Ca2+ signaling is involved in brassinosteroid hormone-mediated gene expression and plant phenotypes.
Abstract
Brassinosteroids (BRs) are hormones that control many aspects of plant growth and development, acting at the cell level to promote division and expansion. BR regulation of plant and plant cell function occurs through altered expression of many genes. Transcriptional reprogramming downstream from cell perception of this hormone is currently known to be mediated by a phosphorylation/dephosphorylation (“phosphorelay”) cascade that alters the stability of two master transcription regulators. Here, we provide evidence that BR perception by their receptor also causes an elevation in cytosolic Ca2+, initiating a Ca2+ signaling cascade in Arabidopsis (Arabidopsis thaliana) cell cytosol. BR-dependent increases in the expression of some genes (INDOLE-3-ACETIC ACID-INDUCIBLE1 and PHYTOCHROME B ACTIVATION-TAGGED SUPPRESSOR1) were impaired in wild-type plants by a Ca2+ channel blocker and also in the defense-no-death (dnd1) mutant, which lacks a functional cyclic GMP-activated cell membrane Ca2+-conducting channel. Alternatively, mutations that impair the BR phosphorelay cascade did not much affect the BR-dependent expression of these genes. Similar effects of the Ca2+ channel blocker and dnd1 mutation were observed on a BR plant growth phenotype, deetiolation of the seedling hypocotyl. Further evidence presented in this report suggests that a BR-dependent elevation in cyclic GMP may be involved in the Ca2+ signaling cascade initiated by this hormone. The work presented here leads to a new model of the molecular steps that mediate some of the cell responses to this plant hormone.
Brassinolide and similar compounds, the brassinosteroids (BRs), are a family of growth-promoting steroidal hormones that are ubiquitous in the plant kingdom. BRs have a positive effect on cell expansion and division; therefore, plants with mutations that impair BR signaling have a dwarf phenotype (Clouse, 2011). BRs regulate a broad range of physiological processes in plants, including reproduction and senescence programs, leaf development, root growth, vascular differentiation, and responses to light as well as other environmental cues, often in an integrated fashion with other hormones (Clouse, 2011; Witthöft and Harter, 2011; Ye et al., 2011). As detailed in a number of recent reviews (Kang et al., 2010; Clouse, 2011, Witthöft and Harter, 2011; Yang et al., 2011), the hormone is perceived at the cell surface upon binding to its receptor, BRASSINOSTEROID INSENSITIVE1 (BRI1). BRI1 is a member of a large family of Leu-rich repeat receptor-like kinases. The global effects of the signaling cascade initiated upon BR binding to the BRI1 receptor on plant growth and development occur through the regulation by the steroid hormone of the expression of a wide array of genes.
Numerous studies, as summarized in the aforementioned reviews, have delineated steps in a protein phosphorylation/dephosphorylation (phosphorelay) cascade as the basis for BR-mediated transcriptional reprogramming. Some of the steps involved in this phosphorelay system include the following. The BR receptor is maintained in an inactive state by binding of cytosolic BRI1 KINASE INHIBITOR1 (BKI1). Hormone binding to BRI1 releases BKI1 and recruits binding of the BRI1 coreceptor BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1), leading to BRI1:BAK1 transphosphorylations. Downstream from receptor phosphorylation, the phosphorelay cascade involves phosphatase-dependent deactivation of the cytosolic kinase BRASSINOSTEROID-INSENSITIVE2 (BIN2), a negative regulator of BR signaling. When activated, BIN2 phosphorylates two master transcription factors (TFs), BRASSINAZOLE RESISTANT1 (BZR1) and BRI1-ETHYL METHANESULFONATE-SUPPRESSOR1 (BES1), preventing their function in the nucleus. In their unphosphorylated, active state (i.e. in the presence of deactivated BIN2), these TFs move to (or are retained in) the nucleus and activate many genes, including other TFs, thus amplifying the BR signaling output.
The signal transduction cascade that links the cell perception of extracellular BR to the control of gene expression, as well as the structure-function relationship of BRI1 and how the receptor acts through autophosphorylation and transphosphorylation to facilitate the BR response cascade, is one of the best-studied signaling pathways in plants (Jaillais et al., 2011). Yet, in the wake of the flood of knowledge developed about BR/BRI1 signaling, some recent reviews point out a striking as-yet-unresolved issue regarding the action of this hormone on plant cells. BR perception at the cell surface involves immediate effects on cell function that suggest a signaling cascade distinct from the phosphorelay system (Witthöft and Harter, 2011; Harter et al., 2012). Furthermore, other recent studies suggest that some BR-dependent plant phenotypes may not be mediated by phosphorelay signaling (Hacham et al., 2011).
Some time ago, Kwezi et al. (2007) identified a guanylyl cyclase (GC) activity associated with the cytosolic domain of Arabidopsis (Arabidopsis thaliana) BRI1; this portion of the receptor (expressed as a recombinant protein in Escherichia coli and affinity purified) generated cyclic GMP (cGMP) from GTP in vitro. Prior studies from this laboratory (Qi et al., 2010) with another Leu-rich-repeat receptor-like kinase (PLANT ELICITOR PEPTIDE RECEPTOR1 [PEPR1]) that has a similar putative GC domain to BRI1 have demonstrated a similar level of in vitro GC activity and, furthermore, provided evidence that PEPR1 signaling involves the activation (possibly due to cGMP generation) of a Ca2+-conducting cyclic nucleotide-gated channel (CNGC) in vivo (Ma et al., 2012). The focus of the work presented here was to test the hypothesis that (some components of) BR:BRI1 signaling involve cytosolic Ca2+ elevation, which is well known to act as a secondary messenger system in all cells (Dodd et al., 2010).
There are few reports of Ca2+ involvement in steps of the BR phosphorelay signaling cascade downstream from BRI1. DWARF1, an enzyme involved in BR synthesis, is activated by Ca2+/calmodulin (CaM); this would suggest that on a long-term basis, the Ca2+ status of cells might impact the generation or steady-state level of the steroidal hormone (Du and Poovaiah, 2005). A report on hormone signaling in wheat (Triticum aestivum; Singla et al., 2006) indirectly suggested that expression of a Ca2+-regulated auxin-responsive gene in wheat orthologous to Arabidopsis INDOLE-3-ACETIC ACID-INDUCIBLE1 (IAA1) is also induced by BR. Working with Arabidopsis, we used genetic and biochemical approaches to evaluate if BR-dependent Ca2+ signaling is involved in the regulation of IAA1 (as well as other BR-responsive gene) expression and plant phenotypes impacted by BR.
RESULTS
Hormone- and Receptor-Dependent Cytosolic Ca2+ Elevation
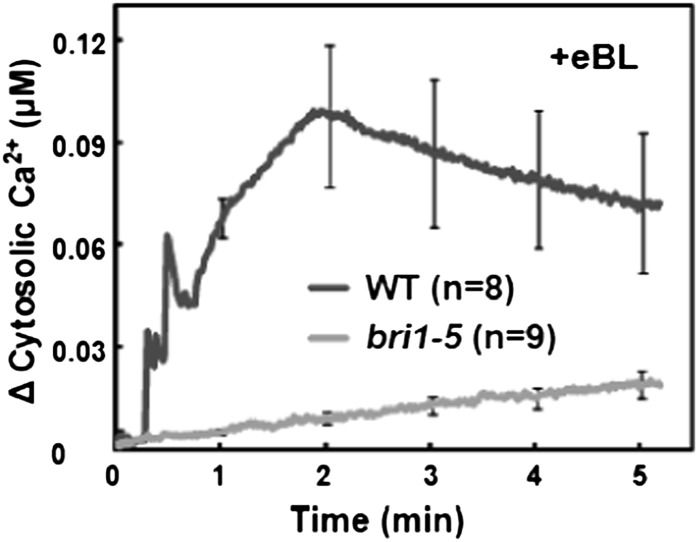
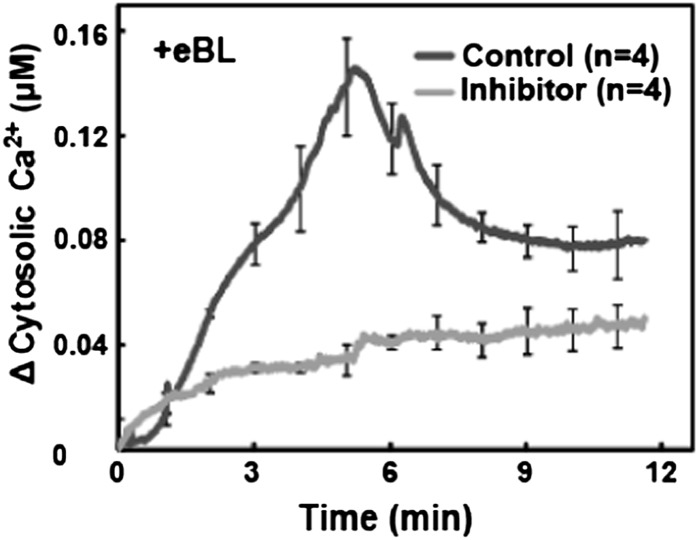
We evaluated BR-induced Ca2+ signaling by monitoring the effect of exogenous epibrassinolide (eBL; see “Materials and Methods”) on the level of cytosolic Ca2+ in detached leaves of Arabidopsis plants expressing the Ca2+ reporter protein apoaequorin. Studies were undertaken using several wild-type Arabidopsis ecotypes (as appropriate, for controls when mutants were used in various ecotype backgrounds) as well as genetic mutants also expressing the gene encoding this Ca2+ reporter protein. As shown in Figure 1, we observed an elevation in cytosolic Ca2+ initiating seconds after adding BR to leaves from ecotype Wassilewskija (Ws-2) wild-type plants. The bri1-5 genotype has reduced sensitivity to BR (Wang et al., 2005). The bri1-5 mutant is the most fertile of the bri1 weak allele mutants and thus is often used (Clouse 2011) for the expression of various transgenes in a genotype with impaired BRI1 receptor signaling. As shown in Figure 1, the cytosolic Ca2+ elevation occurring upon addition of BR to wild-type leaves is impaired in the bri1-5 mutant; generation of the hormone-induced Ca2+ signal requires the function of the BRI1 receptor.
Figure 1.
Exogenous BR-dependent elevation of cytosolic Ca2+ in detached leaves from wild-type Ws-2 (WT) and bri1-5 plants. Ligand was added at time 0; change in Ca2+ was calculated for each replicate by subtracting the value recorded at time 0 from all other measurements. Results are presented as mean values of Ca2+ increase (replicate number in parentheses) ± se shown at 1-min intervals. A similar experimental design and data presentation were used for the experiments shown in Figures 2, 3, and 5. Note that the wild-type plants used for the experiments shown in Figures 2, 3, and 5 were Col. Studies of BR signaling that involve exogenous application of eBL typically use concentrations of 100 nm to 1 µm (Li et al., 2009; Hacham et al., 2011). For the studies of Ca2+ elevation in aequorin-expressing plants shown in this report, we used 100 nm eBL. We found a significantly greater increase in cytosolic Ca2+ when 1 µm was added to leaves of wild-type plants (data not shown).
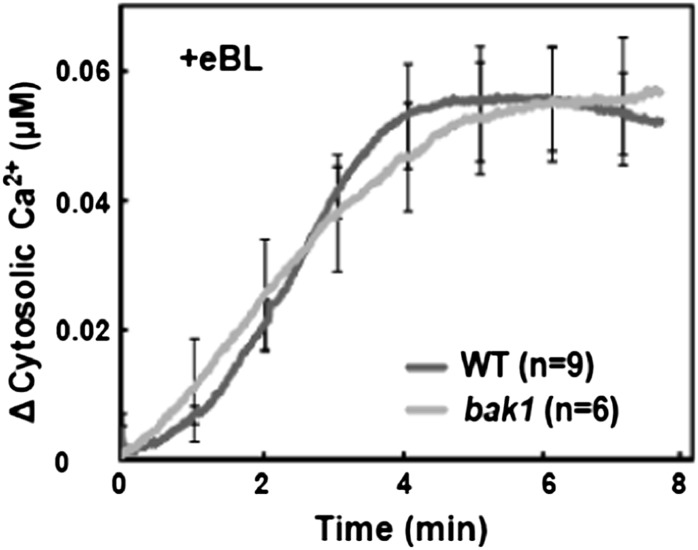
BR signaling through the BRI1 receptor involves physical and functional interactions of BRI1 with its coreceptor BAK1 (Nam and Li, 2002). bak1 mutants that lack the expression of BAK1 mRNA show impairment of some aspects of BR signaling (Chinchilla et al., 2007; Kemmerling et al., 2007). Here, we found no effect of BAK1 mutation on BR/BRI1-dependent Ca2+ signaling; hormone-induced cytosolic Ca2+ elevation was similar in ecotype Columbia (Col) wild-type and bak1 leaves (Fig. 2). Some (but not all) functions of the BAK1 coreceptor involved with BR signaling can be replaced by other members of the BAK1 protein family, such as BAK1-LIKE1 (BKK1; He et al., 2007). The lack of an effect of BAK1 mutation on BR/BRI1 signaling could be due to either (1) the possible replacement of BAK1 function by BKK1 (or a similar protein) in the bak1 mutant or (2) the molecular step(s) leading to BR-dependent Ca2+ signaling involving some aspect of BRI1 function independent of its interaction with a coreceptor. Additional studies suggest that possibility 2 might be the case (i.e. that BR/BRI1-dependent Ca2+ signaling may not require the coreceptor). The work presented in this report includes studies indicating that BR-dependent expression of the genes IAA1 and PHYTOCHROME B ACTIVATION-TAGGED SUPPRESSOR1 (BAS1) requires the generation of a BR-dependent Ca2+ signal (see below). Results shown in Supplemental Figure S1 indicate that BR-dependent IAA1 and BAS1 expression is not compromised in either a bak1 mutant or the bak1/bkk1 double mutant; these results suggest that this component of BR signaling may not involve a coreceptor.
Figure 2.
Exogenous BR-dependent elevation of cytosolic Ca2+ in detached leaves from wild-type Col (WT) and bak1 plants.
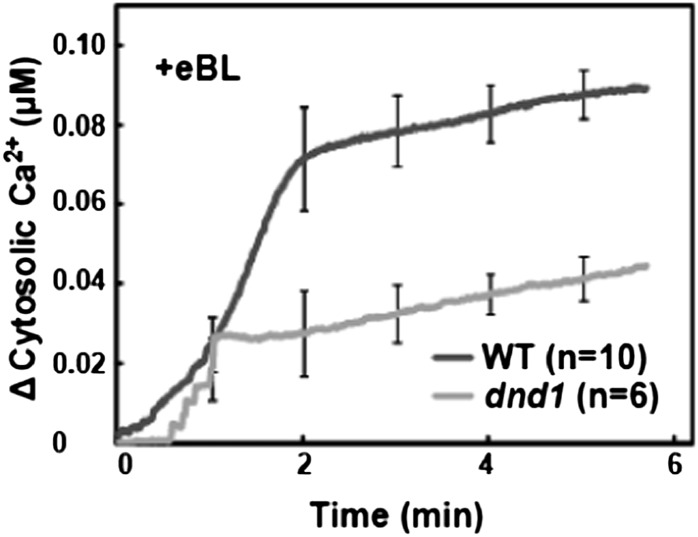
As mentioned above, BRI1 has been shown to have in vitro GC activity independent of the presence of a coreceptor (Kwezi et al., 2007). If BRI1 GC activity is involved in BR-induced cytosolic Ca2+ elevation, the Ca2+ signal could be generated by the opening of cGMP-activated Ca2+-conducting ion channels in the cell membrane upon binding of BR to its receptor. We examined this hypothesis by monitoring BR-dependent cytosolic Ca2+ elevation in the defense-no-death (dnd1) mutant, which lacks a functional CYCLIC NUCLEOTIDE-GATED CHANNEL2 (CNGC2) gene (Clough et al., 2000). CNGC2 is a plasma membrane-localized, inwardly conducting Ca2+-permeable channel (Ali et al., 2007) activated by cGMP (Qi et al., 2010). Genevestigator analysis of the expression patterns of the 20 Arabidopsis CNGCs indicates that CNGC2 is by far the most massively expressed isoform in all tissues (Finka et al., 2012), and CNGC2 has been associated with a number of different plant phenotypes (Chan et al., 2003; Ali et al., 2007; Finka et al., 2012). This prior work supported the rationale of investigating CNGC2 as possibly involved in BR signaling. Results shown in Figure 3 indicate that, in contrast to the lack of an effect of the BAK1 null mutation (Fig. 2), BR-dependent cytosolic Ca2+ elevation is impaired in plants that lack a functional CNGC2 gene.
Figure 3.
Exogenous BR-dependent elevation of cytosolic Ca2+ in detached leaves from wild-type Col (WT) and dnd1 plants.
BR Signaling Involves the Generation of the Secondary Messenger Molecule cGMP
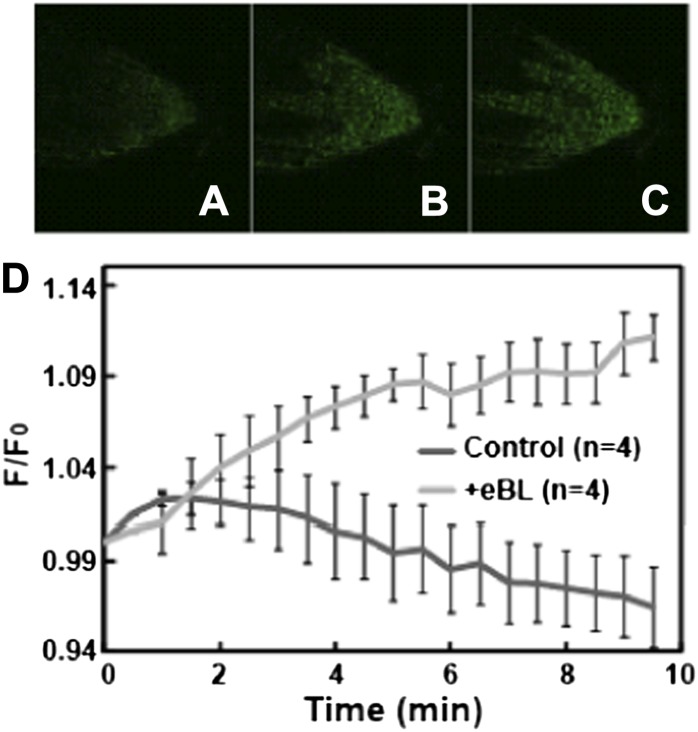
Changes in the level of the cytosolic secondary messenger molecule cGMP during myriad signaling cascades are monitored in animal cells using a non-fluorescence resonance energy transfer-based ratiometric protein, Fluorescence indicator of cyclic GMP (FlincG). Nausch et al. (2008) developed a coding sequence for such a FlincG reporter by fusing the circularly permutated enhanced GFP sequence in tandem with a portion (including the cGMP-binding domain) of the coding sequence for animal type I cGMP-activated protein kinase. Isner and Maathuis (2011) subcloned the FlincG coding sequence into a plant expression plasmid (along with the 35S cauliflower mosaic virus promoter), generated FlincG-expressing Arabidopsis plants, and demonstrated that real-time increases in cytosolic cGMP concentration could be monitored in roots of these plants upon the addition of signaling molecules known to elevate the cyclic nucleotide secondary messenger. Here, we demonstrate that application of BR to seedlings leads to elevation of cGMP at the root tip (in the columella root cap and meristematic zone) that is not evidenced when water alone is added to roots (Fig. 4). This BR-dependent elevation in cytosolic cGMP could be responsible for the CNGC2-mediated, BR-induced Ca2+ signal (Fig. 3).
Figure 4.
Effects of exogenous BR on cell cytosolic cGMP in Arabidopsis seedlings expressing the fluorescent reporter protein FlincG. After application of BR (or water as a control) to one end of the seedling chamber mounted on the stage of a confocal microscope, fluorescence images of the root tip were taken over time. A to C, Images of the root tip at 0, 5, and 10 min after application of BR. Corresponding images of control-treated roots were dark to the eye at all time points. D, Quantitative analysis of change in fluorescence over time is shown for BR- and control (water with 0.001% [v/v] DMSO)-treated roots. Signals are shown as means (biological replication numbers, recorded from different seedlings, are in parentheses) ± se. For each replicate recording, a series of fluorescence ratios at specific times after treatment were generated. The fluorescence ratios were signals recorded at a specific times relative to the signal at time 0 (F/F0; for comparison, see Isner and Maathuis [2011]). The difference in fluorescence change between seedlings treated with BR and water was not due to differences in the initial fluorescence (F0) of the seedlings, which was 374 ± 35 and 397 ± 59 for water- and BR-treated seedlings, respectively.
Results consistent with this model of BR signaling are shown in Figure 5. In this experiment, BR-dependent Ca2+ elevation was monitored in plants exposed to the GC inhibitor 6-anilino-5,8-quinolinedione (LY83583). LY83583 has been used to block cGMP-mediated signaling in a number of plant systems (Salmi et al., 2007; Cousson, 2010; de Montaigu et al., 2010; Teng et al., 2010). BR-induced cytosolic Ca2+ elevation in wild-type plants was blocked in plants treated with the GC inhibitor (Fig. 5). The utility of the GC inhibitor for probing the relationship between BR and Ca2+ signaling is supported by the results of a separate, control experiment. Glu receptors are a second family, besides CNGCs, of Ca2+-conducting channels in plants. Glu receptors conduct Ca2+ in response to this amino acid ligand (Kudla et al., 2010); they are not activated by cGMP. Glu application to wild-type leaves causes an immediate increase in cytosolic Ca2+. Exposure of leaves to the GC inhibitor had no effect on this Glu-dependent Ca2+ signal; the Ca2+ elevation was as great or greater in the presence of the inhibitor as observed when Glu was added to leaves without the inhibitor (Ma et al., 2012). One possible explanation for why the GC inhibitor affected BR-dependent Ca2+ elevation and not Glu-dependent Ca2+ elevation is that BR-dependent generation of cGMP (Fig. 4) is impaired by the inhibitor. Results shown in Supplemental Figure S2 support this contention. Recordings of cGMP-dependent fluorescence were made from wild-type (FlincG-expressing) seedlings exposed to BR after pretreatment with the GC inhibitor or solvent alone (control). Exposure of seedlings to the inhibitor impaired BR-dependent cGMP generation. These results support the conclusion that the well-documented action of LY83583 as a GC inhibitor blocks BR-induced Ca2+ elevation (Figs. 1 and 5) due to the inhibition of BR-induced cGMP rise (Fig. 4; Supplemental Fig. S2), thus impairing the activation of CNGCs (Fig. 3) and not other Ca2+-conducting channels.
Figure 5.
Exogenous BR-dependent elevation of cytosolic Ca2+ in detached leaves from wild-type Col leaves in the absence (control) and presence of a GC inhibitor.
Ca2+ Signaling and BR-Dependent Gene Expression
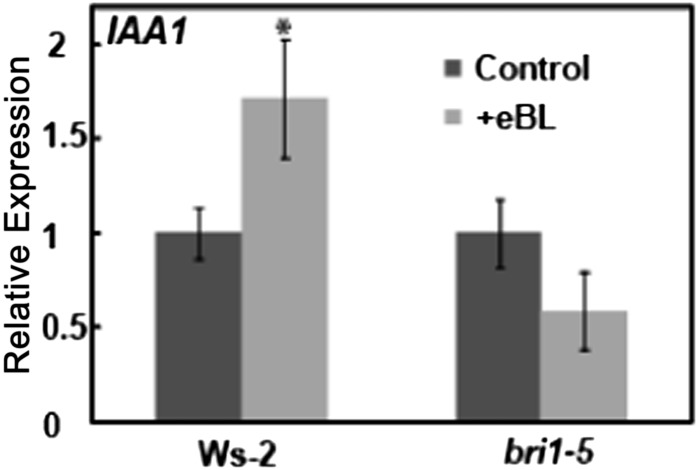
Studies were undertaken to test the hypothesis that BR-dependent Ca2+ elevation impacts gene expression induced by the hormone. In a manner similar to that already demonstrated for orthologous genes in rice (Oryza sativa; Song et al., 2009) and wheat (Singla et al., 2006), we found that BR application to Arabidopsis seedlings increased the expression of IAA1 in Ws-2 wild-type plants (Fig. 6). In contrast to the effects on wild-type plants, application of BR to bri1-5 plants had no significant effect on IAA1 expression.
Figure 6.
Effects of BR application to wild-type Ws-2 and bri1-5 plants on IAA1 expression. Results are presented as means (for a given treatment and genotype) ± se. For each genotype, IAA1 expression in the absence of added BR was normalized to 1. For each genotype, ANOVA analysis was undertaken to evaluate means separation between treatments (presence and absence of exogenous BR application). The asterisk above the bar representing the “+BR” treatment indicates a significant difference (at P < 0.05). A similar data presentation and ANOVA analysis of means separation was used for the experiments shown in Figures 7, 9, and 10 and Supplemental Figures S1 and S4. For this experiment, the expression of IAA1 in the absence of exogenous BR in leaves of bri1-5 plants was 0.39 relative to the level in wild-type plants.
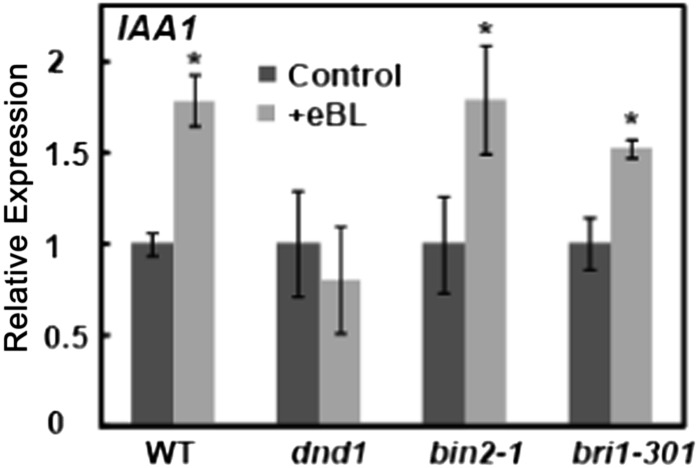
The defective BRI1 receptor in the bri1-5 genotype presumably impairs the signaling cascade linking BR perception to downstream gene (IAA1) expression (Fig. 6). Results presented in Figure 7 suggest that the Ca2+-conducting channel CNGC2 may be involved in this BR signaling cascade. As was the case with the bri1-5 mutant, the BR-dependent IAA1 expression observed in wild-type Col plants is blocked in the dnd1 mutant. This result provides genetic evidence consistent with the possibility that, at least in the case of IAA1, BR-dependent cytosolic Ca2+ elevation (which is impaired in the dnd1 mutant; Fig. 3) can mediate (or at least is associated with) effects of the hormone on gene expression.
Figure 7.
Effects of BR application to wild-type Col (WT), dnd1, bin2-1, and bri1-301 plants on IAA1 expression. The expression of IAA1 in the absence of exogenous BR relative to the level in wild-type plants was 0.73, 0.65, and 0.86 in dnd1, bin2-1, and bri1-301 plants, respectively.
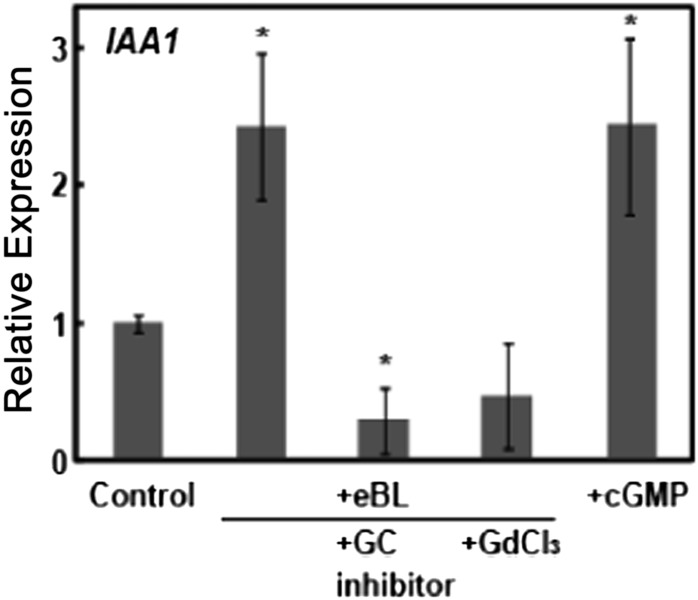
The work presented in this report underlies the development of a novel model of a cellular signaling cascade initiated by perception of the hormone BR. BR application to plants leads to immediate (initiating at less than 1 min) increases in the secondary cytosolic signaling molecules cGMP and Ca2+. Generation of the BR- and BRI1-dependent Ca2+ signal is impaired in the dnd1 mutant. The function of CNGC2 as a component of a Ca2+-conducting channel is required for optimal BR signaling that leads to increased expression of IAA1 (Fig. 7), a gene known to be responsive to BR (Singla et al., 2006; Song et al., 2009). Further experiments examining IAA1 expression are consistent with this model (Fig. 8). Prior studies have shown that elevation of cytosolic cGMP (by application of a lipophilic analog of cGMP to leaves) activates plasma membrane channels composed of CNGC2 subunits, resulting in the elevation of cytosolic Ca2+ (Ali et al., 2007; Qi et al., 2010). Here, we find that addition of a lipophilic analog of cGMP (dibromo-cGMP) to wild-type Col seedlings increased the expression of IAA1 in the absence of added BR (Fig. 8). In contrast to the effect of cGMP addition, application of either a Ca2+ channel blocker or a GC inhibitor completely blocked BR-dependent IAA1 expression (Fig. 8). Consistent with the results shown in Figure 7 using a genetic approach to affect Ca2+-dependent BR signaling (i.e. by monitoring IAA1 expression in the dnd1 mutant), results presented in Figure 8 indicate that the signal transduction cascade linking BR perception by its receptor to downstream gene expression (at least in the case of IAA1) requires the BR-dependent changes in the level of the secondary messenger molecules cGMP and Ca2+. Furthermore, elevation of the secondary messenger molecule cGMP alone mimics the effects of the hormone on IAA1 gene expression.
Figure 8.
Manipulation of the cytosolic secondary messenger molecules Ca2+ and cGMP impacts the effect of exogenous BR on IAA1 expression in wild-type Col plants. The GC inhibitor (LY83583) is added in 0.04% (v/v) DMSO solvent. Therefore, in this experiment, all treatments (including the control) had 0.04% DMSO. Further experiments indicated that in the absence of any DMSO solvent, application of dibromo-cGMP still increased IAA1 expression as compared with the control. Means separation was analyzed by Student's t test comparisons of each treatment individually with the control. Asterisks above the bars representing a treatment indicate significant (at P < 0.05) differences from the control.
We undertook further experiments to compare the relative involvement of Ca2+ signaling and the BR-induced phosphorelay cascade in the effect of the hormone on IAA1 transcript levels. The rationale underlying this work was as follows. Binding of BR to the BRI1 receptor (i.e. initial perception of the hormone) is required to activate both the phosphorelay signal cascade (Jaillais et al., 2011) and the BR-dependent Ca2+ signaling leading to gene expression (Figs. 6–8). We sought to use genotypes that are impaired in steps of the phosphorelay cascade other than mutants that have reduced binding of the hormone to the receptor (e.g. bri1-5). For this work, we used the bin2-1 and bri1-301 mutants. The bin2-1 mutant has a dominant, hypermorphic form of the negative regulator BIN2 (Li et al., 2001; He et al., 2002; Li and Nam, 2002). It has been experimentally verified that BIN2-dependent phosphorylation of BES1 and BZR1 is increased in this genotype, resulting in increased degradation of these master transcription regulators and concomitant impairment of BR signaling through the phosphorelay cascade (He et al., 2002; Li and Nam, 2002; Yin et al., 2002). Mutations that result in a “kinase-dead” BRI1 impair BR signaling, but binding of the hormone to the receptor is not necessarily impacted by such mutations (Wang et al., 2005). The bri1-301 genotype is such a mutant (Xu et al., 2008; Kang et al., 2010). In contrast to the dnd1 mutant, which showed no BR-dependent increase in IAA1 expression, the effect of BR addition on IAA1 expression in these two mutant genotypes is similar to the wild type (Fig. 7).
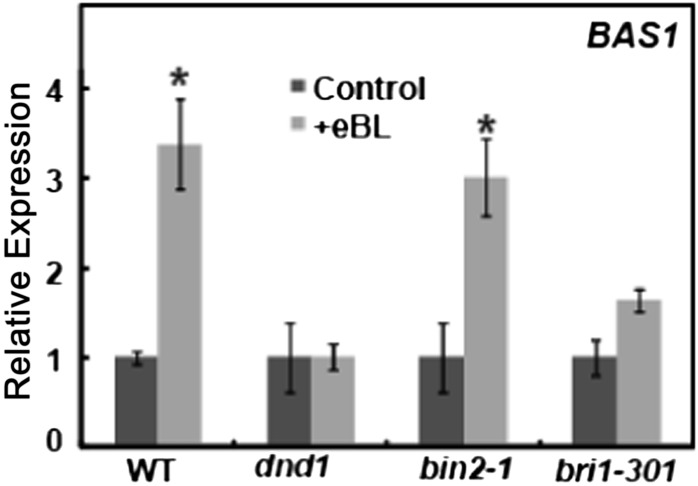
Additional studies were undertaken to further examine the novel finding that impairment of the phosphorelay cascade had little to no effect on BR-dependent gene expression, at least in the case of IAA1. BAS1 encodes a protein involved in BR inactivation/catabolism (Turk et al., 2003, 2005; Thornton et al., 2011), and its expression is increased upon BR perception (Goda et al., 2002) as part of a feedback system that logically regulates the action of this hormone on cells (Tanaka et al., 2005; Turk et al., 2005). We find similar patterns of BR-dependent increases in BAS1 expression among the four genotypes tested as was determined for IAA1 expression. As shown in Figure 9, the BR-dependent increase in BAS1 transcript level found in wild-type plants was absent in dnd1, while exogenous BR application increased BAS1 expression in the bin2-1 and bri1-301 genotypes.
Figure 9.
Effects of BR application to wild-type Col (WT), dnd1, bin2-1, and bri1-301 plants on BAS1 expression. BR application significantly (P < 0.05) increased BAS1 expression in wild-type and bin2-1 plants. BR had no effect on BAS1 level in dnd1 plants. The 60% increase in BAS1 expression observed in bri-301 plants trended toward significance (P < 0.06). The expression of BAS1 in the absence of exogenous BR relative to the level in wild-type plants was 0.72, 0.86, and 1.0 in dnd1, bin2-1, and bri1-301 plants, respectively.
The central tenet of the work presented in our studies is the novel finding that BR perception causes an immediate elevation of cytosolic Ca2+ and that the generation of this Ca2+ signal contributes to at least some component of this hormone’s effects on plant cells. Further experimental evidence supporting this contention is presented in Supplemental Figures S3 and S4. Either chelation of extracellular Ca2+ by prior exposure of Arabidopsis seedlings to EGTA (Aslam et al., 2008) or prevention of cytosolic Ca2+ elevation by application of the lipophilic Ca2+ chelator 1,2-bis-(o-aminophenoxy)-ethane-N,N,N′,N′-tetraacetic acid tetrakis-acetoxymethyl ester (BAPTA-AM; Cousson, 2003; Cruz et al., 2012) prevented the BR-dependent increase in the expression of both IAA (Supplemental Fig. S3A) and BAS1 (Supplemental Fig. S3B).
Additional evidence consistent with the model developed here, that BR perception by the BRI1 receptor is the mechanistic basis for the generation of the Ca2+ signal as well as the downstream effects of the Ca2+ signal on IAA1 and BAS1 expression, is presented in Supplemental Figure S4. Use of the bri1-5 mutant allowed us to develop aequorin-expressing plants with a defective BRI1 receptor (Fig. 1). However, the bri1-5 allele is not a fully null (loss-of-function) mutation. The bri1-701 allele is a bona fide hormone receptor null mutation (Gou et al., 2012). We used the bri1-701 mutant to further evaluate if the BR-evoked signaling that is the focus of our work is dependent on the hormone receptor BRI1. Results shown in Supplemental Figure S4, A and B, demonstrate that BR-dependent increases in the expression of IAA1 and BAS1 are blocked in the br1-701 mutant, as was the case when we tested BR-dependent IAA1 expression in the bri1-5 genotype (Fig. 6).
Ca2+ Signaling and the Phosphorelay Cascade Represent Two Distinct BR Response Pathways
BR perception by its receptor BRI1 is linked to downstream alteration in the expression of at least some BR-responsive genes through a Ca2+ signaling cascade. Results presented in Figures 7 and 9 suggest that BR-responsive genes regulated by Ca2+ signaling might not require activation of the master transcriptional regulators BZR1 and/or BES1 through the BR-dependent phosphorelay cascade. BZR1 and BES1 alter the transcription of BR-responsive genes either directly, by binding to the promoter of specific genes, or indirectly, by modulating the expression of TFs that then alter transcript levels of suites of other genes (Wang et al., 2002; He et al., 2005; Yin et al., 2005; Li et al., 2009).
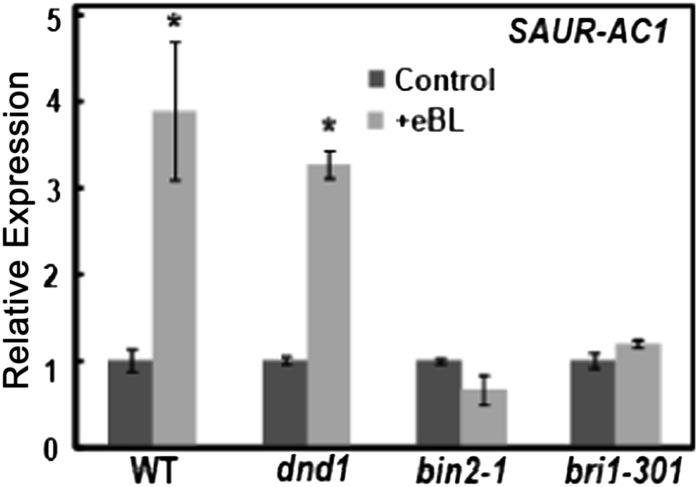
SMALL AUXIN UP RNA1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA (SAUR-AC1) is a BR-responsive gene that has been experimentally confirmed to be activated by direct binding of BES1 to its promoter (Yin et al., 2005). Thus, it is hard to envision that in genotypes that have alterations in steps of the phosphorelay cascade resulting in degradation/inactivation of BES1, BR effects on SAUR-AC1 expression would not be affected. As shown in Figure 10, differences in BR effects on SAUR-AC1 expression among the four genotypes examined were markedly different from those found for either BAS1 (Fig. 9) or IAA1 (Fig. 7). In the case of SAUR-AC1, BR increased the expression level in wild-type and dnd1 plants, while no effect of BR on SAUR-AC1 expression was observed with either the bin2-1 or the bri1-301 genotype. These results suggest that some BR-responsive genes might be regulated by the phosphorelay system and others might be modulated (perhaps independently) by a Ca2+-dependent signaling cascade.
Figure 10.
Effects of BR application to wild-type Col (WT), dnd1, bin2-1, and bri1-301 plants on SAUR-AC1 expression. The expression of SAUR-AC1 in the absence of exogenous BR relative to the level in wild-type plants was 0.91, 0.86, and 0.70 in dnd1, bin2-1, and bri1-301 plants, respectively.
Ca2+ Signaling and BR Phenotypes
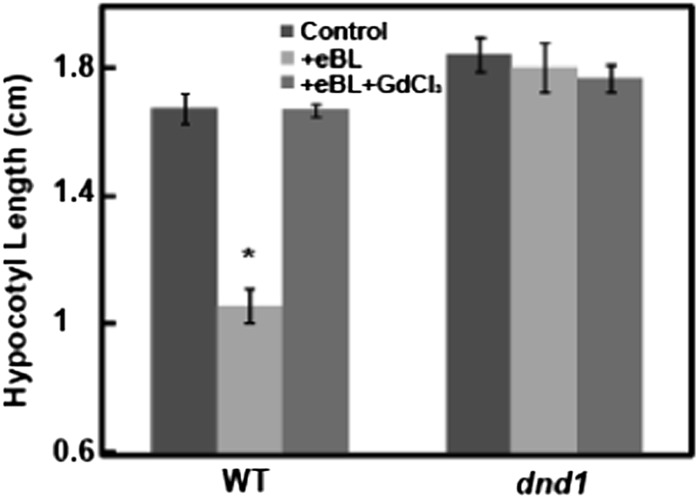
The end point of hormone signaling cascades that involve transcriptional reprogramming is often an altered plant phenotype. This typically involves changes in the growth, form, function, or development of a plant, plant organ, or target cells in response to internal, developmental, or environmental cues. In the work presented here, we have identified a cytosolic Ca2+ elevation as an immediate downstream signal generated upon BR perception by its receptor BRI1 and linked this signal to the transcriptional regulation of several genes by this hormone. As mentioned above, BR signaling results in the potential reprogramming of many genes (Goda et al., 2002; Müssig et al., 2002). The demonstration here of BR-dependent Ca2+ signaling as impacting transcript levels of a few genes does not necessarily link this signaling cascade to BR-dependent phenotypes of plants. Results presented in Figure 11 extend our analysis of BR Ca2+ signaling to plant phenotypes. One documented quantitative measure of the exogenous BR alteration of a plant phenotype is the effect of BR on hypocotyl length of etiolated Arabidopsis seedlings (Xu et al., 2008; Gou et al., 2012). Application of exogenous BR reduces the hypocotyl length of seedlings grown in the dark. This effect is shown with wild-type seedlings in Figure 11. With wild-type seedlings, application of a Ca2+ channel blocker along with BR completely prevented the BR-dependent phenotype. Additional studies were undertaken with dnd1 seedlings. In this case, BR had no effect; hypocotyl length was similar in the presence or absence of BR. Additionally, application of the Ca2+ channel blocker to dnd1 seedlings had no effect. This result suggests that the effect of the Ca2+ channel blocker in reversing the BR-dependent decrease in hypocotyl length is likely not due to some general, inhibitory effect of the channel blocker. Thus, the results of the experiment shown in Figure 11 provide pharmacological and genetic evidence that Ca2+ signaling mediates this BR-related phenotype.
Figure 11.
Ca2+ signaling is involved in the effect of BR on etiolated hypocotyl length of dark-grown Arabidopsis seedlings. Wild-type Col (WT) and dnd1 seedlings were grown on solid medium with water (control), BR, or BR and GdCl3 added. Means separation was analyzed by Student's t test comparisons within each genotype of each treatment individually with the control. The asterisk above the bar representing eBL treatment indicates a significant (P < 0.05) difference from the control (water addition) for that treatment.
DISCUSSION
The work included in this report supports a revised model of BR signaling. The central tenet of this model is that BR perception by its receptor BRI1 leads to an immediate (i.e. less than 1 min) increase in cytosolic Ca2+. At the BR concentration used in our studies of its effect on Ca2+ elevation (100 nm), the BR-dependent Ca2+ signal is of a similar magnitude as that occurring in other cell signaling pathways, such as the plant immune responses evoked by Plant elicitor peptides (Qi et al., 2010) or the ELONGATION FACTOR TU18 peptide (Ranf et al., 2011). In our studies, this BR-dependent Ca2+ signal was demonstrated to be causal to the changed expression of some BR-dependent genes as well as a BR-related phenotype.
There are a number of other assertions that can be formulated based on the experimental results in this report. Exogenous BR application causes in vivo cGMP elevation. This step of the newly delineated BR signaling cascade might occur due to GC activity of the hormone receptor; this point remains speculative and would need to be confirmed by further experiments showing that the putative GC domain of BRI1 is specifically necessary for cGMP generation in planta in response to BR application. We did find that a GC inhibitor blocked BR-dependent Ca2+ elevation, cGMP production, and gene expression and that exogenous application of lipophilic cGMP was found to mimic the effect of the BR hormone on IAA expression; these results are consistent with the involvement of cGMP production in the signaling pathway. Support for the further speculation that BRI1 is responsible for this cGMP production directly could involve the demonstration that the GC catalytic domain of BRI1 (as opposed to the kinase activity of the receptor) is specifically required for initiation of the Ca2+ signaling cascade. Experiments along these lines are not included in this report. However, a recent report (Irving et al., 2012) provided separate evidence linking BR to cGMP generation in vivo using a different experimental approach (ELISA analysis of exogenous BR effects on cGMP in isolated protoplasts) for cGMP analysis than we used (fluorescence of FlincG seedlings). Nonetheless, the putative GC activity of BRI1 has yet to be definitively tested in planta.
The results in this report indicate that BR-dependent Ca2+ signaling and the well-characterized phosphorelay cascade might act independently to activate different BR-responsive genes. However, full elucidation of this point awaits the analysis of more BR-responsive genes.
As mentioned above, there is no BR phosphorelay signaling step downstream from the BRI1 receptor that is currently known to be affected by Ca2+. However, a recent report has demonstrated a physical and functional interaction between BRI1 and the Ca2+-binding protein CaM. Oh et al. (2012) have shown that Arabidopsis CaM7 displays Ca2+-dependent binding to a peptide corresponding to the cytosolic domain of BRI1. The exciting work of Oh et al. (2012) also demonstrated that coexpression of CaM with the cytosolic kinase domain of BRI1 in a heterologous system (E. coli) resulted in an impairment of BRI1 autophosphorylation and transphosphorylation (in this case, of E. coli proteins). This work raises the intriguing possibility that elevation of cytosolic Ca2+ might act, through Ca2+-dependent binding of CaM to BRI1, to shut down the BRI1-dependent phosphorylations that initiate the phosphorelay cascade. Perhaps, then, the BRI1 receptor and activation of a CNGC are the source of the Ca2+ signal that shuts down BRI-dependent phosphorelay signaling. It may also be germane to this evolving model of BR signaling that conductance by CNGCs is down-regulated by CaM (Hua et al., 2003). Thus, we envision a feedback loop that allows for the modulation of both BR signaling pathways.
In prior work from this laboratory characterizing the role of CNGCs in signal transduction pathways (Qi et al., 2010), we speculated that the Ca2+-conducting channel was present along with a receptor protein that generated the ligand that activates the channel as a multienzyme protein complex. In this manner, microdomains may exist where the localized concentration of a cytosolic messenger molecule (such as cyclic nucleotide) is elevated in proximity to the channel that is activated by the molecule. When the recent work of Oh et al. (2012) is considered along with the results presented here, the notion of a multienzyme microdomain could provide an explanation for how BRI1, the Ca2+-conducting CNGC, and CaM act in concert to initiate a finely regulated signal. It is germane to note that in animal cells, CNGCs as well as enzymes that activate the channels and receptors are envisioned to operate as multimolecular complexes existing as membrane “signalosomes” (Trudeau and Zagotta, 2003; Wang and Malbon, 2011; Bankston et al., 2012; Ostrom et al., 2012).
BR-dependent cytosolic Ca2+ elevation could be involved in the immediate responses of cells to BR perception, as mentioned in the introduction. In other words, aside from the well-known Ca2+ signal regulation of gene expression (Finkler et al., 2007; Doherty et al., 2009), a rise in cytosolic Ca2+ could also alter the conductance of K+ and Cl− channels, altering membrane potential (Li et al., 2006). Furthermore, an elevation in cytosolic Ca2+ could have immediate impact on cell function through the activation of Ca2+-dependent protein kinases (Cheng et al., 2002). Of course, these speculations await testing in future studies. Here, the novel effect of BR on cytosolic Ca2+ is documented, raising new questions about cell signaling pathways associated with this hormone.
MATERIALS AND METHODS
Plant Material
All Arabidopsis (Arabidopsis thaliana) lines used in the reported work are in the Col background except bri1-5 (AT4G29400; Noguchi et al., 1999); the bri1-5 mutation is in the Ws-2 background. Col or Ws-2 plants were used as controls as appropriate. The aequorin-expressing lines Col-aeq, Ws-2-aeq, dnd1-aeq (Qi et al., 2010) bri1-5-aeq, and bak1-aeq were used to monitor treatment effects on cytosolic Ca2+ concentration. The bak1-3-aeq line was generated by crossing the bak1-3 mutant with Col-aeq. We chose the bak1-3 allele to generate the aequorin-expressing line because early publications indicated that bak1-3 lacks expression of BAK1 mRNA and this genotype is a BAK1 null mutant (Chinchilla et al., 2007). However, a later publication pointed out that the bak1-3 genotype is a “leaky” null mutant (Gou et al., 2012). Therefore, we used the bak1-4 allele (another transfer DNA insertional mutant) instead of bak1-3 for later experiments. The bak1-4 mutant plants were used for single-mutation gene expression studies (Supplemental Fig. S1). Col plants expressing the cGMP reporter protein δ-FlincG (Isner and Maathuis, 2011) were used for in vivo cGMP measurement. Arabidopsis seeds were surface sterilized by first washing the seeds in 70% (v/v) ethanol, 20% (v/v) bleach, and 0.02% (v/v) Triton X-100, shaking at 300 rpm for 10 min, and then rinsing with 95% (v/v) ethanol three to four times.
For all measurements except the quantitative real-time PCR (qPCR) assay of gene expression, seeds were first planted on one-half-strength Murashige and Skoog (MS) medium (Caisson Labs) supplemented with 1% (w/v) Suc and solidified with 1% (w/v) agar. Seeds were stratified (4°C) for 2 to 3 d after plating to break dormancy. Plants used for cytosolic Ca2+ measurements were grown as follows. Seeds were grown on agar plates for 7 to 10 d at 16 h of light (approximately 100 mol m–2 s–1)/8 h of dark and 25°C and then transplanted into pots containing artificial LP5 mix (Sun Gro). The pots were put in an EGC growth chamber at 12 h of light (approximately 100 mol m–2 s–1)/12 h of dark and 22°C. Healthy, nonsenescing leaves were used after 2 to 3 weeks of growth in pots. For in vivo cGMP measurement, the seeds were set vertically on square plates containing one-half-strength MS medium for 5 d. For measurement of etiolated hypocotyl lengths, square plates containing seeds were placed in 24 h of dark for 14 d at 22°C. For qPCR measurement of gene expression, seeds were grown in tubes containing 3 mL of liquid one-half-strength MS medium (one seed per tube) with shaking (180 rpm) for 14 d at an illumination of 24 h of light (approximately 90 mol m–2 s–1) and 22°C.
Hormone and Reagent Treatment
Over 60 different BR compounds have been identified in plants. Brassinolide and eBL are the most biologically active of these compounds and are typically used in tissue culture applications to study BR effects on plants (Ferrie et al., 2005). In our work, synthetic eBL (Sigma-Aldrich) was applied to plants, seedlings, and detached leaves to test the effects of exogenous BR on plant tissue. Stock solutions of eBL were made in 100% dimethyl sulfoxide (DMSO; Sigma-Aldrich). For all experiments except qPCR studies, the eBL was added at a final concentration of 100 nm. For gene expression analysis, the final concentration of the eBL used was 1 μm (Li et al., 2009).
In some cases, a lipophilic analog of cGMP, 8-bromoguanosine 3′,5′-cyclic monophosphate sodium salt (Sigma-Aldrich), was added to plants (at 150 µm final concentration) in the same manner as the eBL treatment. For these experiments, water (i.e. no solvent) was added to the control treatment. Some experiments involved use of the GC inhibitor LY83583 (Enzolife Sciences) and the cytosolic Ca2+ chelator BAPTA-AM (Sigma-Aldrich). The inhibitors were added to solutions containing leaves or seedlings (20 µm final concentration) 15 min before plants were treated with eBL. For these experiments, control treatments included application of 0.04% (v/v) DMSO in place of the inhibitor. Stock solutions of 50 mm LY83583 and 25 mm BAPTA-AM were made up in 100% DMSO. In some cases, the Ca2+ blocker GdCl3 (at 150 µm final concentration; Sigma-Aldrich) and the extracellular Ca2+ chelator EGTA (Sigma-Aldrich) were used; the channel blocker was added to the plants 15 min before the eBR ligand was applied. Water was used as a control for this work. All experiments were repeated at least twice.
Cytosolic Ca2+ Measurements
The method from Qi et al. (2010) was used with slight modification for cytosolic Ca2+ measurements using aequorin-expressing plants. For details of the methods used to generate the aequorin-expressing genotypes used here (Col-aeq, Ws-2-aeq, dnd1-aeq, and bri1-5-aeq), see Qi et al. (2010). Leaves used for the experiments were cut from 3- to 4-week-old plants expressing cytosol-localized Ca2+-dependent chemiluminescent apoaequorin protein reconstituted with coelenterazine-cp (CTZ-cp; AAT Bioquest), a synthetic derivative of coelenterazine.
Individual detached leaves were placed in a capless 2-mL centrifuge tube containing 500 μL of control buffer (1 mm KCl, 1 mm CaCl2·6H2O, and 10 mm MgCl2·6H2O, adjusted to pH 5.7 with Tris base). For each tube, 1 μL of CTZ-cp was added (10 μm final concentration in 0.2% [v/v] ethanol). The leaf in the control buffer was vacuum infiltrated for 15 s and incubated at room temperature in the dark for 1 to 2 h to allow coelenterazine incorporation into leaves. As the CTZ-cp is a light-sensitive reagent, all preparatory steps after adding the CTZ-cp were carried out in the dark; tubes were covered with foil paper.
A luminometer (TD-20/20; Tuner Design) was used for measurement of the luminescence level. The centrifuge tubes were placed in the luminometer and left for 2 to 3 min to allow leaves to recover from touch-induced Ca2+ spikes induced by handling the tubes. Luminescence level was measured every 1 s, and ligand (eBL) was added only after background luminescence of the leaves was stable. When background luminescence was stable, 500 μL of control buffer containing ligand (at 2× the final concentration) was added to the tubes containing leaves by gently pipetting the solution against the interior wall of the centrifuge tube. For these experiments, eBL (100 nm final concentration) was delivered to the leaves in DMSO solvent (0.001% [v/v] final concentration). The GC inhibitor LY83583 in DMSO (0.04% [v/v] final concentration) was added to the leaves 15 min before adding eBL. Control leaves were treated with 0.1% DMSO 10 min before adding eBL. All the treatments were added at 0 min; results shown in the figures are mean values calculated from a minimum of at least three biological replicates. For each replication, leaves were cut from different plants. After recording luminescence from a treatment replicate, the remaining aequorin (i.e. not bound to Ca2+) in an assay tube was discharged by adding 800 μL of Ca2+-release buffer (2 m CaCl2·6H2O in 30% ethanol) with continued recording for approximately 10 min, until the instantaneous luminescence level dropped below 2,000. Values obtained for aequorin discharge were used to convert luminescence readings to cytosolic Ca2+ concentration for each treatment replicate using an algorithm as described by Qi et al. (2010). Results are presented in the figures as BR-dependent increase in cytosolic Ca2+; this value represents the difference between the Ca2+ level recorded at any time point and the basal level of Ca2+ measured at time 0 (i.e. prior to BR application). The basal level of cytosolic Ca2+ for the experiments reported here ranged from 0.20 to 0.24 µm and averaged 0.220 ± 0.004 µm.
In Vivo cGMP Measurements
Individual 7-d-old seedlings (Col plants expressing the cGMP reporter FlincG) were placed in 60 μL of water on a 24- × 40-mm cover glass and covered with a 22- × 22-mm coverslip. The small coverslip was taped (on two opposite sides) onto the larger one (forming a chamber), and then the chamber with a seedling inside was taped onto the stage of a confocal microscope (Nikon A1R). Measurements of fluorescence were made prior to the addition of ligand; seedlings with roots having high levels of GFP expression (i.e. GFP fluorescence with just water in the chamber) were chosen for use. The root tip was located under bright-field illumination, observation of background GFP fluorescence was made, and after adjustment of the perfect focus, 150 μL of water containing eBL in 0.001% (v/v) DMSO was delivered to one open end of the chamber at 0 min. The control “water-only” treatment contained just DMSO. A filter paper was kept in contact with the chamber solution at the opposite side of the chamber to wick water as the ligand was delivered. Time-lapse changes of fluorescence were captured every 30 s using 480/20-nm excitation wavelengths using the microscope’s high-performance optical offset (Perfect Focus System) to facilitate real-time correction of focus drift. Fluorescence intensity at the root tip was quantified as relative brightness (within a defined range of 256 shades of gray per unit area) using NIH ImageJ processing/analysis software.
qPCR Analysis
qPCR analyses were performed to study genotype and treatment effects on the expression levels of some BR-responsive genes. The growth of Arabidopsis seedlings in one-half-strength MS liquid medium was determined according to Qi et al. (2010). The seedlings were grown in separate tubes containing 3 mL of liquid medium for 14 d on a shaker (180 rpm) with 24 h of illumination (approximately 90 mol m–2 s–1) at 22°C. The ligand eBL (or lipophilic cGMP) was added directly to the growth medium, and the seedlings were incubated for a further 8 h. Water containing 0.001% (v/v) DMSO was given to the seedling for the control treatment. For experiments involving the addition of inhibitors, LY83583 (20 μm), GdCl3 (150 μm), EGTA (20 mm), and BAPTA-AM (10 μm) were added to the growth medium containing plants 15 min prior to the addition of eBL. Seedlings were kept on the shaker in the light during inhibitor pretreatment and exposure to eBL. After incubation, the seedlings were collected and frozen immediately with liquid N2 and stored at −80°C for future use. Total RNA were extracted from whole liquid-grown seedlings using the Plant RNA Extraction Kit (Macherey-Nagel). During the RNA extraction process, tissue extracted was treated with rDNase (Macherey-Nagel) to remove potential genomic DNA from samples.
After extraction, 500 ng of total RNA was used for reverse transcription by using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The synthesized complementary DNA was diluted 1:10 (v/v) in water, and 1 µL of the diluted complementary DNA was used for each qPCR. qPCR assays were performed using the ABI 7900 HT Real-Time PCR system with the SYBR Green gene expression assay (Applied Biosystems). Treatment effects on the expression levels of IAA1 (AT4G14560), BAS1 (AT2G26710), and SAUR-AC1 (AT4G38850) were examined. 18s rRNA was used as an endogenous control. The primers used for these analyses were as follows: IAA1 (forward), 5′-AGTCACCAATGGGCTTAACC-3′; IAA1 (reverse), 5′-CTGTTGAGTCGTTGTTCTTGC-3′; BAS1 (forward), 5′-CCCGTTGGCTTCATACCG T-3′; BAS1 (reverse), 5′-TTACAGCGAGTGTCAATTTGGC-3′; SAUR-AC1 (forward), 5′-AGGAGTTTCTTGGGTGCTAAG-3′; SAUR-AC1 (reverse), 5′-CATAGACCGCCATGAATCCTC-3′; 18s rRNA (forward), 5′-CGGCTACCACATCCAAGGAA-3′; 18s rRNA (reverse), 5′-GCTGGAATTACCGCGGCT-3′.
For each analysis, three mechanical replications were tested on one plate, and each treatment mean was generated from analysis of at least three biological replications from separate RNAs isolated from different individual seedlings. ANOVA of corresponding threshold cycle values was used for the evaluation of means separation among treatments in an experiment (as noted in the figure legends) and to generate se values of the means for control treatments (Schmittgen and Livak, 2008). In many cases, experiments involving qPCR analysis of gene expression involved the evaluation of eBL effects on different genotypes. In these cases, gene expression in the absence of added eBL was normalized to 1 for each genotype in a given experiment (as noted in the figures). In these cases, the relative amount of gene expression in the absence of eBL for each mutant line used in an experiment (i.e. in a given figure) is compared with the expression level in wild-type plants for that experiment. This information is provided in the corresponding figure legend.
Measurement of Etiolated Hypocotyl Lengths
This assay of a BR-dependent plant phenotype allows for quantitative evaluation of exogenously added BR effects on seedlings and was adopted from Xu et al. (2008) and Müssig et al. (2002). Arabidopsis seedlings were grown vertically on one-half-strength MS medium on square plates containing 1% (w/v) agar at 22°C in the dark for 14 d. eBL (100 nm) and/or GdCl3 (150 μm) were added to the solid growth medium before pouring the medium onto the petri plates. For the control treatment, plants were grown on medium containing 0.001% (v/v) DMSO. The hypocotyl lengths of seedlings were measured after 14 d.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effects of BR application to wild-type (Col), bak1, and bak1/bkk plants on IAA1 and BAS1 expression.
Supplemental Figure S2. Effects of GC inhibitor on BR-induced cGMP elevation in wild type.
Supplemental Figure S3. Effects of Ca2+ chelator on BR-induced IAA1 and BAS1 gene expression.
Supplemental Figure S4. Effects of BR application to the wild type (Col) and the BR null mutant bri1-701 on IAA1 and BAS1 expression.
Acknowledgments
We thank Dr. Frans J.M. Maathuis (York University) for the FlincG-expressing Arabidopsis seeds, Dr. Jianmin Li (University of Michigan) for bin2-1 seeds, Dr. Yonghong Wang (Institute of Genetics, Chinese Academy of Science) for bri1-301 seeds, and Dr. S.C. Huber (University of Illinois) for bak1/bkk1 seeds.
Glossary
- BR
brassinosteroid
- TFs
transcription factors
- GC
guanylyl cyclase
- cGMP
cyclic GMP
- CNGC
cyclic nucleotide-gated channel
- CaM
calmodulin
- eBL
epibrassinolide
- Ws-2
Wassilewskija
- Col
Columbia
- LY83583
6-anilino-5,8-quinolinedione
- FlincG
Fluorescence indicator of cyclic GMP
- BAPTA-AM
1,2-bis-(o-aminophenoxy)-ethane-N,N,N′,N′-tetraacetic acid tetrakis-acetoxymethyl ester
- qPCR
quantitative real-time PCR
- MS
Murashige and Skoog
- DMSO
dimethyl sulfoxide
- CTZ-cp
coelenterazine-cp
References
- Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, Berkowitz GA. (2007) Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam SN, Newman MA, Erbs G, Morrissey KL, Chinchilla D, Boller T, Jensen TT, De Castro C, Ierano T, Molinaro A, et al. (2008) Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr Biol 18: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Bankston JR, Camp SS, DiMaio F, Lewis AS, Chetkovich DM, Zagotta WN. (2012) Structure and stoichiometry of an accessory subunit TRIP8b interaction with hyperpolarization-activated cyclic nucleotide-gated channels. Proc Natl Acad Sci USA 109: 7899–7904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CW, Schorrak LM, Smith RK, Jr, Bent AF, Sussman MR. (2003) A cyclic nucleotide-gated ion channel, CNGC2, is crucial for plant development and adaptation to calcium stress. Plant Physiol 132: 728–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J. (2002) Calcium signaling through protein kinases: the Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129: 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RK, Jr, Bent AF. (2000) The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA 97: 9323–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD. (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousson A. (2003) Two potential Ca2+-mobilizing processes depend on the abscisic acid concentration and growth temperature in the Arabidopsis stomatal guard cell. J Plant Physiol 160: 493–501 [DOI] [PubMed] [Google Scholar]
- Cousson A. (2010) Indolyl-3-butyric acid-induced Arabidopsis stomatal opening mediated by 3′,5′-cyclic guanosine-monophosphate. Plant Physiol Biochem 48: 977–986 [DOI] [PubMed] [Google Scholar]
- Cruz LF, Cobine PA, De LA, Fuente L. (2012) Calcium increases Xylella fastidiosa surface attachment, biofilm formation, and twitching motility. Appl Environ Microbiol 78: 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Montaigu A, Sanz-Luque E, Galván A, Fernández E (2010) A soluble guanylate cyclase mediates negative signaling by ammonium on expression of nitrate reductase in Chlamydomonas. Plant Cell 22: 1532–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D. (2010) The language of calcium signaling. Annu Rev Plant Biol 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Poovaiah BW. (2005) Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature 437: 741–745 [DOI] [PubMed] [Google Scholar]
- Ferrie AMR, Dirpaul J, Krishna P, Krochko J, Keller WA. (2005) Effects of brassinosteroids on microspore embryogenesis in Brassica species. In Vitro Cell Dev Biol Plant 41: 742–745 [Google Scholar]
- Finka A, Cuendet AF, Maathuis FJ, Saidi Y, Goloubinoff P. (2012) Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 24: 3333–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkler A, Ashery-Padan R, Fromm H. (2007) CAMTAs: calmodulin-binding transcription activators from plants to human. FEBS Lett 581: 3893–3898 [DOI] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J. (2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. (2011) Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harter K, Meixner AJ, Schleifenbaum F. (2012) Spectro-microscopy of living plant cells. Mol Plant 5: 14–26 [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY. (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, Russell SD, Li J. (2007) BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol 17: 1109–1115 [DOI] [PubMed] [Google Scholar]
- Hua BG, Mercier WR, Zinlieski RE, Berkowitz GA. (2003) Functional interaction of calmodulin with a plant cyclic nucleotide gated cation channel. Plant Physiol Biochem 41: 945–954 [Google Scholar]
- Irving HR, Kwezi L, Wheeler J, Gehring C. (2012) Moonlighting kinases with guanylate cyclase activity can tune regulatory signal networks. Plant Signal Behav 7: 201–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isner JC, Maathuis FJ. (2011) Measurement of cellular cGMP in plant cells and tissues using the endogenous fluorescent reporter FlincG. Plant J 65: 329–334 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Hothorn M, Belkhadir Y, Dabi T, Nimchuk ZL, Meyerowitz EM, Chory J. (2011) Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev 25: 232–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B, Wang H, Nam KH, Li J, Li J. (2010) Activation-tagged suppressors of a weak brassinosteroid receptor mutant. Mol Plant 3: 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerling B, Schwedt A, Rodriguez P, Mazzotta S, Frank M, Qamar SA, Mengiste T, Betsuyaku S, Parker JE, Müssig C, et al. (2007) The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol 17: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K. (2010) Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwezi L, Meier S, Mungur L, Ruzvidzo O, Irving H, Gehring C. (2007) The Arabidopsis thaliana brassinosteroid receptor (AtBRI1) contains a domain that functions as a guanylyl cyclase in vitro. PLoS ONE 2: e449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nam KH. (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. (2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kim BG, Cheong YH, Pandey GK, Luan S. (2006) A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc Natl Acad Sci USA 103: 12625–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y. (2009) Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J 58: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Walker RK, Zhao Y, Berkowitz GA. (2012) Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc Natl Acad Sci USA 109: 19852–19857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig C, Fischer S, Altmann T. (2002) Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Nausch LW, Ledoux J, Bonev AD, Nelson MT, Dostmann WR. (2008) Differential patterning of cGMP in vascular smooth muscle cells revealed by single GFP-linked biosensors. Proc Natl Acad Sci USA 105: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MH, Kim HS, Wu X, Clouse SD, Zielinski RE, Huber SC. (2012) Calcium/calmodulin inhibition of the Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase provides a possible link between calcium and brassinosteroid signalling. Biochem J 443: 515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom RS, Bogard AS, Gros R, Feldman RD. (2012) Choreographing the adenylyl cyclase signalosome: sorting out the partners and the steps. Naunyn Schmiedebergs Arch Pharmacol 385: 5–12 [DOI] [PubMed] [Google Scholar]
- Qi Z, Verma R, Gehring C, Yamaguchi Y, Zhao Y, Ryan CA, Berkowitz GA. (2010) Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc Natl Acad Sci USA 107: 21193–21198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D. (2011) Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J 68: 100–113 [DOI] [PubMed] [Google Scholar]
- Salmi ML, Morris KE, Roux SJ, Porterfield DM. (2007) Nitric oxide and cGMP signaling in calcium-dependent development of cell polarity in Ceratopteris richardii. Plant Physiol 144: 94–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Singla B, Chugh A, Khurana JP, Khurana P. (2006) An early auxin-responsive Aux/IAA gene from wheat (Triticum aestivum) is induced by epibrassinolide and differentially regulated by light and calcium. J Exp Bot 57: 4059–4070 [DOI] [PubMed] [Google Scholar]
- Song Y, You J, Xiong L. (2009) Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol Biol 70: 297–309 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S. (2005) Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol 138: 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Xu W, Ma M. (2010) cGMP is required for seed germination in Arabidopsis thaliana. J Plant Physiol 167: 885–889 [DOI] [PubMed] [Google Scholar]
- Thornton LE, Peng H, Neff MM. (2011) Rice CYP734A cytochrome P450s inactivate brassinosteroids in Arabidopsis. Planta 234: 1151–1162 [DOI] [PubMed] [Google Scholar]
- Trudeau MC, Zagotta WN. (2003) Calcium/calmodulin modulation of olfactory and rod cyclic nucleotide-gated ion channels. J Biol Chem 278: 18705–18708 [DOI] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Denzel MA, Torres QI, Neff MM. (2003) CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol 133: 1643–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Wang H, Torres QI, Ward JM, Murthy G, et al. (2005) BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J 42: 23–34 [DOI] [PubMed] [Google Scholar]
- Wang HY, Malbon CC. (2011) Probing the physical nature and composition of signalsomes. J Mol Signal 6: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD. (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Witthöft J, Harter K. (2011) Latest news on Arabidopsis brassinosteroid perception and signaling. Front Plant Sci 2: 58–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Huang J, Li B, Li J, Wang Y. (2008) Is kinase activity essential for biological functions of BRI1? Cell Res 18: 472–478 [DOI] [PubMed] [Google Scholar]
- Yang CJ, Zhang C, Lu YN, Jin JQ, Wang XL. (2011) The mechanisms of brassinosteroids’ action: from signal transduction to plant development. Mol Plant 4: 588–600 [DOI] [PubMed] [Google Scholar]
- Ye H, Li L, Yin Y. (2011) Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J Integr Plant Biol 53: 455–468 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]