Abstract
Plant cell nuclei can generate calcium responses to a variety of inputs, tantamount among them the response to signaling molecules from symbiotic microorganisms.
An astonishing variety of plant and animal cellular functions are coordinated by intracellular calcium (Ca2+; Berridge et al., 2000; White and Broadley, 2003). Although Ca2+ is a common second messenger, it is translated into specific developmental processes, implying specificity of recognition of individual Ca2+ responses (Dolmetsch et al., 1997, 1998). Such specificity in Ca2+ signaling is a function of its mechanism of activation, the spatial nature of its release, and the developmental context within which the Ca2+ response occurs (Berridge et al., 2000; Evans et al., 2001; Ng and McAinsh, 2003; Di Capite et al., 2009). Together, the frequency, amplitude, and spatial location of the Ca2+ release can differ, and these lead to variations in Ca2+ responses, commonly known as the Ca2+ signature (McAinsh and Pittman, 2009). The spatial differences across Ca2+ responses derive from Ca2+ release from diverse stores, with differential activation of Ca2+ channels that occur in restricted locations within the cell (Berridge et al., 2000; Di Capite et al., 2009; McAinsh and Pittman, 2009).
It has been hotly debated whether the nucleus of animal cells can act as an independent Ca2+ compartment from the rest of the cell. In animal cells, the generation of nucleoplasmic Ca2+ signals has been shown to be essential to regulate specific processes, including transcription, cell growth, and proliferation (Bootman et al., 2009), but the origin of nucleoplasmic Ca2+ increase in animal cells remains contentious. The Ca2+ channels inositol-1,4,5-trisphosphate and ryanodine receptors, as well as Ca2+-ATPases, are all present on the inner nuclear envelope (Bootman et al., 2009), highlighting the potential of the nucleus to independently generate Ca2+ signals. In contrast to the animal field, it is widely accepted that the nuclei of plant cells can produce an autonomous Ca2+ response (Pauly et al., 2000, 2001; Mazars et al., 2009), and these nuclear Ca2+ events have distinct biological roles from those regulated by cytosolic Ca2+ release. For example, transient nuclear Ca2+ events are required for wind-induced calmodulin expression (van Der Luit et al., 1999) and for sphingolipid-induced programmed cell death (Lachaud et al., 2010). Nuclear Ca2+, whether autonomous or of cytoplasmic origin, functions as a second messenger to stimulate numerous Ca2+-sensitive processes, notably transcriptional regulation (Kaplan et al., 2006; Whalley et al., 2011), by binding to Ca2+-sensing proteins such as calmodulin, transcription factors, kinases, or phosphatases (Galon et al., 2010; Reddy et al., 2011). In this review, we discuss the latest knowledge on nuclear Ca2+ signaling in plants. Among of the best plant models for nuclear Ca2+ signaling are the Ca2+ oscillations that occur during symbiotic signaling (Ehrhardt et al., 1996; Miwa et al., 2006; Sieberer et al., 2009; Chabaud et al., 2011), and while not exclusive, this review will focus on this signaling process. Over the past decade, tremendous progress has been made in understanding the mechanisms of encoding and decoding Ca2+ oscillations during symbiotic signaling, and this provides a platform for understanding nuclear Ca2+ signaling more broadly in plants.
MEASURING Ca2+ IN PLANT NUCLEI
A number of approaches have been used to measure Ca2+ responses in plants and, in particular, in the nucleus. Initially, microinjection with Ca2+-responsive dyes was used, and this revealed nuclear Ca2+ responses, such as the Ca2+ oscillations induced in legume root hair cells in response to the rhizobial signaling molecule Nod factor (Ehrhardt et al., 1996). Such dyes are not restricted to the nucleus; thus, the nuclear changes could only be surmised by measuring fluorescence changes within the nuclear region. Concurrent with the use of Ca2+-responsive dyes has been the development of Ca2+-responsive proteins, whether naturally occurring such as aequorin or synthetic Ca2+ reporters such as cameleon (Allen et al., 1999; Knight et al., 1991; van Der Luit et al., 1999). The advantage of such protein reporters is the ability to target these proteins to different cellular compartments, including the nucleus (Mithöfer and Mazars, 2002; Krebs et al., 2012; Mehlmer et al., 2012). This has provided conclusive proof of Ca2+ changes within the nucleoplasm of plant cells as well as providing the ease of measurement of nuclear Ca2+ events in response to a variety of different stimuli, such as sphingolipids (Lachaud et al., 2010), wind or cold stresses (van Der Luit et al., 1999), mastoparan (Pauly et al., 2001), osmotic shocks (Pauly et al., 2000), pathogen elicitors (Lecourieux et al., 2005), jasmonic acid (Walter et al., 2007), and symbiotic signals (Sieberer et al., 2009; Chabaud et al., 2011).
LINKING SIGNAL PERCEPTION TO THE ACTIVATION OF NUCLEAR Ca2+ RELEASE
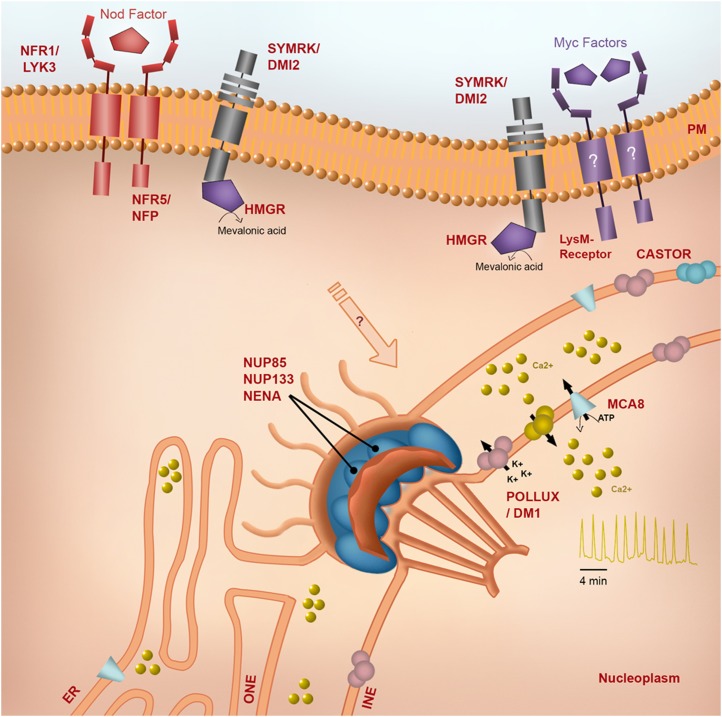
The best-studied inducers of nuclear Ca2+ responses are the symbiotic signals produced by rhizobial bacteria and arbuscular mycorrhizal (AM) fungi. Such Nod factors and Myc factors are lipochitooligosaccharides (LCOs) with a variety of modifications, dependent on the producing organism (Lerouge et al., 1990; Maillet et al., 2011; Genre et al., 2013). Recognition of these LCO signals involves plasma membrane LysM receptor-like kinases (Fig. 1; Amor et al., 2003; Madsen et al., 2003, 2011; Radutoiu et al., 2003; Smit et al., 2007; Antolín-Llovera et al., 2012) that, at least for the Nod factor receptors, have been shown to bind directly to the appropriate LCO signal (Broghammer et al., 2012). Recognition of rhizobia, AM fungi, and their LCO signals leads to the induction of Ca2+ oscillations in the nucleus (Sieberer et al., 2009; Genre et al., 2013) and, considering their recognition by plasma membrane-localized receptors, implies the production of diffusible secondary messengers that can link recognition at the plasma membrane to the Ca2+ changes in the nucleus. While the precise nature of these secondary messengers remains elusive, a number of clues provide indications of their potential structure.
Figure 1.
The symbiotic machinery required to induce nuclear-localized Ca2+ oscillations. In L. japonicus (Lj) and M. truncatula (Mt), the Nod factor receptors LjNFR1/MtLYK3 and LjNFR5/MtNFP are associated with the recognition of Nod factor (Amor et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Smit et al., 2007; Broghammer et al., 2012). SYMRK is positioned at the cross point between Nod factor and AM perception (Endre et al., 2002; Stracke et al., 2002) and is proposed to form a complex with the Nod factor receptors and the yet to be identified AM receptors (Antolín-Llovera et al., 2012). SYRMK interacts with 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), which is involved in the production of mevalonic acid, a precursor for many potential second signals (Stermer et al., 1994; Kevei et al., 2007). Downstream of SYMRK are the nucleoporins NUP85, NUP133, and NENA, the ion channels LjPOLLUX (MtDMI1) and LjCASTOR, as well as the Ca2+ pump MCA8 (Kanamori et al., 2006; Saito et al., 2007; Charpentier et al., 2008; Groth et al., 2010; Capoen et al., 2011). In L. japonicus, the function of DMI1 is divided between both POLLUX and CASTOR. The selectivity filter of DMI1 differs from CASTOR and POLLUX by one amino acid, which seems sufficient to fulfill both functions (Venkateshwaran et al., 2012), making MtCASTOR redundant. ER, Endoplasmic reticulum; PM, plasma membrane.
The G-protein agonist mastoparan and its synthetic analog Mas7 were shown to activate Ca2+ oscillations in a manner analogous to Nod factor-induced responses (Charron et al., 2004; Sun et al., 2007). Furthermore, inhibitors of phospholipase D and phospholipase C block Nod factor-induced Ca2+ oscillations (Engstrom et al., 2002; Charron et al., 2004). Together, these data indicate that G-proteins may induce nuclear Ca2+ oscillations through their regulation of phospholipases. Inositol phosphates, which are products of phospholipase C, can coordinate Ca2+ responses in plant cells (Gilroy et al., 1990), and they are major activators of Ca2+ channels in animal cells (Bootman et al., 2009). However, there is currently no direct link demonstrated between the Nod factor receptors and the phospholipases or G-proteins; furthermore, there is little evidence for inositol phosphates functioning in symbiosis signaling. At the plasma membrane, the symbiosis receptor-like kinase, SYMRK, is hypothesized to complex with the Nod and Myc factor receptors (Antolín-Llovera et al., 2012) and to be associated with the production of the secondary messenger that activates nucleoplasmic Ca2+ oscillations. Proteins that interact with SYMRK include a plant mitogen-activated protein kinase kinase (Chen et al., 2012), and a 3-hydroxy-3-methylglutaryl-CoA reductase, HMGR1 (Kevei et al., 2007), and both of these were found to positively regulate the rhizobial association (Kevei et al., 2007; Chen et al., 2012). 3-Hydroxy-3-methylglutaryl-CoA reductase, an enzyme involved in lipid signaling via mevalonate production, and mitogen-activated protein kinase kinase could both be involved in downstream signaling through the generation of secondary messengers and phosphorylation cascades, respectively (Stermer et al., 1994; Taj et al., 2010). Both could be associated directly or indirectly in the activation or modulation of the symbiotic Ca2+ channel.
THE NUCLEAR MACHINERY REQUIRED FOR SYMBIOTIC Ca2+ RESPONSES
The genetic dissection of plant symbioses has led to the identification of a number of proteins present in the nucleus that have roles in the generation of Ca2+ oscillations. Three nucleoporins, NUP85, NUP133, and NENA (Kanamori et al., 2006; Saito et al., 2007; Groth et al., 2010), are all part of the nucleopore scaffold (Alber et al., 2007) and are required for symbiotic Ca2+ oscillations (Fig. 1). The main function of the nucleopore complex is to mediate macromolecular transport, such as mRNA export and protein import across the nuclear envelope (Alber et al., 2007). The role of these nucleoporins in the generation of Ca2+ oscillations could be associated with the diffusion of a symbiotic signal from the plasma membrane to the nucleus in order to activate nucleoplasmic Ca2+ oscillations. However, an alternative explanation is the transport of specific integral membrane proteins to the inner nuclear membrane, and in Saccharomyces cerevisiae, nucleoporins are required for this function (Deng and Hochstrasser, 2006; King et al., 2006). The specific yeast nucleoporins required are Nup188 and Nup170, which, like NUP85, NUP133, and NENA, locate at the nucleopore scaffold (Alber et al., 2007). These observations suggest that the nucleopore scaffold could play a role in translocating proteins to the inner nuclear membrane that are essential for the generation of Ca2+ oscillations. In Medicago truncatula, the ion channel DMI1 (Lotus japonicus homolog POLLUX) and the SERCA-type Ca2+-ATPase MCA8 are essential for nucleoplasmic Ca2+ oscillations (Capoen et al., 2011; Venkateshwaran et al., 2012), and both localize to the nuclear membranes (refer to Fig. 1; Riely et al., 2007; Capoen et al., 2011). However, in contrast to MCA8, DMI1 was shown to preferentially localize to the inner nuclear membrane (Capoen et al., 2011), and the targeting of this protein may be at least one of the roles of the nuclear pore scaffold in the generation of symbiotic Ca2+ oscillations.
The localization of ion channels and a Ca2+-ATPase at the nuclear envelope (Fig. 1; Riely et al., 2007; Charpentier et al., 2008; Capoen et al., 2011), as well as the spatiotemporal analyses showing the emergence of Ca2+ oscillations predominantly at the periphery of the nucleus (Sieberer et al., 2009; Capoen et al., 2011), strongly suggest that the lumen of the nuclear envelope contiguous with the endoplasmic reticulum constitutes the Ca2+ store for symbiotic Ca2+ signaling. This observation suggests that the components localized at the nuclear envelope/endoplasmic reticulum are primarily involved in controlling the release of Ca2+. The nuclear-localized ion channel DMI1 (L. japonicus POLLUX), which permeates potassium, seems unlikely to be directly responsible for the Ca2+ release (Charpentier et al., 2008; Venkateshwaran et al., 2012). Indeed, pharmacological and yeast expression analyses highlight that DMI1 might be a tight regulator of the yet unidentified symbiotic Ca2+ channel (Peiter et al., 2007). In agreement with this observation, mathematical modeling reveals that the association of three components (DMI1, a putative voltage/ligand-activated Ca2+ channel, and a Ca2+ pump) is sufficient to produce the symbiotic Ca2+ oscillations (Granqvist et al., 2012). This mathematical modeling suggests that DMI1 functions to regulate the Ca2+ channel as a counter ion channel and a modulator of membrane potential in two steps (Charpentier et al., 2013). First, activation of DMI1 generates a potassium current that facilitates an initial, limited Ca2+ release via a partially activated Ca2+ channel. This Ca2+ release provides a positive feedback, via a predicated Ca2+-binding pocket in DMI1 (Edwards et al., 2007), that fully activates DMI1, whose potassium influx hyperpolarizes the membrane to open a putative voltage-gated Ca2+ channel. The Ca2+ released is then pumped back into the store via the Ca2+-ATPase. In this mathematical model, the positive Ca2+ feedback and the voltage fluctuation of the nuclear envelope play a major role in sustaining the Ca2+ oscillations. Recent studies that have demonstrated the Ca2+ modulation of the nuclear envelope potential to induce Ca2+ bursts in neurons (Yamashita, 2011) and that have shown the expression of DMI1 in human embryonic kidney cells sufficient to activate Ca2+ oscillations upon Ca2+ stimulation (Venkateshwaran et al., 2012) provide support for the mathematical modeling.
IS INFORMATION ENCODED IN THE NUCLEAR Ca2+ SIGNATURE?
Intrinsic to the Ca2+ signature hypothesis is the idea that information is encoded in the structure of the Ca2+ response (McAinsh and Pittman, 2009). In mammalian cells, it is well established that the amplitude and frequency of the Ca2+ oscillations can encode the specificity of the response (Dolmetsch et al., 1998). Thus, in T lymphocyte cells, rapid and irregular Ca2+ oscillations activate different Ca2+-sensitive transcription factors, leading to specific gene expression patterns (Dolmetsch et al., 1998). In plants, evidence for information encoding came from studies in guard cells, where enforced Ca2+ oscillations of different structures gave different long-term effects for stomatal closure (Allen et al., 1999).
The nucleoplasmic Ca2+ oscillations induced by symbionts are cell autonomous, as nonsynchronous Ca2+ oscillations occur between adjacent cells (Sieberer et al., 2009; Chabaud et al., 2011). Furthermore, the structures of the oscillations differ between cells (Ehrhardt et al., 1996; Sieberer et al., 2009; Chabaud et al., 2011), providing the basis for hypothesizing information encoding within the Ca2+ response. Rhizobia and AM fungi utilize the same symbiosis signaling pathway for the activation of and response to the Ca2+ oscillations (Parniske, 2008). Despite these similarities in signaling, specificity must be maintained to ensure an appropriate response to each symbiont. Such specificity of signaling could be encoded within the Ca2+ oscillations, could be a function of as yet unknown parallel signaling, or could be associated with the differential cell types that the symbionts use for initial colonization: Rhizobia colonize through root hair cells, while AM fungi colonize atrichoblasts (refer to Fig. 2).
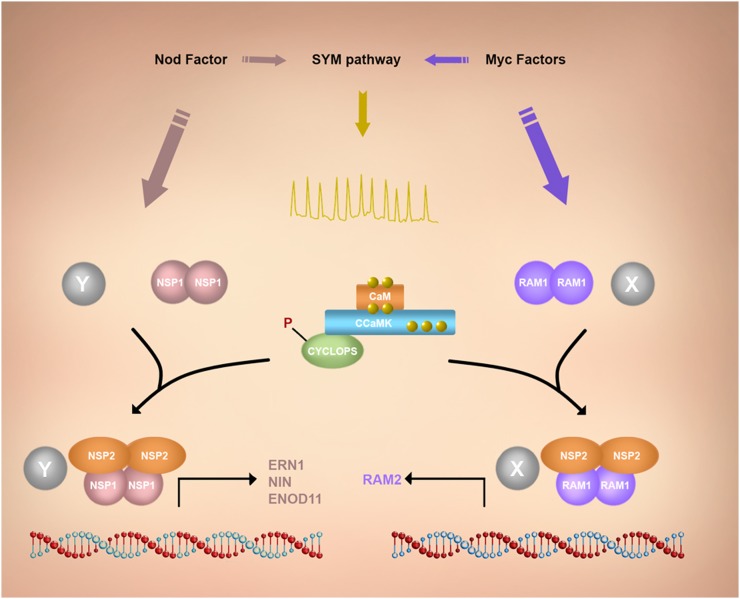
Figure 2.
Symbiotic signaling downstream of Ca2+ oscillations. The generation of Ca2+ oscillations requires components essential for both rhizobia and AM symbioses, collectively referred to as the common symbiosis (SYM) signaling pathway. CCaMK is responsible for decoding the Ca2+ oscillations through its association with Ca2+ and calmodulin (CaM; Singh and Parniske, 2012). CCaMK associates with and phosphorylates CYCLOPS (Yano et al., 2008). Downstream of CCaMK and CYCLOPS, a suit of GRAS transcription factors are required to activate nodulation or AM programs (Oldroyd, 2013). NODULATION SIGNALING PATHWAY2 (NSP2) associates with both the nodulation-specific GRAS transcription factor NSP1 (Hirsch et al., 2009) and the AM-specific GRAS transcription factor REQUIRED FOR ARBUSCULAR MYCORRHIZATION1 (RAM1; Gobbato et al., 2012). The complex NSP2/NSP1 is required for the expression of nodulation genes (ERN1, NIN, and ENOD11), while the complex NSP2/RAM1 modulates the expression of AM-specific genes such as RAM2 (Oldroyd, 2013). In this model, we hypothesize that upon symbiont stimulation (Myc or Nod factors), the transcription factor complexes are recruited by specific unknown AM (X) or nodulation (Y) components. The CCaMK/CYCLOPS complex is activated via Ca2+ oscillations and activates symbiotic gene expression, either independently or in combination with the GRAS protein complexes.
In the root hair cell, nucleoplasmic Ca2+ oscillations are induced after a lag of 6 to 20 min following Nod factor addition and reach a period of regular frequency (with approximately 100 s between each spike) after an initial short burst of high-frequency oscillations comprising three to six spikes (Ehrhardt et al., 1996; Miwa et al., 2006; Sieberer et al., 2009). Each spike produced is characterized by an asymmetric shape, resulting from an initial rapid release of Ca2+ followed by a much slower Ca2+ reuptake (Sieberer et al., 2009; Fig. 1). Although mathematical modeling suggests that the short burst of oscillations can be generated by a different buffering capacity of the cell (Granqvist et al., 2012), the biological relevance of this initial rapid burst of Ca2+ spikes remains to be determined.
Initial studies of Ca2+ oscillations induced at early stages of AM-fungal interactions before contact with the epidermal cell indicated differences to the Ca2+ oscillations activated by Nod factor, providing the basis for hypothesizing that the specificity of symbiosis signaling may be encoded within the structure of the Ca2+ response (Kosuta et al., 2008; Chabaud et al., 2011). Interestingly, chitooligosaccharides, which are produced by AM fungi in addition to LCOs, activate similar irregular and low-frequency Ca2+ oscillations (Genre et al., 2013). However, upon contact with the fungal hyphopodium, the nuclear Ca2+ oscillations induced are similar in shape and frequency to the rhizobial bacteria-induced Ca2+ oscillations (Chabaud et al., 2011; Sieberer et al., 2012). Furthermore, a direct comparison of cortical cells colonized by AM fungi and rhizobial bacteria indicates that both symbionts induced similar nuclear Ca2+ oscillations of low frequency prior to colonization and of high frequency upon colonization (Sieberer et al., 2012). These observations suggest that the Myc and Nod factors delivered by AM fungi and rhizobial bacteria might trigger diverse Ca2+ signaling responses, with the periodicity and shape of the Ca2+ response being dependent upon the concentration of symbiotic factors and probably the mix of Myc factors.
During symbiont colonization, the path of infection through the root is predicted by a preinfection structure that predicts the route of the invading AM fungus or rhizobia-colonized infection thread (van Brussel et al., 1992; Genre et al., 2005). This predicted path of infection is always directed by the cells where nuclei exhibit high-frequency Ca2+ oscillations (Sieberer et al., 2012). During rhizobial infection of root cortical cells, this high-frequency Ca2+ oscillation is sustained for 40 to 55 min, which corresponds to 35 to 45 spikes and attenuates synchronously with the infection progression (Sieberer et al., 2012). Although it is unclear whether low- and high-frequency Ca2+ oscillations induce different posttranslational or transcriptional changes, previous studies suggested that a minimum of 36 spikes were required to induce a symbiotic marker in response to Nod factor (Miwa et al., 2006). While low-frequency Ca2+ oscillations could induce the symbiotic marker, its expression was considerably delayed (Miwa et al., 2006). We propose that although the Ca2+ oscillations do not encode specific information for rhizobial or AM fungal colonization, they do encode information regarding the nature and concentration of the symbiotic signals perceived by the cell: only those cells perceiving the appropriate mix of factors at the right concentration support robust and sustained Ca2+ oscillations. Such cells undergo the appropriate programming for symbiont colonization, and these are the cells that undergo the developmental changes associated with prepenetration. It is possible that the irregular Ca2+ oscillations observed prior to symbiont colonization may play a role in the initial stages of priming the cell for symbiotic associations, or they may simply reflect cellular signaling that is insufficient to sustain a symbiotic response.
DECODING THE NUCLEOPLASMIC Ca2+ RESPONSES
The direct sensing of Ca2+ requires Ca2+-binding proteins (Batistič and Kudla, 2012), and a number of Ca2+ sensors, such as calmodulin-domain protein kinases and calmodulins, are predicted to be present in the nucleus (Biro et al., 1984; Rodríguez-Concepción et al., 1999; Reddy et al., 2011). The nuclear location of such Ca2+-decoding proteins implies that the nucleus itself has the capability to independently respond to Ca2+ signals. Clearly, in addition to such autonomous Ca2+ signaling, most cellular signaling, including Ca2+ signaling, will transduce to the nucleus; however, for simplicity in this review, we focus only on autonomous nuclear Ca2+ signaling events. Again, the best model for understanding the decoding of Ca2+ signals in the nucleus is in symbiotic signaling that utilizes a nuclear-localized Ca2+- and calmodulin-dependent Ser/Thr protein kinase (CCaMK; Lévy et al., 2004; Mitra et al., 2004; refer to Fig. 2). Gain-of-function mutations of CCaMK are sufficient to induce symbiotic processes, such as spontaneous nodulation in the absence of rhizobia (Gleason et al., 2006; Tirichine et al., 2006), and the promotion of prepenetration structures that are associated with AM colonization (Takeda et al., 2012). Moreover, in the presence of the autoactive CCaMK, the symbiotic signaling components upstream of Ca2+ oscillations become dispensable for nodulation and mycorrhization (Hayashi et al., 2010; Madsen et al., 2010). These observations indicate that the main role of the upstream signaling components is to generate Ca2+ oscillations whose predominant function is the activation of CCaMK.
CCaMK can bind Ca2+ either directly via three C-terminal EF hand domains or indirectly via a Ca2+/calmodulin-binding domain (Sathyanarayanan et al., 2000; Gleason et al., 2006; Tirichine et al., 2006; Swainsbury et al., 2012). This dual Ca2+-binding capability of CCaMK is unique compared with Ca2+-binding proteins in both animals and plants (Hrabak et al., 2003) and underlines the mechanistic complexity of this Ca2+-sensing kinase. Several studies combining homology modeling with the animal Ca2+/calmodulin-dependent protein kinase II and mutational analyses have highlighted the importance of the kinase domain and the autophosphorylation state of CCaMK to positively or negatively regulate its activity (Hayashi et al., 2010; Liao et al., 2012; Shimoda et al., 2012; Singh and Parniske, 2012; Takeda et al., 2012). The autophosphorylation of CCaMK is dependent on Ca2+ binding to the EF hand domains, while substrate phosphorylation is promoted by calmodulin binding to CCaMK (Sathyanarayanan et al., 2000; Shimoda et al., 2012). Many of the specifics of the CCaMK mode of action have been studied, and for a more detailed description of CCaMK activation, see the review by Du and Poovaiah (2013) in this edition.
CCaMK interacts with and phosphorylates the nuclear-localized CYCLOPS (Messinese et al., 2007; Yano et al., 2008; Horváth et al., 2011; refer to Fig. 2). CYCLOPS encodes a coiled-coil protein required for both AM and rhizobial infection (Yano et al., 2008; Horváth et al., 2011). Interestingly, the mutation to Asp of two of the CYCLOPS Ser residues that are phosphorylated by CCaMK creates a gain of function in CYCLOPS that leads to spontaneous nodulation when this mutant is transformed into legume roots (M. Parniske, personal communication). This observation suggests that the activation of the core complex CCaMK/CYCLOPS is sufficient to trigger downstream signaling associated with nodule organogenesis. CYCLOPS might function directly with downstream nuclear-localized GRAS family transcriptional regulators to coordinate the transcriptional events associated with rhizobial and AM invasion (Fig. 2; Kaló et al., 2005; Smit et al., 2005; Gleason et al., 2006; Hirsch et al., 2009; Gobbato et al., 2012).
CONCLUSION
The plant cell nucleus has the capability to mount an autonomous Ca2+ response, and such nuclear Ca2+ signaling is likely to be associated with a variety of processes. Because of its ease of genetic dissection, the symbiotic signaling pathway has emerged as the best model for studying nuclear Ca2+ signaling in plants. The perception of symbiotic signals leads to the generation of nucleoplasmic Ca2+ oscillations, with high-frequency Ca2+ oscillations associated with cellular programming that defines the pathway within the root for symbiont colonization. The establishment of nuclear Ca2+ oscillations involves a potassium-permeable channel, a Ca2+-ATPase, and a hypothetical Ca2+ channel that are located on the nuclear membranes. These are predicted to function in combination to sustain Ca2+ oscillations, following activation by an as yet unknown secondary messenger. One of the main purposes of the Ca2+ oscillations appears to be the activation of CCaMK and its phosphorylation of CYCLOPS. Subsequent transcriptional reprogramming to permit either AM or rhizobial colonization is dependent on the Ca2+-decoding complex defined by CCaMK/CYCLOPS and a suite of GRAS domain transcription factors (refer to Fig. 2). How the specificity of symbiosis signaling is encoded has yet to be defined, but it appears that the robustness of the Ca2+ oscillations is important to define the cells that will ultimately house the invading symbionts. While genetic dissection of symbiotic signaling has provided a framework to understand nuclear Ca2+ signaling, questions remain, particularly with regard to the nature of the secondary messenger(s) that link signal recognition at the plasma membrane to the activation of Ca2+ responses in the nucleus as well as the structure of the nuclear Ca2+ channels that coordinate this process.
Glossary
- Ca2+
intracellular calcium
- AM
arbuscular mycorrhizal
- LCO
lipochitooligosaccharide
- CCaMK
Ca2+- and calmodulin-dependent Ser/Thr protein kinase
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. (2007) The molecular architecture of the nuclear pore complex. Nature 450: 695–701 [DOI] [PubMed] [Google Scholar]
- Allen GJ, Kwak JM, Chu SP, Llopis J, Tsien RY, Harper JF, Schroeder JI. (1999) Cameleon calcium indicator reports cytoplasmic calcium dynamics in Arabidopsis guard cells. Plant J 19: 735–747 [DOI] [PubMed] [Google Scholar]
- Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C. (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Antolín-Llovera M, Ried MK, Binder A, Parniske M. (2012) Receptor kinase signaling pathways in plant-microbe interactions. Annu Rev Phytopathol 50: 451–473 [DOI] [PubMed] [Google Scholar]
- Batistič O, Kudla J. (2012) Analysis of calcium signaling pathways in plants. Biochim Biophys Acta 1820: 1283–1293 [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Lipp P, Bootman MD. (2000) The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol 1: 11–21 [DOI] [PubMed] [Google Scholar]
- Biro RL, Daye S, Serlin BS, Terry ME, Datta N, Sopory SK, Roux SJ. (1984) Characterization of oat calmodulin and radioimmunoassay of its subcellular distribution. Plant Physiol 75: 382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootman MD, Fearnley C, Smyrnias I, MacDonald F, Roderick HL. (2009) An update on nuclear calcium signalling. J Cell Sci 122: 2337–2350 [DOI] [PubMed] [Google Scholar]
- Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ, et al. (2012) Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA 109: 13859–13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoen W, Sun J, Wysham D, Otegui MS, Venkateshwaran M, Hirsch S, Miwa H, Downie JA, Morris RJ, Ané JM, et al. (2011) Nuclear membranes control symbiotic calcium signaling of legumes. Proc Natl Acad Sci USA 108: 14348–14353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M, Genre A, Sieberer BJ, Faccio A, Fournier J, Novero M, Barker DG, Bonfante P. (2011) Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol 189: 347–355 [DOI] [PubMed] [Google Scholar]
- Charpentier M, Bredemeier R, Wanner G, Takeda N, Schleiff E, Parniske M. (2008) Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20: 3467–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Vaz Martins T, Granqvist E, Oldroyd GE, Morris RJ. (2013) The role of DMI1 in establishing Ca2+ oscillations in legume symbioses. Plant Signal Behav 8: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron D, Pingret JL, Chabaud M, Journet EP, Barker DG. (2004) Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol 136: 3582–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Zhu H, Ke D, Cai K, Wang C, Gou H, Hong Z, Zhang Z. (2012) A MAP kinase kinase interacts with SymRK and regulates nodule organogenesis in Lotus japonicus. Plant Cell 24: 823–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Hochstrasser M. (2006) Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature 443: 827–831 [DOI] [PubMed] [Google Scholar]
- Di Capite J, Ng SW, Parekh AB. (2009) Decoding of cytoplasmic Ca2+ oscillations through the spatial signature drives gene expression. Curr Biol 19: 853–858 [DOI] [PubMed] [Google Scholar]
- Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. (1997) Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 386: 855–858 [DOI] [PubMed] [Google Scholar]
- Dolmetsch RE, Xu K, Lewis RS. (1998) Calcium oscillations increase the efficiency and specificity of gene expression. Nature 392: 933–936 [DOI] [PubMed] [Google Scholar]
- Edwards A, Heckmann AB, Yousafzai F, Duc G, Downie JA. (2007) Structural implications of mutations in the pea SYM8 symbiosis gene, the DMI1 ortholog, encoding a predicted ion channel. Mol Plant Microbe Interact 20: 1183–1191 [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR. (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kaló P, Kiss GB. (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Engstrom EM, Ehrhardt DW, Mitra RM, Long SR. (2002) Pharmacological analysis of Nod factor-induced calcium spiking in Medicago truncatula: evidence for the requirement of type IIA calcium pumps and phosphoinositide signaling. Plant Physiol 128: 1390–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans NH, McAinsh MR, Hetherington AM. (2001) Calcium oscillations in higher plants. Curr Opin Plant Biol 4: 415–420 [DOI] [PubMed] [Google Scholar]
- Galon Y, Finkler A, Fromm H. (2010) Calcium-regulated transcription in plants. Mol Plant 3: 653–669 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Balzergue C, Puech-Pagès V, Novero M, Rey T, Fournier J, Rochange S, Bécard G, Bonfante P, et al. (2013) Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol 198: 190–202 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG. (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17: 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Read ND, Trewavas AJ. (1990) Elevation of cytoplasmic calcium by caged calcium or caged inositol triphosphate initiates stomatal closure. Nature 346: 769–771 [DOI] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Muñoz A, Poovaiah BW, Oldroyd GE. (2006) Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Gobbato E, Marsh JF, Vernié T, Wang E, Maillet F, Kim J, Miller JB, Sun J, Bano SA, Ratet P, et al. (2012) A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr Biol 22: 2236–2241 [DOI] [PubMed] [Google Scholar]
- Granqvist E, Wysham D, Hazledine S, Kozlowski W, Sun J, Charpentier M, Martins TV, Haleux P, Tsaneva-Atanasova K, Downie JA, et al. (2012) Buffering capacity explains signal variation in symbiotic calcium oscillations. Plant Physiol 160: 2300–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M, Takeda N, Perry J, Uchida H, Dräxl S, Brachmann A, Sato S, Tabata S, Kawaguchi M, Wang TL, et al. (2010) NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Banba M, Shimoda Y, Kouchi H, Hayashi M, Imaizumi-Anraku H. (2010) A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts. Plant J 63: 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Kim J, Muñoz A, Heckmann AB, Downie JA, Oldroyd GE. (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth B, Yeun LH, Domonkos A, Halász G, Gobbato E, Ayaydin F, Miró K, Hirsch S, Sun J, Tadege M, et al. (2011) Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol Plant Microbe Interact 24: 1345–1358 [DOI] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EM, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, et al. (2006) A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA 103: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan B, Davydov O, Knight H, Galon Y, Knight MR, Fluhr R, Fromm H. (2006) Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18: 2733–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevei Z, Lougnon G, Mergaert P, Horváth GV, Kereszt A, Jayaraman D, Zaman N, Marcel F, Regulski K, Kiss GB, et al. (2007) 3-Hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 19: 3974–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G. (2006) Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 442: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ. (1991) Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Kosuta S, Hazledine S, Sun J, Miwa H, Morris RJ, Downie JA, Oldroyd GE. (2008) Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc Natl Acad Sci USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M, Held K, Binder A, Hashimoto K, Den Herder G, Parniske M, Kudla J, Schumacher K. (2012) FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J 69: 181–192 [DOI] [PubMed] [Google Scholar]
- Lachaud C, Da Silva D, Cotelle V, Thuleau P, Xiong TC, Jauneau A, Brière C, Graziana A, Bellec Y, Faure JD, et al. (2010) Nuclear calcium controls the apoptotic-like cell death induced by d-erythro-sphinganine in tobacco cells. Cell Calcium 47: 92–100 [DOI] [PubMed] [Google Scholar]
- Lecourieux D, Lamotte O, Bourque S, Wendehenne D, Mazars C, Ranjeva R, Pugin A. (2005) Proteinaceous and oligosaccharidic elicitors induce different calcium signatures in the nucleus of tobacco cells. Cell Calcium 38: 527–538 [DOI] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J. (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784 [DOI] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Liao J, Singh S, Hossain MS, Andersen SU, Ross L, Bonetta D, Zhou Y, Sato S, Tabata S, Stougaard J, et al. (2012) Negative regulation of CCaMK is essential for symbiotic infection. Plant J 72: 572–584 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Antolín-Llovera M, Grossmann C, Ye J, Vieweg S, Broghammer A, Krusell L, Radutoiu S, Jensen ON, Stougaard J, et al. (2011) Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J 65: 404–417 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Mazars C, Bourque S, Mithöfer A, Pugin A, Ranjeva R. (2009) Calcium homeostasis in plant cell nuclei. New Phytol 181: 261–274 [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Pittman JK. (2009) Shaping the calcium signature. New Phytol 181: 275–294 [DOI] [PubMed] [Google Scholar]
- Mehlmer N, Parvin N, Hurst CH, Knight MR, Teige M, Vothknecht UC. (2012) A toolset of aequorin expression vectors for in planta studies of subcellular calcium concentrations in Arabidopsis thaliana. J Exp Bot 63: 1751–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messinese E, Mun JH, Yeun LH, Jayaraman D, Rougé P, Barre A, Lougnon G, Schornack S, Bono JJ, Cook DR, et al. (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact 20: 912–921 [DOI] [PubMed] [Google Scholar]
- Mithöfer A, Mazars C. (2002) Aequorin-based measurements of intracellular Ca2+-signatures in plant cells. Biol Proced Online 4: 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR. (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GE, Downie JA. (2006) Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J 48: 883–894 [DOI] [PubMed] [Google Scholar]
- Ng CK, McAinsh MR. (2003) Encoding specificity in plant calcium signalling: hot-spotting the ups and downs and waves. Ann Bot (Lond) 92: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Pauly N, Knight MR, Thuleau P, Graziana A, Muto S, Ranjeva R, Mazars C. (2001) The nucleus together with the cytosol generates patterns of specific cellular calcium signatures in tobacco suspension culture cells. Cell Calcium 30: 413–421 [DOI] [PubMed] [Google Scholar]
- Pauly N, Knight MR, Thuleau P, van der Luit AH, Moreau M, Trewavas AJ, Ranjeva R, Mazars C. (2000) Control of free calcium in plant cell nuclei. Nature 405: 754–755 [DOI] [PubMed] [Google Scholar]
- Peiter E, Sun J, Heckmann AB, Venkateshwaran M, Riely BK, Otegui MS, Edwards A, Freshour G, Hahn MG, Cook DR, et al. (2007) The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol 145: 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah BW, Du L, Wang H, Yang T. (2013) Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant:microbe interactions. Plant Phys 163: 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Reddy AS, Ali GS, Celesnik H, Day IS. (2011) Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23: 2010–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely BK, Lougnon G, Ané JM, Cook DR. (2007) The symbiotic ion channel homolog DMI1 is localized in the nuclear membrane of Medicago truncatula roots. Plant J 49: 208–216 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Yalovsky S, Zik M, Fromm H, Gruissem W. (1999) The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J 18: 1996–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yoshikawa M, Yano K, Miwa H, Uchida H, Asamizu E, Sato S, Tabata S, Imaizumi-Anraku H, Umehara Y, et al. (2007) NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19: 610–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayanan PV, Cremo CR, Poovaiah BW. (2000) Plant chimeric Ca2+/calmodulin-dependent protein kinase: role of the neural visinin-like domain in regulating autophosphorylation and calmodulin affinity. J Biol Chem 275: 30417–30422 [DOI] [PubMed] [Google Scholar]
- Shimoda Y, Han L, Yamazaki T, Suzuki R, Hayashi M, Imaizumi-Anraku H. (2012) Rhizobial and fungal symbioses show different requirements for calmodulin binding to calcium calmodulin-dependent protein kinase in Lotus japonicus. Plant Cell 24: 304–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Fournier J, Timmers AC, Barker DG. (2012) A switch in Ca2+ spiking signature is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula. Plant J 69: 822–830 [DOI] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Timmers AC, Monin A, Fournier J, Barker DG. (2009) A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol 151: 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Parniske M. (2012) Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr Opin Plant Biol 15: 444–453 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T. (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R. (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Stermer BA, Bianchini GM, Korth KL. (1994) Regulation of HMG-CoA reductase activity in plants. J Lipid Res 35: 1133–1140 [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al. (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Sun J, Miwa H, Downie JA, Oldroyd GE. (2007) Mastoparan activates calcium spiking analogous to Nod factor-induced responses in Medicago truncatula root hair cells. Plant Physiol 144: 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainsbury DJ, Zhou L, Oldroyd GE, Bornemann S. (2012) Calcium ion binding properties of Medicago truncatula calcium/calmodulin-dependent protein kinase. Biochemistry 51: 6895–6907 [DOI] [PubMed] [Google Scholar]
- Taj G, Agarwal P, Grant M, Kumar A. (2010) MAPK machinery in plants: recognition and response to different stresses through multiple signal transduction pathways. Plant Signal Behav 5: 1370–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Maekawa T, Hayashi M. (2012) Nuclear-localized and deregulated calcium- and calmodulin-dependent protein kinase activates rhizobial and mycorrhizal responses in Lotus japonicus. Plant Cell 24: 810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, et al. (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- van Brussel AA, Bakhuizen R, van Spronsen PC, Spaink HP, Tak T, Lugtenberg BJ, Kijne JW. (1992) Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science 257: 70–72 [DOI] [PubMed] [Google Scholar]
- van Der Luit AH, Olivari C, Haley A, Knight MR, Trewavas AJ. (1999) Distinct calcium signaling pathways regulate calmodulin gene expression in tobacco. Plant Physiol 121: 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateshwaran M, Cosme A, Han L, Banba M, Satyshur KA, Schleiff E, Parniske M, Imaizumi-Anraku H, Ané JM. (2012) The recent evolution of a symbiotic ion channel in the legume family altered ion conductance and improved functionality in calcium signaling. Plant Cell 24: 2528–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A, Mazars C, Maitrejean M, Hopke J, Ranjeva R, Boland W, Mithöfer A. (2007) Structural requirements of jasmonates and synthetic analogues as inducers of Ca2+ signals in the nucleus and the cytosol of plant cells. Angew Chem Int Ed Engl 46: 4783–4785 [DOI] [PubMed] [Google Scholar]
- Whalley HJ, Sargeant AW, Steele JF, Lacoere T, Lamb R, Saunders NJ, Knight H, Knight MR. (2011) Transcriptomic analysis reveals calcium regulation of specific promoter motifs in Arabidopsis. Plant Cell 23: 4079–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, Broadley MR. (2003) Calcium in plants. Ann Bot (Lond) 92: 487–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M. (2011) Fluctuations in nuclear envelope’s potential mediate synchronization of early neural activity. Biochem Biophys Res Commun 406: 107–111 [DOI] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]