Abstract
Calcium/calmodulin-mediated signaling contributes in diverse roles in plant growth, development, and response to environmental stimuli.
During calcium (Ca2+) signaling, decoding the stimulus-response coupling involves a set of Ca2+ sensor proteins or Ca2+-binding proteins (DeFalco et al., 2010a; Kudla et al., 2010). These proteins usually possess one or more classical helix-loop-helix elongation factor (EF) hand motifs. Three major types of Ca2+-sensor proteins in plants are calmodulin (CaM)/CaM-like proteins, calcium-dependent protein kinases (CDPKs), and calcineurin B-like proteins. As compared with animals, plant genomes encode more diversified Ca2+ sensors; with the exception of canonic CaM, all other types of Ca2+ sensors (CaM-like proteins, CDPKs, and calcineurin B-like proteins) are plant specific. The large population and unique structural composition of Ca2+-binding proteins and the diversity of the target proteins regulated by the Ca2+ sensors reflect the complexity of Ca2+ signaling, which helps plants adapt to the changing environment. This update will be limited primarily to discussions on CaM and CaM-binding proteins and the recent advances in Ca2+/CaM-mediated signaling.
CaM is a conserved Ca2+-binding protein found in all eukaryotes. The discovery of CaM can be traced back to the 1970s. An activator of cyclic nucleotide phosphodiesterase was shown to be involved in the regulation of cAMP concentration, which was stimulated by Ca2+ (Kakiuchi and Yamazaki, 1970; Cheung, 1971). The activator was found to bind Ca2+ and was eventually named “calmodulin,” an abbreviation of Ca2+-modulated protein. Since its discovery over 40 years ago, CaM has been regarded as a model Ca2+-binding protein and has been subjected to intensive studies in biochemistry, cell biology, and molecular biology because of its importance in almost all aspects of cellular regulation (Poovaiah and Reddy, 1987, 1993; Bouche et al., 2005; DeFalco et al., 2010a; Du et al., 2011; Reddy et al., 2011b). Disruption or depletion of the single copy of the CaM gene in yeast (Saccharomyces cerevisiae) results in a recessive lethal mutation (Davis et al., 1986), suggesting that CaM has a critical role in eukaryotic cells.
The structure of CaM has been well studied, and the prototype of CaM found in all eukaryotes has 149 amino acids with two globular domains, each containing two EF hands connected by a long flexible helix (Meador et al., 1993; Zhang et al., 1995; Yun et al., 2004; Ishida et al., 2009). As more and more genomes are sequenced, it is becoming clear that CaM belongs to a small gene family in plants. In the model plant Arabidopsis (Arabidopsis thaliana), seven CaM genes encode for four highly conserved isoforms (CaM1/4, CaM2/3/5, CaM6, and CaM7) that differ in only one to five amino acid residues. Loss-of-function mutations of individual CaMs indicate that the different CaMs may have overlapping yet different functions. For example, a loss of function in Arabidopsis AtCaM2 affects pollen germination (Landoni et al., 2010). Phenotypic analysis showed that in normal growth conditions, atcam2-2 plants were indistinguishable from the wild type, while genetic analysis showed a reduced transmission of the atcam2-2 allele through the male gametophyte, and in vitro pollen germination revealed a reduced level of germination in comparison with the wild type. However, the atcam3 knockout mutant showed a clear reduction in thermotolerance after heat treatment at 45°C for 50 min (Zhang et al., 2009). Overexpression of AtCaM3 in either the atcam3 knockout or wild-type background significantly rescued or increased the thermotolerance, respectively. Further analysis of individual CaM mutants under different stress conditions should reveal more on the functional significance of individual CaM genes.
STRATEGIES TO IDENTIFY CaM-BINDING PROTEINS
CaM has no inherent catalytic activity, but its activity is reflected in modulating the function of the target proteins by physically interacting with them (Rhoads and Friedberg, 1997; Hoeflich and Ikura, 2002). The CaM-binding domain (CaMBD)/motifs in the target proteins are not conserved. However, the target peptides usually form a basic amphipathic α-helix, which contains hydrophobic residues on one side and basic residues on the other side. The hydrophobic portion of the target peptide is often held in the hydrophobic pocket of CaM to anchor the target peptide, and the acidic clusters of CaM then interact with the basic portion of the target peptide. The remarkable flexibility of the central linker and the exceptionally large numbers of Met residues in the hydrophobic pocket give CaM conformational plasticity to adjust to a variety of target peptides (Ikura and Ames, 2006). However, the variation in the primary structure of CaMBDs makes it very difficult to identify CaM targets just based on the amino acid sequences of proteins.
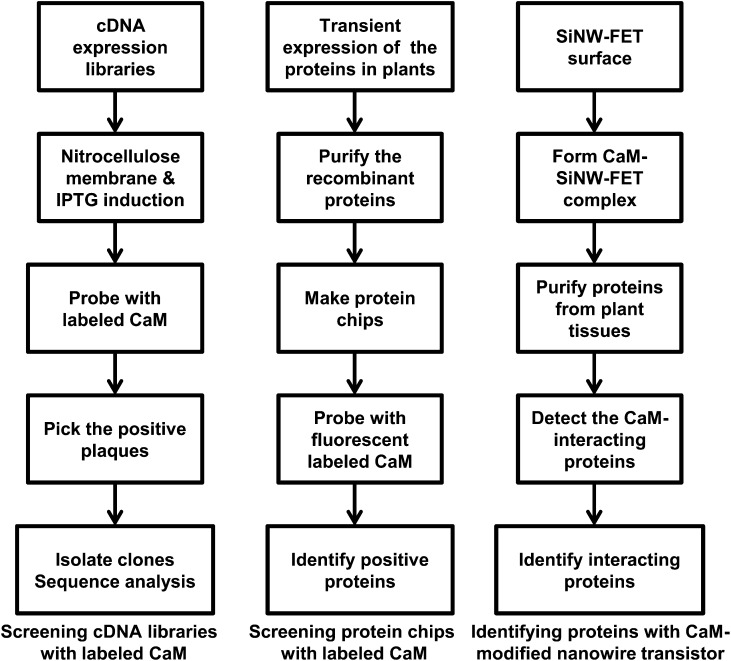
Yeast hybrid systems, based on the reconstitution of functional transcription factors, are the most commonly used approaches to isolate interacting proteins. However, these approaches are not effective for obtaining CaM-binding proteins, possibly because CaM does not undergo Ca2+-induced conformational changes in yeast cells under normal conditions. Coimmunoprecipitation and purification with a CaM-Sepharose column are also not very effective. So far, the majority of CaM-binding proteins in plants have been identified by screening complementary DNA (cDNA) expression libraries with radiolabeled or biotinylated CaM. The most useful probe is 35S-labeled CaM (Fromm and Chua, 1992; Reddy et al., 1993; Yang and Poovaiah, 2000b), and a general scheme for this strategy is shown in Figure 1. The cDNA expression libraries are grown on plates until the plaques appear. The isopropyl β-d-1-thiogalactopyranoside-absorbed nitrocellulose membranes are overlaid onto the plaques to induce the expression of protein. Then, those membranes are incubated with the labeled CaM. The positive plaques are picked up and verified.
Figure 1.
Illustration of the three main approaches used to identify CaM-binding proteins. IPTG, Isopropyl β-d-1-thiogalactopyranoside; SINW-FET, silicon nanowire field-effect transmitter.
Screening cDNA libraries is easy and straightforward. However, many false-positive clones can be picked up, and researchers could miss the real targets because eukaryotic proteins expressed in bacteria are often misfolded. Popescu et al. (2007) developed a protein microarray approach in which proteins expressed in plants were purified and used to make protein chips. The initial cost for preparing the protein chips with a large collection is high, because each protein needs to be expressed and purified from plants. However, once the system is established, this could be used as a high-throughput approach to find the targets for different CaM isoforms in an entire genome (Fig. 1). So far, over 80 plant CaM-target proteins have been characterized using these approaches. However, it is believed that there are many more putative CaM-target proteins yet to be discovered. The current estimate of CaM targets in the Arabidopsis genome is about 500 (T. Yang and B.W. Poovaiah, unpublished data). These target proteins are involved in almost all aspects of plant growth and development as well as in responses to abiotic and biotic stresses. Table I summarizes the characterized CaM-binding proteins and their CaM-binding motifs.
Table I. Plant CaM-binding proteins with defined CaMBDs.
|
CaM-Binding Proteins |
References |
|---|---|
| Protein phosphorylation/dephosphorylation | |
| Chimeric Ca2+/CaM-dependent protein kinase (CCaMK) | Patil et al. (1995); Takezawa et al. (1996) |
| Diacylglycerol kinase (LeCBDGK) | Snedden and Blumwald (2000) |
| NAD kinase (NAD2) | Turner et al. (2004) |
| Cytoplasmic receptor-like kinase (CRCK1) | Yang et al. (2004) |
| Nicotiana tobacum Ca2+/calmodulin-dependent protein kinase 1 (NtCaMK1) | Ma et al. (2004) |
| PP2C-like phosphatase | Takezawa (2003) |
| NPK phosphatase (NtMKP1) | Yamakawa et al. (2004); Ishida et al. (2009) |
| Receptor-like kinase (CRLK1) | Yang et al. (2010) |
| Nuclear proteins/transcription regulators | |
| SRs/CaMTAs | Yang and Poovaiah (2002a); Du et al. (2009) |
| AtBT | Du and Poovaiah (2004) |
| Auxin-induced protein ZmSAUR1 | Yang and Poovaiah (2000) |
| IQD1 | Levy et al. (2005) |
| AtMYB2 | Yoo et al. (2005) |
| CBP60g | Wang et al. (2009) |
| AtGT2L | Xi et al. (2012) |
| Nuclear protein PCBP | Reddy et al. (2002) |
| WRKY group IId AtWRKY7 | Park et al. (2005) |
| ASYMMETRIC LEAVES1 (AS1) | Han et al. (2012) |
| Metabolism | |
| Glu decarboxylase | Baum et al. (1993, 1996) |
| Catalase | Yang and Poovaiah (2002b) |
| FAD-dependent oxidoreductase, DWF1 | Du and Poovaiah (2005) |
| Membrane proteins/ion channels/transporters | |
| Disease resistance gene MLO | Kim et al. (2002a, 2002b) |
| Cyclic nucleotide-gated cation channels (CNCG) | Arazi et al. (1999); Kohler et al. (1999); Ali et al. (2007) |
| Vacuolar Ca2+-ATPase | Malmstrom et al. (2000) |
| Endoplasmic reticulum Ca2+-ATPase | Hong et al. (1999) |
| Plasma membrane Ca2+-ATPase** | Chung et al. (2000) |
| AAA family CIP111 | Zielinski (2002) |
| AAA+-ATPase AFG1L1 | Bussemer et al. (2009) |
| Apyrase | Hsieh et al. (2000); Steinebrunner et al. (2003) |
| Others | |
| Kinesin-like protein | Reddy et al. (1996); Wang et al. (1996) |
| Pollen-specific protein (MPCBP, NPG1) | Safadi et al. (2000) |
| Chaperonin10 | Yang and Poovaiah (2000c) |
| DRL1 | Nelissen et al. (2003) |
| NADPH-dependent dehydrogenase Tic32 | Chigri et al. (2006) |
| Ubiquitin-specific protease6 (AtUBP6) | Moon et al. (2005) |
Recently, an mRNA display technique and a CaM-modified nanowire transistor method have been developed for isolating CaM-binding proteins in humans (Shen et al., 2005; Lin et al., 2010). The mRNA display technique to identify Ca2+/CaM-binding proteins is to screen the mRNA-displayed proteome libraries with biotinylated CaM. Covalent fusions between an mRNA and the peptide or protein that it encodes can be generated by in vitro translation of synthetic mRNAs that carry puromycin. The mRNA display provides a powerful means for reading and amplifying a protein sequence after it has been selected from large libraries (Shen et al., 2005). The CaM-modified nanowire transistor (biosensor) approach is to use a highly sensitive and reusable silicon nanowire field-effect transistor for the detection of protein-protein interactions (Fig. 1). The reusable device is made possible by the reversible association of glutathione S-transferase-tagged CaM with a glutathione-modified transistor (Lin et al., 2010). The minimum concentration of Ca2+ required to activate CaM is 1 μm, and this sensitive nanowire transistor can serve as a high-throughput biosensor and substitute for immunoprecipitation methods used in the identification of interacting proteins. These approaches will be useful for the further identification and characterization of true plant CaM-target proteins, especially those proteins triggered by different stimuli.
Ca2+/CaM-MEDIATED REGULATION OF PROTEIN PHOSPHORYLATION
Ca2+/CaM-stimulated phosphorylation in plants was first observed in the 1980s (Veluthambi and Poovaiah, 1984), long before the cloning of the first CaM-binding kinase from apple (Malus domestica; Watillon et al., 1993). Plant CaM-binding kinases can be classified into four distinct subgroups based on their structural features (Fig. 2). The first subgroup is the CaM-binding protein kinases similar to mammalian CaMK, such as apple calmodulin-binding peptide 1, Nicotiana tobacum Ca2+/calmodulin-dependent protein kinase, maize Ca2+/calmodulin-dependent protein kinase, and Arabidopsis calmodulin binding protein kinase 1 (AtCBK1) to AtCBK3. These kinases are sometimes called CDPK-related kinases (CRKs, also called CBKs) because they carry a kinase domain in the N terminus and degenerated, nonfunctional EF hands in their C terminus (Zhang et al., 2002; Zhang and Lu, 2003; Hua et al., 2004; Wang et al., 2004). The second subgroup is Ca2+ and calcium/calmodulin-dependent protein kinases (CCaMKs), found in most of the higher plants (Patil et al., 1995). CCaMK is a plant-specific protein kinase that carries a Ser/Thr kinase domain in the N-terminal portion, a CaM-binding autoinhibitory domain, and a visinin-like domain (VLD) with three distinct Ca2+-binding EF hands in the C terminus. The third subgroup belongs to receptor-like kinases (RLKs); hence, they are called CaM-binding receptor-like kinases. Plant genomes carry a relatively large RLK family that shares homology with animal receptor kinases, with an extracellular domain, a transmembrane domain, and a kinase domain. Some RLKs from plants contain only a kinase domain and thus are named receptor-like cytoplasmic kinases. Some receptor-like cytoplasmic kinases, such as CRCK1 and homologs (Yang et al., 2004), and RLKs, such as SRK, CaM-binding receptor-like kinase, AtCaMRLK, AtCRLK1, and BRI1, are CaM-binding proteins (Vanoosthuyse et al., 2003; Charpenteau et al., 2004; Kim et al., 2009a; DeFalco et al., 2010b; Yang et al., 2010; Oh et al., 2012). Last but not least, some of the mitogen-activated protein kinases (MAPKs), specifically members of subgroup D of MAPK in Arabidopsis and rice (Oryza sativa), were reported to be regulated by Ca2+/CaM (Ding et al., 2009; Takahashi et al., 2011). Structural features of CaM-regulated protein kinases are summarized in Figure 2. Although the regulation of some of these kinases by CaM remains to be confirmed, accumulated evidence indicates that Ca2+-mediated signals could regulate a broad range of physiological activities related to plant growth, development, and responses to environmental stimuli (Zhang et al., 2002; Vanoosthuyse et al., 2003; Charpenteau et al., 2004; Hua et al., 2004; Liu et al., 2008; Kim et al., 2009a; Yang et al., 2010; Oh et al., 2012). Among all these CaM-regulated kinases, CCaMK has been widely studied because of its role in symbioses, developmental processes, and stress responses.
Figure 2.
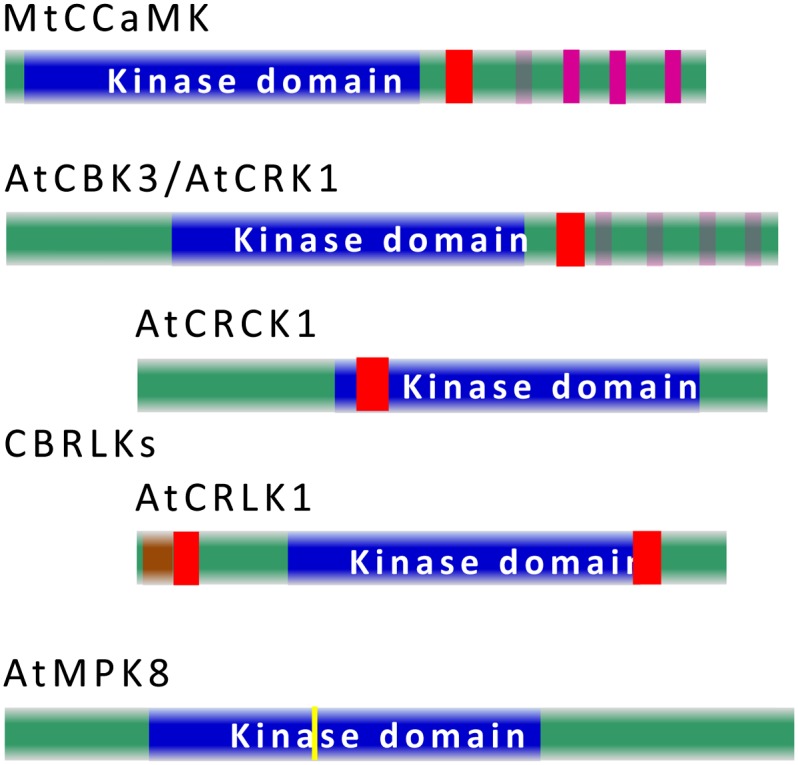
Schematic presentation of the domain structure of Ca2+/CaM-regulated protein kinases. One example of each class of CaM-regulated kinases, MtCCaMK (UniProt Q6RET7), AtCBK3/AtCRK1 (UniProt O80673), AtCRCK1 (UniProt Q9FIL7), AtCRLK1 (UniProt Q9FIU5), and AtMPK8 (UniProt Q9LM33), is presented. The kinase domain is in blue, CaMBD is in red, functional EF hands are in bright purple and degenerated EF hands are in light purple, the transmembrane domain is in light brown, and the yellow bar in MAPK8 represents the conserved TDY motif recognized and phosphorylated by MAPK kinases. CaMBDs in the MAPK D subgroup, including MPK8, are not currently resolved.
ROLE OF Ca2+/CaM-REGULATED KINASE IN PLANT-MICROBE INTERACTIONS
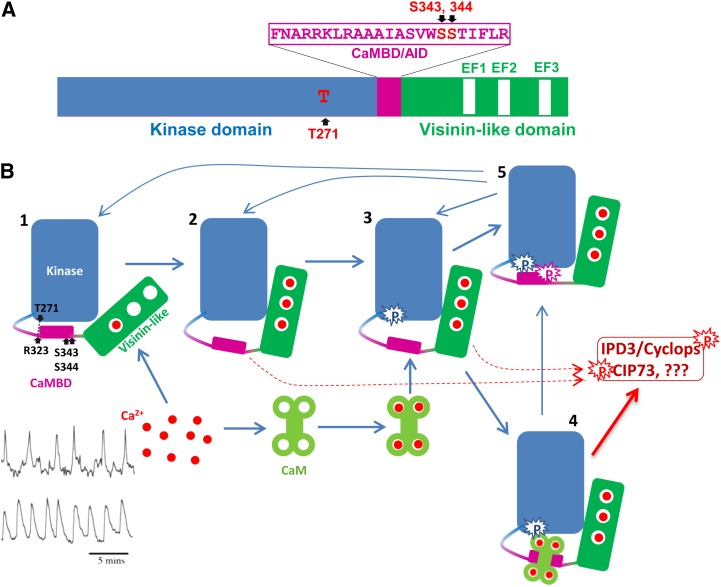
Ca2+ spiking in the nucleus and perinuclear region of root hair cells has been documented as one of the earliest cellular responses after the perception of symbionts by host plants (Ehrhardt et al., 1996; Wais et al., 2000; Walker et al., 2000; Kosuta et al., 2008). Accumulating results have revealed a common symbiotic pathway composed of eight components, SYMRK/DMI2, POLLUX/DMI1, CASTOR, NENA, NUP85, NUP133, CCaMK, and CYCLOPS/IPD3, which are all required for the normal establishment of both root nodulation symbiosis (RNS) and arbuscular mycorrhizal symbiosis (AMS; Singh and Parniske, 2012; Oldroyd, 2013). CCaMK, which carries structural features enabling it to interact with Ca2+ and Ca2+/CaM, acts as a decoder of the encrypted Ca2+ signal (Levy et al., 2004; Mitra et al., 2004). An activation mechanism of CCaMK is proposed in the next section of this update (Fig. 3). CCaMK was first identified and cloned in lily (Lilium spp.), and biochemical studies on lily CCaMK showed that its kinase activities are regulated by both Ca2+ and Ca2+/CaM (Patil et al., 1995; Takezawa et al., 1996). CCaMK alone is inactive, with little or no kinase activity. Its autophosphorylation, predominantly at the Thr-271 residue, is significantly stimulated when Ca2+ levels increase (Takezawa et al., 1996). Autophosphorylation at this position drastically increases its affinity for CaM, a phenomenon called CaM trapping (Sathyanarayanan et al., 2000), and also increases its substrate phosphorylation activity (Takezawa et al., 1996). The kinase activities of CCaMK in leguminous plants are closely related to its function in supporting the establishment of RNS and AMS. Site mutations T271A of MtCCaMK and T265I of LjCCaMK resulted in deregulated and constitutive substrate phosphorylation activity and produced spontaneous nodulation when used for the complementation of ccamk null mutations in both Medicago truncatula and Lotus japonicus (Ramachandiran et al., 1997; Gleason et al., 2006; Tirichine et al., 2006). A recent study on L. japonicus CCaMK indicated that Thr-265 in its native state interacts with other amino acids along with the positively charged Arg-317 in the CaMBD/autoinhibitory domain (AID) region, and this interaction is disrupted when Thr-265 is mutated to Ala or Ile (Shimoda et al., 2012). Together with the aforementioned biochemical data, this suggests that the native Thr-265 (Thr-271 in M. truncatula) is critical in maintaining an ideal intramolecular interaction between the autoinhibitory domain and the kinase domain that keeps CCaMK inactive. Changing the structure at this position through autophosphorylation provides a regulatory mechanism for the substrate phosphorylation capacity of CCaMK and the associated physiological function in supporting plant-microbe symbioses.
Figure 3.
A, Schematic diagram of MtCCaMK showing the domain structure and autophosphorylation sites. B, Activation of CCaMK by Ca2+ and Ca2+/CaM. State 1, CCaMK most likely with Ca2+ loaded in the first EF hand. CCaMK is inactive in this state. State 2, CCaMK loaded with Ca2+ in its three EF hands in the VLD. Because of the Ca2+-induced changes in the tertiary structure of CCaMK, the kinase is active and Thr-271 is ready for autophosphorylation. State 3, Thr-271 is autophosphorylated and CCaMK shows a dramatic increase in its affinity for CaM, a phenomenon called CaM trapping. State 4, Ca2+ and Ca2+/CaM-loaded CCaMK. The kinase is fully active and should be able to phosphorylate substrates as well as CCaMK itself. As long as the CCaMK is bound by Ca2+/CaM, it should remain active. State 5, Ser-343 and/or Ser-344 are phosphorylated, CCaMK can no longer interact with CaM, and its kinase activity is also turned off, whether the Ca2+ concentration is high enough to load in its EF hands. Mycorrhizal and Nod factor-induced Ca2+ spikings (bottom left corner: mycorrhizal is at top and Nod factor is at bottom) are adapted from Kosuta et al. (2008).
Very recently, Ser-337 in LjCCaMK (corresponding to Ser-343 in MtCCaMK) and Ser-344 in MtCCaMK (corresponding to Ser-338 in LjCCaMK) in the CaMBD were reported to be novel autophosphorylation sites in CCaMK (Liao et al., 2012). Phosphorylation at Ser-337 of LjCCaMK and Ser-344 of MtCCaMK both negatively regulate their interaction with CaM and their kinase activities. Furthermore, the phosphomimicking mutation of Ser-337 of LjCCaMK and Ser-344 of MtCCaMK failed to complement null mutants of CCaMK in both RNS and AMS. These results indicate that autophosphorylation at Ser-337 of LjCCaMK and Ser-344 of MtCCaMK has negative regulatory functions in LjCCaMK and MtCCaMK at both the biochemical and physiological levels. Interestingly, the negative regulation through the autophosphorylation of Ser-337 of LjCCaMK is required for the proper progression and establishment of both RNS and AMS (Liao et al., 2012; Routray et al., 2013), but autophosphorylation at Ser-344 acts solely as a negative control to switch off the activated MtCCaMK (Routray et al., 2013), implying a complex regulation of CCaMK through autophosphorylation.
The well-characterized CaMKII, the closest homolog of CCaMK in animals, also has several autophosphorylation sites in its autoinhibitory/CaMBD, and its activities are delicately regulated through differential autophosphorylation at different sites (Hanson and Schulman, 1992). Similar to CCaMK, phosphorylation of CaMKII at its autophosphorylation sites in the CaMBD also interferes with its interaction with CaM and negatively regulates its kinase activity (Colbran and Soderling, 1990; Hanson and Schulman, 1992). A high sequence homology of 79% between CaMKII and CCaMK around the CaM/autoinhibitory region (Colbran and Soderling, 1990; Patil et al., 1995), the presence of autophosphorylation sites in both of the kinases, and their similar impact on CaM-binding properties indicate that CCaMK could be inactivated in a similar way to CaMKII, and this hypothesis has now been confirmed (Liao et al., 2012; Routray et al., 2013). Although CCaMK is very similar to CaMKII, especially in its kinase domain and CaMBD/AID region, CCaMK is drastically different from its cousin in the C terminus and acquired the ability to receive Ca2+ signals using both the CaMBD and the Ca2+-binding VLD. Hence, it is not surprising to see that its mode of activation is different from that of CaMKII.
Although CCaMK was reported to be a major regulator of plant-microbe symbioses, evidence also suggests that CCaMK is involved in other aspects of plant life. Tobacco (Nicotiana tabacum) CCaMK was indicated to play a role in controlling the development of anther (Poovaiah et al., 1999), and pea (Pisum sativum) CCaMK was up-regulated in roots in response to low temperature and salt treatment (Pandey et al., 2002). Recent results showed that CCaMK from maize (Zea mays; ZmCCaMK) and rice (OsCCaMK) both play important roles in abscisic acid-induced antioxidant protection (Ma et al., 2012; Shi et al., 2012). Together, these results suggest that CCaMK acts as a multifunctional regulatory protein in plants.
PROPOSED ACTIVATION MECHANISM OF CCaMK
Based on published data, we propose an activation mechanism of CCaMK (Fig. 3). The first EF hand of CCaMK has a very high affinity for Ca2+ (Swainsbury et al., 2012). Hence, in the resting condition, it is likely that CCaMK is loaded with Ca2+ in its EF1 site, and at least one of the other two sites must be unloaded to keep CCaMK responsive to Ca2+ spiking. It could also be postulated that in the resting condition (state 1), the CaMBD/AID interacts with the kinase activity center and keeps the kinase at an inactive or low-activity status (Takezawa et al., 1996). The hydrogen bond-based interaction between Thr-271 and Arg-323 is predicted to be critical for maintaining this intramolecular interaction to seal the kinase activity of CCaMK under this condition (Shimoda et al., 2012). Alteration in the status of Thr-271 or changes in the Ca2+-loading status of VLD could break this inhibition of the kinase activity. Ca2+ binding to the three EF hands in the VLD induces a change in the tertiary structure of CCaMK, activates its kinase activity, and also makes the Thr-271 accessible for autophosphorylation (state 2, transient and active, may phosphorylate substrates; Sathyanarayanan et al., 2000, 2001; Gleason et al., 2006; Swainsbury et al., 2012). Thr-271 was reported to be the preferred and likely the first autophosphorylated site of CCaMK (Sathyanarayanan et al., 2001; Routray et al., 2013), and autophosphorylation at this site induces an increase in its affinity for CaM by about 200-fold, a phenomenon called CaM trapping (state 3, active and may phosphorylate substrate, transient, and CaM trapping; Sathyanarayanan et al., 2001; Gleason et al., 2006; Tirichine et al., 2006). Once the CCaMK reached state 3, CaM in the vicinity was attracted to CCaMK, even though the Ca2+ concentration could have already decreased to a level lower than that in the stage of Ca2+ loading to EF hands in the VLD. Once bound by CaM in the CaMBD/AID region, CCaMK is fully activated and can phosphorylate substrates such as IPD3/Cyclops, CIP73, and itself (state 4, CCaMK fully loaded with Ca2+ and Ca2+/CaM and highly active; Messinese et al., 2007; Yano et al., 2008; Kang et al., 2011). In addition, CaM binding could protect Ser-343 and Ser-344 from being phosphorylated by CCaMK, which is supported by the in vitro phosphorylation assays that demonstrated that CaM binding decreases the autophosphorylation level of CCaMK (Takezawa et al., 1996).
Recent results showed that, if Ser-343 and/or Ser-344 are autophosphorylated, CCaMK will no longer interact with Ca2+/CaM and its kinase activity will be turned off (Liao et al., 2012; Routray et al., 2013). Hence, another impact of Ca2+/CaM binding to CCaMK could be to maintain its activity for a prolonged period even after Ca2+ decreases to some extent. Since the interaction between Ca2+/CaM and CCaMK is dynamic, the fully activated CCaMK will still have a chance to be autophosphorylated at Ser-343 and Ser-344 in its CaMBD; this will cap the CCaMK from further CaM binding (a phenomenon called CaM capping) and also inactivate itself (state 5; Liao et al., 2012; Routray et al., 2013). This inactivation can be postponed by decreasing Ca2+ concentration. If CCaMK is regulated to state 3 and there is no CaM available in the vicinity of CCaMK, the Thr-271-phosphorylated active kinase could immediately phosphorylate the unprotected Ser-343 and Ser-344 in its CaMBD and inactivate the kinase activity rapidly (state 5). This was observed to occur in vitro within 30 s (Sathyanarayanan and Poovaiah, 2002). Hence, the phosphorylation of Thr-271 acts as a “binary logic switch” in the decision-making process; it could quickly activate or quickly turn off the kinase activity of CCaMK depending on whether CaM is available once the Thr-271 is phosphorylated. Logically, this inactivation via the autophosphorylation of Ser-343 and/or Ser-344 in the CaMBD could be mitigated or avoided by decreases in Ca2+ concentration; this could be the reason why this kinase is activated in vivo by Ca2+ spiking (Oldroyd and Downie, 2004). After losing the phosphate groups at Thr-271, Ser-343, and/or Ser-344, the CCaMK at state 5 could be changed back to state 1, 2, or 3. Different from CaMKs in animals, which need to form a dodecamer to read the oscillative Ca2+ signals (Hudmon and Schulman, 2002), the dually regulated CCaMK, which receives Ca2+ signals from both VLD and CaMBD, could sense and respond to oscillative Ca2+ signals in a monomer format. Recent empirical data obtained from truncated versions of CCaMK also support that CCaMK may not form multimers in the presence or absence of Ca2+ (Swainsbury et al., 2012).
Ca2+/CaM-MEDIATED REGULATION OF TRANSCRIPTIONAL CONTROL IN PLANTS
Transcriptional control is an end result of many signal transduction pathways, including Ca2+/CaM-mediated signaling. CaM interacts with a variety of DNA-binding proteins/transcription factors. Over 90 CaM-binding proteins (CBPs) are DNA-binding proteins that fit into several families of known transcription factors, including CAMTAs (also known as AtSRs), WRKY IID, bZIP, MYB, Trihelix, NAC, CBP60, MADS, and GRAS (Reddy et al., 2011a), and the CaMBDs in some of the newly identified CaM-binding transcription factors remain to be determined (Popescu et al., 2007). In this section, we will focus only on recent developments in the direct regulation of Ca2+/CaM on transcriptional machinery through the actions of CAMTAs/SRs.
The Best Characterized CaM-Regulated Transcription Factors: CAMTAs
NtER1 was the first member of the CAMTA family reported to be a CaM-binding protein (Yang and Poovaiah, 2000a). Follow-up studies showed that CAMTAs belong to a conserved transcription factor family that exists in all the examined multicellular eukaryotes (Reddy et al., 2000; Bouche et al., 2002; Yang and Poovaiah, 2002a; Finkler et al., 2007). The expression of CAMTAs is developmentally regulated (Yang et al., 2012) and responds to different abiotic and biotic signals (Yang and Poovaiah, 2002a; Yang et al., 2013). Target cis-elements for this family are (A/C/G)CGCG(T/C/G) (Yang and Poovaiah 2002a) and (A/C)CGTGT (Choi et al., 2005; Doherty et al., 2009; Du et al., 2009; Kim et al., 2009b; Galon et al., 2010b). All members of the CAMTA family carry a CG-1 DNA-binding domain in the N terminus, followed by a TIG domain, ankyrin repeats, a Ca2+-dependent CaMBD, and tandem repeats of the IQ motif that interact with CaM in a Ca2+-independent manner (Bouche et al., 2002; Yang and Poovaiah, 2002a; Finkler et al., 2007; Du et al., 2009). The functions of AtSRs/CaMTAs were found to be dependent on their interaction with Ca2+/CaM (Choi et al., 2005; Du et al., 2009).
Loss-of-function mutants of AtSR1/CAMTA3 were reported to have pleiotropic, temperature-dependent, constitutive disease-resistant phenotypes, including compromised growth, spontaneous leaf chlorosis with autonomous lesions, constitutive expression of pathogenesis-related genes, and elevated resistance against both virulent and avirulent strains of Pseudomonas syringae pv tomato DC3000 (Galon et al., 2008; Du et al., 2009). These phenotypes were correlated with higher levels of endogenous salicylic acid (SA), demonstrating that AtSR1/CAMTA3 is a negative regulator of SA-mediated defense responses (Du et al., 2009). It was also shown that AtSR1/CAMTA3 interacts with a CGCG box motif in the −1-kb promoter region of EDS1 both in vivo and in vitro and suppresses the transcription of EDS1, a critical player in the SA activation loop and toll interleukin 1 receptor-nucleotide binding domain (NB)-leucine rich repeat (LRR)-type R gene-mediated defense in Arabidopsis (Du et al., 2009). Recently, AtSR1/CAMTA3 was also shown to negatively regulate ethylene-mediated senescence by recognizing a CGCG box in the promoters of EIN3 and disease resistance by interacting with a CGCG box in the promoters of NDR1, a key signaling component required for coiled-coil-NB-LRR-type R gene-mediated plant immunity (Nie et al., 2012). Similar to the function of CAMTA3, OsCBT, a CAMTA member from rice, also plays a negative role in regulating rice defense against both the bacterial pathogen Xanthomonas oryzae pv oryzae and the rice blast fungus Magnaporthe grisea (Koo et al., 2009). Very recently, CAMTA3 was shown to play a critical role in plant defense against insect herbivory (Laluk et al., 2012; Qiu et al., 2012). atsr1/camta3 null mutants are more vulnerable to herbivore attack, and CaM binding was shown to be required for AtSR1/CAMTA3 in maintaining normal levels of plant resistance to herbivore attack. In addition, it was observed that elevated SA levels in atsr1 mutant plants have a negative impact on both basal and induced biosynthesis of jasmonates, a critical hormone-mediated wounding response in plants (Qiu et al., 2012). Furthermore, compared with the wild type, the atsr1 mutant accumulates less of the insect repellent metabolite glucosinolate, and this coincides with the altered expression of several genes involved in glucosinolate metabolism, such as MYB51, AtST5α, and IQD1 (Laluk et al., 2012). In a different line of research, CAMTA3 was shown to recognize the conserved DNA motif 2 (CM2, CCGCGT) in the promoter of CBF2, a critical transcription factor required for Arabidopsis cold acclimation and the subsequent establishment of freezing tolerance as well as the regulation of cold-induced gene expression (Doherty et al., 2009). Very recently, results showed that CAMTA1 and CAMTA2 share some functional redundancy with CAMTA3 in suppressing SA biosynthesis and the quick induction of CBF1, CBF2, and CBF3 transcription (Kim et al., 2013). AtCAMTA1 was reported to be an auxin-responsive gene, and hypocotyl elongation of AtCAMTA1 knockout or repression lines is hyperresponsive to exogenous application of auxin, indicating that CAMTA1 could regulate plant growth through the action of auxin (Galon et al., 2010a).
Recent Breakthrough in Ca2+/CaM-Mediated Transcriptional Control of Plant Immunity
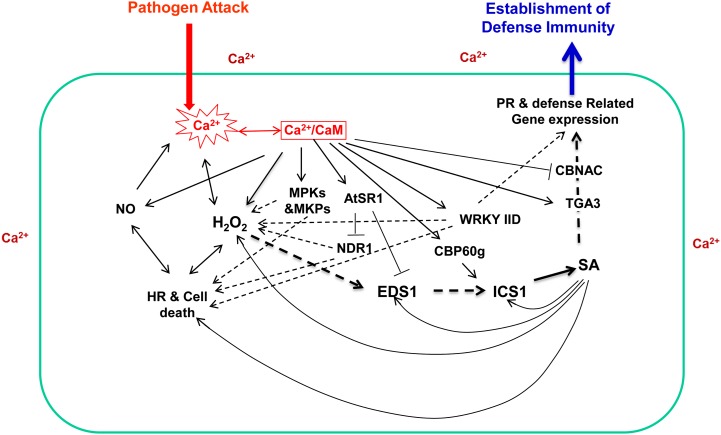
Ca2+/CaM-mediated signaling has been documented to be involved in almost every aspect of a plant’s life, including plant growth and development, as well as plant responses to biotic and abiotic stresses (Du and Poovaiah, 2005; Lecourieux et al., 2006; Yang et al., 2007; DeFalco et al., 2010a; Du et al., 2011). In the past several years, mechanisms by which Ca2+/CaM regulate plant defenses against pathogenic microbes have been revealed at an impressively rapid pace. Figure 4 summarizes the accumulated information of how Ca2+/CaM regulates the functions of target proteins involved in plant responses to pathogen attack. Rapid increases in intracellular Ca2+ concentration, oxidative burst, nitric oxide (NO) production, hypersensitive responses and the associated cell death, accumulation of SA, induced expression of pathogenesis-related genes, and the establishment of local and systemic resistances are common defense responses that occur at different stages during the establishment of immune responses after plants are challenged with pathogens (Nimchuk et al., 2003; Lecourieux et al., 2006; Fu and Dong, 2013). These changes are presented in Figure 4 in a rough temporal order. The key defense-related events/signaling components, such as hydrogen peroxide (H2O2), NO, and SA, are well known to induce the production of each other and, hence, are positioned directly or indirectly in various feed-forward amplification loops and act as positive regulators of hypersensitive and defense responses (Lecourieux et al., 2006; Ali et al., 2007; Fu and Dong, 2013). Accumulated data revealed that Ca2+/CaM-mediated regulations oversee and coordinate the progression of the entire plant immune system.
Figure 4.
Multiple controls of plant immunity by Ca2+/CaM. Empirically confirmed regulations are presented with solid lines, regulations not directly confirmed are presented with dashed lines, arrowheads indicate positive control, and T-heads indicate negative control. HR, Hypersensitive response; PR, pathogenesis related.
Properly controlled production of NO with the involvement of Ca2+, CaM, and NO synthase after the perception of lipopolysaccharides or avirulent pathogen and the balanced action of NO and H2O2 contribute to the appropriate progression of the hypersensitive reaction and the establishment of plant defenses (Ali et al., 2007). On the other hand, NO has also been reported to be a powerful stimulator of intracellular Ca2+ in plants, providing a path of feedback to the Ca2+ signaling system (Besson-Bard et al., 2008). While well known as a regulator of endogenous Ca2+ levels, H2O2 production during the oxidative burst requires Ca2+ influx, which activates the plasma membrane-localized NADPH oxidase (Keller et al., 1998). Furthermore, Ca2+/CaM has been proposed to increase H2O2 generation through Ca2+/CaM-dependent NAD kinase, which affects the concentration of available NADPH during the assembly and activation of NADPH oxidase (Harding et al., 1997; Turner et al., 2004). Moreover, Ca2+/CaM binds to plant catalase and enhances its activity (Yang and Poovaiah, 2002b). Recently, Ca2+/CaM was also reported to regulate H2O2-mediated defense responses by regulating the MAPK cascade through the action of MAPKs and MAPK phosphatases (Lee et al., 2008; Bartels et al., 2009; Takahashi et al., 2011).
SA has been generally accepted as a defense hormone that controls plant defense against biotrophic pathogens (Nimchuk et al., 2003; Fu and Dong, 2013). The SA activation pathway is under extensive regulation by Ca2+/CaM-mediated signaling. Transcription of the SA biosynthesis gene ICS1/SID2 is controlled by the Ca2+/CaM-binding transcription factor CBP60g (Wang et al., 2009, 2011; Zhang et al., 2010), providing a venue for the Ca2+ signal to activate the production of SA. The transcription of two critical genes, EDS1 and NDR1, required for the activation of both toll interleukin 1 receptor-NB-LRR and coiled-coil-NB-LRR R gene-activated immunities, is negatively controlled by CaM-regulated AtSR1/CAMTA3, enabling tight control over the synthesis of SA and providing an effective approach to avoid the misactivation of effector-triggered immunity as well as pathogen-associated molecular pattern-triggered immunity, since SA is critical for both (Nimchuk et al., 2003; Fu and Dong, 2013). In addition, Ca2+/CaM could also provide both positive and negative controls through WRKY7, WRKY11, WRKY17, and WRKY53 (Park et al., 2005; Journot-Catalino et al., 2006; Kim et al., 2006; Murray et al., 2007; Popescu et al., 2007), although their direct downstream target genes and their regulation by CaM remain unknown.
Defense-related gene expression after the accumulation of SA also seems to be regulated by Ca2+/CaM-mediated signaling. CaM binding to TGA3 enhances its interaction with the target promoter (Szymanski et al., 1996); furthermore, TGA3 interacts with NPR1, a critical transcription cofactor involved in SA perception and the expression of a broad spectrum of defense-related genes (Fu and Dong, 2013), providing a possible option to regulate the output of defensive reactions. Furthermore, CaM could also repress the expression of pathogenesis-related genes through the action of the transcription factor CBNAC (Kim et al., 2012). It is very interesting to see that Ca2+/CaM can exert a well-balanced control even at the final stage of defense responses after the induced accumulation of SA.
Defense comes with a price; mutant plants with misactivation or constitutive activation of defense responses usually suffer significantly in their growth and development or can even die (Heil and Baldwin, 2002). The deployment of multiple positive and negative regulatory controls on different progression stages during the establishment of plant immunity demonstrates the critical importance of balancing these immune responses. As summarized in this update, Ca2+, the universal messenger in eukaryotes, including plants, could act as a competent conductor in orchestrating these powerful physiological activities, which could protect plants from pathogen attack or cause them to commit “suicide.”
CONCLUSION
Since the discovery of CaM over 40 years ago, and especially during the last decade, tremendous progress has been made in the isolation and characterization of Ca2+/CaM and its target proteins. A complex Ca2+/CaM-regulated network is beginning to emerge, but it is far from complete. While accepting the unique roles of Ca2+ as a messenger, we are also realizing the complexities and the challenges of Ca2+/CaM-mediated signaling and its cross talk with different signal transduction pathways in plants. As more genomes are sequenced and more high-throughput tools are developed, the significance and the scope of this network are being realized at an unprecedented rate. However, there are several issues that still need to be resolved. First, we believe that many CaM-binding proteins remain to be identified, not to mention the target proteins of CaM-like proteins. Hence, the identification of novel CaM-binding proteins will still be one of the most important tasks for plant scientists. Classic approaches to identify targets of CaM through protein-protein interaction-based expression library screening may still be used, but improved approaches, such as the CaM-modified nanowire transistor method, are highly desirable. Second, the functional significance of these target proteins in terms of biochemical, molecular, and physiological activities needs to be adequately studied. Third, we need to determine how Ca2+/CaM and its interactors respond to upstream signals and how they regulate various downstream signal transduction pathways. For example, scientists need to determine whether and how Ca2+/CaM helps CCaMK to initiate RNS or AMS based on the subtle differences in Ca2+ spiking induced by Nod or Myc factors. Fourth, we need to understand the significance and complexity of Ca2+/CaM-mediated regulations, their roles as both positive and negative regulators to balance a particular signaling pathway, and how these regulatory roles are coordinated. To cite a specific example: when, where, and how Ca2+/CaM helps plants to produce a positive or negative regulation of the SA-mediated immune response to ensure appropriate protection against a potential pathogen and avoid the negative consequences of overreaction during defense. Progress in these areas will significantly improve our understanding of Ca2+/CaM-mediated signaling in plants.
Glossary
- EF
elongation factor
- Ca2+
calcium
- CaM
calmodulin
- CDPK
calcium-dependent protein kinase
- CaMBD
calmodulin-binding domain
- cDNA
complementary DNA
- CCaMK
calcium/calmodulin-dependent protein kinase
- RLK
receptor-like kinase
- MAPK
mitogen-activated protein kinase
- RNS
root nodulation symbiosis
- AMS
arbuscular mycorrhizal symbiosis
- AID
autoinhibitory domain
- VLD
visinin-like domain
- SA
salicylic acid
- H2O2
hydrogen peroxide
- NO
nitric oxide
References
- Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, Berkowitz GA. (2007) Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arazi T, Sunkar R, Kaplan B, Fromm H. (1999) A tobacco plasma membrane calmodulin-binding transporter confers Ni2+ tolerance and Pb2+ hypersensitivity in transgenic plants. Plant J 20: 171–182 [DOI] [PubMed] [Google Scholar]
- Bartels S, Anderson JC, González Besteiro MA, Carreri A, Hirt H, Buchala A, Métraux JP, Peck SC, Ulm R. (2009) MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21: 2884–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum G, Chen Y, Arazi T, Takatsuji H, Fromm H. (1993) A plant glutamate decarboxylase containing a calmodulin binding domain. Cloning, sequence, and functional analysis. J Biol Chem 268: 19610–19617 [PubMed] [Google Scholar]
- Baum G, Lev-Yadun S, Fridmann Y, Arazi T, Katsnelson H, Zik M, Fromm H. (1996) Calmodulin binding to glutamate decarboxylase is required for regulation of glutamate and GABA metabolism and normal development in plants. EMBO J 15: 2988–2996 [PMC free article] [PubMed] [Google Scholar]
- Besson-Bard A, Pugin A, Wendehenne D. (2008) New insights into nitric oxide signaling in plants. Annu Rev Plant Biol 59: 21–39 [DOI] [PubMed] [Google Scholar]
- Bouché N, Scharlat A, Snedden W, Bouchez D, Fromm H. (2002) A novel family of calmodulin-binding transcription activators in multicellular organisms. J Biol Chem 277: 21851–21861 [DOI] [PubMed] [Google Scholar]
- Bouché N, Yellin A, Snedden WA, Fromm H. (2005) Plant-specific calmodulin-binding proteins. Annu Rev Plant Biol 56: 435–466 [DOI] [PubMed] [Google Scholar]
- Bussemer J, Chigri F, Vothknecht UC. (2009) Arabidopsis ATPase family gene 1-like protein 1 is a calmodulin-binding AAA+-ATPase with a dual localization in chloroplasts and mitochondria. FEBS J 276: 3870–3880 [DOI] [PubMed] [Google Scholar]
- Charpenteau M, Jaworski K, Ramirez BC, Tretyn A, Ranjeva R, Ranty B. (2004) A receptor-like kinase from Arabidopsis thaliana is a calmodulin-binding protein. Biochem J 379: 841–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung WY. (1971) Cyclic 3′,5′-nucleotide phosphodiesterase. Evidence for and properties of a protein activator. J Biol Chem 246: 2859–2869 [PubMed] [Google Scholar]
- Chigri F, Hormann F, Stamp A, Stammers DK, Bolter B, Soll J, Vothknecht UC. (2006) Calcium regulation of chloroplast protein translocation is mediated by calmodulin binding to Tic32. Proc Natl Acad Sci USA 103: 16051–16056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi MS, Kim MC, Yoo JH, Moon BC, Koo SC, Park BO, Lee JH, Koo YD, Han HJ, Lee SY, et al. (2005) Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J Biol Chem 280: 40820–40831 [DOI] [PubMed] [Google Scholar]
- Chung WS, Lee SH, Kim JC, Heo WD, Kim MC, Park CY, Park HC, Lim CO, Kim WB, Harper JF, et al. (2000) Identification of a calmodulin-regulated soybean Ca(2+)-ATPase (SCA1) that is located in the plasma membrane. Plant Cell 12: 1393–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbran RJ, Soderling TR. (1990) Calcium/calmodulin-independent autophosphorylation sites of calcium/calmodulin-dependent protein kinase II: studies on the effect of phosphorylation of threonine 305/306 and serine 314 on calmodulin binding using synthetic peptides. J Biol Chem 265: 11213–11219 [PubMed] [Google Scholar]
- Davis TN, Urdea MS, Masiarz FR, Thorner J. (1986) Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell 47: 423–431 [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Bender KW, Snedden WA. (2010a) Breaking the code: Ca2+ sensors in plant signalling. Biochem J 425: 27–40 [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Chiasson D, Munro K, Kaiser BN, Snedden WA. (2010b) Characterization of GmCaMK1, a member of a soybean calmodulin-binding receptor-like kinase family. FEBS Lett 584: 4717–4724 [DOI] [PubMed] [Google Scholar]
- Ding X, Richter T, Chen M, Fujii H, Seo YS, Xie M, Zheng X, Kanrar S, Stevenson RA, Dardick C, Li Y, et al. (2009) A rice kinase-protein interaction map. Plant Physiol 149: 1478–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Ali GS, Simons KA, Hou J, Yang T, Reddy ASN, Poovaiah BW. (2009) Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457: 1154–1158 [DOI] [PubMed] [Google Scholar]
- Du L, Poovaiah BW. (2004) A Novel Family of Ca2+/Calmodulin-Binding Proteins Involved in Transcriptional Regulation: Interaction with fsh/Ring3 Class Transcription Activators. Plant Mol Biol 54: 549–569 [DOI] [PubMed] [Google Scholar]
- Du L, Poovaiah BW. (2005) Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature 437: 741–745 [DOI] [PubMed] [Google Scholar]
- Du L, Yang T, Puthanveettil SV, Poovaiah BW, Luan S (2011) Decoding of calcium signal through calmodulin: calmodulin-binding proteins in plants. In S Luan, ed, Coding and Decoding of Calcium Signals in Plants. Springer, Berlin, pp 177–233 [Google Scholar]
- Ehrhardt DW, Wais R, Long SR. (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Finkler A, Ashery-Padan R, Fromm H. (2007) CAMTAs: calmodulin-binding transcription activators from plants to human. FEBS Lett 581: 3893–3898 [DOI] [PubMed] [Google Scholar]
- Fromm H, Chua NH. (1992) Cloning of plant cDNAs encoding calmodulin-binding proteins using 35S-labeled recombinant calmodulin as a probe. Plant Mol Biol Rep 10: 199–206 [Google Scholar]
- Fu ZQ, Dong X. (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Galon Y, Aloni R, Nachmias D, Snir O, Feldmesser E, Scrase-Field S, Boyce JM, Bouché N, Knight MR, Fromm H. (2010a) Calmodulin-binding transcription activator 1 mediates auxin signaling and responds to stresses in Arabidopsis. Planta 232: 165–178 [DOI] [PubMed] [Google Scholar]
- Galon Y, Finkler A, Fromm H. (2010b) Calcium-regulated transcription in plants. Mol Plant 3: 653–669 [DOI] [PubMed] [Google Scholar]
- Galon Y, Nave R, Boyce JM, Nachmias D, Knight MR, Fromm H. (2008) Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett 582: 943–948 [DOI] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Muñoz A, Poovaiah BW, Oldroyd GE. (2006) Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Han HJ, Park HC, Byun HJ, Lee SM, Kim HS, Yun DJ, Cho MJ, Chung WS. (2012) The transcriptional repressor activity of ASYMMETRIC LEAVES1 is inhibited by direct interaction with calmodulin in Arabidopsis. Plant Cell Environ 35: 1969–1982 [DOI] [PubMed] [Google Scholar]
- Hanson PI, Schulman H. (1992) Inhibitory autophosphorylation of multifunctional Ca2+/calmodulin-dependent protein kinase analyzed by site-directed mutagenesis. J Biol Chem 267: 17216–17224 [PubMed] [Google Scholar]
- Harding SA, Oh SH, Roberts DM. (1997) Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J 16: 1137–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, Baldwin IT. (2002) Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci 7: 61–67 [DOI] [PubMed] [Google Scholar]
- Hoeflich KP, Ikura M. (2002) Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 108: 739–742 [DOI] [PubMed] [Google Scholar]
- Hong B, Ichida A, Wang Y, Gens JS, Pickard BG, Harper JF. (1999) Identification of a calmodulin-regulated Ca2+-ATPase in the endoplasmic reticulum. Plant Physiol 119: 1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh HL, Song CJ, Roux SJ. (2000) Regulation of a recombinant pea nuclear apyrase by calmodulin and casein kinase II. Biochim Biophys Acta 1494: 248–255 [DOI] [PubMed] [Google Scholar]
- Hua W, Zhang L, Liang S, Jones RL, Lu YT. (2004) A tobacco calcium/calmodulin-binding protein kinase functions as a negative regulator of flowering. J Biol Chem 279: 31483–31494 [DOI] [PubMed] [Google Scholar]
- Hudmon A, Schulman H. (2002). Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem J 364: 593–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura M, Ames JB. (2006) Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily: two ways to promote multifunctionality. Proc Natl Acad Sci USA 103: 1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H, Rainaldi M, Vogel HJ. (2009) Structural studies of soybean calmodulin isoform 4 bound to the calmodulin-binding domain of tobacco mitogen-activated protein kinase phosphatase-1 provide insights into a sequential target binding mode. J Biol Chem 284: 28292–28305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journot-Catalino N, Somssich IE, Roby D, Kroj T. (2006) The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiuchi S, Yamazaki R. (1970) Calcium dependent phosphodiesterase activity and its activating factor (PAF) from brain studies on cyclic 3′,5′-nucleotide phosphodiesterase (3). Biochem Biophys Res Commun 41: 1104–1110 [DOI] [PubMed] [Google Scholar]
- Kang H, Zhu H, Chu X, Yang Z, Yuan S, Yu D, Wang C, Hong Z, Zhang Z. (2011) A novel interaction between CCaMK and a protein containing the Scythe_N ubiquitin-like domain in Lotus japonicus. Plant Physiol 155: 1312–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Damude HG, Werner D, Doerner P, Dixon RA, Lamb C. (1998) A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell 10: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Jung MS, Lee K, Kim KE, Yoo JH, Kim MC, Kim DH, Cho MJ, Chung WS. (2009a) An S-locus receptor-like kinase in plasma membrane interacts with calmodulin in Arabidopsis. FEBS Lett 583: 36–42 [DOI] [PubMed] [Google Scholar]
- Kim HS, Park HC, Kim KE, Jung MS, Han HJ, Kim SH, Kwon YS, Bahk S, An J, Bae DW, et al. (2012) A NAC transcription factor and SNI1 cooperatively suppress basal pathogen resistance in Arabidopsis thaliana. Nucleic Acids Res 40: 9182–9192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Fan B, Chen Z. (2006) Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol 142: 1180–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MC, Chung WS, Yun DJ, Cho MJ. (2009b) Calcium and calmodulin-mediated regulation of gene expression in plants. Mol Plant 2: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MC, Lee SH, Kim JK, Chun HJ, Choi MS, Chung WS, Moon BC, Kang CH, Park CY, Yoo JH, et al. (2002) Mlo, a Modulator of Plant Defense and Cell Death, Is a Novel Calmodulin-binding Protein. J Biol Chem 277: 19304–19314 [DOI] [PubMed] [Google Scholar]
- Kim MC, Panstruga R, Elliott C, Muller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P. (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416: 447–451 [DOI] [PubMed] [Google Scholar]
- Kim Y, Park S, Gilmour SJ, Thomashow MF. (2013) Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J 75: 364–376 [DOI] [PubMed] [Google Scholar]
- Kohler C, Merkle T, Neuhaus G. (1999) Characterisation of a novel gene family of putative cyclic nucleotide- and calmodulin-regulated ion channels in Arabidopsis thaliana. Plant J 18: 97–104 [DOI] [PubMed] [Google Scholar]
- Koo SC, Choi MS, Chun HJ, Shin DB, Park BS, Kim YH, Park HM, Seo HS, Song JT, Kang KY, et al. (2009) The calmodulin-binding transcription factor OsCBT suppresses defense responses to pathogens in rice. Mol Cells 27: 563–570 [DOI] [PubMed] [Google Scholar]
- Kosuta S, Hazledine S, Sun J, Miwa H, Morris RJ, Downie JA, Oldroyd GE. (2008) Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc Natl Acad Sci USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K. (2010) Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K, Prasad KV, Savchenko T, Celesnik H, Dehesh K, Levy M, Mitchell-Olds T, Reddy AS. (2012) The calmodulin-binding transcription factor SIGNAL RESPONSIVE1 is a novel regulator of glucosinolate metabolism and herbivory tolerance in Arabidopsis. Plant Cell Physiol 53: 2008–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landoni M, De Francesco A, Galbiati M, Tonelli C. (2010) A loss-of-function mutation in Calmodulin2 gene affects pollen germination in Arabidopsis thaliana. Plant Mol Biol 74: 235–247 [DOI] [PubMed] [Google Scholar]
- Lecourieux D, Ranjeva R, Pugin A. (2006) Calcium in plant defence-signalling pathways. New Phytol 171: 249–269 [DOI] [PubMed] [Google Scholar]
- Lee K, Song EH, Kim HS, Yoo JH, Han HJ, Jung MS, Lee SM, Kim KE, Kim MC, Cho MJ, et al. (2008) Regulation of MAPK phosphatase 1 (AtMKP1) by calmodulin in Arabidopsis. J Biol Chem 283: 23581–23588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Levy M, Wang QM, Kaspi R, Parrella MP, Abel S. (2005) Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J 43: 79–96 [DOI] [PubMed] [Google Scholar]
- Liao J, Singh S, Hossain MS, Andersen SU, Ross L, Bonetta D, Zhou Y, Sato S, Tabata S, Stougaard J, et al. (2012) Negative regulation of CCaMK is essential for symbiotic infection. Plant J 72: 572–584 [DOI] [PubMed] [Google Scholar]
- Lin TW, Hsieh PJ, Lin CL, Fang YY, Yang JX, Tsai CC, Chiang PL, Pan CY, Chen YT. (2010) Label-free detection of protein-protein interactions using a calmodulin-modified nanowire transistor. Proc Natl Acad Sci USA 107: 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HT, Gao F, Li GL, Han JL, Liu DL, Sun DY, Zhou RG. (2008) The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. Plant J 55: 760–773 [DOI] [PubMed] [Google Scholar]
- Ma F, Lu R, Liu H, Shi B, Zhang J, Tan M, Zhang A, Jiang M. (2012) Nitric oxide-activated calcium/calmodulin-dependent protein kinase regulates the abscisic acid-induced antioxidant defence in maize. J Exp Bot 63: 4835–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Liang SP, Jones RL, Lu YT. (2004) Characterization of a novel calcium/calmodulin-dependent protein kinase from tobacco. Plant Physiol 135: 1280–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom S, Akerlund HE, Askerlund P. (2000) Regulatory role of the N terminus of the vacuolar calcium-ATPase in cauliflower. Plant Physiol 122: 517–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador WE, Means AR, Quiocho FA. (1993) Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science 262: 1718–1721 [DOI] [PubMed] [Google Scholar]
- Messinese E, Mun JH, Yeun LH, Jayaraman D, Rougé P, Barre A, Lougnon G, Schornack S, Bono JJ, Cook DR, et al. (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact 20: 912–921 [DOI] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GED, Long SR. (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon BC, Choi MS, Kang YH, Kim MC, Cheong MS, Park CY, Yoo JH, Koo SC, Lee SM, Lim CO, et al. (2005) Arabidopsis ubiquitin-specific protease 6 (AtUBP6) interacts with calmodulin. FEBS Lett 579: 3885–3890 [DOI] [PubMed] [Google Scholar]
- Murray SL, Ingle RA, Petersen LN, Denby KJ. (2007) Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol Plant Microbe Interact 20: 1431–1438 [DOI] [PubMed] [Google Scholar]
- Nelissen H, Clarke JH, De Block M, De Block S, Vanderhaeghen R, Zielinski RE, Dyer T, Lusta S, Inze D, Van Lijsebettens M. (2003) DRL1, a homolog of the yeast TOT4/KT112 protein, has a function in meristem activity and organ growth in plants. Plant Cell 15: 639–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie H, Zhao C, Wu G, Wu Y, Chen Y, Tang D. (2012) SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol 158: 1847–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk Z, Eulgem T, Holt BF, III, Dangl JL. (2003) Recognition and response in the plant immune system. Annu Rev Genet 37: 579–609 [DOI] [PubMed] [Google Scholar]
- Oh MH, Kim HS, Wu X, Clouse SD, Zielinski RE, Huber SC. (2012) Calcium/calmodulin inhibition of the Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase provides a possible link between calcium and brassinosteroid signalling. Biochem J 443: 515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA. (2004) Calcium, kinases and nodulation signalling in legumes. Nat Rev Mol Cell Biol 5: 566–576 [DOI] [PubMed] [Google Scholar]
- Pandey S, Tiwari SB, Tyagi W, Reddy MK, Upadhyaya KC, Sopory SK. (2002) A Ca2+/CaM-dependent kinase from pea is stress regulated and in vitro phosphorylates a protein that binds to AtCaM5 promoter. Eur J Biochem 269: 3193–3204 [DOI] [PubMed] [Google Scholar]
- Park CY, Lee JH, Yoo JH, Moon BC, Choi MS, Kang YH, Lee SM, Kim HS, Kang KY, Chung WS, et al. (2005) WRKY group IId transcription factors interact with calmodulin. FEBS Lett 579: 1545–1550 [DOI] [PubMed] [Google Scholar]
- Patil S, Takezawa D, Poovaiah BW. (1995) Chimeric plant calcium/calmodulin-dependent protein kinase gene with a neural visinin-like calcium-binding domain. Proc Natl Acad Sci USA 92: 4897–4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah BW, Reddy ASN. (1987) Calcium messenger system in plants. CRC Crit Rev Plant Sci 6: 47–103 [DOI] [PubMed] [Google Scholar]
- Poovaiah BW, Reddy ASN. (1993) Calcium and signal transduction in plants. CRC Crit Rev Plant Sci 12: 185–211 [DOI] [PubMed] [Google Scholar]
- Poovaiah BW, Xia M, Liu Z, Wang W, Yang T, Sathyanarayanan PV, Franceschi VR. (1999) Developmental regulation of the gene for chimeric calcium/calmodulin-dependent protein kinase in anthers. Planta 209: 161–171 [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Seay M, Gerstein M, Snyder M, Dinesh-Kumar SP. (2007) Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc Natl Acad Sci USA 104: 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu YJ, Xi J, Du LQ, Suttle JC, Poovaiah BW. (2012) Coupling calcium/calmodulin-mediated signaling and herbivore-induced plant response through calmodulin-binding transcription factor AtSR1/CAMTA3. Plant Mol Biol 79: 89–99 [DOI] [PubMed] [Google Scholar]
- Ramachandiran S, Takezawa D, Wang W, Poovaiah BW. (1997) Functional domains of plant chimeric calcium/calmodulin-dependent protein kinase: regulation by autoinhibitory and visinin-like domains. J Biochem 121: 984–990 [DOI] [PubMed] [Google Scholar]
- Reddy AS, Ali GS, Celesnik H, Day IS. (2011a) Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23: 2010–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Day IS, Narasimhulu SB, Safadi F, Reddy VS, Golovkin M, Harnly MJ. (2002) Isolation and characterization of a novel calmodulin-binding protein from potato. J Biol Chem 277: 4206–4214 [DOI] [PubMed] [Google Scholar]
- Reddy AS, Reddy VS, Golovkin M. (2000) A calmodulin binding protein from Arabidopsis is induced by ethylene and contains a DNA-binding motif. Biochem Biophys Res Commun 279: 762–769 [DOI] [PubMed] [Google Scholar]
- Reddy AS, Safadi F, Narasimhulu SB, Golovkin M, Hu X. (1996) A novel plant calmodulin-binding protein with a kinesin heavy chain motor domain. J Biol Chem 271: 7052–7060 [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Ben-Hur A, Day IS. (2011b) Experimental and computational approaches for the study of calmodulin interactions. Phytochemistry 72: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Takezawa D, Fromm H, Poovaiah BW. (1993) Isolation and characterization of 2 cDNAs that encode for calmodulin-binding proteins from corn root-tips. Plant Sci 94: 109–117 [Google Scholar]
- Rhoads AR, Friedberg F. (1997) Sequence motifs for calmodulin recognition. FASEB J 11: 331–340 [DOI] [PubMed] [Google Scholar]
- Routray P, Miller JB, Du L, Oldroyd G, Poovaiah BW. (July 19, 2013) Phosphorylation of S344 in the calmodulin-binding domain negatively affects CCaMK function during bacterial and fungal symbioses. Plant J http://dx.doi.org/10.1111/tpj.12288 [DOI] [PubMed] [Google Scholar]
- Safadi F, Reddy VS, Reddy AS. (2000) A pollen-specific novel calmodulin-binding protein with tetratricopeptide repeats. J Biol Chem 275: 35457–35470 [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan PV, Cremo CR, Poovaiah BW. (2000) Plant chimeric Ca2+/Calmodulin-dependent protein kinase. Role of the neural visinin-like domain in regulating autophosphorylation and calmodulin affinity. J Biol Chem 275: 30417–30422 [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan PV, Poovaiah BW. (2002) Autophosphorylation-dependent inactivation of plant chimeric calcium/calmodulin-dependent protein kinase. Eur J Biochem 269: 2457–2463 [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan PV, Siems WF, Jones JP, Poovaiah BW. (2001) Calcium-stimulated autophosphorylation site of plant chimeric calcium/calmodulin-dependent protein kinase. J Biol Chem 276: 32940–32947 [DOI] [PubMed] [Google Scholar]
- Shen XC, Valencia CA, Szostak JW, Dong B, Liu RH. (2005) Scanning the human proteome for calmodulin-binding proteins. Proc Natl Acad Sci USA 102: 5969–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Ni L, Zhang A, Cao J, Zhang H, Qin T, Tan M, Zhang J, Jiang M. (2012) OsDMI3 is a novel component of abscisic acid signaling in the induction of antioxidant defense in leaves of rice. Mol Plant 5: 1359–1374 [DOI] [PubMed] [Google Scholar]
- Shimoda Y, Han L, Yamazaki T, Suzuki R, Hayashi M, Imaizumi-Anraku H. (2012) Rhizobial and fungal symbioses show different requirements for calmodulin binding to calcium calmodulin-dependent protein kinase in Lotus japonicus. Plant Cell 24: 304–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Parniske M. (2012) Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr Opin Plant Biol 15: 444–453 [DOI] [PubMed] [Google Scholar]
- Snedden WA, Blumwald E. (2000) Alternative splicing of a novel diacylglycerol kinase in tomato leads to a calmodulin-binding isoform. Plant J 24: 317–326 [DOI] [PubMed] [Google Scholar]
- Steinebrunner I, Wu J, Sun Y, Corbett A, Roux SJ. (2003) Disruption of apyrases inhibits pollen germination in Arabidopsis. Plant Physiol 131: 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainsbury DJ, Zhou L, Oldroyd GE, Bornemann S. (2012) Calcium ion binding properties of Medicago truncatula calcium/calmodulin-dependent protein kinase. Biochemistry 51: 6895–6907 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Liao B, Zielinski RE. (1996) Calmodulin isoforms differentially enhance the binding of cauliflower nuclear proteins and recombinant TGA3 to a region derived from the Arabidopsis Cam-3 promoter. Plant Cell 8: 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F, Mizoguchi T, Yoshida R, Ichimura K, Shinozaki K. (2011) Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol Cell 41: 649–660 [DOI] [PubMed] [Google Scholar]
- Takezawa D. (2003) Characterization of a novel plant PP2C-like protein Ser/Thr phosphatase as a calmodulin-binding protein. J Biol Chem 278: 38076–38083 [DOI] [PubMed] [Google Scholar]
- Takezawa D, Ramachandiran S, Paranjape V, Poovaiah BW. (1996) Dual regulation of a chimeric plant serine/threonine kinase by calcium and calcium/calmodulin. J Biol Chem 271: 8126–8132 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, et al. (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Turner WL, Waller JC, Vanderbeld B, Snedden WA. (2004) Cloning and characterization of two NAD kinases from Arabidopsis: identification of a calmodulin binding isoform. Plant Physiol 135: 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V, Tichtinsky G, Dumas C, Gaude T, Cock JM. (2003) Interaction of calmodulin, a sorting nexin and kinase-associated protein phosphatase with the Brassica oleracea S locus receptor kinase. Plant Physiol 133: 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluthambi K, Poovaiah BW. (1984) Calcium-promoted protein phosphorylation in plants. Science 223: 167–169 [DOI] [PubMed] [Google Scholar]
- Wais RJ, Galera C, Oldroyd G, Catoira R, Penmetsa RV, Cook D, Gough C, Denarié J, Long SR. (2000) Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc Natl Acad Sci USA 97: 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SA, Viprey V, Downie JA. (2000) Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by nod factors and chitin oligomers. Proc Natl Acad Sci USA 97: 13413–13418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, Glazebrook J. (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog 5: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Truman W, Sato M, Nguyen V, Katagiri F, Glazebrook J. (2011) CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J 67: 1029–1041 [DOI] [PubMed] [Google Scholar]
- Wang W, Takezawa D, Narasimhulu SB, Reddy AS, Poovaiah BW. (1996) A novel kinesin-like protein with a calmodulin-binding domain. Plant Mol Biol 31: 87–100 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liang S, Xie QG, Lu YT. (2004) Characterization of a calmodulin-regulated Ca2+-dependent-protein-kinase-related protein kinase, AtCRK1, from Arabidopsis. Biochem J 383: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watillon B, Kettmann R, Boxus P, Burny A. (1993) A calcium/calmodulin-binding serine/threonine protein kinase homologous to the mammalian type II calcium/calmodulin-dependent protein kinase is expressed in plant cells. Plant Physiol 101: 1381–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J, Qiu YJ, Du LQ, Poovaiah BW. (2012) Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Sci 185: 274–280 [DOI] [PubMed] [Google Scholar]
- Yamakawa H, Katou S, Seo S, Mitsuhara I, Kamada H, Ohashi Y. (2004) Plant MAPK Phosphatase Interacts with Calmodulins. J Biol Chem 279: 928–936 [DOI] [PubMed] [Google Scholar]
- Yang T, Chaudhuri S, Yang L, Chen Y, Poovaiah BW. (2004) Calcium/calmodulin up-regulates a cytoplasmic receptor-like kinase in plants. J Biol Chem 279: 42552–42559 [DOI] [PubMed] [Google Scholar]
- Yang T, Chaudhuri S, Yang L, Du L, Poovaiah BW. (2010) A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J Biol Chem 285: 7119–7126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Du L, Poovaiah BW. (2007) Concept of redesigning proteins by manipulating calcium/calmodulin-binding domains to engineer plants with altered traits. Funct Plant Biol 34: 343–352 [DOI] [PubMed] [Google Scholar]
- Yang T, Peng H, Whitaker BD, Conway WS. (2012) Characterization of a calcium/calmodulin-regulated SR/CAMTA gene family during tomato fruit development and ripening. BMC Plant Biol 12: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Peng H, Whitaker BD, Jurick WM. (2013) Differential expression of calcium/calmodulin-regulated SlSRs in response to abiotic and biotic stresses in tomato fruit. Physiol Plant 148: 445–455 [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW. (2000a) An early ethylene up-regulated gene encoding a calmodulin-binding protein involved in plant senescence and death. J Biol Chem 275: 38467–38473 [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW. (2000b) Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J Biol Chem 275: 3137–3143 [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW. (2000c) Arabidopsis chloroplast chaperonin 10 is a calmodulin-binding protein. Biochem Biophys Res Commun 275: 601–607 [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW. (2002a) A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J Biol Chem 277: 45049–45058 [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW. (2002b) Hydrogen peroxide homeostasis: activation of plant catalase by calcium/calmodulin. Proc Natl Acad Sci USA 99: 4097–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo JH, Park CY, Kim JC, Heo WD, Cheong MS, Park HC, Kim MC, Moon BC, Choi MS, Kang YH, et al. (2005) Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J Biol Chem 280: 3697–3706 [DOI] [PubMed] [Google Scholar]
- Yun CH, Bai J, Sun DY, Cui DF, Chang WR, Liang DC. (2004) Structure of potato calmodulin PCM6: the first report of the three-dimensional structure of a plant calmodulin. Acta Crystallogr D Biol Crystallogr 60: 1214–1219 [DOI] [PubMed] [Google Scholar]
- Zhang L, Liu BF, Liang S, Jones RL, Lu YT. (2002) Molecular and biochemical characterization of a calcium/calmodulin-binding protein kinase from rice. Biochem J 368: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu YT. (2003) Calmodulin-binding protein kinases in plants. Trends Plant Sci 8: 123–127 [DOI] [PubMed] [Google Scholar]
- Zhang M, Tanaka T, Ikura M. (1995) Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat Struct Biol 2: 758–767 [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhou RG, Gao YJ, Zheng SZ, Xu P, Zhang SQ, Sun DY. (2009) Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol 149: 1773–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu S, Ding P, Wang D, Cheng YT, He J, Gao M, Xu F, Li Y, Zhu Z, et al. (2010) Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107: 18220–18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski RE. (2002) Characterization of three new members of the Arabidopsis thaliana calmodulin gene family: conserved and highly diverged members of the gene family functionally complement a yeast calmodulin null. Planta 214: 446–455 [DOI] [PubMed] [Google Scholar]