The bioinformatic tool, ROSMETER, differentiates different reactive oxygen species, their subcellular origin, to give new insight into senescence and biotic and abiotic stresses.
Abstract
The chemical identity of the reactive oxygen species (ROS) and its subcellular origin will leave a specific imprint on the transcriptome response. In order to facilitate the appreciation of ROS signaling, we developed a tool that is tuned to qualify this imprint. Transcriptome data from experiments in Arabidopsis (Arabidopsis thaliana) for which the ROS type and organelle origin are known were compiled into indices and made accessible by a Web-based interface called ROSMETER. The ROSMETER algorithm uses a vector-based algorithm to portray the ROS signature for a given transcriptome. The ROSMETER platform was applied to identify the ROS signatures profiles in transcriptomes of senescing plants and of those exposed to abiotic and biotic stresses. An unexpected highly significant ROS transcriptome signature of mitochondrial stress was detected during the early presymptomatic stages of leaf senescence, which was accompanied by the specific oxidation of mitochondria-targeted redox-sensitive green fluorescent protein probe. The ROSMETER analysis of diverse stresses revealed both commonalties and prominent differences between various abiotic stress conditions, such as salt, cold, ultraviolet light, drought, heat, and pathogens. Interestingly, early responses to the various abiotic stresses clustered together, independent of later responses, and exhibited negative correlations to several ROS indices. In general, the ROS transcriptome signature of abiotic stresses showed limited correlation to a few indices, while biotic stresses showed broad correlation with multiple indices. The ROSMETER platform can assist in formulating hypotheses to delineate the role of ROS in plant acclimation to environmental stress conditions and to elucidate the molecular mechanisms of the oxidative stress response in plants.
During the normal course of plant development and during stress, reactive oxygen species (ROS) production may occur in diverse subcellular compartments, including membranes, chloroplasts, mitochondria, peroxisomes, and cytoplasm. In the light, plastids are a major source of ROS, but in the dark, or in nongreen tissues, the mitochondria are major source of ROS (Rhoads et al., 2006). One electron transfer to oxygen will create the superoxide anion, O2.−. Superoxide anions are generated in multiple ways and at different sites. For example, one source is the respiratory burst oxidases multigene family (Rboh; NADPH oxidases) localized to the plasmalemma (Sagi and Fluhr, 2006) that can be internalized on vesicles (Leshem et al., 2006; Ashtamker et al., 2007). The production of O2.−, possibly by xanthine oxidase in the peroxisome or peroxisome membrane proteins, has also been demonstrated (Nyathi and Baker, 2006). Other sites of superoxide production are in PSI by the photoreduction of oxygen (Asada, 2006) and in complexes I and III of the mitochondrial electron transport chain (Møller, 2001; Rhoads et al., 2006). The charged superoxides can spontaneously or catalyzed by superoxide dismutase (SOD) be dismutated to oxygen and hydrogen peroxide (H2O2). H2O2 can also be formed by photorespiration in the peroxisome as a by-product of glycolate oxidation and during the metabolism of ureides (Corpas et al., 2001). H2O2 will react with the soluble ferrous iron in a Fenton reaction to produce the hydroxyl radical OH· (Halliwell and Gutteridge 2007). Singlet oxygen 1O2 can be generated photodynamically in PSII in the light through energy transfer from photoactivated pigments such as chlorophyll, through the activity of peroxidases, or by the decomposition of membrane hydroperoxides formed by other ROS (Miyamoto et al., 2007).
ROS play a role in cell death and in cellular signaling (Wagner et al., 2004; Foyer and Noctor, 2005; Mittler et al., 2011; Suzuki et al., 2012). While high ROS concentrations are harmful to essential biological processes, sublethal doses of ROS act as secondary messengers that can be sensed by specific redox-sensitive proteins responsible for the activation of a signal transduction culminating in altered gene expression. Different ROS have different activities (Halliwell and Gutteridge, 2007) and hence elicit different protein modifications, which might be manifested in eliciting different gene expression (Møller and Sweetlove, 2010). Moreover, the subcellular site in which the modification in ROS/oxidation state occurs can also serve as a specific signal of a cellular redox network (Foyer and Noctor, 2003; Møller and Sweetlove, 2010; König et al., 2012).
To assess the specificity of ROS-driven transcript expression, Gadjev et al. (2006) compared transcriptome data generated from ROS-related microarray experiments in which the specific identity of the ROS and its subcellular site of production were known. A set of general oxidative ROS-related genes was identified. More importantly, sets of genes that were highly induced in response to H2O2, superoxide, or singlet oxygen emanating from a specific organelles were also identified. By examining the expression of these ROS-related genes in mutant plants and during senescence, it was possible to identify the involvement of ROS in signaling (Jing et al., 2008; Rosenwasser et al., 2011). However, a simplified platform is desirable that will take into consideration both up- and down-regulated genes and, in a comparative manner, identify ROS footprints. In this manner, the chemical identity and subcellular production of the ROS can be discerned using a more rigorous statistical analysis. A bioinformatic tool, ROSMETER, was developed to provide an organelle/type-dependent ROS-related transcriptomic signature. It integrates diverse, large-scale data sets of ROS-induced genes into a framework that predicts and describes the specificity of ROS transcriptomic signature. We corroborated the ROSMETER predictions by an analysis of additional transcriptome data for which ROS measurements were available. In addition, we apply ROSMETER to analyze various environmental states, leading to novel hypotheses.
RESULTS
The ROSMETER Platform
To assess the appearance of the ROS-related transcriptomic signature in a given large-scale gene expression data set, we first compiled a set of indices, each composed of a list of significantly changed genes that represent a specific ROS type or ROS emanating from a specific organelle. This approach, in which large gene expression profiles are compared with a set of genes extracted from a whole gene list, representing a specific biological response, is less susceptible to experimental noise in comparison with common clustering methods in which all the array is compared (Sasaki et al., 2011).
The indices were compiled from transcriptome data of defined mutations or of direct chemical applications that lead to increases in ROS production. Each of the selected experiments has the advantage of ROS accumulation thought to originate in a specific organelle, and in most cases the specific ROS type is known (Supplemental Table S1). The indices include the following: (1) accumulation of H2O2 in the cytoplasm in plants mutated in cytosolic ascorbate peroxidase exposed to high light (KO-APX1 + HL; Davletova et al., 2005a); (2) apoplastic multiple ROS, the result of ozone treatment (A. Shirras http://affymetrix.arabidopsis.info/narrays/experimentpage.pl?experimentid=26); (3) production of singlet oxygen in the flu mutant in chloroplasts due to the accumulation of photodynamic chlorophyll precursors (op den Camp et al., 2003); (4) direct application of 20 mm H2O2 to Arabidopsis (Arabidopsis thaliana) seedlings (Davletova et al., 2005b); (5) oxidative stress brought on by a mutation in mitochondrial alternative oxidase that is exposed to mild light and drought stress (TDNA-AOX1a-MLD; Giraud et al., 2008); (6) down-regulation by antisense AOX1a (AS-AOX1; Umbach et al., 2005); (7) oxidative stress brought on by the application of rotenone, an inhibitor of mitochondrial complex I (Garmier et al., 2008); (8) accumulation of H2O2 in the peroxisomes in catalase2 mutants exposed to high light (CAT2HP1 + HL; Vanderauwera et al., 2005); (9) treatment with 3-aminotriazole (AT), a catalase inhibitor spray, which lead to increasing H2O2 in peroxisome (Gechev et al., 2005); (10) production of superoxide in chloroplasts lacking chloroplastic superoxide dismutase (KD-SOD; Rizhsky et al., 2003); (11) superoxide formation in the chloroplast and mitochondria after application of methyl viologen (MV; Kilian et al., 2007). For each experiment, signal intensities were normalized and compared in fold change-transformed values relative to the control sample (e.g. the wild type or mock treatment). Additionally, statistical significance (P values) based on a two-tailed Student’s t test was calculated for each gene. Statistically significant (P < 0.05) most changed (induced or repressed) transcripts were extracted from each of the above ROS-related experiments and used to define the indices of ROSMETER.
The similarity between the transcripts in the indices and data of interest is established by using vector-based correlation, which is thoroughly described by Kuruvilla et al. (2002). In accordance with the vector-based correlation, each microarray experiment is represented by a high-dimensional vector in which each gene is of different dimension. The similarity between the vector of the experiment and the indices was assessed by comparing the cosine of the angle between the vectors (Kuruvilla et al., 2002), which is essentially analogous to the Pearson correlation coefficient. Due to the stringent statistical requirement, some comparisons may yield low numbers of genes; therefore, only correlation values generated from the comparison of at least 45 genes are presented. Complete correlation is indicated by the numeral 1, and the highest possible negative correlation is indicated by the numeral −1. The numeral 0 indicates no correlation. Finally, the results are summarized in a table that includes for each comparison the correlation values and the number of genes that participated. The correlation data are then illustrated by a heat map in which red presents positive correlation, green presents negative correlation, and black stands for no correlation. The experiments examined are clustered by nearest neighbor correlation. This approach was recently adopted for discerning between hormone-related transcription signatures in Arabidopsis (Volodarsky et al., 2009).
In addition to the ROS signature analysis, ROSMETER also returns to the user lists of genes that participated in each correlation analysis and, therefore, enables downstream functional analysis. Users can analyze their own transcriptome data by accessing the ROSMETER Web-based site http://app.agri.gov.il/noa/ROSMETER.php.
Discerning Footprints of ROS-Related Experiments
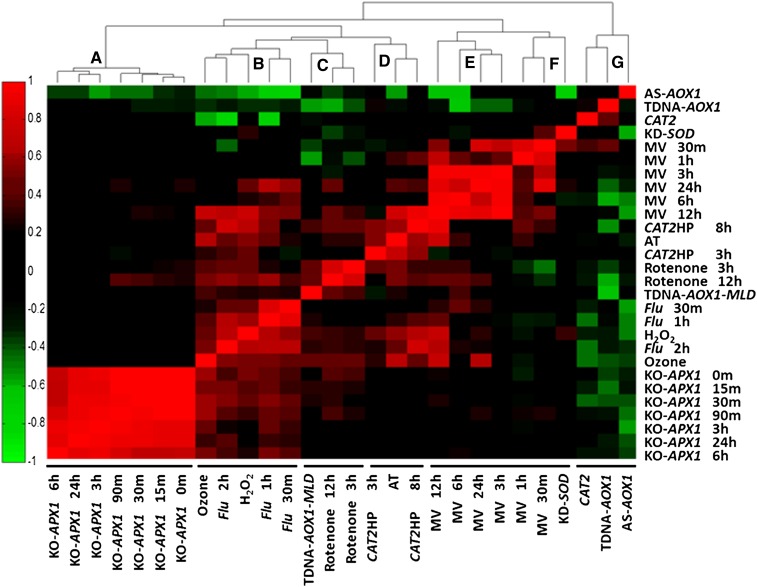
To obtain a global viewpoint about the specificity of the different ROS indices, ROSMETER was applied to the indexed transcriptomes themselves. The ROS indices are shown on the abscissa (x axis), and the microarray experiments are shown on the ordinate (y axis; Fig. 1). The positions of the indices on the abscissa are determined by clustergram of nearest neighbor correlation. Subsequently, in this figure, the indices were compared with their own transcriptomes and with the transcriptomes of the other indices. The order at the y axis is arranged according to the order of the indices on the x axis (Fig. 1; for correlation values, see Supplemental Table S2).
Figure 1.
Correlations between ROS indices and the ROS-related microarray data that were used to build the indices. The ROS indices are listed on the abscissa and are clustered by nearest neighbor correlation, and the ROS experiments are shown on the ordinate. The color-coded results of correlations for each index are shown as a heat map. Correlation values are between 1 (complete positive correlation; red) and −1 (highest negative correlation; green), where 0 indicates no correlation (black). The experiments are listed in Supplemental Table S1, and the numerical values of the correlation are given in Supplemental Table S2. [See online article for color version of this figure.]
The analysis revealed seven distinct nearest neighbor clusters that can be grouped, by and large, according to the type of organelle stress. Cluster A includes the knockout of cytoplasmic ascorbate peroxidase (KO-APX1) experiments, which are thought to represent cytoplasmic H2O2. This cluster shows the highest degree of self-correlation. Cluster B is a mix of H2O2, flu (0.5, 1, and 2 h), and ozone experiments. Interestingly, the later time point of flu (2 h) is more similar to H2O2 and ozone than to flu 30 min, suggesting that the early time point of the flu experiment represent a unique response to singlet oxygen while the later time points might represent less specific ROS responses, possibly due to oxidative damage. Cluster C consists of rotenone treatments (3 and 12 h) and of mild light and drought stress applied to the alternative oxidase knockout (TDNA-AOX1-MLD). Both experiments are associated with a mitochondrial stress but have not been shown to directly generate ROS. Cluster D comprises AT, an inhibitor of catalase and of Catalase2-deficient plants (CAT2HP1) exposed to high light for 3 and 8 h (CAT2HP3h and CAT2HP8h). CAT2 is a part of peroxisome ROS detoxification. The fact that AT treatment is correlated with CAT2HP3h and CAT2HP8h gene expression indicates that this cluster can represent peroxisome stress.
Clusters E and F consist of the MV late (3–24 h) and early (0.5 and 1 h) time points, respectively, and cluster F also contains KD-SOD. The late MV time points (3–24 h) tend to show more cross reactivity with other ROS, particularly H2O2, and may indicate secondary stress effects. The earliest time points of MV and KD-SOD may indicate an overproduction of superoxide in the chloroplast. Interestingly, cluster G includes the indices of CAT2, TDNA-AOX1, and AS-AOX1. This cluster comprises transcriptome data from plants that are mutated in essential antioxidant enzymes that were not exposed to any stress conditions. The observation that these indices exhibit negative correlations, instead of simply no correlation with most of the data in Figure 1, may indicate that in such mutant plants a compensatory scavenging mechanism has been activated.
If ROS responses were well defined and restricted, one would have expected a strict correlation value of 1 mainly along the diagonal. However, inspection of Figure 1 reveals a much more complicated picture. For example, the indices of ozone, flu, and H2O2 were particularly promiscuous and share responses with MV, CAT2HP, AT, and also with rotenone treatment of 12 h and to some degree with KO-APX1 exposed to light. Furthermore, these indices show negative correlation with AS-AOX1. In addition, the indices of 12-h MV, CAT2HP8h, and AT show correlations with ozone, flu, and H2O2 experiments. In summary, the global view of transcriptome analysis through the ROSMETER tool, as reflected in the indices of different ROS types and cellular sources, portrays specificity and its limits.
Examination of ROSMETER Predictions Using Experiments in Which the Source or Type of Oxidative Stress Is Known
In order to assess the range of correlation values that have biological meaning, we estimated both the lowest correlation values that need to be considered as well as values that are expected to be significant. For the lower end of the estimate, data were randomized and analyzed for correlation to the ROSMETER indices. In this case, we expect a correlation value of 0. The expression data of flu 2 h, which shows a strong transcription response in terms of the number of transcripts that changed, was randomized and the result compared with the indices. The resultant average correlation score to all the indices was near zero (average = 0.03, σ = 0.026). Thus, if correlation values of a particular experiment are distributed normally, values beyond 3.2906σ sd (i.e. 0.09) have only a 0.1% chance of being random. Thus, only values below −0.13 or above 0.13 need be considered (Volodarsky et al., 2009).
ROSMETER was first evaluated by queries of transcriptome data from experiments with predictable results. Thus, data involving rotenone application for 3 h (40 µm) that were not used in the ROSMETER indices were analyzed (Clifton et al., 2005; Fig. 2A; Supplemental Table S3). In this case, the highest correlation, as expected, was to rotenone 3 h and somewhat less to rotenone 12 h (above 0.45 and 0.39, respectively). Oligomycin is a proton channel blocker, inhibiting mitochondrial electron transport by blocking ATP synthase activity (Schwarzländer et al., 2012). Analysis of transcriptome data following this treatment can be seen to correlate with multiple indices (Fig. 2A). However, significant correlations are, as expected, with mitochondrial functions (i.e. rotenone [0.48] and TDNA-AOX1-MLD [0.38]). The data also show correlation with MV 6 h, which is expected, as rotenone and MV 6 h show a degree of overlap (Fig. 1). These examples demonstrate the ability of ROSMETER analysis to identify cellular transcription responses to specific ROS stress and suggest that correlation values above 0.4 can be considered as significant correlation values providing biological insights.
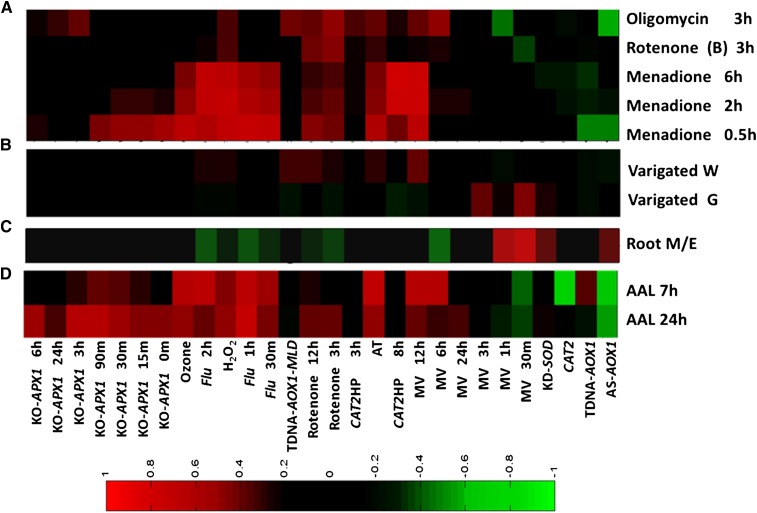
Figure 2.
ROSMETER analysis of gene expression of oligomycin, rotenone, and menadione (A), green and white sectors of the variegated mutant var2 (B), root development (C), and AAL toxin (D). The correlation values between the transcriptome data and ROS indices are shown as a color-coded heat map as described in Figure 1. The ROS indices are shown on the abscissa. Expression data for rotenone, oligomycin treatments, and AAL toxin were obtained from Clifton et al. (2005), Schwarzländer et al. (2012), and Gechev et al. (2004), respectively; expression data for var2 and root development were downloaded from GEO data sets (http://www.ncbi.nlm.nih.gov/gds/). Expression values in the var2 mutant either in the green or white sector were normalized to wild-type plants. Variegated W and variegated G are white and green sectors, respectively, of var2 (AT2G30950). For root development, data of the wild-type meristematic zone (M) were normalized to the wild-type elongation zone (E). [See online article for color version of this figure.]
To further attach significance to the correlation scores made by ROSMETER, we examined data from experiments in which the ROS identity or ROS/redox changes have been demonstrated. The treatment of root tissue with menadione was shown to induce a rapid oxidation of the redox-sensitive GFP (roGFP) probe in multiple cellular compartments (Lehmann et al., 2009). Analysis by ROSMETER shows rapid and transient correlation with early KO-APX1 indices and a strong correlation with late flu 2 h (average of 0.67 for 3 time points), ozone (average of 0.52), H2O2 (average of 0.69), and MV 12 h (average of 0.72) and a weaker correlation to rotenone indices (Fig. 2A; Supplemental Table S3). Hence, ROSMETER analysis shows that application of menadione leads to the formation of a pervasive ROS signal that is consistent with the reported change in oxidation state in the cytoplasm, chloroplast, and mitochondria.
Another example is in the case of the variegated (var) mutant. This mutant develops variegated green and white sectors during development, and nitroblue tetrazolium (NBT) staining representing superoxides, accumulated in the green tissue of the var mutant well above the wild-type levels, while no NBT staining was detected in the white sectors (Miura et al., 2010). ROSMETER analysis of the transcriptome data revealed in the green tissue a high correlation to MV-related signals (greater than 0.4; Fig. 2B, variegated G; Supplemental Table S3), consistent with the excess staining observed for superoxide. However, in the white sectors, the correlation was to mitochondrial and peroxisomal stress (Fig. 2B, variegated W; Supplemental Table S3; i.e. rotenone, TDNA-AOX-MLD, and AT). The correlations with rotenone and TDNA-AOX-MLD may indicate that white sectors are more reliant on mitochondrial functions. In addition, white tissue shows correlation to MV 12 h, and as noted above, the MV 12-h index shows additional components of general H2O2 stress (Fig. 1).
The predictive value of using ROSMETER can also be ascertained from an experiment where changes in ROS type have been shown to be correlated with the transition from proliferation to differentiation in roots (Tsukagoshi et al., 2010). Superoxide as measured by NBT was shown to accumulate in the meristematic zone, containing actively dividing cells, whereas H2O2 accumulated in the elongation zone, as determined by dihydroethidium. The expression data of the meristematic zone of wild-type plants was normalized to that from the elongation zone, and the data were analyzed by ROSMETER (Fig. 2C; Supplemental Table S3). In accordance with the increase in superoxide level in the meristematic zone, ROSMETER yielded positive correlations for superoxide-related signatures (0.46 for MV 30 min, 0.39 for MV 1 h, and 0.34 for KD-SOD).
Increased H2O2 level and, consequently, activation of programmed cell death have been demonstrated in Arabidopsis plants mutated in ceramid synthase LAG One Homolog2 (LOH2) and treated with Alternaria Alternata (AAL) fungal toxin (Gechev et al., 2004). The ROSMETER analysis was performed on the expression data of AAL toxin-induced cell death 7 and 24 h post toxin application. In accordance with increased H2O2 level as measured by 3,3′-diaminobenzidine staining, we found positive correlation to indices representing the overproduction of H2O2 in cytoplasm (KO-APX), peroxisome (AT), and external application of H2O2 (Fig. 2D; Supplemental Table S3). Positive correlations were also found to indices of the flu mutant. Thus, gene expression profiles from experiments in which the type and origin of ROS have been established help to calibrate the correlation values computed by ROSMETER and data of ROS or roGFP corroborate the predictive capability of ROSMETER.
Analysis of Dark-Induced and Developmental Senescence in Leaves
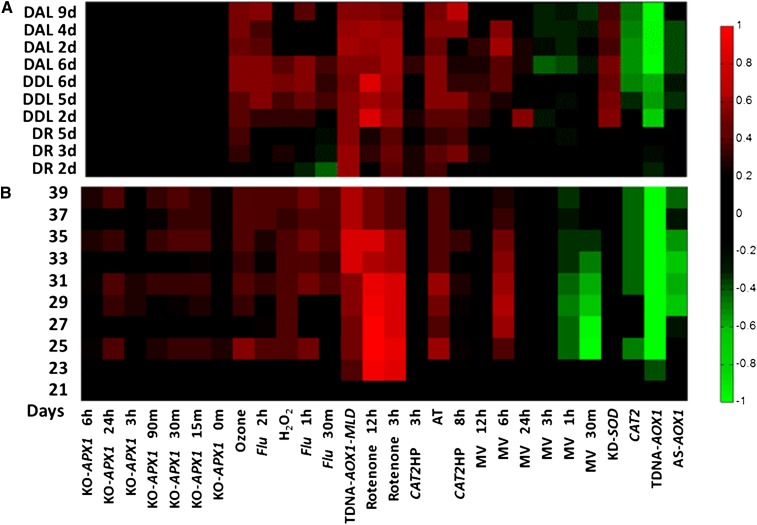
Recent analysis of data from darkened detached rosettes showed elevation of the mitochondria-related ROS transcriptome (Rosenwasser et al., 2011; Fig. 3A). Here, we extend this analysis to darkened detached leaves and to darkened attached leaves (van der Graaff et al., 2006; Fig. 3A; Supplemental Table S4). As shown, in all dark-induced systems, there were high correlations to rotenone and TDNA-AOX1-MLD (i.e. mitochondrial stress).
Figure 3.
ROSMETER analysis of developmental and dark-induced senescence transcriptomes. A, Dark-induced senescence. The transcriptomes of dark detached rosette (DR) from Rosenwasser et al. (2011) were compared with those of darkened attached leaf (DAL) or darkened detached leaf (DDL) obtained from van der Graaff et al. (2006). B, Developmental senescence. The microarray data set of leaf development from 19 to 39 DAS is from Breeze et al. (2011). The averaged data of each day were analyzed. [See online article for color version of this figure.]
A high-resolution time-course profile of gene expression during development of a single leaf (19–39 d after sowing [DAS]) and grown in a 16/8-h light/dark cycle was recently compiled (Breeze et al., 2011). Analysis of the data by the ROSMETER platform shows neutral correlation with all the ROS indices on day 21 (Fig. 3B; Supplemental Table S4). Remarkably, from day 23 on, high correlation values were observed, particularly to mitochondria-related ROS footprints (rotenone and TDNA-AOX1-MLD). Interestingly, on this day, plants commenced to bolt and the leaves reached a state of full expansion. However, the overt visual sign of senescence (i.e. yellowing at the tip of the leaves) was observed only around 31 DAS (Breeze et al., 2011). This analysis suggests a role of mitochondria stress at early presymptomatic stages of leaf senescence. Interestingly, negative correlation values to the MV (early response) footprints were found starting at day 25; however, such chloroplast-dependent footprints were by and large absent in dark-induced senescence.
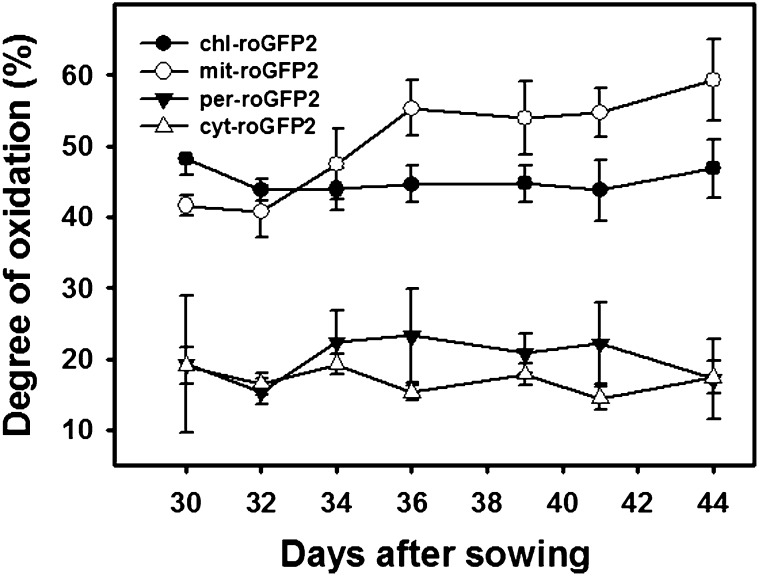
In order to assess if the redox state of cells undergoing senescence reflects that portrayed by ROSMETER analysis, we followed the subcellular changes in roGFP oxidation. The fluorescence of the roGFP probe reflects the local glutathione redox potential (Meyer et al., 2007). Measurements were carried out in Arabidopsis transgenic lines, in which the probe was localized to mitochondria (mit-roGFP2), plastids (pla-roGFP2), cytoplasm (cyt-GRX1-roGFP2), or peroxisomes (per-GRX1-roGFP2). The degree of probe oxidation in mitochondria reporter lines (mit-roGFP2) showed an increase following 32 DAS that was maintained at high levels (Fig. 4). In contrast, the peroxisomal reporter lines showed an initial increase at this time that was followed by a decrease to the initial levels 12 d later (Fig. 4). In this experiment, bolting started 32 DAS; hence, the increase in mitochondria oxidation was correlated with bolting. Note that the degree of roGFP oxidation in plastids or mitochondria is similar to the values reported by us previously (Rosenwasser et al., 2011) but higher than the values reported by Schwarzländer et al. (2008). It is possible that the lower basal redox state of the roGFP, observed by others, is due to the use of younger plants and lower light intensity used for their growth. In all, these results are consistent with the mitochondrial stress footprint displayed by ROSMETER.
Figure 4.
Changes in the degree of oxidation of roGFP in various cellular compartments during developmental senescence. The degree of roGFP oxidation in transgenic Arabidopsis lines expressing roGFP2 in plastids, mitochondria cytoplasm, and peroxisomes was determined following measurements of fluorescence at 515 nm resulting from excitation at 400 or 485 nm using a fluorometer as described in “Materials and Methods.”
Analysis of ROS Signatures in Abiotic Stresses Using ROSMETER
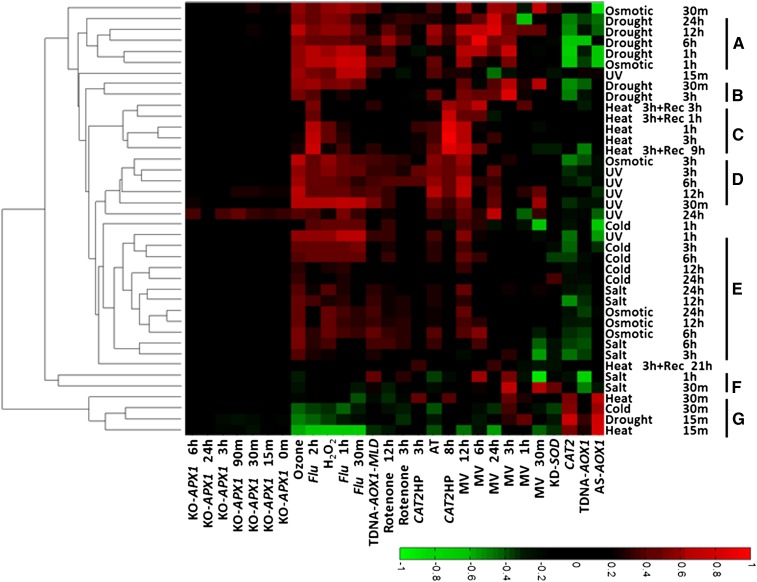
Increased levels of ROS have been suggested to have dual roles during abiotic stresses, both as a signal for mounting protective functions and as an agent causing oxidative damage (Suzuki et al., 2012). It was of interest to compare the ROS signatures of several stresses using ROSMETER. The AtGenExpress global stress data set consists of transcriptomic data generated by the Affymetrix ATH1 platform of hydroponically grown Arabidopsis plants subjected to different environmental stresses. These include UV-B (15 min, 1.18 W m−2), drought (10% weight loss in air for 15 min and then recovery), heat (3 h at 38°C and then recovery), cold (4°C for the indicated times), salt (150 mm for the indicated times), and high osmolarity (300 mm mannitol for the indicated times; Kilian et al., 2007). The data were analyzed using the ROSMETER platform as shown in Figure 5 and Supplemental Table S5.
Figure 5.
ROSMETER analysis of various abiotic stresses. The expression data of Arabidopsis plants exposed to various abiotic stress conditions (Kilian et al., 2007) are from GEO data sets (http://www.ncbi.nlm.nih.gov/gds/). They include salt (GSE5623), osmotic (GSE5622), cold (GSE5621), UV-B (GSE5626), heat (GSE5628), and drought (GSE5624) stresses. Only data from shoot tissue were analyzed. For each experiment, fold change values were normalized to the control series (GSE5620). The indices are listed on the abscissa, and the abiotic stress experiments are listed on the ordinate and clustered according to the similarity of their profiles represented in clusters A to G. [See online article for color version of this figure.]
Strikingly, robust correlations corresponding to flu, H2O2, ozone, and AT signatures and the MV 12-h indices were obtained, but only neutral correlations were seen with cytoplasm-based KO-APX1 (Fig. 5). Furthermore, a bimodal response to stress can be observed where the early responses (clusters G and F; Fig. 5) clustered separately from the late responses of the corresponding stresses. Hence, cluster F of salt treatment (30 min and 1 h) exhibits low correlations to the indices, except to MV 3 h and negative correlation to TDNA-AOX1 and MV 30 min. Cluster G contains the early responses to cold (30 min), drought (15 min), and heat (15 and 30 min), and interestingly, the response negatively correlated with the indices of general ROS (H2O2 or ozone) and flu. However, other early treatments (e.g. UV-B at 15 min or osmotic stress) clearly stimulated correlations with multiple ROS responses.
In general, it is evident that the abiotic stress experiments showed positive correlations to the various indices and that clustering was generally according to the nature of the stress. Clusters A and B, containing mainly the data of drought responses, show high correlations by and large to late MV, AT, and indices of general ROS. Heat stress from 1 and 3 h shows a distinctive signature of peroxisomal H2O2 production that persisted during recovery of 1 to 9 h (cluster C) but nearly disappeared at 21 h. For the late time points of UV stress (cluster D; 3, 6, and 12 h and 30 min), high correlations were found especially to ozone, AT, and MV 12-h indices. Cluster E includes cold (3, 6, 12, and 24 h), salt (3, 6, 12, and 24 h), and the response to change in osmolarity (6, 12, and 24 h). It is characterized by relatively lower correlations to the indices, indicating less of a ROS response under the experimental conditions used. Interestingly, the early time points of osmotic stress were interspersed with drought and heat, but the later time points of osmotic stress were more similar to salt stress. In conclusion, the differences in ROS transcriptomic signatures of the various abiotic stresses suggest that each stress provides a unique response.
The AtGenExpress global stress also contains expression data generated from the roots of the stressed plant. In order to compare the root and shoot responses, we examined the ROS signatures in shoot versus root following treatment with either heat or UV and recovery from heat (Supplemental Fig. S1). Under UV stress, where only the aerial parts of the plants were irradiated, distinct ROS signatures were observed in the shoot and the roots. For example, in the shoots, the footprints of flu, H2O2, ozone, and MV 12 h dominated, but those of rotenone were found in roots, suggesting a different kind of ROS-related transcriptome expression involved in root versus shoot responses to UV illumination. In contrast, in heat stress, where the whole plant was exposed to the treatment, ROS signatures of most time points show similarity between roots and shoot (Supplemental Fig. S1A). Thus, the analysis by the ROSMETER platform indicates that the responses of two different tissues are similar.
Analysis of ROS Signatures in Biotic Stresses Using ROSMETER
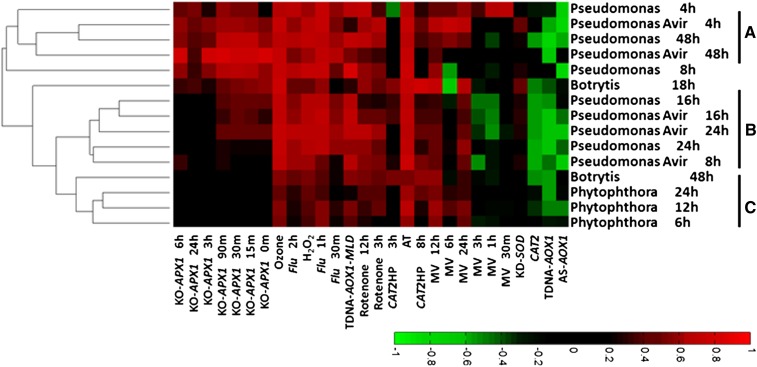
ROS signaling pathways have been suggested to play an important role in plant responses to biotic stresses (Grant and Loake, 2000; Sagi and Fluhr, 2001). In an avirulent interaction, the recognition of a pathogen by plant cells initiates a response that includes the production of H2O2, leading eventually to cell death (Torres, 2010). The ROSMETER platform was applied to analyze the available transcriptome data of Arabidopsis plants exposed to the following pathogens: the fungal biotroph Phytophthora infestans, the fungal necrotroph Botrytis cinerea, and the bacterial biotroph Pseudomonas syringae with and without avirulent factor (Fig. 6; Supplemental Table S6).
Figure 6.
ROSMETER analysis of various biotic stresses. The expression data of Arabidopsis plants exposed to biotic stress conditions were downloaded from GEO data sets (http://www.ncbi.nlm.nih.gov/gds/). They include P. infestans (GSE5616), B. cinerea (GSE5684), and P. syringae (GSE5685). For each experiment, fold change values were normalized to the untreated plants. The biotic stress experiments are listed on the ordinate, and the ROS indices are listed on the abscissa. Groups A to C represent different clusters. [See online article for color version of this figure.]
Interestingly, the correlations of P. syringae (4 and 48 h; cluster A) and B. cinerea (18 h) and, to a lesser degree, P. syringae (8, 16, and 24 h; cluster B) but not P. infestans (cluster C) showed high correlations with APX1-related indices. This is in contrast to the signatures for abiotic stress data (Fig. 5). Furthermore, the results of P. syringae (clusters A and B) show that as far as the ROS signature is concerned, virulent and avirulent strains yield similar correlations. This indicates that the major ROS signatures are not related to the hypersensitive response. Generally, the analysis of the response to the diverged pathogen types (necrotrophic and biotrophic fungi and virulent and avirulent bacteria) yields very high correlation values between the biotic stress expression data and several ROS indices such as H2O2, ozone, AT, and flu related. High correlations were also found to the mitochondrial indices, but minimal correlations were found to MV indices (except to MV 12 and 24 h). This suggests that the plant response to pathogens involves mitochondrial stress and other ROS but not stress caused by the production of superoxide in chloroplasts.
DISCUSSION
Specificity in the ROS Response
In this study, we show how the ROSMETER platform facilitates the characterization of ROS transcriptome signatures. Its algorithm uses a rigorous comparative approach based on vector correlation that takes into account the direction and size of all significant changes of transcript levels across the transcriptome (Volodarsky et al., 2009). The ROSMETER tool is based on the assumption that the subcellular source of the ROS and its distinct chemical identity will determine the transcriptome signature. Indices were created for a set of experiments for which the ROS type and its organelle location have been defined. The indices show a robust degree of specific correlation with their own transcriptome data (Fig. 1). Moreover, cluster analysis of the ROS indices revealed that they cluster according to the subcellular localization of ROS production (Fig. 1), and ROS transcriptomic footprints have been identified for ROS in cytoplasm, mitochondria, peroxisomes, and plastids. Hence, this analysis supports the hypothesis that transcriptome signatures can reflect ROS origin and type and is consistent with the observation that each ROS is more highly correlated to itself along the diagonal (Fig. 1). However, a significant degree of cross correlation can be observed that, in itself, is not unexpected. The cross reactivity may be due to one or more of the following reasons: spontaneous or enzymatic interconversion of ROS, movement of ROS to a different location, collateral general damage that induces further ROS, or parallel pathways of gene activation. This makes it challenging to associate a distinct response to a particular type or ROS/organelle.
Interconversion of ROS is very common. For example, hydroxyl radical and superoxides convert into H2O2 via the Fenton reaction and SOD, respectively (Halliwell and Gutteridge, 2007). It is well established that superoxides spontaneously, or enzymatically, dismutate by SOD to H2O2, and indeed, the RBOH class of NADPH oxidases release superoxide that is readily detected as H2O2 (Ashtamker et al., 2007). The charged superoxides will not readily cross the membranes; however, H2O2 can readily diffuse via aquaporin to other cellular sites (Bienert et al., 2007).
The singlet oxygen-like response is very notable in many transcriptomes. It remains to be shown if singlet oxygen is a true intermediate. Singlet oxygen is short lived, since it rapidly dissipates by interaction with cellular components. Singlet oxygens, which are produced in the flu mutant upon the transition of the mutant from darkness to light, are thought to be released within plastids (op den Camp et al., 2003). However, singlet oxygen can rapidly convert to superoxide (Saito et al., 1981), and they can also interact with cellular component membranes in type II-mediated lipid peroxidation (Triantaphylidès et al., 2008). Indeed, fatty acid peroxidation was observed to occur in the flu mutant (Przybyla et al., 2008). Thus, cellular responses that produce hydroxyl radical, singlet oxygen, or superoxide could overlap. Indeed, although about 50 genes encoding putative transcription factors in the case of flu were shown to be rapidly up-regulated upon the generation of singlet oxygen, it is difficult to point to a purely singlet oxygen-induced transcript (Kim et al., 2008). It is possible that the difference in the response of the flu mutant to early (0.5 and 1 h) and later (2 h) light exposure, which clustered with H2O2 and ozone (Fig. 1, cluster B), represents this scenario, where the early response to singlet oxygen overproduction might be associated with singlet oxygen while the later response might be associated with lipid peroxidation.
Cross reactivity was predominant in three disparate ROS indices: flu, H2O2, and ozone. In addition, they appear to be coregulated together by a multitude of stresses of abiotic and biotic origin (Figs. 1, cluster B, and 3–5). It is not clear if singlet oxygen is a common intermediate, or if flu, H2O2, and ozone stresses converge to impact on a common pathway. Within the indices themselves, H2O2 is an important source of singlet oxygen and can disproportionate to singlet oxygen in the presence of ions like Mo6+ or Ca2+ (Wahlen et al., 2005). Ozone was shown to readily generate singlet oxygen during interaction with the large amounts of ascorbate that plant cells maintain (Kanofsky and Sima, 1995). Thus, each of the indices can, in theory, generate singlet oxygen, and the results of the ROSMETER analysis are consistent with this possibility. However, only the application of real-time visualization technology for singlet oxygen can resolve the issue.
Another case of cross reactivity is apparent in the MV 12-h treatment, which shares high similarity to flu, H2O2, and ozone indices (Fig. 1). This time point is exceptional, as superoxide produced by MV would reach a peak in the 16-h-light/8-h-dark cycle in which the plants were maintained (Kilian et al., 2007) and that can be dismutated to H2O2. It is also possible that, at this time point, the excess light via singlet oxygen would lead to the depletion of carotenoids, which are major scavenging compounds (Ramel et al., 2012). Thus, either superoxides or singlet formation may be the reason that the flu, H2O2, ozone, and MV 12-h indices tend to participate in cross correlation (Fig. 1).
Nonetheless, several indices exhibit specific responses. For example, rotenone treatment is correlated with that of TDNA-AOX1 exposed to mild light stress (Fig. 1) and also to oligomycin, another inhibitor of mitochondrial metabolism (Fig. 2A). Rotenone inhibits the transfer of electrons from iron-sulfur centers in complex I to ubiquinone, and that inhibition can produce ROS in animal mitochondria (Li et al., 2003). Although in Arabidopsis, rotenone treatment does not impact the mitochondrial roGFP signal (Schwarzländer et al., 2009), it does significantly reduce ascorbate levels (Millar et al., 2003) without changing reduced glutathione levels (Garmier et al., 2008). As roGFP equilibrates with reduced glutathione (Meyer et al., 2007), no prominent change would develop in roGFP; however, a specific mitochondrial ROS footprint might develop.
Irrespective of the specificity of the indices, the output of the ROSMETER tool was corroborated in several cases by measurement of either ROS or roGFP oxidation, as demonstrated for menadione (Lehmann et al., 2009), the var mutant (Miura et al., 2010), AAL treatment (Gechev et al., 2004), root meristem (Tsukagoshi et al., 2010; Fig. 2), dark-induced senescence (Rosenwasser et al., 2011; Fig. 3A), and developmental senescence (Figs. 3B and 4).
Novel Insights Provided by ROSMETER Analysis
The ROSMETER platform analysis of consecutive days during bolting and senescence showed a clear correlation to the mitochondrial ROS footprint and a negative correlation to the chloroplast ROS footprint (Fig. 3B). This result was corroborated by the direct measurement of roGFP oxidation in mitochondria (Fig. 4). Positive correlations to mitochondrial stress were also found in dark-induced senescence (Rosenwasser et al., 2011; Fig. 3A), suggesting that developmental and induced senescence yield similar ROS signatures. The dramatic increase of both roGFP oxidation and the mitochondria-related transcriptomic signature corresponds to bolting, and this developmental step was shown previously to be associated with an increase in H2O2 from whole plants (Miao et al., 2004). Hence, the use of the ROSMETER platform has shown that mitochondrial oxidative stress is an important signaling event during developmental senescence.
ROSMETER also shows positive correlation to the indices representing peroxisomal stress (AT treatment; Fig. 3), but the correlation values to those indices were lower than for the mitochondrial indices. Similarly, a slight increase in roGFP oxidation was found in peroxisomes (Fig. 4). These results are in agreement with the finding that H2O2 levels and lipid peroxidation rate increased significantly in peroxisomes during senescence (del Rio et al., 1998). Taken together, it is suggested that ROS stresses emanating from the mitochondria and peroxisomes occur early during natural senescence. Hence, mitochondria may play a central role as subcellular sensors of senescence and trigger senescence-activated genes.
In general, the ROSMETER analysis shows that the indices of group B (Fig. 1; flu, H2O2, and ozone) are activated in most of the stresses except in cold (6–24 h), osmotic (6–24 h), and salt treatments, further emphasizing different pathways for abiotic stresses. ROSMETER analysis also shows that early times of stress responses (cold, drought, and heat) clustered together, while the later time responses tended to cluster according to the type of stress (Fig. 5). This may suggest a bimodal response to these stresses. Significant negative correlations were found between early responses to heat, drought, and cold and indices of the H2O2, ozone, and flu. However, at later time points, drought stress exhibits major positive correlations with these indices in addition to those of MV. Water deficiency may lead to the disruption of photosynthesis, possibly by the overproduction of superoxide ions at PSI by the Mehler reaction, which can be further dismutated to H2O2 by SOD. This is because, under water stress, CO2 availability is reduced, due to stomata closure, leading to overreduction of the electron transport chain (Asada, 2006; Miller et al., 2010). Singlet oxygen typical of a flu mutant can also be generated under water stress conditions at PSII by excited triplet-state chlorophyll at the P680 reaction center and in the light-harvesting complex when the electron transport chain is overreduced (Asada, 2006; Miller et al., 2010).
Heat stress is characterized by the most unusual indices showing correlation to seemingly disparate indices, flu, CAT2HP8h, and MV 12 h, that generally do not group together. This is possible because heat can generate two distinct signals correlating with different gene sets. Indeed, only 11 of the selected genes overlapped with flu, CAT2HP8h, and MV 12 h (Supplemental Table S7). In comparison with abiotic stresses, the biotic stresses, especially that of virulent and avirulent P. syringae, yield correlations to most of the indices, including those of ROS stress related to the cytoplasm (represented by KO-APX1). This may be indicative of a massive and prolonged ROS response. Curiously, little difference is seen between virulent and avirulent responses as far as the ROS signature is concerned. This indicates that the hypersensitive response during avirulence does not generate specific detectable ROS beyond that seen in the virulent responses. Hence, the ROS signals seen here are not involved in mediating the resistance response or are masked by other major R gene-independent ROS processes.
The insight gained by the ROSMETER analysis offers a new portal for formulating hypotheses in ROS-related research. The ROSMETER tool can be accessed through http://app.agri.gov.il/Noa/ROSMETER.php.
CONCLUSION
It is demonstrated that a newly developed bioinformatic tool, ROSMETER, can discern between the responses of different ROS types and their subcellular origins. This tool can be used to characterize the ROS response of various stresses and mutants for the purpose of formulating new hypotheses related to the involvement of ROS. In this study, this tool shows new insight into senescence and biotic and abiotic stresses.
MATERIALS AND METHODS
The ROSMETER Platform
ROS-related experiments used for ROSMETER and the details of the experiments are described in Supplemental Table S1. Affymetrix CEL files were downloaded from the following sites: (1) from the Gene Expression Omnibus (GEO) databases, flu mutant (GSE10876), AS-AOX1 (GDS1532), ozone treatment (GSE5722), H2O2 treatment (GSE5530; http://www.ncbi.nlm.nih.gov/geo/); (2) from the ArrayExpress database, TDNA-AOX1 plants grown under normal conditions or with moderate light and drought treatment (E-ATMX-32), rotenone treatment (E-MEXP-1797), CAT2HP1 (E-MEXP-449; http://www.ebi.ac.uk/microarray-as/ae/); (3) from The Arabidopsis Information Resource database: AtGenExpress oxidative stress in response to MV treatment (http://arabidopsis.org/servlets/TairObject?type=expression_set&id=1007966941). CEL files of KD-SOD and KO-APX1 experiments were obtained from Gadjev et al. (2006). Expression data (fold change and P values) of AT-treated plants were obtained from Gechev et al. (2005).
All CEL files were normalized by robust multiarray average, and fold change was calculated for all time points of all experiments using the average expression values of the treatments/mutants relative to the control. Statistical significance (P values) based on two-tailed Student’s t test was calculated per each gene. CEL file processing and statistical analyses were done using the ROBIN software (Lohse et al., 2010). In the case of flu mutant data, only one replicate for each time point was available. In that case, the 1,000 genes that show the highest fold change values were selected for indexing. Only genes that appear in the Affymetrix ATH1 were included in the analysis. The conditions of the treatments are summarized in Supplemental Table S1.
Gene response indices based on these ROS-related experiments were compiled using the same methods used for building HORMONOMETER (Volodarsky et al., 2009). Gene expression fold change values of different treatments/mutants versus control samples were extracted from each of the ROS-related experiments. A Perl script was used to extract from each experiment a gene expression list composed of 1,000 highest absolute fold change values that have P < 0.05. This constitutes the index of that particular experiment. We limited the number of genes in each index to 1,000, since, in general, the maximum number of detectable modified genes is less than 1,000 for most treatments. Moreover, by computing the average vector correlation values using different size indices, Volodarsky et al. (2009) showed that the values rapidly converged and hardly changed between index sizes of 500 and 1,000. Following the extraction of significantly changed transcripts, the script extracts the fold change value for each transcript in the ROS indices and in the examined gene expression data and uses an algebraic vector-based comparison to compare each index with the examined experiment. The resulting correlation values are between −1 and 1. The closer the two vectors are, the higher the result. Two opposite vectors will result in a correlation value of −1. To avoid high correlation values resulting from small numbers of genes, only correlation values generated from the comparison of at least 45 genes were presented. The correlation values were represented as a heat map by using MATLAB code.
ROSMETER can be accessed through user interface by submission of data in a “comma separated values” file format that includes fold change and P values for each gene in the transcriptome to be examined (detailed instructions can be found on the Web site and in Supplemental Instruction S1). The user receives the computed correlation ranks for each of the indices in a table and a clustergram that arranges the experiments according to their similarity to each other in terms of the ranks.
Microarray Data of Stress-Related Experiments
Supplemental Table S8 summarizes the microarray experiments that were analyzed using ROSMETER. For experiments in which the Affymetrix platform were used, the CEL files containing Affymetrix expression data were downloaded from GEO databases (http://www.ncbi.nlm.nih.gov/projects/geo/). Robust multiarray average-normalized data, fold change values, and P values were obtained using ROBIN software for each experiment (Lohse et al., 2010). Expression data of rotenone and oligomycin treatments were made by Clifton et al. (2005) and were downloaded from Schwarzländer et al. (2012). Agilent microarray data for the var mutant were analyzed using Partek software. Agilent platform Arabidopsis (Arabidopsis thaliana) data for the AAL experiments were supplied by Gechev et al. (2004). Data for the menadione treatment (60 μm) were obtained from the Arabidopsis oligonucleotide microarrays version 3.0 platform (http://ag.arizona.edu/microarray/) and downloaded from Lehmann et al. (2009). Since the ROSMETER was made based on the Affymetrix ATH1 chip, where expression data were obtained using other platforms (Agilent, CATMA), only genes that also appear in the Affymetrix platform were used for the analysis.
Senescence data of darkened detached leaves and darkened attached leaves were obtained from van der Graaff et al. (2006), and for comparison, we added the dark-induced senescence data of detached rosettes (Rosenwasser et al., 2011). The developmental senescence data were obtained from Breeze et al. (2011). Data of the mean values for eight biological replicates for each of 11 d were used, and all data were normalized to the first day (19 DAS). Expression data were downloaded from the CATdb Web site (http://urgv.evry.inra.fr/cgi-in/projects/CATdb/catdb_index.pl). A total of 10,125 genes that appear in both the CATMA microarray platform and the Affymetrix chip (ATH1) were used.
Measurements of roGFP Oxidation/Reduction
Arabidopsis wild-type plants or those expressing either roGFP2 in the mitochondria (mit-roGFP2) and plastids (pla-roGFP2) or GRX1-roGFP2 in cytoplasm (cyt-GRX1-roGFP2) and peroxisomes (per-GRX1-roGFP2) were grown at 21°C under a 12-h-day/12-h-night cycle and illuminated at 80 μmol m−2 s−1. Leaf discs were excised from the seventh leaf at consecutive days starting from 30 DAS. Bolting occurred at 32 DAS. The roGFP degree of oxidation was determined by the fluorometer Synergy TM2 (BioTek Instruments), as described (Rosenwasser et al., 2010). Leaf discs were excited by using 400- ± 15- and 485- ± 10-nm filters, and fluorescence values were measured using a 525- ± 10-nm emission filter. Fluorescence was first measured in the resting state in each disc. This was followed by the consecutive addition of 50 mm dithiothreitol followed by 1 m H2O2 (Rosenwasser et al., 2010). The latter treatments gave maximal reduced or oxidized states of roGFP fluorescence, respectively. Background emission intensities were obtained for 16 leaf discs of wild-type plants exposed to the same excitation wavelengths under the same conditions, and these values were averaged and subtracted from the roGFP values. The degree of oxidation of roGFP was calculated according to Schwarzländer et al. (2008).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ROS signatures in shoot versus root following treatment with heat or UV.
Supplemental Table S1. Summary of the 11 ROS-related experiments included in the ROSMETER platform.
Supplemental Table S2. Table of correlation values for Figure 1.
Supplemental Table S3. Table of correlation values for Figure 2.
Supplemental Table S4. Table of correlation values for Figure 3.
Supplemental Table S5. Table of correlation values for Figure 5.
Supplemental Table S6. Table of correlation values for Figure 6.
Supplemental Table S7. Comparison between flu, CAT2HP8h, and MV 12-h indices in heat stress.
Supplemental Table S8. A list of experiments that were analyzed for ROS signature using ROSMETER.
Supplemental Instruction S1. Instructions for submission to ROSMETER site.
Glossary
- ROS
reactive oxygen species
- SOD
superoxide dismutase
- H2O2
hydrogen peroxide
- AT
3-aminotriazole
- MV
methyl viologen
- roGFP
redox-sensitive GFP
- NBT
nitroblue tetrazolium
- DAS
days after sowing
- GEO
Gene Expression Omnibus
References
- Asada K. (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashtamker C, Kiss V, Sagi M, Davydov O, Fluhr R. (2007) Diverse subcellular locations of cryptogein-induced reactive oxygen species production in tobacco Bright Yellow-2 cells. Plant Physiol 143: 1817–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert GP, Møller ALB, Kristiansen KA, Schulz A, Møller IM, Schjoerring JK, Jahn TP. (2007) Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J Biol Chem 282: 1183–1192 [DOI] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton R, Lister R, Parker KL, Sappl PG, Elhafez D, Millar AH, Day DA, Whelan J. (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol Biol 58: 193–212 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, del Río LA. (2001) Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci 6: 145–150 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. (2005a) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. (2005b) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio LA, Pastori GM, Palma JM, Sandalio LM, Sevilla F, Corpas FJ, Jimenez A, Lopez-Huertas E, Hernandez JA. (1998) The activated oxygen role of peroxisomes in senescence. Plant Physiol 116: 1195–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2003) Redox sensing and signaling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant 119: 355–364 [Google Scholar]
- Foyer CH, Noctor G. (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F. (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmier M, Carroll AJ, Delannoy E, Vallet C, Day DA, Small ID, Millar AH. (2008) Complex I dysfunction redirects cellular and mitochondrial metabolism in Arabidopsis. Plant Physiol 148: 1324–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Gadjev IZ, Hille J. (2004) An extensive microarray analysis of AAL-toxin-induced cell death in Arabidopsis thaliana brings new insights into the complexity of programmed cell death in plants. Cell Mol Life Sci 61: 1185–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Minkov IN, Hille J. (2005) Hydrogen peroxide-induced cell death in Arabidopsis: transcriptional and mutant analysis reveals a role of an oxoglutarate-dependent dioxygenase gene in the cell death process. IUBMB Life 57: 181–188 [DOI] [PubMed] [Google Scholar]
- Giraud E, Ho LHM, Clifton R, Carroll A, Estavillo G, Tan Y-F, Howell KA, Ivanova A, Pogson BJ, Millar AH, et al (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JJ, Loake GJ. (2000) Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol 124: 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC (2007) Free Radicles in Biology and Medicine. Oxford University Press, New York [Google Scholar]
- Jing HC, Hebeler R, Oeljeklaus S, Sitek B, Stühler K, Meyer HE, Sturre MJ, Hille J, Warscheid B, Dijkwel PP. (2008) Early leaf senescence is associated with an altered cellular redox balance in Arabidopsis cpr5/old1 mutants. Plant Biol (Stuttg) (Suppl 1) 10: 85–98 [DOI] [PubMed] [Google Scholar]
- Kanofsky JR, Sima PD. (1995) Singlet oxygen generation from the reaction of ozone with plant leaves. J Biol Chem 270: 7850–7852 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kim C, Meskauskiene R, Apel K, Laloi C. (2008) No single way to understand singlet oxygen signalling in plants. EMBO Rep 9: 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Muthuramalingam M, Dietz K-J. (2012) Mechanisms and dynamics in the thiol/disulfide redox regulatory network: transmitters, sensors and targets. Curr Opin Plant Biol 15: 261–268 [DOI] [PubMed] [Google Scholar]
- Kuruvilla FG, Shamji AF, Sternson SM, Hergenrother PJ, Schreiber SL. (2002) Dissecting glucose signalling with diversity-oriented synthesis and small-molecule microarrays. Nature 416: 653–657 [DOI] [PubMed] [Google Scholar]
- Lehmann M, Schwarzländer M, Obata T, Sirikantaramas S, Burow M, Olsen CE, Tohge T, Fricker MD, Møller BL, Fernie AR, et al (2009) The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol Plant 2: 390–406 [DOI] [PubMed] [Google Scholar]
- Leshem Y, Melamed-Book N, Cagnac O, Ronen G, Nishri Y, Solomon M, Cohen G, Levine A. (2006) Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proc Natl Acad Sci USA 103: 18008–18013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, Robinson JP. (2003) Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem 278: 8516–8525 [DOI] [PubMed] [Google Scholar]
- Lohse M, Nunes-Nesi A, Krüger P, Nagel A, Hannemann J, Giorgi FM, Childs L, Osorio S, Walther D, Selbig J, et al. (2010) Robin: an intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol 153: 642–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R. (2007) Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52: 973–986 [DOI] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U. (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Millar AH, Mittova V, Kiddle G, Heazlewood JL, Bartoli CG, Theodoulou FL, Foyer CH. (2003) Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol 133: 443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Miura E, Kato Y, Sakamoto W. (2010) Comparative transcriptome analysis of green/white variegated sectors in Arabidopsis yellow variegated2: responses to oxidative and other stresses in white sectors. J Exp Bot 61: 2433–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Ronsein GE, Prado FM, Uemi M, Corrêa TC, Toma IN, Bertolucci A, Oliveira MCB, Motta FD, Medeiros MHG, et al (2007) Biological hydroperoxides and singlet molecular oxygen generation. IUBMB Life 59: 322–331 [DOI] [PubMed] [Google Scholar]
- Møller IM. (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- Møller IM, Sweetlove LJ. (2010) ROS signalling: specificity is required. Trends Plant Sci 15: 370–374 [DOI] [PubMed] [Google Scholar]
- Nyathi Y, Baker A. (2006) Plant peroxisomes as a source of signalling molecules. Biochim Biophys Acta 1763: 1478–1495 [DOI] [PubMed] [Google Scholar]
- op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Gobel C, Feussner I, et al (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybyla D, Göbel C, Imboden A, Hamberg M, Feussner I, Apel K. (2008) Enzymatic, but not non-enzymatic, 1O2-mediated peroxidation of polyunsaturated fatty acids forms part of the EXECUTER1-dependent stress response program in the flu mutant of Arabidopsis thaliana. Plant J 54: 236–248 [DOI] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Cuiné S, Triantaphylidès C, Ravanat J-L, Havaux M. (2012) Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiol 158: 1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DM, Umbach AL, Subbaiah CC, Siedow JN. (2006) Mitochondrial reactive oxygen species: contribution to oxidative stress and interorganellar signaling. Plant Physiol 141: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R. (2003) The water-water cycle is essential for chloroplast protection in the absence of stress. J Biol Chem 278: 38921–38925 [DOI] [PubMed] [Google Scholar]
- Rosenwasser S, Rot I, Meyer AJ, Feldman L, Jiang K, Friedman H. (2010) A fluorometer-based method for monitoring oxidation of redox-sensitive GFP (roGFP) during development and extended dark stress. Physiol Plant 138: 493–502 [DOI] [PubMed] [Google Scholar]
- Rosenwasser S, Rot I, Sollner E, Meyer AJ, Smith Y, Leviatan N, Fluhr R, Friedman H. (2011) Organelles contribute differentially to reactive oxygen species-related events during extended darkness. Plant Physiol 156: 185–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi M, Fluhr R. (2001) Superoxide production by plant homologues of the gp91(phox) NADPH oxidase: modulation of activity by calcium and by tobacco mosaic virus infection. Plant Physiol 126: 1281–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi M, Fluhr R. (2006) Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol 141: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito I, Matsuura T, Inoue K. (1981) Formation of superoxide ion from singlet oxygen: use of a water-soluble singlet oxygen source. J Am Chem Soc 103: 188–190 [Google Scholar]
- Sasaki E, Takahashi C, Asami T, Shimada Y. (2011) AtCAST, a tool for exploring gene expression similarities among DNA microarray experiments using networks. Plant Cell Physiol 52: 169–180 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M, Fricker MD, Müller C, Marty L, Brach T, Novak J, Sweetlove LJ, Hell R, Meyer AJ. (2008) Confocal imaging of glutathione redox potential in living plant cells. J Microsc 231: 299–316 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M, Fricker MD, Sweetlove LJ. (2009) Monitoring the in vivo redox state of plant mitochondria: effect of respiratory inhibitors, abiotic stress and assessment of recovery from oxidative challenge. Biochim Biophys Acta 1787: 468–475 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M, König A-C, Sweetlove LJ, Finkemeier I. (2012) The impact of impaired mitochondrial function on retrograde signalling: a meta-analysis of transcriptomic responses. J Exp Bot 63: 1735–1750 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G. (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35: 259–270 [DOI] [PubMed] [Google Scholar]
- Torres MA. (2010) ROS in biotic interactions. Physiol Plant 138: 414–429 [DOI] [PubMed] [Google Scholar]
- Triantaphylidès C, Krischke M, Hoeberichts FA, Ksas B, Gresser G, Havaux M, Van Breusegem F, Mueller MJ. (2008) Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol 148: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Fiorani F, Siedow JN. (2005) Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol 139: 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inzé D, Van Breusegem F. (2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139: 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge U-I, Kunze R. (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volodarsky D, Leviatan N, Otcheretianski A, Fluhr R. (2009) HORMONOMETER: a tool for discerning transcript signatures of hormone action in the Arabidopsis transcriptome. Plant Physiol 150: 1796–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Przybyla D, op den Camp R, Kim C, Landgraf F, Lee KP, Würsch M, Laloi C, Nater M, Hideg E, et al (2004) The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]
- Wahlen J, De Hertogh S, De Vos DE, Nardello V, Bogaert S, Aubry J-M, Alsters PL, Jacobs PA. (2005) Disproportionation of hydrogen peroxide into singlet oxygen catalyzed by lanthanum-exchanged zeolites. J Catal 233: 422–433 [Google Scholar]