Abstract
These kinases are identified as integrators in plant signaling, with distinct as well as shared phosphorylation substrates mediating pathway specificity.
Calcium ions (Ca2+) have long been recognized as a major, conserved second messenger in eukaryotic signal transduction. In plants, the multigene family of calcium-dependent protein kinases (CDPKs) encodes structurally conserved, unimolecular calcium sensor/protein kinase effector proteins. CDPKs are ideal candidates for perceiving intracellular changes in Ca2+ concentration and translating them into specific phosphorylation events to initiate further downstream signaling processes. Accordingly, CDPKs have been predominantly characterized in rapid abiotic stress and immune signaling responses, with little focus upon CDPK function in long-term adaptive processes or plant development. The long-awaited identification of in vivo/in planta CDPK targets, including their respective phosphorylation sites, allows the elucidation of CDPK function on a molecular and biochemical level. Moreover, the resolution of kinase-specific phosphorylation patterns within a target protein provides mechanistic evidence suggesting that CDPKs act as signaling hubs in plant stress signaling and development. The CDPK gene family comprises Ser/Thr protein kinases organized in four subgroups (Harmon et al., 2000; Cheng et al., 2002); families of varying size have been characterized in Arabidopsis (Arabidopsis thaliana; 34), rice (Oryza sativa; 29), wheat (Triticum aestivum; 20), maize (Zea mays; 35), and poplar (Populus trichocarpa; 20; Harmon et al., 2000; Asano et al., 2005; Li et al., 2008; Ma et al., 2013; Zuo et al., 2013). CDPKs have a conserved molecular structure, consisting of a variable N-terminal domain, fused to a Ser/Thr kinase domain, and a CDPK activation domain (CAD). The CAD is formed from a pseudosubstrate region (sometimes referred to as the “inhibitory junction domain”) and a Ca2+-binding domain, highly homologous to calmodulin (also described as a calmodulin-like domain; Fig. 1A). The calcium-binding domain of a canonical CDPK contains up to four elongation factor (EF) hand motifs. These are organized in N-terminal and C-terminal EF lobes and may display different Ca2+-binding affinities (Harper et al., 2004; Harper and Harmon, 2005; Boudsocq and Sheen, 2013; Liese and Romeis, 2013).
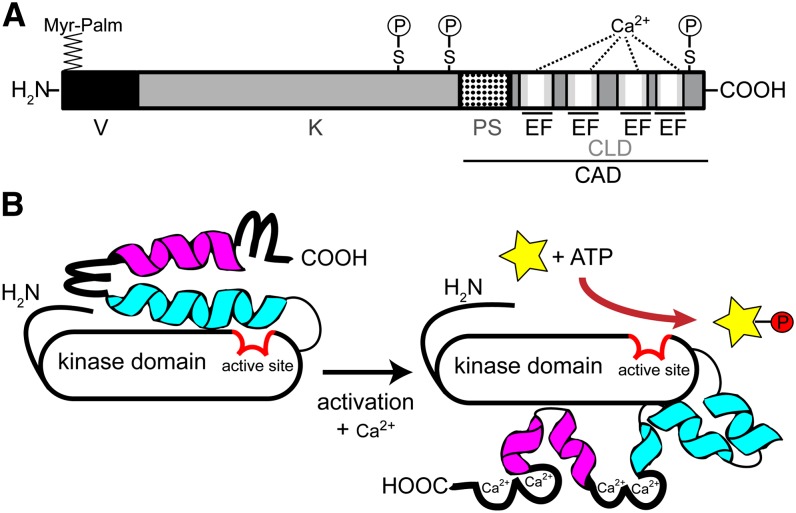
Figure 1.
Scheme of CDPK modular structure and activation. A, Domain structure of Ca2+-dependent protein kinase, drawn to scale for AtCPK28, consisting of 523 amino acids. V, N-terminal variable domain; K, kinase domain; PS, pseudosubstrate segment; CLD, calmodulin-like domain; EF, EF hand. The four white bars within the EF hand motifs represent the EF hand Ca2+-binding loop. The position of myristoylation and palmitoylation (Myr-Palm) and the in vivo autophosphorylation sites (P) at Ser (S) residues are depicted. B, Model for CDPK activation (modified after Wernimont et al., 2010). Calcium binding to the EF hand motifs (loop) induces a conformational change, in which helix 1 (blue) and helix 2 (magenta) break into segments, rotating the calmodulin-like domain around the kinase domain. This rotation increases the accessibility of the N-terminal variable domain as well as of the active site (red) in the kinase domain (black), allowing phosphorylation activity toward its substrate proteins (yellow star, P).
Based on this modular structure, a model for CDPK activation was proposed in which Ca2+ binding to the CAD induces conformational changes, leading to the release of the pseudosubstrate segment from the active site (Fig. 1B). This results in kinase activity, enabling CDPK autophosphorylation as well as the phosphorylation of target substrates (Liese and Romeis, 2013). With the recent availability of crystal structures from apicomplexan CDPKs, encompassing the kinase and CAD, this model has been refined (Wernimont et al., 2010, 2011). In the absence of Ca2+, the autoinhibitory region and the N-terminal lobe of the Ca2+-binding domain form two extended α-helices that block the active site, and likely bury the N-terminal part, of the kinase. In addition, one may speculate that the N-terminal variable domain (which is absent in the crystal structure of apicomplexan CDPKs) may be hidden, so that it has no access to the CDPK target. Upon Ca2+ binding, the structure of the helices is broken down, inducing a rotation of the entire regulatory domain around the kinase domain, probably releasing the N-terminal variable domain. It has been proposed that both the apo- and calcium-bound forms of the enzyme are stabilized by distinct contact sites between the kinase and regulatory domains (Wernimont et al., 2010). It is tempting to speculate that, in particular, some of the amino acids involved in forming these contact sites may be subjected to posttranslational modifications during the activation process (Liese and Romeis, 2013), modulating the stability of the active or inactive form.
CDPKs have been shown to function in many different aspects of plant biology, from environmental stress signaling upon the perception of abiotic and biotic stress stimuli to hormone-regulated processes in development (Cheng et al., 2002; Ludwig et al., 2004; Asano et al., 2012; Boudsocq and Sheen, 2013). While previously, CDPK characterization often relied on the biochemical activation proteins or the transcriptional induction of the respective genes in a given signaling response, recent studies draw a more comprehensive picture of CDPK function, including in vivo/in planta data concerning kinase/substrate interactions. Despite this significant progress, elucidating the impact of biochemical regulation on the in vivo function of a distinct enzyme in a specific intracellular response remains a key challenge in CDPK research. The reasons for this challenge are, first, the lack of mechanistic understanding of in planta kinase activation (e.g. distinct stimulus- and/or pathway-specific posttranslational modifications of the kinase may exist) and, second, putative functional redundancy (Mori et al., 2006); no obvious, single cdpk mutant phenotype has been described for a long time, probably due to close homology within the CDPK subfamilies.
Here, we report on the current understanding of how single CDPK domains may provide the basis for CDPK isoform response specificity. We also summarize the latest progress in CDPK function, with a focus on in vivo substrate identification.
THE N-TERMINAL VARIABLE DOMAIN: SOLELY FOR LOCALIZATION?
The N-terminal variable domain differs in length and amino acid sequence even within the same species (Ito et al., 2010). With the majority of CDPKs harboring a predicted N-myristoylation and N-palmitoylation site, the N terminus was thought to be necessary for membrane association (Harmon et al., 2000; Cheng et al., 2002). In support of this idea, CDPK variants that carry amino acid substitutions G2A (myristoylation) and/or C4/5A (palmitoylation) were defective in the membrane association demonstrated for AtCPK2 (Lu and Hrabak, 2002), AtCPK6, AtCPK9, and AtCPK13 (Benetka et al., 2008), AtCPK3 (Mehlmer et al., 2010), AtCPK5 (Lu and Hrabak, 2013), and AtCPK16 (Stael et al., 2011) from Arabidopsis, NtCDPK2/NtCDPK3 from Nicotiana tabacum (Witte et al., 2010), and StCDPK4/StCDPK5 from potato (Solanum tuberosum; Asai et al., 2013).
Additionally, the N-terminal variable domain is also subjected to in vivo phosphorylation. Both constitutively phosphorylated amino acids and stress stimulus-induced in vivo phosphorylation events, catalyzed either by the enzyme itself or by upstream kinases, were reported. In domain-swap experiments, in which the N-terminal variable domains were exchanged between NtCDPK2 and NtCDPK3 in Nicotiana benthamiana, the observed in vivo phosphorylation pattern was solely determined by the N-terminal variable domain, irrespective of the protein kinase domain used (Witte et al., 2010). This indicates that in vivo phosphorylation of CDPKs cannot simply be deduced by sequence alignment, either among close homologs within the same species or among orthologs from different species. Furthermore, introduction of the G2A amino acid substitution in NtCDPK2 resulted not only in a loss of membrane localization but, concomitantly, prevented phosphorylation by upstream kinases at sites within the N-terminal variable domain (Witte et al., 2010). Likewise, a requirement for correct CDPK localization for CDPK function was shown for potato StCDPK5 phosphorylating substrate protein NADPH oxidase StRBOHB (for respiratory burst oxidase homolog B) in plant immune signaling. A chimeric protein containing the variable domain, including myristoylation and palmitoylation sites, of tomato (Solanum lycopersicum) SlCDPK2 and the kinase domain and CAD from potato StCDPK5 is localized to the trans-Golgi system (as is SlCDPK2 itself) and is not able to phosphorylate StRBOHB (Asai et al., 2013). The requirement for the variable domain in directing target specificity was further shown by interspecies domain-swap experiments between NtCDPK1 and AtCPK9 (Ito et al., 2010).
The challenge: it is conceivable that a single CDPK isoform, which participates in various and independent stress signaling pathways, may show a differential acylation/phosphorylation pattern within its respective variable domain. However, only a very careful analysis of (stimulus-induced) variable domain-dependent in vivo localization and interaction with potential partner proteins, ideally in the absence of ectopic overexpression of kinase and/or substrate proteins, will elucidate the role of the variable domain for substrate specificity.
THE PROTEIN KINASE DOMAIN: WHAT REGULATES ACTIVITY TOWARD A SUBSTRATE IN VIVO?
Standard biochemical assays characterizing CDPK in vitro kinase activity have been conducted either with purified or recombinant proteins synthesized in Escherichia coli and often using the synthetic peptide, syntide2, or histone protein as a model substrate. Kinase activity has been determined in the absence or presence of different Ca2+ concentrations, and the effect of further regulatory components such as lipids (Dixit and Jayabaskaran, 2012) or 14-3-3 proteins (Lachaud et al., 2013; Latz et al., 2013) was investigated. With the identification of stress- or pathway-specific CDPK isoforms, the biochemical characterization is now extended to in vivo kinase activation, mediated by stimulus-induced, posttranslational modification/phosphorylation of the CDPK, which may occur at all enzyme domains (Liese and Romeis, 2013). Kinase properties have been determined by in-gel kinase or immune complex kinase assays, in the absence or presence of model substrates or presumed in vivo targets, and autophosphorylation or substrate phosphorylation was visualized by autoradiography. CDPKs were shown to be activated as rapidly as 1 to 2 min after stress exposure; activation often occurred transiently, and activation and response duration were dependent on the isoform as well as the biological process under investigation (Romeis et al., 2000; Ludwig et al., 2005; Boudsocq et al., 2010). For example, in plant immune signaling, the bacterial 22-amino acid, flagellin-derived peptide flg22 elicited a transient activation for AtCPK5 and AtCPK6, peaking at 5 to 15 min (Boudsocq et al., 2010; Dubiella et al., 2013), while enhanced activation was observed between 2 and 3 h after the expression of the bacterial effector proteins AvrRpm1 and AvrRpt2 (Gao et al., 2013).
The rapid transient activation of CDPK kinase activity points to concerted action involving (upstream) protein kinases, phosphatases, and respective substrate proteins. Indeed, joint regulation of Slow Anion Channel1 (SLAC1) activity in the context of stomatal aperture has been demonstrated for the kinases Open Stomata1 (OST1) and AtCPK23/AtCPK21 and the phosphatase Abscisic Acid Insensitive1 (ABI1; Geiger et al., 2010, 2011; Brandt et al., 2012; Scherzer et al., 2012).
The identification of CDPK in vivo activity has long been hampered by the lack of bona fide in vivo substrate proteins. It became evident that some CDPKs, when purified or as recombinant enzymes, promiscuously phosphorylate proteins in vitro at many different Ser and Thr residues and far beyond the previously proposed CDPK phosphorylation motifs (Harper and Harmon, 2005). Selection based on protein-protein interaction screens such as the yeast two-hybrid assay has so far yielded a low success rate for the identification of in vivo CPDK targets, probably due to the complex mechanism of CDPK activation, combined with a transient kinase-substrate interaction. Significant progress in the identification of signaling pathways modulated by CDPKs was made by the ectopic expression of truncated, constitutively active CDPK variants, which lack the entire regulatory CAD (Sheen, 1996; Romeis et al., 2001; Ludwig et al., 2005). With respect to immune signaling, transient expression of these gain-of-function variants identified candidates AtCPK4, AtCPK5, AtCPK6, and AtCPK11 as triggering pathogen-associated molecular pattern-related transcriptional reprogramming in Arabidopsis leaf mesophyll protoplasts (Boudsocq et al., 2010) and AtCPK5 and AtCPK6 as inducing a reactive oxygen species (ROS)-mediated cell death response in Nicotiana benthamiana leaves (Dubiella et al., 2013). Putative pathway and response specificity was confirmed in transgenic lines overexpressing the kinase and/or in loss-of-function mutants lacking (multiple) CDPK gene(s). The tool of transient ectopic expression has been developed even further, to directly investigate CDPK kinase/phosphorylation substrate relationships in vivo. Abscisic acid (ABA)-related anion channel activity was assessed in Xenopus laevis oocytes (Geiger et al., 2010, 2011), pollen tube growth-related cation channel activity in COS-7 cells (Zhao et al., 2013), and plant immune signaling-related responses in protoplasts and/or leaves of wild-type and mutant plants (Boudsocq et al., 2010; Dubiella et al., 2013; Gao et al., 2013). Despite this significant progress, however, one should remember that CDPKs may have been functionally assessed under conditions of overexpression (potentially promoting substrate promiscuity) and in the absence of the tissue- and cell type-specific localization.
Recently, state-of-the-art phosphoproteomics was shown to represent the next level in in vivo analysis of CDPK function. In particular, selected reaction monitoring mass spectrometry (SRM-MS) enabled the in vivo identification and quantification of kinase isoform-specific phosphorylation patterns in given substrate proteins. For example, six flg22-induced, AtCPK5-dependent, differentially phosphorylated amino acids in the N terminus of the NADPH oxidase RBOHD were identified and quantified, from both Arabidopsis plants and protoplasts (Dubiella et al., 2013). Amino acids Ser-39, Ser-148, Ser-163, and Ser-347 showed increased, and Ser-162 and Ser-343 showed reduced or unaltered, phosphorylation, which correlated with AtCPK5 activity. While all identified Ser residues were required for RBOHD-catalyzed ROS production, these data provide an unambiguous in vivo signature for AtCPK5 activity in a specific signaling response involving a distinct target protein. These data also indicate a functional differentiation (or hierarchy) at neighboring amino acids, suggesting that the RBOHD protein may be targeted by additional protein kinases in vivo. These kinases may be CDPKs, further calcium-regulated kinases such as CIPKs, or kinases from different classes. Interestingly, Ser-184 and Ser-185 in RBOHF are homologous to Ser-162 and Ser-163 in RBOHD (of which only Ser-163 is directly phosphorylated by CPK5; Dubiella et al., 2013). Ser-184 of RBOHF can be phosphorylated in vitro by the recombinant ABA-signaling-related SnRKII (for Snf1-related protein kinase)-type kinase OST1 (Sirichandra et al., 2009). Pending an in vivo proof, these data provide evidence for exquisite signaling pathway-specific regulation of identical substrate proteins through phosphorylation at neighboring amino acids by different protein kinases.
The challenge: modern mass spectrometry analysis combined with CDPK gain- and loss-of-function approaches will allow the identification of CDPK- and pathway-specific substrate proteins. Future research is also expected to address the role and hierarchy of multiple in vivo phosphorylation events within a single target protein, which may be catalyzed by different protein kinases. In addition, SRM-MS may help to dissect sites subjected to CDPK autophosphorylation and to assess their contribution to stimulus-specific activating/inactivating modifications at the enzyme itself.
THE CDPK ACTIVATION DOMAIN: ARE ALL CDPKs REGULATED BY CALCIUM IN VIVO?
CDPKs were primarily perceived as enzymes that swap between a Ca2+-free inactive and a Ca2+-bound active enzyme form. While still valid, this model has been refined (Fig. 1). Experimental evidence indicates a differential contribution of N-terminal and C-terminal EF lobes within the CAD in either stabilizing or releasing the pseudosubstrate region (Christodoulou et al., 2004). Furthermore, CDPK variants, harboring site-specific amino acid substitutions in single or double EF hand motifs, demonstrated a differential requirement of the N- and C-terminal EF hand pairs in standard in vitro protein kinase assays (Zhao et al., 1994; Franz et al., 2011). Remarkably, a comparison between Ca2+ binding and phosphorylation activity identified Arabidopsis CDPKs that displayed apparent Ca2+-insensitive kinase activity. These isoforms lacked up to three consensus EF hand motifs but were still able to bind Ca2+ in vitro, while lacking all four EF hand motifs like AtCPK25 abolished Ca2+ binding (Boudsocq et al., 2012). So far, it is entirely unknown how in vivo sequence variations in single EF hand motifs or different calcium-binding domains influence the translation of Ca2+ concentration changes into CDPK activation in a stimulus- and spatiotemporal-specific manner. With the availability of in vivo substrate proteins, a functional assessment of isoform-intrinsic Ca2+ dependency now appears more feasible. For example, the phosphorylation of SLAC1 by AtCPK21 displays activation kinetics corresponding with four Ca2+-binding sites. Accordingly, constitutive phosphorylation coinciding with functional ion channel activation was only observed for truncated AtCPK21-VK (only variable and kinase domain) lacking the CAD. In contrast, AtCPK23 appears to be Ca2+ insensitive, and slow, constitutive SLAC1 activation was observed even with the full-length enzyme (Geiger et al., 2010). AtCPK11 and AtCPK24 have been identified as functional negative regulators of Arabidopsis pollen tube growth. AtCPK11, but not AtCPK24, displays Ca2+ sensitivity in in vitro kinase assays, and phosphorylates recombinant AtCPK24, but not vice versa. Furthermore, enhanced expression of AtCPK24, but not AtCPK11, was observed during pollen tube growth (Zhao et al., 2013). Taken together, in spite of the current lack of in vivo proof for the phosphorylation of either kinase and potential cation channel substrate, these results provide the first evidence for a fascinating scenario in CDPK regulation: AtCPK11 perceives alternating changes in Ca2+ concentration, which may be relayed, through phosphorylation, to a second CDPK, AtCPK24, which finally phosphorylates the effector substrate, here probably a Shaker-type K+ channel.
The challenge: under the hypothesis of distinct CDPKs being allocated to various biological processes and signaling pathways, and considering the increasing number of corresponding, even overlapping, in vivo phosphorylation substrates known for CDPKs, future research will be needed to determine Ca2+-binding affinities and compare the calcium activation kinetics of CDPK isoforms in vivo.
CDPKs IN PLANT DEVELOPMENT
A major role for CDPK function in plant development has emerged in pollen tube growth, distinguished by an oscillating Ca2+ gradient. Twelve members of the Arabidopsis CDPK gene family are predominantly expressed in pollen (Myers et al., 2009). For AtCPK17/AtCPK34 as well as AtCPK11/AtCPK24, a role in pollen tube growth and fitness has been experimentally validated in double mutant lines (Myers et al., 2009; Zhao et al., 2013).
Furthermore, CDPKs are associated with the activity of phytohormones such as GA3. A well-documented example of a CDPK modulating GA3 homeostasis is NtCDPK1 interacting in vivo with the bZIP transcription factor REPRESSOR OF SHOOT GROWTH, which directly regulates GA3 biosynthesis and is inactivated by NtCDPK1-dependent phosphorylation (Ishida et al., 2008). Recently, a developmental phenotype in stem elongation and vascular development was described for a single CDPK (AtCPK28) loss-of-function mutant in Arabidopsis. Even more remarkably, this phenotype was strictly growth phase dependent. Specifically upon the transition to the generative growth phase, cpk28 plants were characterized by shorter stems, increased secondary growth, and shortened petioles with enhanced anthocyanin production (Matschi et al., 2013). The stem phenotype correlated with reduced transcript levels of GA3 biosynthesis genes and could be partially rescued by exogenous application of GA3. The wild-type phenotype could be fully restored in cpk28 upon complementation with AtCPK28 protein expressed under the control of native AtCPK28, but also under the control of tissue-specific promoters, providing evidence for the participation of a mobile signal in mediating AtCPK28 protein function. Interestingly, transient silencing of the orthologous gene pair NaCDPK4 and NaCDPK5 in Nicotiana attenuata likewise resulted in plants of reduced height, based on compromised GA3-mediated stem elongation (Heinrich et al., 2013), accompanied by increased levels of the biologically active phytohormone, jasmonic acid, conjugate JA-Ile (Yang et al., 2012). Consequently, these silenced plants displayed enhanced resistance toward the herbivore Manduca sexta. One may speculate that CPK28 and its orthologs form a branching point between GA3 and JA-Ile, with the CDPK modulating the activity of these two phytohormone signaling pathways in order to balance plant growth and defense. The pleiotropic phenotypes raise the question of whether hormonal imbalance is an immediate consequence of reduced CDPK activity or a result of an as yet unknown underlying developmental process affected by these CDPKs. AtCPK28 was shown to display Ca2+-dependent kinase activity in vitro, suggesting an important role of Ca2+ sensing in developmental processes.
The challenge: future research is expected to identify in vivo features of developmentally induced changes in Ca2+ concentrations, including the nature of the inducing stimulus and the source of Ca2+ ions; for example, in the case of AtCPK28, activation correlating with the transition to the generative growth phase. Subsequently, the mechanism of the translation of Ca2+/CDPK activity toward a specific CDPK target into phytohormone-regulated growth processes remains to be elucidated.
CDPKs IN ABIOTIC STRESS SIGNALING
CDPKs have been known for many years to participate in Ca2+-related signal transduction induced by abiotic stress stimuli in the context of salinity, drought, and cold; in particular, isoforms were implicated in ABA-mediated signaling. CDPK-dependent changes in ion fluxes, metabolic changes, or alterations in gene expression were addressed. CDPKs were often identified as positive regulators of abiotic stress responses, and the overexpression of the respective kinase resulted in plants with enhanced stress tolerance (Asano et al., 2012; Boudsocq and Sheen, 2013).
Interestingly, with respect to abiotic stress signaling in Arabidopsis, CDPK substrates have so far been identified predominantly among either ABA Response Factor (ABF) transcriptional regulators or (guard cell-expressed) ion channel proteins and transporters. A positive regulatory effect of CDPKs in drought stress signaling may be explained by the enhanced expression of ABA-responsive genes (Xu et al., 2010). Consistently, in vivo interactions were shown between ABF2 and AtCPK4, AtCPK7, AtCPK10, and AtCPK30 (Lu et al., 2013), as well as between ABF1 and AtCPK11 (Lynch et al., 2012), using split-yellow fluorescent protein studies. Furthermore, in in vitro kinase assays, AtCPK4 and AtCPK11 were shown to phosphorylate ABF1 and ABF4 (Zhu et al., 2007; Lynch et al., 2012). Notably, other kinases, such as members of the SnRK gene families, also are able to interact with and to phosphorylate ABF2 in an ABA-dependent manner in vivo (Umezawa et al., 2013). In addition, CDPK phosphorylation substrates were found among cation and anion channels. CPK3 was reported as phosphorylating a two-pore K+ channel (TPK1), enabling 14-3-3 protein-mediated channel activation (Latz et al., 2013). AtCPK6, AtCPK21, and AtCPK23 functionally interact with and phosphorylate the slow voltage-gated anion channel, SLAC1, as a prerequisite for channel activation (Geiger et al., 2010, 2011; Brandt et al., 2012; Demir et al., 2013), and anion currents mediated by SLAC1 and SLAC1 Homolog3 (SLAH3) were shown to be essential for ABA-mediated stomatal opening and closure (Hedrich, 2012). In addition, ABA-dependent, in planta interaction and colocalization in lipid rafts were shown for SLAH3 and AtCPK21 (Demir et al., 2013). While AtCPK21 was able to phosphorylate SLAH3 in vivo, response activation was suppressed in the presence of phosphatase ABI1 (Geiger et al., 2011).
The challenge: interestingly, single CDPK isoforms appear to participate in rapid short-term signaling reactions (e.g. AtCPK21 and AtCPK23 proteins function as positive regulators in ABA-induced stomatal closure) as well as in the regulation of long-term adaptive processes (e.g. cpk21 and cpk23 mutant lines display an accumulation of stress-related metabolite and marker genes, concomitant with an increase in stress tolerance; Ma and Wu, 2007; Franz et al., 2011). While some molecular CDPK targets in rapid signaling responses have been identified, the nature of the tissue- and/or cell type-specific CDPK target proteins mediating long-term responses still has to be determined.
CDPKs IN BIOTIC STRESS SIGNALING
CDPKs were recognized early as key players participating in the translation of pathogen signal-induced changes in the Ca2+ concentration into plant defense reactions. These reactions include the synthesis of ROS, altered gene expression, changes in phytohormone synthesis and signaling, and cell death (Tena et al., 2011; Boudsocq and Sheen, 2013). The rapid biochemical activation of CDPKs by pathogen stress and subsequent deactivation within 15 to 120 min (Romeis et al., 2000) suggests a role of CDPKs in the onset of plant immune reactions. Such rapid in vivo activation was first described for Nicotiana tabaccum NtCDPK2, triggered by the fungal elicitor Avr9 in plants that harbored the corresponding tomato disease resistance gene Cf-9 (Romeis et al., 2000, 2001; Ludwig et al., 2005). Likewise, potato StCDPK4 and StCDPK5 were identified as modulators of early defense reactions, including StRBOHB-mediated ROS production in response to a Phytophthora infestans elicitor (Kobayashi et al., 2007, 2012). In Arabidopsis leaf mesophyll protoplasts, flg22-induced biochemical activation was shown for AtCPK5 and AtCPK6 (orthologs to StCDPK4 and StCDPK5; Dubiella et al., 2013) and for AtCPK4 and AtCPK11 (Boudsocq et al., 2010), whereas AtCPK1 and AtCPK2 (orthologs to NtCDPK2) were implicated predominantly during effector-triggered immunity (Gao et al., 2013). Functional studies also demonstrated a differing, partially overlapping pattern of defense gene expression triggered by these enzymes (Boudsocq et al., 2010). In addition, the NADPH oxidase isoform RBOHD, as well as the WRKY transcriptional regulators WRKY8, WRKY28, and WRKY48, were reported as in vitro substrates phosphorylated by a subset of these CDPKs. Mutants lacking multiple CDPK or WRKY genes were unable to mount a full plant defense mediating resistance to bacteria (Gao et al., 2013). A recent study in Arabidopsis demonstrated AtCPK5-mediated changes in defense gene expression as well as long-lasting salicylic acid-dependent enhanced bacterial resistance. AtCPK5 was shown to rapidly phosphorylate specific Ser residues of RBOHD in vivo in response to bacterial flg22. Furthermore, AtCPK5 and AtRBOHD were shown to constitute a self-propagating mutual activation circuit, which provides the basis for ROS-mediated signal transduction from local to distal plant sites, resulting in the biochemical activation of AtCPK5 and AtCPK5-mediated defense responses in distal cells, ultimately driving resistance to pathogens (Dubiella et al., 2013). Interestingly, both AtCDPK5 and RBOHD contain EF hand Ca2+-binding motifs and undergo posttranslational regulation, not only by phosphorylation but also by Ca2+ binding. While the precise role of Ca2+ within this mutual activation process has still to be elucidated, CDPKs seem to participate in a Ca2+- and ROS-driven mechanism for cell-to-cell immune signal propagation, complementing a model that has been postulated previously (Torres et al., 2005; Miller et al., 2009).
The challenge: during the onset of the plant immune response, single CDPKs have been shown to activate both substrate proteins in the plasma membrane (e.g. proteins involved in rapid ROS-based signal propagation) and transcriptional regulators functioning in the nucleus (e.g. those responsible for long-lasting changes in gene expression). While subcellular localization has been reported for some CDPKs, a detailed cell biological analysis of CDPK localization or potential immune stimulus-dependent relocalization in a native plant system without overexpression or (heterologous) ectopic expression is still missing.
CDPKs AS SIGNALING HUBS?
The progress in recent CDPK research, and in particular the identification of in vivo substrates, points to two hallmark features of CDPK signaling: (1) a single enzyme family and even a single isoform may integrate signals triggered by different biological processes, suggesting a role of CDPKs as hubs in stress signaling and development; and (2) multiple CDPK isoforms not only share the same signaling pathway but even target an identical substrate protein.
A distinct CDPK may not only contribute to rapid and long-term responses within one signaling pathway but may control responses even in different signaling pathways. For example, guard cell-expressed AtCPK6 is implicated in ABA signaling, and AtCPK6-mediated ROS generation and direct phosphorylation of SLAC1 was reported (Mori et al., 2006; Munemasa et al., 2011). When expressed in leaf protoplasts, AtCPK6 undergoes rapid flg22-induced biochemical activation and induces transcriptional reprogramming reminiscent of plant innate immune signaling (Gao et al., 2013). An even more stunning example is CPK11, reported to participate in plant immune and abiotic stress signaling (affecting transcriptional regulators) as well as in pollen tube growth (regulating a K+ channel; Zhu et al., 2007; Zhao et al., 2013).
Furthermore, CDPKs apparently engage in joint actions. Arabidopsis AtCPK4, AtCPK5, AtCPK6, and AtCPK11, and recently also AtCPK1/AtCPK2, have been implicated in pathogen-induced plant defense signaling, with partially overlapping in vitro phosphorylation targets identified (Gao et al., 2013). AtCPK6, AtCPK21, and AtCPK23 participate in the activation of the abiotic stress-related anion channel SLAC1, which is also targeted by OST1 (Brandt et al., 2012). Although these data, in part, rely on ectopic expression, recombinant proteins, or heterologous experimental systems, the demand arises for a model outlining the molecular mechanism of such apparent multiple enzyme regulation. With the prerequisite of native coexpression in the same cell, does functional CDPK isoform specificity with respect to phosphorylation and subsequent intracellular response exist at all, and if yes, how can it be mechanistically explained?
One requirement is the identification of a distinct, stimulus-specific phosphorylation pattern for a defined in vivo substrate protein for each individual CDPK. This has been shown by SRM-MS analysis in the case of RBOHD phosphorylation by AtCPK5. Also, differential, site-specific in vitro phosphorylation of SLAC1 was reported for the kinases AtCPK23 and OST1. Such concerted regulation of phosphorylation by different protein kinases and dephosphorylation by phosphatases may be correlated with a positive or negative effect on the CDPK substrate (activity) itself as well as on induced downstream responses.
A second key may be CDPK regulation by Ca2+ binding within the CAD. It is conceivable that CDPK isoforms within a single cell may have distinct Ca2+-binding affinities. Thus, depending on the subcellular localization with respect to a (stimulus-dependent) local change in the intracellular Ca2+ concentration, a distinct isoform may become activated or remain in its inactive form.
While technical advances in mass spectrometry facilitated the resolution of CDPK isoform-specific phosphorylation in vivo at the biochemical level, the deciphering of differential in vivo CDPK Ca2+-binding affinities, as well as their requirement for triggering pathway-specific CDPK function, remains one of the biggest immediate challenges to be tackled in the field of CDPK research.
Acknowledgments
We thank Dr. Helen Fones for critical reading of the manuscript.
Glossary
- CDPK
calcium-dependent protein kinase
- CAD
CDPK activation domain
- EF
elongation factor
- ROS
reactive oxygen species
- ABA
abscisic acid
- SRM-MS
selected reaction monitoring mass spectrometry
References
- Asai S, Ichikawa T, Nomura H, Kobayashi M, Kamiyoshihara Y, Mori H, Kadota Y, Zipfel C, Jones JDG, Yoshioka H. (2013) The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J Biol Chem 288: 14332–14340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Hayashi N, Kikuchi S, Ohsugi R. (2012) CDPK-mediated abiotic stress signaling. Plant Signal Behav 7: 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Tanaka N, Yang G, Hayashi N, Komatsu S. (2005) Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol 46: 356–366 [DOI] [PubMed] [Google Scholar]
- Benetka W, Mehlmer N, Maurer-Stroh S, Sammer M, Koranda M, Neumüller R, Betschinger J, Knoblich JA, Teige M, Eisenhaber F. (2008) Experimental testing of predicted myristoylation targets involved in asymmetric cell division and calcium-dependent signalling. Cell Cycle 7: 3709–3719 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Droillard MJ, Regad L, Laurière C. (2012) Characterization of Arabidopsis calcium-dependent protein kinases: activated or not by calcium? Biochem J 447: 291–299 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Sheen J. (2013) CDPKs in immune and stress signaling. Trends Plant Sci 18: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng S-H, Sheen J. (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B, Brodsky DE, Xue S, Negi J, Iba K, Kangasjärvi J, Ghassemian M, Stephan AB, Hu H, Schroeder JI. (2012) Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc Natl Acad Sci USA 109: 10593–10598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J. (2002) Calcium signaling through protein kinases: the Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129: 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou J, Malmendal A, Harper JF, Chazin WJ. (2004) Evidence for differing roles for each lobe of the calmodulin-like domain in a calcium-dependent protein kinase. J Biol Chem 279: 29092–29100 [DOI] [PubMed] [Google Scholar]
- Demir F, Horntrich C, Blachutzik JO, Scherzer S, Reinders Y, Kierszniowska S, Schulze WX, Harms GS, Hedrich R, Geiger D, et al. (2013) Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc Natl Acad Sci USA 110: 8296–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit AK, Jayabaskaran C. (2012) Phospholipid mediated activation of calcium dependent protein kinase 1 (CaCDPK1) from chickpea: a new paradigm of regulation. PLoS ONE 7: e51591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T. (2013) Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz S, Ehlert B, Liese A, Kurth J, Cazalé AC, Romeis T. (2011) Calcium-dependent protein kinase CPK21 functions in abiotic stress response in Arabidopsis thaliana. Mol Plant 4: 83–96 [DOI] [PubMed] [Google Scholar]
- Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J, et al. (2013) Bifurcation of Arabidopsis NLR immune signaling via Ca²⁺-dependent protein kinases. PLoS Pathog 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Maierhofer T, Al-Rasheid KAS, Scherzer S, Mumm P, Liese A, Ache P, Wellmann C, Marten I, Grill E, et al. (2011) Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci Signal 4: ra32. [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KAS, Grill E, et al. (2010) Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA 107: 8023–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Harper JF. (2000) CDPKs: a kinase for every Ca2+ signal? Trends Plant Sci 5: 154–159 [DOI] [PubMed] [Google Scholar]
- Harper JF, Breton G, Harmon A. (2004) Decoding Ca2+ signals through plant protein kinases. Annu Rev Plant Biol 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Harper JF, Harmon A. (2005) Plants, symbiosis and parasites: a calcium signalling connection. Nat Rev Mol Cell Biol 6: 555–566 [DOI] [PubMed] [Google Scholar]
- Hedrich R. (2012) Ion channels in plants. Physiol Rev 92: 1777–1811 [DOI] [PubMed] [Google Scholar]
- Heinrich M, Hettenhausen C, Lange T, Wünsche H, Fang J, Baldwin IT, Wu J. (2013) High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of Nicotiana attenuata stems. Plant J 73: 591–606 [DOI] [PubMed] [Google Scholar]
- Ishida S, Yuasa T, Nakata M, Takahashi Y. (2008) A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 20: 3273–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y. (2010) Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca2+-dependent protein kinase is important for substrate recognition. Plant Cell 22: 1592–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, Shimamoto K, Doke N, Yoshioka H. (2007) Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Yoshioka M, Asai S, Nomura H, Kuchimura K, Mori H, Doke N, Yoshioka H. (2012) StCDPK5 confers resistance to late blight pathogen but increases susceptibility to early blight pathogen in potato via reactive oxygen species burst. New Phytol 196: 223–237 [DOI] [PubMed] [Google Scholar]
- Lachaud C, Prigent E, Thuleau P, Grat S, Da Silva D, Brière C, Mazars C, Cotelle V. (2013) 14-3-3-regulated Ca2+-dependent protein kinase CPK3 is required for sphingolipid-induced cell death in Arabidopsis. Cell Death Differ 20: 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz A, Mehlmer N, Zapf S, Mueller TD, Wurzinger B, Pfister B, Csaszar E, Hedrich R, Teige M, Becker D. (2013) Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol Plant 6: 1274–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AL, Zhu YF, Tan XM, Wang X, Wei B, Guo HZ, Zhang ZL, Chen XB, Zhao GY, Kong XY, et al. (2008) Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol Biol 66: 429–443 [DOI] [PubMed] [Google Scholar]
- Liese A, Romeis T. (2013) Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim Biophys Acta 1833: 1582–1589 [DOI] [PubMed] [Google Scholar]
- Lu SX, Hrabak EM. (2002) An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol 128: 1008–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Hrabak EM. (2013) The myristoylated amino-terminus of an Arabidopsis calcium-dependent protein kinase mediates plasma membrane localization. Plant Mol Biol 82: 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Chen X, Wu Y, Wang Y, He Y, Wu Y. (2013) Directly transforming PCR-amplified DNA fragments into plant cells is a versatile system that facilitates the transient expression assay. PLoS ONE 8: e57171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig AA, Romeis T, Jones JDG. (2004) CDPK-mediated signalling pathways: specificity and cross-talk. J Exp Bot 55: 181–188 [DOI] [PubMed] [Google Scholar]
- Ludwig AA, Saitoh H, Felix G, Freymark G, Miersch O, Wasternack C, Boller T, Jones JDG, Romeis T. (2005) Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc Natl Acad Sci USA 102: 10736–10741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch T, Erickson BJ, Finkelstein RR. (2012) Direct interactions of ABA-insensitive(ABI)-clade protein phosphatase(PP)2Cs with calcium-dependent protein kinases and ABA response element-binding bZIPs may contribute to turning off ABA response. Plant Mol Biol 80: 647–658 [DOI] [PubMed] [Google Scholar]
- Ma P, Liu J, Yang X, Ma R. (2013) Genome-wide identification of the maize calcium-dependent protein kinase gene family. Appl Biochem Biotechnol 169: 2111–2125 [DOI] [PubMed] [Google Scholar]
- Ma SY, Wu WH. (2007) AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol Biol 65: 511–518 [DOI] [PubMed] [Google Scholar]
- Matschi S, Werner S, Schulze WX, Legen J, Hilger HH, Romeis T. (2013) Function of calcium-dependent protein kinase CPK28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J 73: 883–896 [DOI] [PubMed] [Google Scholar]
- Mehlmer N, Wurzinger B, Stael S, Hofmann-Rodrigues D, Csaszar E, Pfister B, Bayer R, Teige M. (2010) The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J 63: 484–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal 2: ra45. [DOI] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al. (2006) CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Hossain MA, Nakamura Y, Mori IC, Murata Y. (2011) The Arabidopsis calcium-dependent protein kinase, CPK6, functions as a positive regulator of methyl jasmonate signaling in guard cells. Plant Physiol 155: 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers C, Romanowsky SM, Barron YD, Garg S, Azuse CL, Curran A, Davis RM, Hatton J, Harmon AC, Harper JF. (2009) Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J 59: 528–539 [DOI] [PubMed] [Google Scholar]
- Romeis T, Ludwig AA, Martin R, Jones JD. (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20: 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Jones JD. (2000) Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell 12: 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer S, Maierhofer T, Al-Rasheid KAS, Geiger D, Hedrich R. (2012) Multiple calcium-dependent kinases modulate ABA-activated guard cell anion channels. Mol Plant 5: 1409–1412 [DOI] [PubMed] [Google Scholar]
- Sheen J. (1996) Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274: 1900–1902 [DOI] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, et al. (2009) Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett 583: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Stael S, Bayer RG, Mehlmer N, Teige M. (2011) Protein N-acylation overrides differing targeting signals. FEBS Lett 585: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J. (2011) Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL. (2005) Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K. (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal 6: rs8. [DOI] [PubMed] [Google Scholar]
- Wernimont AK, Amani M, Qiu W, Pizarro JC, Artz JD, Lin YH, Lew J, Hutchinson A, Hui R. (2011) Structures of parasitic CDPK domains point to a common mechanism of activation. Proteins 79: 803–820 [DOI] [PubMed] [Google Scholar]
- Wernimont AK, Artz JD, Finerty P, Jr, Lin YH, Amani M, Allali-Hassani A, Senisterra G, Vedadi M, Tempel W, Mackenzie F, et al. (2010) Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat Struct Mol Biol 17: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte CP, Keinath N, Dubiella U, Demoulière R, Seal A, Romeis T. (2010) Tobacco calcium-dependent protein kinases are differentially phosphorylated in vivo as part of a kinase cascade that regulates stress response. J Biol Chem 285: 9740–9748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Tian YS, Peng RH, Xiong AS, Zhu B, Jin XF, Gao F, Fu XY, Hou XL, Yao QH. (2010) AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta 231: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Yang DH, Hettenhausen C, Baldwin IT, Wu J. (2012) Silencing Nicotiana attenuata calcium-dependent protein kinases, CDPK4 and CDPK5, strongly up-regulates wound- and herbivory-induced jasmonic acid accumulations. Plant Physiol 159: 1591–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LN, Shen LK, Zhang WZ, Zhang W, Wang Y, Wu WH. (2013) Ca2+-dependent protein kinase11 and 24 modulate the activity of the inward rectifying K+ channels in Arabidopsis pollen tubes. Plant Cell 25: 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Pokutta S, Maurer P, Lindt M, Franklin RM, Kappes B. (1994) Calcium-binding properties of a calcium-dependent protein kinase from Plasmodium falciparum and the significance of individual calcium-binding sites for kinase activation. Biochemistry 33: 3714–3721 [DOI] [PubMed] [Google Scholar]
- Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang X-F, Wu F-Q, et al. (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo R, Hu R, Chai G, Xu M, Qi G, Kong Y, Zhou G. (2013) Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa). Mol Biol Rep 40: 2645–2662 [DOI] [PubMed] [Google Scholar]