud release shortly following apex decapitation of chickpea (Cicer arietinum) seedlings. Physiol Plant 119: 295–308 [Google Scholar]
- Peng S, Khush GS, Cassman KG (1994) Evolution of the new plant ideotype for increased yield potential. In KG Cassman, ed, Breaking the Yield Barrier: Proceedings of a Workshop on Rice Yield Potential in Favorable Environments. International Rice Research Institute, Los Banos, Philippines, pp 5–20 [Google Scholar]
- Pierik R, De Wit M, Voesenek LACJ. (2011) Growth-mediated stress escape: convergence of signal transduction pathways activated upon exposure to two different environmental stresses. New Phytol 189: 122–134 [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Crawford S, Smith RS, Ljung K, Bennett T, Ongaro V, Leyser O. (2009) Control of bud activation by an auxin transport switch. Proc Natl Acad Sci USA 106: 17431–17436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin CH, Hay MJM, Newton PCD, Greer DH. (1994) Effect of light quality (red:far-red ratio) at the apical bud of the main stolon on morphogenesis of Trifolium repens L. Ann Bot (Lond) 74: 119–123 [Google Scholar]
- Sellaro R, Yanovsky MJ, Casal JJ. (2011) Repression of shade-avoidance reactions by sunfleck induction of HY5 expression in Arabidopsis. Plant J 68: 919–928 [DOI] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I. (2005) A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Ram H, Abbas N, Chattopadhyay S. (2012) Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabidopsis thaliana. J Biol Chem 287: 25995–26009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. (1995) Physiological and ecological function within the phytochrome family. Annu Rev Plant Physiol Plant Mol Biol 46: 289–315 [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235–4245 [DOI] [PubMed] [Google Scholar]
- Su H, Abernathy SD, White RH, Finlayson SA. (2011) Photosynthetic photon flux density and phytochrome B interact to regulate branching in Arabidopsis. Plant Cell Environ 34: 1986–1998 [DOI] [PubMed] [Google Scholar]
- Tamas IA, Ozbun JL, Wallace DH, Powell LE, Engels CJ. (1979) Effect of fruits on dormancy and abscisic acid concentration in the axillary buds of Phaseolus vulgaris L. Plant Physiol 64: 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. (2006) Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J 45: 1028–1036 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker DJ. (1977) Effects of far-red light on lateral bud outgrowth in decapitated tomato plants and associated changes in levels of auxin and abscisic acid. Plant Sci Lett 8: 339–344 [Google Scholar]
- Tucker DJ, Mansfield TA. (1972) Effects of light quality on apical dominance in Xanthium strumarium and associated changes in endogenous levels of abscisic acid and cytokinins. Planta 102: 140–151 [DOI] [PubMed] [Google Scholar]
- Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, Sakurai N, Suzuki H, Saito K, Shibata D, Kobayashi M, et al. (2009) Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J 57: 1065–1078 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y. (2009) Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol 150: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker L, Jacomet S, Korner C. (2002) Trends in biomass fractionation in wheat and barley from wild ancestors to modern cultivars. Plant Biol 4: 258–265 [Google Scholar]
- Waldie T, Hayward A, Beveridge CA. (2010) Axillary bud outgrowth in herbaceous shoots: how do strigolactones fit into the picture? Plant Mol Biol 73: 27–36 [DOI] [PubMed] [Google Scholar]
- Wan C, Sosebee RE. (1998) Tillering responses to red:far-red light ratio during different phenological stages in Eragrostis curvula. Environ Exp Bot 40: 247–254 [Google Scholar]
- Weinig C, Stinchcombe JR, Schmitt J. (2003) Evolutionary genetics of resistance and tolerance to natural herbivory in Arabidopsis thaliana. Evolution 57: 1270–1280 [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP. (2011) grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci USA 108: E506–E512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrough KM, Nelson CJ, Coutts JH. (1983) Relationship between tillering and forage yield of tall fescue. I. Yield. Crop Sci 23: 333–337 [Google Scholar]
- Zhao DL, Atlin GN, Bastiaans L, Spiertz JHJ. (2006) Developing selection protocols for weed competitiveness in aerobic rice. Field Crops Res 97: 272–285 [Google Scholar]