The biosynthesis of β-citraurin from β-cryptoxanthin and zeaxanthin contribute to the reddish peel color of citrus fruits.
Abstract
In this study, the pathway of β-citraurin biosynthesis, carotenoid contents and the expression of genes related to carotenoid metabolism were investigated in two varieties of Satsuma mandarin (Citrus unshiu), Yamashitabeni-wase, which accumulates β-citraurin predominantly, and Miyagawa-wase, which does not accumulate β-citraurin. The results suggested that CitCCD4 (for Carotenoid Cleavage Dioxygenase4) was a key gene contributing to the biosynthesis of β-citraurin. In the flavedo of Yamashitabeni-wase, the expression of CitCCD4 increased rapidly from September, which was consistent with the accumulation of β-citraurin. In the flavedo of Miyagawa-wase, the expression of CitCCD4 remained at an extremely low level during the ripening process, which was consistent with the absence of β-citraurin. Functional analysis showed that the CitCCD4 enzyme exhibited substrate specificity. It cleaved β-cryptoxanthin and zeaxanthin at the 7,8 or 7′,8′ position. But other carotenoids tested in this study (lycopene, α-carotene, β-carotene, all-trans-violaxanthin, and 9-cis-violaxanthin) were not cleaved by the CitCCD4 enzyme. The cleavage of β-cryptoxanthin and zeaxanthin by CitCCD4 led to the formation of β-citraurin. Additionally, with ethylene and red light-emitting diode light treatments, the gene expression of CitCCD4 was up-regulated in the flavedo of Yamashitabeni-wase. These increases in the expression of CitCCD4 were consistent with the accumulation of β-citraurin in the two treatments. These results might provide new strategies to improve the carotenoid contents and compositions of citrus fruits.
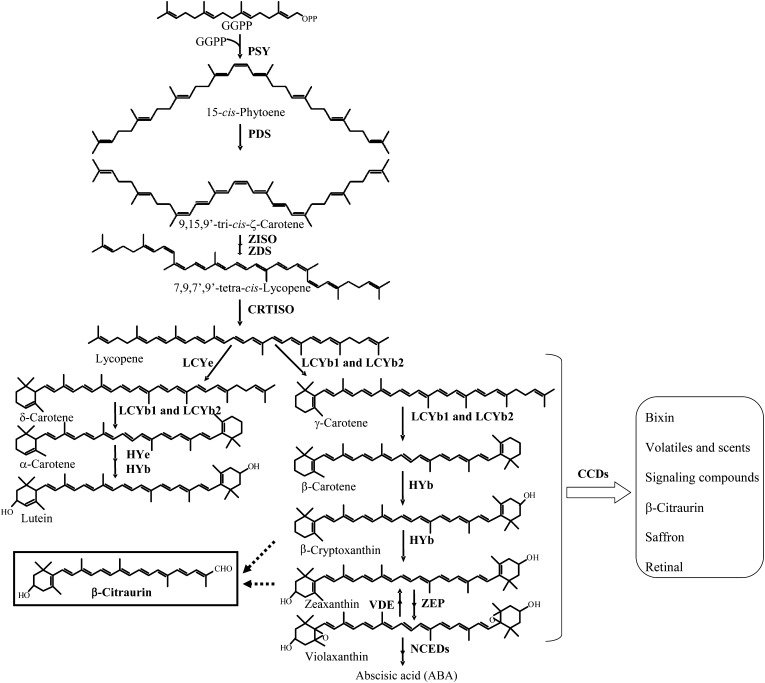
Carotenoids, a diverse group of pigments widely distributed in nature, fulfill a variety of important functions in plants and play a critical role in human nutrition and health (Schwartz et al., 1997; Cunningham and Gantt, 1998; Havaux, 1998; Krinsky et al., 2003; Ledford and Niyogi, 2005). The pathway of carotenoid biosynthesis has been well documented in various plant species, including Arabidopsis (Arabidopsis thaliana; Park et al., 2002), tomato (Lycopersicon esculentum; Isaacson et al., 2002), pepper (Capsicum annuum; Bouvier et al., 1998), citrus (Citrus spp.; Kato et al., 2004, 2006; Rodrigo et al., 2004; Rodrigo and Zacarías, 2007; Kato, 2012; Zhang et al., 2012a), and apricot (Prunus armenaica; Kita et al., 2007). Genes encoding the enzymes in the carotenoid biosynthetic pathway have been cloned, and their expression profiles have also been characterized (Fig. 1). As carotenoids contain a series of conjugated double bonds in the central chain, they can be oxidatively cleaved in a site-specific manner (Mein et al., 2011). The oxidative cleavage of carotenoids not only regulates their accumulation but also produces a range of apocarotenoids (Walter et al., 2010). In higher plants, many different apocarotenoids derive from the cleavage of carotenoids and have important metabolic functions, such as plant hormones, pigments, aroma and scent compounds, as well as signaling compounds (Fig. 1). A well-known example is abscisic acid, which is a C15 compound derived from the cleavage of the 11,12 double bond of 9-cis-violaxanthin and 9′-cis-neoxanthin (Schwartz et al., 1997; Tan et al., 1997; Cutler and Krochko, 1999; Chernys and Zeevaart, 2000; Giuliano et al., 2003).
Figure 1.
Carotenoid and apocarotenoid metabolic pathway in plants. GGPP, Geranylgeranyl diphosphate. Enzymes, listed here from top to bottom, are named according to the designation of their genes: PSY, phytoene synthase; PDS, Phytoene desaturase; ZDS, ζ-carotene desaturase; ZISO, 15-cis-ζ-carotene isomerase; CRTISO, carotenoid isomerase; LCYb, lycopene β-cyclase; LCYe, lycopene ε-cyclase; HYe, ε-ring hydroxylase; HYb, β-ring hydroxylase; ZEP, zeaxanthin epoxidase; VDE, violaxanthin deepoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase.
Carotenoid cleavage dioxygenases (CCDs) are a group of enzymes that catalyze the oxidative cleavage of carotenoids (Ryle and Hausinger, 2002). CCDs are nonheme iron enzymes present in plants, bacteria, and animals. In plants, CCDs belong to an ancient and highly heterogenous family (CCD1, CCD4, CCD7, CCD8, and 9-cis-epoxycarotenoid dioxygenases [NCEDs]). The similarity among the different members is very low apart from four strictly conserved His residues and a few Glu residues (Kloer and Schulz, 2006; Walter et al., 2010). In Arabidopsis, the CCD family contains nine members (CCD1, NCED2, NCED3, CCD4, NCED5, NCED6, CCD7, CCD8, and NCED9), and orthologs in other plant species are typically named according to their homology with an Arabidopsis CCD (Huang et al., 2009). In our previous study, the functions of CitCCD1, CitNCED2, and CitNCED3 were investigated in citrus fruits (Kato et al., 2006). The recombinant CitCCD1 protein cleaved β-cryptoxanthin, zeaxanthin, and all-trans-violaxanthin at the 9,10 and 9′,10′ positions and 9-cis-violaxanthin at the 9′,10′ position. The recombinant CitNCED2 and CitNCED3 proteins cleaved 9-cis-violaxanthin at the 11,12 position to form xanthoxin, a precursor of abscisic acid (Kato et al., 2006). To date, information on the functions of other CCDs in citrus fruits remains limited, while the functions of CCD7 and CCD8, as well as NCED5, NCED6, and NCED9, in Arabidopsis have been characterized (Kloer and Schulz, 2006; Walter et al., 2010). In Arabidopsis, CCD7 cleaves all-trans-β-carotene at the 9′,10′ position to form all-trans-β-apo-10′-carotenal. All-trans-β-apo-10′-carotenal is further shortened by AtCCD8 at the 13,14 position to produce β-apo-13-carotenone (Alder et al., 2012). NCED5, NCED6, and NCED9 cleave 9-cis-violaxanthin at the 11,12 position to form xanthoxin (Tan et al., 2003). Compared with other CCDs, the function of CCD4 is poorly understood. In Chrysanthemum morifolium, CmCCD4a contributed to the white color formation by cleaving carotenoids into colorless compounds (Ohmiya et al., 2006). Recently, it has been reported that CsCCD4, CmCCD4a, and MdCCD4 could cleave β-carotene to yield β-ionone (Rubio et al., 2008; Huang et al., 2009).
β-Citraurin, a C30 apocarotenoid, is a color-imparting pigment responsible for the reddish color of citrus fruits (Farin et al., 1983). In 1936, it was first discovered in Sicilian oranges (Cual, 1965). In citrus fruits, the accumulation of β-citraurin is not a common event; it is only observed in the flavedos of some varieties during fruit ripening. The citrus varieties accumulating β-citraurin are considered more attractive because of their red-orange color (Ríos et al., 2010). Although more than 70 years have passed since β-citraurin was first identified, the pathway of its biosynthesis is still unknown. As its structure is similar to that of β-cryptoxanthin and zeaxanthin, β-citraurin was presumed to be a degradation product of β-cryptoxanthin or zeaxanthin (Oberholster et al., 2001; Rodrigo et al., 2004; Ríos et al., 2010; Fig. 1). To date, however, the specific cleavage reaction producing β-citraurin has not been elucidated. In this study, we found that the CitCCD4 gene was involved in the synthesis of β-citraurin, using two citrus varieties of Satsuma mandarin (Citrus unshiu), Yamashitabeni-wase, which accumulates β-citraurin predominantly, and Miyagawa-wase, which does not accumulate β-citraurin. To confirm the role of the CitCCD4 gene further, functional analyses of the CitCCD4 enzyme were performed in vivo and in vitro. Additionally, the regulation of β-citraurin content and CitCCD4 gene expression in response to ethylene and red light-emitting diode (LED) light treatments was also examined. This study, to our knowledge, is the first to investigate the biosynthesis of β-citraurin in citrus fruits. The results might provide new strategies to enhance the nutritional and commercial qualities of citrus fruits.
RESULTS
Isolation and Identification of β-Citraurin
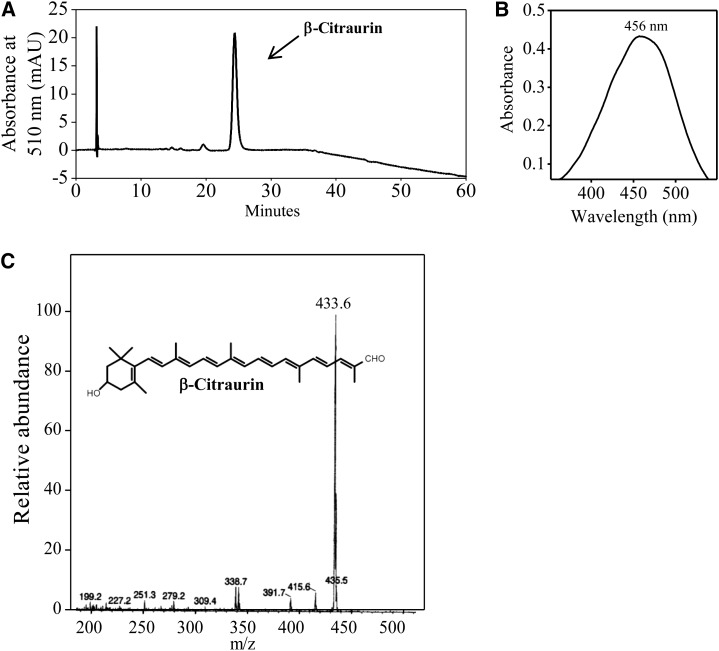
The crude extract of carotenoids from the flavedo of Yamashitabeni-wase was loaded on a column of silica gel. The red pigment was collected and analyzed by HPLC. As shown in Figure 2, the peak that eluted at 24 min was isolated. The mass spectrum of the eluent showed a molecular ion at mass-to-charge ratio 433 ([M+H+]). The absorption maximum of the eluent was 456 nm in ethanol. The mass spectrum and absorption maximum of the eluent were consistent with those of β-citraurin (Farin et al., 1983; Agócs et al., 2007). Therefore, the peak that eluted at 24 min was identified as β-citraurin.
Figure 2.
Isolation and identification of β-citraurin from the flavedo of Yamashitabeni-wase. A, HPLC analysis of β-citraurin. mAU, Milliabsorbance units. B, UV-visible light spectrum of β-citraurin. C, Fast atom bombardment mass spectrometry spectrum of β-citraurin.
Changes in the Contents and Compositions of Carotenoids in the Flavedos
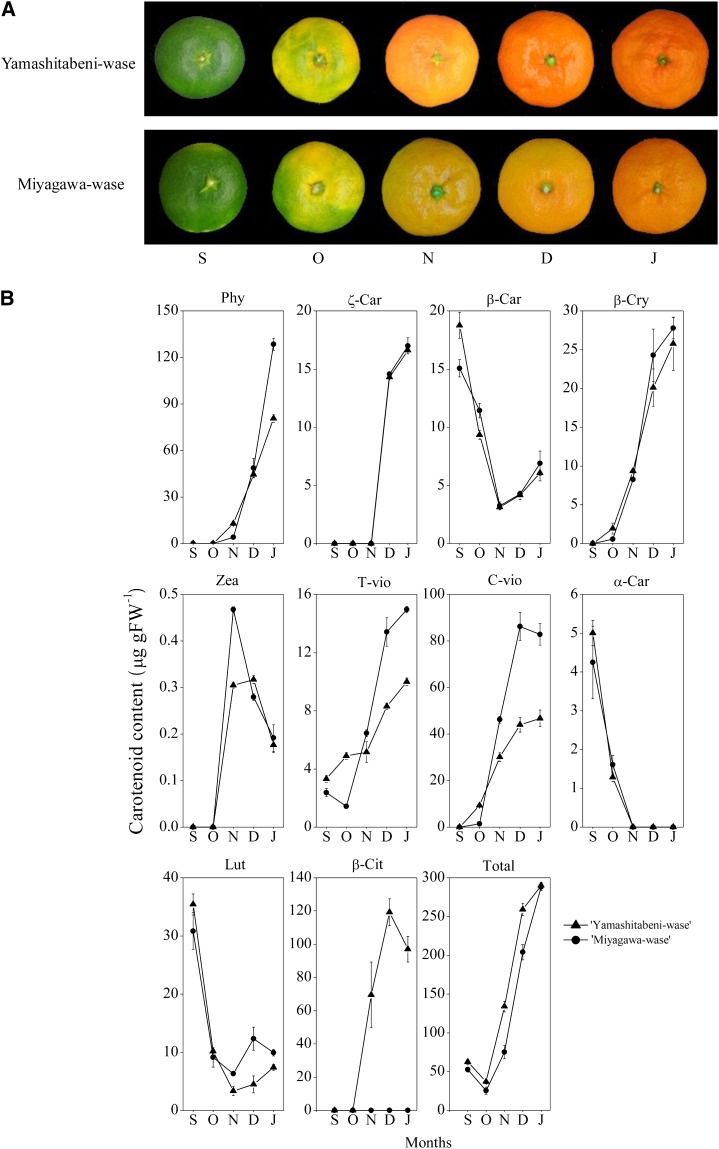
According to changes in the color of the flavedo, the ripening process of citrus fruits can be divided into three stages: a green stage (September), a transition stage (from October to November), and a mature stage (from December to January; Kato, 2012). As shown in Figure 3A, two varieties of Satsuma mandarin, Yamashitabeni-wase and Miyagawa-wase, showed a color break gradually from the transition stage. In the mature stage, Yamashitabeni-wase displayed a reddish color in peel, while Miyagawa-wase displayed a yellowish color in peel.
Figure 3.
Carotenoid accumulation in flavedos of Yamashitabeni-wase and Miyagawa-wase during the ripening process. A, Changes in the external colors of Yamashitabeni-wase and Miyagawa-wase during the ripening process. B, Changes in the carotenoid contents in flavedos of Yamashitabeni-wase and Miyagawa-wase during the ripening process. The results shown are means ± se for triplicate samples. S, September; O, October; N, November; D, December; J, January; Phy, phytoene; ζ-Car, ζ-carotene; β-Car, β-carotene; β-Cry, β-cryptoxanthin; Zea, zeaxanthin; T-vio, all-trans-violaxanthin; C vio, 9-cis-violaxanthin; α-Car, α-carotene; Lut, lutein; β-Cit, β-citraurin; Total, total carotenoids; FW, Fresh weight.
The changes in the contents and compositions of carotenoids were examined in the flavedos of Yamashitabeni-wase and Miyagawa-wase during fruit ripening. As shown in Figure 3B, a significant difference in the β-citraurin content was observed between the two varieties. In Yamashitabeni-wase, β-citraurin content increased rapidly from October. In contrast, β-citraurin was undetectable in Miyagawa-wase throughout the ripening process. During fruit ripening, the contents of β,ε-carotenoids, lutein, and α-carotene decreased rapidly to a low level from September, while massive accumulation of β,β-xanthophylls (β-cryptoxanthin, zeaxanthin, all-trans-violaxanthin, and 9-cis-violaxanthin) occurred in the two varieties during the ripening process (Fig. 3B). The contents of β-cryptoxanthin and zeaxanthin increased rapidly from October in Yamashitabeni-wase and Miyagawa-wase. The content of β-cryptoxanthin was similar between the two varieties throughout the ripening process (Fig. 3B). The content of zeaxanthin was higher in Miyagawa-wase than in Yamashitabeni-wase in November. The contents of all-trans-violaxanthin and 9-cis-violaxanthin increased gradually from September in Yamashitabeni-wase. In Miyagawa-wase, the contents of all-trans-violaxanthin and 9-cis-violaxanthin increased significantly from October, and their contents were much higher than those in Yamashitabeni-wase from November to January (Fig. 3B).
Changes in the Expression of Genes Related to Carotenoid Metabolism
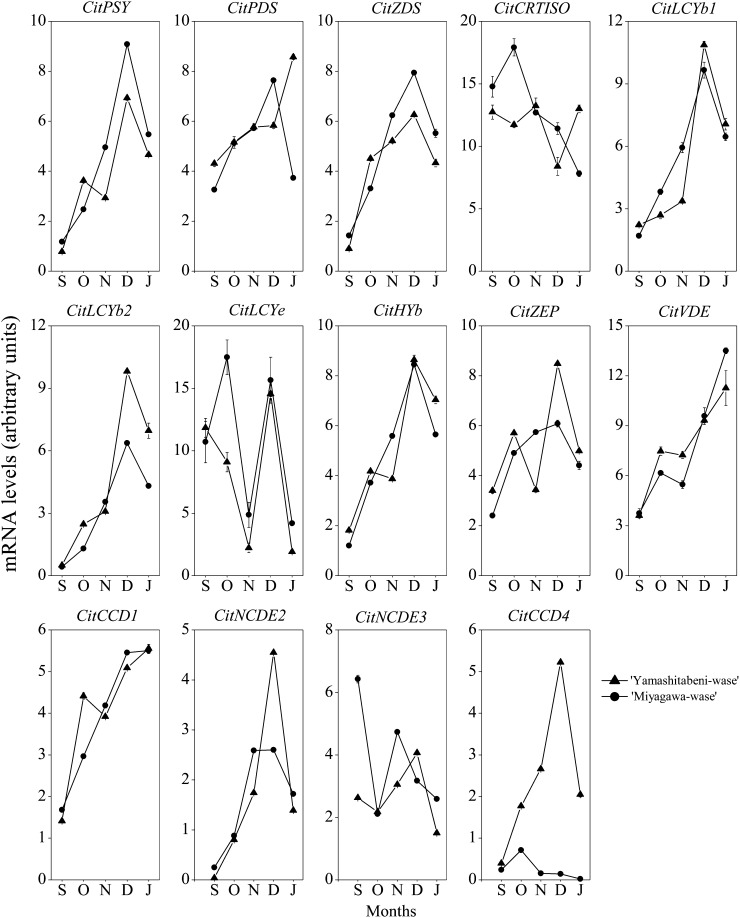
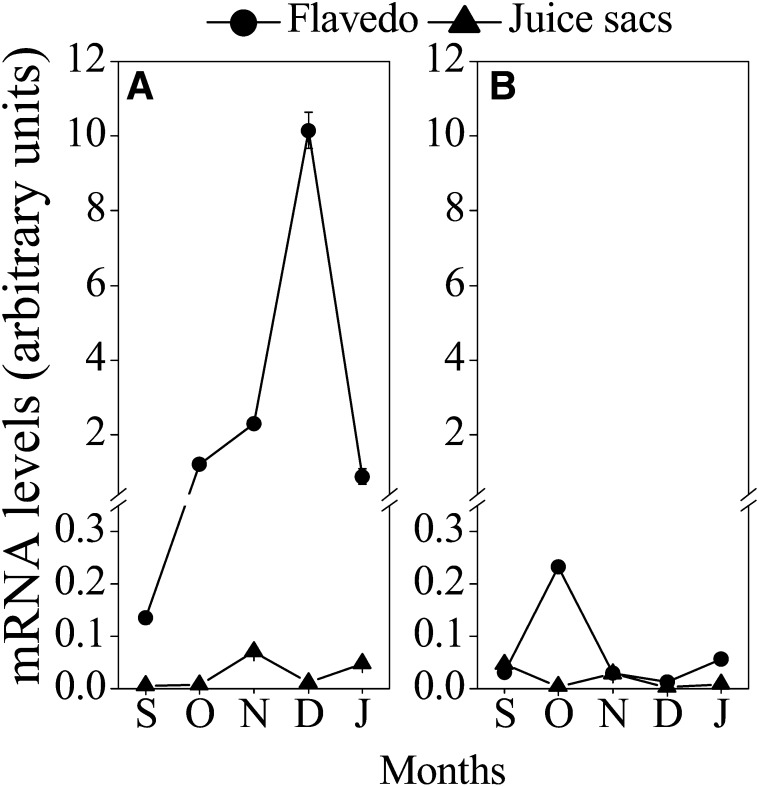
The expression of a set of genes related to carotenoid metabolism, phytoene synthase (PSY), phytoene desaturase (PDS), z-carotene desaturase (ZDS), carotenoid isomerase (CRTISO), lycopene b-cyclase (LCYb), lycopene e-cyclase (LCYe), β-ring hydroxylase (HYb), zeaxanthin epoxidase (ZEP), violaxanthin de-epoxidase (VDE), NCED2, and CCD1, increased from September to December in the flavedos of Yamashitabeni-wase and Miyagawa-wase. Interestingly, we found that the expression of CitCCD4 increased significantly, with a peak in December, in the flavedo of Yamashitabeni-wase; however, it remained at a low level in the flavedo of Miyagawa-wase during the ripening process (Fig. 4). Moreover, in the juice sacs, where β-citraurin was not accumulated, the expression level of CitCCD4 was extremely low in the two varieties during the ripening process (Fig. 5; Supplemental Fig. S1). These results indicated that CitCCD4 might be involved in the biosynthesis of β-citraurin in the flavedo of Yamashitabeni-wase.
Figure 4.
Changes in the expression of genes related to carotenoid metabolism in flavedos of Yamashitabeni-wase and Miyagawa-wase during the ripening process. The results shown are means ± se for triplicate samples. S, September; O, October; N, November; D, December; J, January. The mRNA levels were analyzed by TaqMan real-time quantitative RT-PCR. Real-time RT-PCR amplification of 18S rRNA was used to normalize the expression of the genes under identical conditions.
Figure 5.
Changes in the expression of CitCCD4 in flavedos and juice sacs of Yamashitabeni-wase (A) and Miyagawa-wase (B) during the ripening process. The results shown are means ± se for triplicate samples. S, September; O, October; N, November; D, December; J, January. The mRNA levels were analyzed by TaqMan real-time quantitative RT-PCR. Real-time RT-PCR amplification of 18S rRNA was used to normalize the expression of the genes under identical conditions.
Isolation of Full-Length Complementary DNA of CitCCD4
In this study, full-length complementary DNAs (cDNAs) of the CitCCD4 gene were isolated from Yamashitabeni-wase and Miyagawa-wase, and their sequences were analyzed. The sequences of CitCCD4 in Yamashitabeni-wase and Miyagawa-wase were identical at the nucleic acid level and the amino acid level (Supplemental Fig. S2). The nucleotide sequence of CitCCD4 contained 1,692 bp and encoded a putative protein of 563 amino acids with an estimated molecular mass of 63 kD. CitCCD4 protein was predicted to localize in cytoplasm (utility: 8.0) or chloroplast (utility: 5.0) by WoLF PSORT (Horton et al., 2007; http://wolfpsort.org/). Using the online prediction server TMpred, the CitCCD4 protein was predicted to have two transmembrane helices, which indicated that it was an integral membrane protein (Supplemental Fig. S2). In the N-terminal region of the protein encoded by CitCCD4, no characteristic transit peptide was detected by TargetP.
A BLAST search in Citrus Genome Database (http://www.citrusgenomedb.org) revealed that the cDNA sequence of CitCCD4 was identical to scaffold_128:294016:295707 of the citrus gene clementine0.9_006329m.g. The gene structure of clementine0.9_006329m.g included a 5′ untranslated region, a coding sequence, and a 3′ untranslated region. To further identify the genome organization of CitCCD4, the genomic DNAs of CitCCD4 were amplified by PCR from the start codon to the stop codon and the sequences were analyzed in Yamashitabeni-wase and Miyagawa-wase. The sequence analysis revealed that the CitCCD4 gene did not contain introns in either Yamashitabeni-wase or Miyagawa-wase (Supplemental Fig. S3).
Subcellular Localization of the CitCCD4-GFP Fusion Protein Expressed in Tobacco Leaves
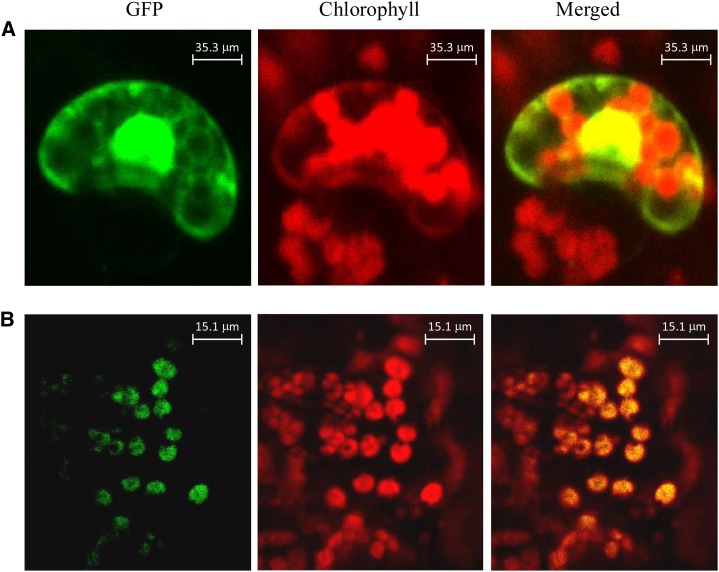
To determine the subcellular localization of CitCCD4, the cDNA of CitCCD4 without the stop codon was fused with a GFP reporter gene under the control of the cauliflower mosaic virus 35S promoter and bombarded in tobacco (Nicotiana tabacum) leaves. After 16 to 18 h, the localization of GFP fusion proteins was observed by confocal laser-scanning microscopy. Confocal imaging of GFP fluorescence showed that the CitCCD4 fusion protein was located in the chloroplast (Fig. 6).
Figure 6.
Subcellular localization of the CitCCD4-GFP fusion protein. A, Tobacco SR1 leaves were transfected with the control vector (35S-GFP). B, Tobacco SR1 leaves were transfected with the recombinant vector (35S-CitCCD4-GFP).
Functional Analyses of Recombinant CitCCD4 Protein
In this study, the functions of CitCCD4 were investigated in vivo and in vitro. Lycopene, α-carotene, β-carotene, and zeaxanthin were used as substrates to assay the enzyme activity of CitCCD4 in vivo. β-Cryptoxanthin, zeaxanthin, all-trans-violaxanthin, and 9-cis-violaxanthin were used as substrates to assay the enzyme activity of CitCCD4 in vitro.
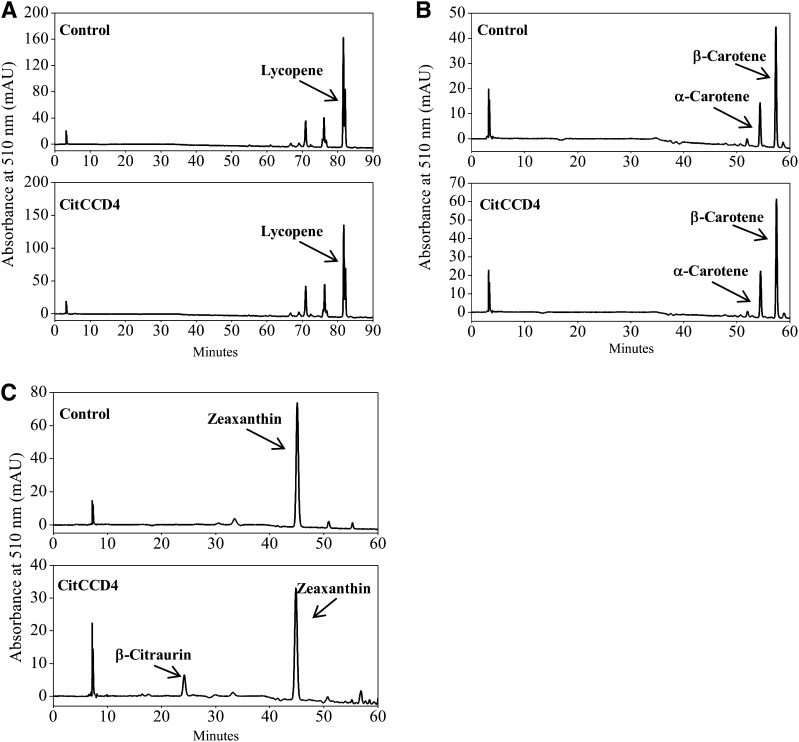
To examine the enzyme activity of CitCCD4 in vivo, the cDNA of CitCCD4 isolated from the flavedo of Yamashitabeni-wase was cloned into the pRSF-2 Ek/LIC vector. The recombinant plasmid was transformed to the lycopene-accumulating Escherichia coli BL21 (DE3) cells, α-carotene- and β-carotene-accumulating E. coli BL21 (DE3) cells, as well as zeaxanthin-accumulating E. coli BL21 (DE3) cells. Carotenoids were extracted from bacteria, and their contents and compositions were analyzed by HPLC. No cleaved products were observed when CitCCD4 was expressed in E. coli BL21 (DE3) cells accumulating lycopene, α-carotene, and β-carotene (Fig. 7, A and B). Thus, the CitCCD4 enzyme cannot cleave lycopene, α-carotene, and β-carotene. When CitCCD4 was expressed in E. coli BL21 (DE3) cells accumulating zeaxanthin, the peak for β-citraurin, which eluted at 24 min, was observed (Fig. 7C).
Figure 7.
Functional analysis of the CitCCD4 enzyme in vivo. A, HPLC analysis of the cleavage products in lycopene-accumulating E. coli BL21 (DE3) cells transformed with pRSF-2 Ek/LIC-CitCCD4. B, HPLC analysis of the cleavage products in α-carotene- and β-carotene-accumulating E. coli BL21 (DE3) cells transformed with pRSF-2 Ek/LIC-CitCCD4. C, HPLC analysis of the cleavage products in zeaxanthin-accumulating E. coli BL21 (DE3) cells transformed with pRSF-2 Ek/LIC-CitCCD4. mAU, Milliabsorbance units.
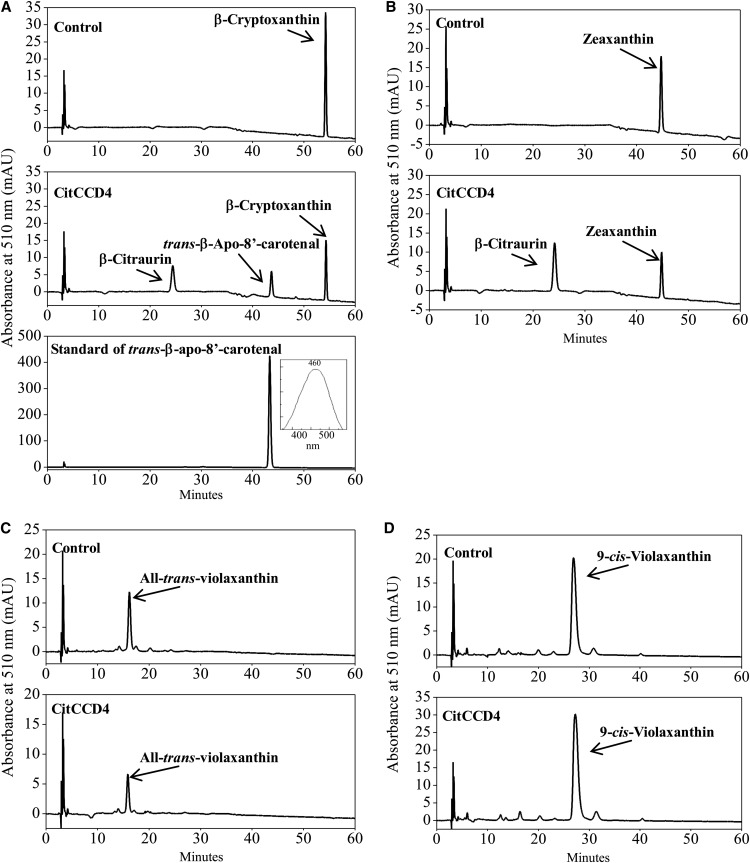
To examine the enzyme activity of CitCCD4 in vitro, the cDNA of CitCCD4 isolated from the flavedo of Yamashitabeni-wase was cloned into the pCold I vector. The recombinant protein expressed in E. coli cells was extracted and confirmed by SDS-PAGE. When all-trans-violaxanthin and 9-cis-violaxanthin were used as substrates for the cleavage reaction of the recombinant CitCCD4 enzyme, no cleavage product was detected (Fig. 8, C and D). When β-cryptoxanthin and zeaxanthin were used as substrates, however, the peak for β-citraurin, which eluted at 24 min, was observed (Fig. 8, A and B). In the case of β-cryptoxanthin, the other cleaved product, which eluted at 43 min, was also detected (Fig. 8A). The absorption maximum of the peak at 43 min was 460 nm. Both the elution time and absorption maximum of this eluent were identical to those of standard trans-β-apo-8′-carotenal (Fig. 8A). Therefore, the eluent at 43 min was identified as trans-β-apo-8′-carotenal. These results suggested that CitCCD4 enzyme could cleave β-cryptoxanthin and zeaxanthin at the 7,8 or 7′,8′ position, and the cleavage of β-cryptoxanthin and zeaxanthin contributed to the biosynthesis of β-citraurin.
Figure 8.
Functional analysis of the CitCCD4 enzyme in vitro. A, HPLC analysis of the cleavage products from the incubation of β-cryptoxanthin with recombinant CitCCD4. B, HPLC analysis of the cleavage products from the incubation of zeaxanthin with recombinant CitCCD4. C, HPLC analysis of the cleavage products from the incubation of all-trans-violaxanthin with recombinant CitCCD4. D, HPLC analysis of the cleavage products from the incubation of 9-cis-violaxanthin with recombinant CitCCD4. mAU, Milliabsorbance units.
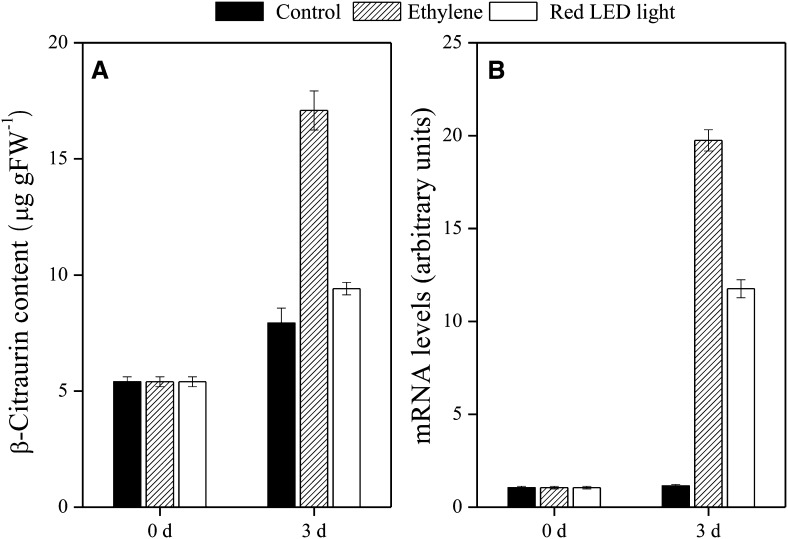
Effects of Ethylene and Red LED Light on β-Citraurin Content and CitCCD4 Expression
To enhance the content of β-citraurin, the effects of ethylene and red LED light (660 nm) on β-citraurin content and the expression of CitCCD4 were investigated using the fruits of Yamashitabeni-wase harvested in October. During the experimental periods, the accumulation of β-citraurin was observed in the control as well as the ethylene- and red light-treated groups (Fig. 9). Compared with the control, the content of β-citraurin was significantly increased by ethylene treatment. The expression of CitCCD4 was up-regulated by ethylene and red LED light treatments (Fig. 9). These increases in the expression of CitCCD4 were well consistent with the accumulation of β-citraurin in the two treatments.
Figure 9.
Effects of ethylene and red LED light on β-citraurin content (A) and gene expression of CitCCD4 (B) in flavedo of Yamashitabeni-wase. The results shown are means ± se for triplicate samples. The mRNA levels were analyzed by TaqMan real-time quantitative RT-PCR. Real-time RT-PCR amplification of 18S rRNA was used to normalize the expression of the genes under identical conditions. FW, Fresh weight.
DISCUSSION
Accumulation of β-Citraurin in the Flavedos of Citrus Fruits
The accumulation of β-citraurin, a red pigment, has been detected in only a few citrus varieties (Xu et al., 2011). Ríos et al. (2010) reported that β-citraurin was observed in the flavedo of a citrus clementine (Citrus clementina) mutant, 39B3, while it was absent in the flavedo of the other citrus clementine mutant, 39E7. The defective synthesis of β-citraurin was proposed to cause the yellowish color of fully ripe 39E7 flavedo. Because β-citraurin is a characteristic pigment, it can be isolated in pure form among other carotenoids from the flavedos of citrus fruits (Agócs et al., 2007). In this study, we isolated β-citraurin from the flavedo of Yamashitabeni-wase using a silica gel column and HPLC. The mass spectrum and absorption maximum of β-citraurin were consistent with those reported previously (Farin et al., 1983; Agócs et al., 2007; Fig. 2). In Yamashitabeni-wase, the content of β-citraurin increased significantly from October. To the best of our knowledge, this is the first report to show that β-citraurin is a main carotenoid that accumulates in Satsuma mandarin Yamashitabeni-wase. In the other Satsuma mandarin variety, Miyagawa-wase, β-citraurin was undetectable throughout the ripening process (Fig. 3B). This difference led to different peel colors between the two varieties. The accumulation of β-citraurin contributed to the reddish peel in Yamashitabeni-wase, while the complete absence of β-citraurin led to the yellowish peel in Miyagawa-wase (Fig. 3A). Although more than 70 years have passed since β-citraurin was first identified in citrus fruits, the pathway of β-citraurin biosynthesis has yet to be elucidated (Cual, 1965). In this study, the contents of β-cryptoxanthin, zeaxanthin, and β-citraurin rapidly increased in Yamashitabeni-wase from October. The concomitant increases in β-cryptoxanthin, zeaxanthin, and β-citraurin indicated that β-citraurin may be a breakdown product of β-cryptoxanthin and zeaxanthin. Additionally, β-citraurin did not accumulate in Miyagawa-wase and zeaxanthin was further converted into all-trans-violaxanthin and 9-cis-violaxanthin; as a result, the contents of all-trans-violaxanthin and 9-cis-violaxanthin were higher than those in Yamashitabeni-wase.
Isolation and Sequence Analysis of CitCCD4
To find the enzyme responsible for β-citraurin biosynthesis, we compared the expression of genes related to carotenoid metabolism between the two varieties of Satsuma mandarin, Yamashitabeni-wase and Miyagawa-wase. In the flavedo, a significant difference in the expression of CitCCD4 was observed between the two varieties. In Yamashitabeni-wase, the expression of CitCCD4 increased rapidly, with a peak in December, which was consistent with the accumulation of β-citraurin (Fig. 4). In Miyagawa-wase, the expression of CitCCD4 remained at a low level during the ripening process, which was consistent with the absence of β-citraurin in the flavedo (Fig. 4). Moreover, in the juice sacs, where β-citraurin was not accumulated, the expression of CitCCD4 was extremely low in both Yamashitabeni-wase and Miyagawa-wase (Fig. 5; Supplemental Fig. S1). These results suggested CitCCD4 to be a key factor regulating β-citraurin biosynthesis.
In this study, full-length cDNAs of the CitCCD4 gene were isolated from Yamashitabeni-wase and Miyagawa-wase, and their sequences were analyzed. The homology of CitCCD4 between Yamashitabeni-wase and Miyagawa-wase was 100% identity at the nucleic acid and amino acid levels (Supplemental Fig. S2). In clementine, two types of CCD4, CcCCD4a and CcCCD4b, were isolated (Agustí et al., 2007). The identities of CitCCD4 investigated herein between CcCCD4a and CcCCD4b at the amino acid level were 52% and 99%, respectively (Supplemental Fig. S4). The nucleotide sequence of CitCCD4 contained 1,692 bp and encoded a putative protein of 563 amino acids with an estimated molecular mass of 63 kD. The nucleotide sequence of CcCCD4b contained 1,683 bp and encoded a putative protein of 560 amino acids. The nucleotide sequence of CitCCD4 was not identical to CcCCD4a or CcCCD4b. In addition, we isolated the 2,000-bp genomic DNA sequences of CitCCD4 promoters from Yamashitabeni-wase and Miyagawa-wase, and no significant difference was observed between the two varieties (data not shown). Ríos et al. (2010) reported that CcGCC1, a transcriptional regulator, played an important role in regulating the color break in citrus fruits. Thus, it is possible that some transcriptional regulators are involved in controlling the expression of CitCCD4 in the flavedos of Yamashitabeni-wase and Miyagawa-wase. Further research is needed to confirm this.
In contrast to CCD1, CCD7, and CCD8, where the introns were conserved, in CCD4 the intron-exon structure was highly dynamic (Mein et al., 2011). In this study, no intron was observed in CitCCD4 in Yamashitabeni-wase or Miyagawa-wase (Supplemental Fig. S3). Huang et al. (2009) reported that RdCCD4 and AtCCD4 contained no intron, while MdCCD4, OfCCD4, and CmCCD4 contained introns. The absence or presence of introns was related to the different expression patterns and biochemical functions of these CCD4 genes (Huang et al., 2009).
Unlike CCD1 enzymes, which were located in the cytoplasm, most CCD4 enzymes were located in plastids (Ytterberg et al., 2006; Rubio et al., 2008). In the N-terminal region of the protein encoded by CitCCD4, no characteristic transit peptide was predicted. However, the subcellular localization of the CitCCD4-GFP fusion protein expressed in tobacco leaves suggested that CitCCD4 fusion protein was imported into plastids (Fig. 6). A similar phenomenon was also observed in the CsZCD enzyme. Bouvier et al. (2003) found that CsZCD was compartmentalized to the plastid, although it did not contain a typical cleavable transit peptide. In plants, carotenoids are synthesized and stored in plastids (Kloer and Schulz, 2006). The location of CitCCD4 within plastids allowed it to access its carotenoid substrates, such as β-cryptoxanthin and zeaxanthin.
Functional Analyses of Recombinant CitCCD4 Enzyme
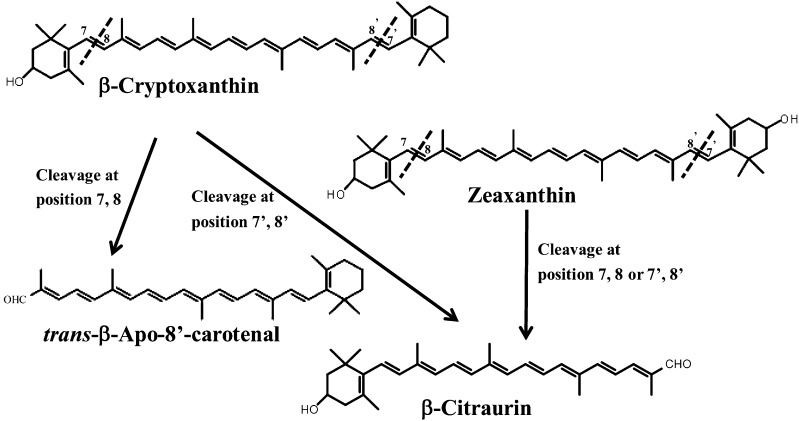
Compared with other CCDs, information about the functions of CCD4 is limited. In potato (Solanum tuberosum), the down-regulation of CCD4 gene expression using RNA interference resulted in increased violaxanthin content (Campbell et al., 2010). In C. morifolium, CmCCD4a contributed to the white color formation by cleaving carotenoids into colorless compounds (Ohmiya et al., 2006). A common feature of CCD4 identified in several recent studies is a 9,10 or 9′,10′ cleavage activity to yield β-ionone (Rubio et al., 2008; Huang et al., 2009). However, the 7,8 or 7′,8′ cleavage activity of CCD4 remains to be confirmed. Bouvier et al. (2003) reported that CsZCD specifically catalyzed the cleavage of zeaxanthin at the 7,8 and 7′,8′ positions of the chromophore and initiated the formation of saffron (Crocus sativus) secondary metabolites. In this study, we found that the CitCCD4 enzyme exhibited substrate specificity. It cleaved β-cryptoxanthin and zeaxanthin at the 7,8 or 7′,8′ position (Figs. 7 and 8). But other carotenoids tested in this study (lycopene, α-carotene, β-carotene, all-trans-violaxanthin, and 9-cis-violaxanthin) were not cleaved by CitCCD4 (Figs. 7 and 8). Moreover, the cleavage of β-cryptoxanthin and zeaxanthin by the CitCCD4 enzyme led to the formation of β-citraurin. In addition, when β-cryptoxanthin and zeaxanthin were added together in the same reaction solution, the contents of β-cryptoxanthin and zeaxanthin decreased simultaneously along with the biosynthesis of β-citraurin (data not shown). The specific cleavage reaction of CitCCD4 presented here further confirmed the previous speculations that β-citraurin was a breakdown product of β-cryptoxanthin and zeaxanthin (Fig. 10). In addition, when β-cryptoxanthin was used as the substrate for the cleavage reaction of the recombinant CitCCD4 enzyme, trans-β-apo-8′-carotenal, which eluted at 43 min, was also detected except for β-citraurin (Fig. 10). In clementine, extremely low content of trans-β-apo-8′-carotenal was detected in the fruits harvested in November, while in the fully ripened fruits, it was absent (Ríos et al., 2010). In our study, we were unable to detect trans-β-apo-8′-carotenal in the flavedo or juice sacs of Yamashitabeni-wase. It is possible that trans-β-apo-8′-carotenal may be further cleaved by other CCDs in Yamashitabeni-wase.
Figure 10.
The pathway from β-cryptoxanthin and zeaxanthin to β-citraurin catalyzed by CitCCD4 in citrus fruits.
Effects of Ethylene and Red LED Light on β-Citraurin Content and CitCCD4 Expression
It has been reported that ethylene treatment increased the contents of carotenoids; as a result, the degreening process of citrus fruits was accelerated (Rodrigo and Zacarías, 2007). We previously found that red LED light was effective in enhancing carotenoid contents, especially the content of β-cryptoxanthin, while blue LED light had no significant effect on the carotenoid content in the flavedo of Satsuma mandarin (Ma et al., 2012). In this study, to investigate the regulatory effects of ethylene and red LED light on β-citraurin accumulation, fruits harvested in October were used, as the changes in β-citraurin content were most significant at this stage. As shown in Figure 9, the content of β-citraurin was increased by the ethylene and red LED light treatments. Additionally, with ethylene and red LED light treatments, the gene expression of CitCCD4 was up-regulated in the flavedo of Yamashitabeni-wase. These increases in the expression of CitCCD4 were well consistent with the accumulation of β-citraurin in the two treatments. The results presented here provide more insights into the regulatory mechanism of β-citraurin metabolism in citrus fruits.
CONCLUSION
In this study, the biosynthetic pathway of β-citraurin was investigated using two citrus varieties of Satsuma mandarin, Yamashitabeni-wase, which accumulates β-citraurin predominantly, and Miyagawa-wase, which does not accumulate β-citraurin. The results suggested that CitCCD4 was a key gene regulating the biosynthesis of β-citraurin. The cleavage of β-cryptoxanthin and zeaxanthin by the CitCCD4 enzyme led to the formation of β-citraurin. In addition, the ethylene and red light treatments were effective in enhancing the content of β-citraurin by up-regulating the expression of CitCCD4 in the flavedo of Yamashitabeni-wase. These results might contribute to elucidate the mechanism of β-citraurin accumulation in citrus fruits, which could facilitate improvement in citrus nutritional and commercial qualities.
MATERIALS AND METHODS
Plant Material
Two varieties of Satsuma mandarin (Citrus unshiu), Yamashitabeni-wase and Miyagawa-wase, cultivated at the Fujieda Farm of Shizuoka University, were used as materials. Fruit samples were collected periodically from September to January. The flavedos and juice sacs were separated from sampled fruits, immediately frozen in liquid nitrogen, and kept at −80°C until used.
Extraction and Determination of Carotenoids
The identification and quantification of carotenoids were conducted according to the methods described by Kato et al. (2004). Pigments were extracted from the samples using a hexane:acetone:ethanol (2:1:1, v/v) solution containing 0.1% (w/v) 2,6-di-tert-butyl-4-methylphenol and 10% (w/v) magnesium carbonate basic. After the organic solvents had been completely evaporated, the extracts containing carotenoids esterified to fatty acids were saponified with 20% (w/v) methanolic KOH. Water-soluble extracts were then removed by adding NaCl-saturated water. The pigments repartitioned into the diethyl ether phase were recovered and evaporated to dryness. Subsequently, the residue was redissolved in a methyl tert-butyl ether:methanol (1:1, v/v) solution. An aliquot (20 μL) was separated by a reverse-phase HPLC system (Jasco) fitted with a YMC Carotenoid S-5 column of 250 × 4.6 mm i.d. (Waters) at a flow rate of 1 mL min−1. The eluent was monitored by a photodiode array detector (MD-2015; Jasco). The standard for each carotenoid was prepared according to the methods described by Kato et al. (2004). The carotenoid concentration was estimated by the standard curves and expressed as milligrams per gram fresh weight. Total carotenoid is the sum of the content of various carotenoids identified in this study. Carotenoid quantification was performed in three replicates.
Isolation and Identification of β-Citraurin
The carotenoids extracted from the flavedo of Yamashitabeni-wase were loaded on a column of silica gel (2 cm diameter and 52 cm length) using a hexane:ethyl ether:isopropyl alcohol (7:3:1, v/v) solution as eluent. The red pigment was collected and evaporated to dryness. The collection was subjected to HPLC and identified by spectrophotometry and mass spectrometry. The UV-visible spectra were taken with a spectrophotometer. Fast atom bombardment mass spectrometry analysis was performed with the API 2000 triple-stage quadrupole mass spectrometer (Applied Biosystems).
Isolation and Sequence Analysis of CitCCD4
Total RNA was extracted from the flavedos of Yamashitabeni-wase and Miyagawa-wase according to the method described by Ikoma et al. (1996). First-strand cDNA was synthesized from 2 μg of total RNA using TaqMan Reverse Transcription Reagents (Applied Biosystems). The cDNA fragment of CitCCD4 was amplified by PCR using the degenerate PCR primers designed according to the common sequences that have been reported previously (Supplemental Table S1). The amplified cDNAs were sequenced using the BigDye Terminator Version 3.1 Cycle Sequencing Kit (Applied Biosystems) with an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). After sequencing, RACE-PCR was performed using the SMART RACE cDNA Amplification Kit (Clontech Laboratories; Supplemental Table S1). The amplified cDNAs of the 5′ and 3′ ends for CitCCD4 were cloned and sequenced. End-to-end PCR was performed with the cDNAs of Yamashitabeni-wase and Miyagawa-wase using the primers designed from the cDNA sequences amplified by RACE-PCR (Supplemental Table S1).
The alignment of CitCCD4 between Yamashitabeni-wase and Miyagawa-wase was created using the Genetyx Analysis Program (Genetyx). Information regarding gene structure was obtained from the Citrus Genome Database (http://www.citrusgenomedb.org). The sublocation of CitCCD4 was predicated by WoLF PSORT (Horton et al., 2007; http://wolfpsort.org/). Predictions of transit peptides of CitCCD4 were carried out using TargetP. The transmembrane helices of CitCCD4 were predicted using the online prediction server TMpred (Hofmann and Stoffel, 1993).
Isolation of CitCCD4 Genomic DNA
Genomic DNA was isolated from leaf tissues of Yamashitabeni-wase and Miyagawa-wase using a Qiagen DNA Mini Kit. A genomic DNA fragment from the start codon to the stop codon of CitCCD4 was amplified by PCR. The PCR program for CitCCD4 was as described above. The amplified DNA was separated by electrophoresis on 1.5% agarose gels and sequenced using the BigDye Terminator Version 3.1 Cycle Sequencing Kit (Applied Biosystems) with an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
Total RNA Extraction and Real-Time Quantitative Reverse Transcription-PCR
Total RNA was extracted from the flavedos and juice sacs of Yamashitabeni-wase and Miyagawa-wase at different stages according to the method described by Ikoma et al. (1996). Total RNA was cleaned up using the RNeasy Mini Kit (Qiagen) with on-column DNase digestion. The reverse transcription (RT) reaction was performed with 2 μg of purified RNA and a random hexamer at 37°C for 60 min using TaqMan Reverse Transcription Reagents (Applied Biosystems).
TaqMan Minor Groove Binder probes and sets of primers for genes related to carotenoid metabolism (CitPSY, CitPDS, CitZDS, CitLCYb1, CitLCYb2, CitLCYe, CitHYb, CitZEP, CitCRTISO, CitVDE, CitCCD1, CitNCED2, CitNCED3, and CitCCD4) were designed on the basis of sequences conserved between the two varieties for each gene with Primer Express software (Supplemental Table S2). For the endogenous control, the TaqMan rRNA Control Reagents VIC Probe (Applied Biosystems) was used. TaqMan real-time PCR was carried out with the TaqMan Universal PCR Master Mix (Applied Biosystems) using ABI PRISM 7300 (Applied Biosystems) according to the manufacturer’s instructions. Each reaction contained 900 nm of the primers, 250 nm TaqMan MGB Probe, and template cDNA. The thermal cycling conditions were 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The levels of gene expression were analyzed with ABI PRISM 7300 Sequence Detection System Software (Applied Biosystems) and normalized with the results of 18S ribosomal RNA. Real-time quantitative RT-PCR was performed in three replicates for each sample.
Transient Expression of the CitCCD4-GFP Fusion Protein in Tobacco Leaves
The full-length CitCCD4 without the stop codon was added to the 4-bp sequence (CACC) on the N terminus and subcloned into pENTR/D-TOPO (Invitrogen). The CitCCD4 sequences were subcloned into new pUGW5 consisting of a synthetic GFP under the control of the cauliflower mosaic virus 35S promoter by using the Gateway LR recombination reaction system (Invitrogen). Plasmid DNA (0.8 μg) was introduced into tobacco (Nicotiana tabacum) SR1 leaves using a pneumatic particle gun (PDS-1000/He; Bio-Rad). The conditions for bombardment were a vacuum of 28 inches of mercury, a helium pressure of 1,300 p.s.i., and a 6-cm target distance using 1-μm gold microcarriers. After the bombardment, the tobacco leaves were incubated for 16 to 18 h at room temperature. Green fluorescence detected with a band-pass 500- to 530-nm filter and chlorophyll autofluorescence detected with a long-pass 560-nm filter were observed using a Leica TCS-SL confocal laser scanning microscope (Leica Microsystems).
Functional Analysis of the CitCCD4 Enzyme in Vivo
The CitCCD4 cDNA from Yamashitabeni-wase was cloned into the pRSF-2 Ek/LIC vector. The recombinant plasmid was transformed into lycopene-accumulating Escherichia coli BL21 (DE3) cells, α-carotene- and β-carotene-accumulating E. coli BL21 (DE3) cells, as well as zeaxanthin-accumulating E. coli BL21 (DE3) cells (Misawa and Shimada, 1997; Zhang et al., 2012b). The transformants were plated in Luria-Bertani medium supplemented with chloramphenicol (50 μg mL−1), carbenicillin (50 μg mL−1), and kanamycin (50 μg mL−1) and incubated at 37°C for 20 h. The colonies were incubated in 100 mL of 2×yeast-tryptone (YT) medium with chloramphenicol (50 μg mL−1), carbenicillin (50 μg mL−1), and kanamycin (50 μg mL−1) at 37°C for 16 h. Then, 2 mL of culture solution was inoculated into 200 mL of 2×YT medium with chloramphenicol (50 μg mL−1), carbenicillin (50 μg mL−1), and kanamycin (50 μg mL−1). After 8 h at 27°C, 200 μL of 0.1 m isopropyl β-d-thiogalactoside was added and cultured overnight at 27°C.
Cultures of E. coli cells were centrifuged at 5,000g for 10 min, and the bacterial pellet was washed twice with Tris-HCl (pH 8.0). The pellet was dried using vacuum freeze drying and stored at −20°C until the HPLC analysis. The freeze-ground material was extracted with a mixture of chloroform and methanol (2:1, v/v) until all the color was removed from the E. coli cells. The carotenoid extracts were reduced to dryness by rotary evaporation and then dissolved in the methyl tert-butyl ether:methanol (1:1, v/v) solution containing 0.1% butylated hydroxyl toluene. The identification and quantification of carotenoids were conducted according to the methods described by Kato et al. (2004).
Functional Analysis of CitCCD4 Enzyme in Vitro
The CitCCD4 cDNA from Yamashitabeni-wase was cloned into the pCold I vector. The recombinant plasmid was transformed into XL1-Blue cells. The transformants were plated in Luria-Bertani medium supplemented with carbenicillin (50 μg mL−1) and incubated at 37°C for 20 h. The colonies were incubated in 100 mL of 2×YT medium with carbenicillin (50 μg mL−1) at 37°C for 16 h. Then, 2 mL of culture solution was inoculated into 200 mL of 2×YT medium with carbenicillin (50 μg mL−1). Cultures were grown at 37°C until an optical density at 600 nm of 0.7 was reached. The culture solution was quickly refrigerated at 15°C and left to stand for 30 min. The expression of proteins was induced by the addition of 200 μL of 500 mm isopropyl β-d-thiogalactoside, and the cultures were grown at 15°C for an additional 24 h. The cells were harvested by centrifugation, frozen in liquid nitrogen, and then resuspended in 2.5 mL of extraction buffer (0.1 m Tris-HCl, pH 7.2, 30 mm sodium ascorbate, 5 mm dithiothreitol, 10% [v/v] glycerol, and 0.05% [v/v] Triton X-100). After sonication and centrifugation, the complex was subjected to gel filtration using a PD-10 (Pharmacia LKB Biotechnology) column. The recombinant proteins were analyzed by SDS-PAGE with a 12% (w/v) polyacrylamide gel. The concentration of recombinant proteins was determined using the Bradford assay. Five micrograms of the recombinant protein was used for each cleavage reaction.
For the substrates of carotenoids, β-cryptoxanthin and zeaxanthin were obtained from Extrasynthese, and all-trans-violaxanthin and 9-cis-violaxanthin were prepared from the flavedo of Valencia orange (Citrus sinensis; Kato et al., 2006). The enzymatic activities of the recombinant CitCCD4 protein were assayed in a reaction mixture consisting of 0.1 m Tris-HCl, pH 7.2, 30 mm sodium ascorbate, 50 μm FeSO4, 20 μg of catalase, 0.05% (v/v) Triton X-100, 20% (v/v) glycerol, 1 mm carotenoid substrate (β-cryptoxanthin, zeaxanthin, all-trans-violaxanthin, and 9-cis-violaxanthin), and 5 μg of the recombinant protein in a total volume of 200 μL at 27°C for 3 h. After the incubation, 1 mL of water was added to the reaction mixture. The reaction products were partitioned three times into 1.2 mL of ethyl acetate, evaporated to dryness, and dissolved in methanol. An aliquot (20 μL) was separated by a reverse-phase HPLC system (Jasco) fitted with a YMC Carotenoid S-5 column of 250 mm × 34.6 mm i.d. (Waters) at a flow rate of 1 mL min−1. The eluent was detected with a photodiode array detector (MD-2015; Jasco). The product was analyzed using the following elution schedule: 20% (v/v) methanol and 80% (v/v) water (0–5 min), a linear gradient to 100% (v/v) methanol (5–25 min), and 100% (v/v) methanol (25–40 min). For the identification of trans-β-apo-8′-carotenal, a purchased standard was used (Sigma).
Treatment with Ethylene and Red LED Light
Fruits of Yamashitabeni-wase harvested in October were used. For the ethylene treatment, fruits were treated with 50 μL L−1 ethylene for 3 d at 20°C. For the red LED light treatment, fruits were irradiated with red LED light (660 nm) at an intensity of 150 μmol m−2 s−1 for 3 d at 20°C. Fruits stored at 20°C (relative humidity, 75%) in the dark were used as the control. After each treatment, the flavedos were immediately frozen in liquid nitrogen and kept at −80°C until used.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AB114648, CitPSY; AB114649, CitPDS; AB114650, CitZDS; AB114652, CitLCYb1; AB719392, CitLCYb2; AB114655, CitLCYe; AB114653, CitHYb; AB114654, CitZEP; AB114651, CitCRTISO; AB781688, CitVDE; AB219164, CitCCD1; AB219169, CitNCED2; AB219174, CitNCED3; and AB781691, CitCCD4.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Changes in the carotenoid contents in juice sacs of Yamashitabeni-wase and Miyagawa-wase during the ripening process.
Supplemental Figure S2. Alignment of deduced amino acid sequences of CitCCD4 in Yamashitabeni-wase and Miyagawa-wase.
Supplemental Figure S3. Ethidium bromide gel showing PCR amplification of CitCCD4 from genomic DNA and cDNA of Yamashitabeni-wase and Miyagawa-wase.
Supplemental Figure S4. Alignment of deduced amino acid sequences of CitCCD4, CcCCD4a, and CcCCD4b.
Supplemental Table S1. Primer sequences used for the amplification of CitCCD4.
Supplemental Table S2. Primer sequences and TaqMan MGB probes used for quantitative RT-PCR of the genes related to carotenoid metabolism.
Acknowledgments
We thank Prof. Norihiko Misawa (Research Institute for Bioresources and Biotechnology, Ishikawa Prefectural University) for providing the pACCRT-EIB plasmid. We also thank Prof. Tsuyoshi Nakagawa (Center for Integrated Research in Science, Shimane University) for the gift of pUGW5 plasmid.
Glossary
- CCD
carotenoid cleavage dioxygenase
- LED
light-emitting diode
- cDNA
complementary DNA
- RT
reverse transcription
References
- Agócs A, Nagy V, Szabó Z, Márk L, Ohmacht R, Deli J. (2007) Comparative study on the carotenoid composition of the peel and the pulp of different citrus species. Innov Food Sci Emerg Technol 8: 390–394 [Google Scholar]
- Agustí J, Zapater M, Iglesias DJ, Cercós M, Tadeo FR, Talón M. (2007) Differential expression of putative 9-cis-epoxycarotenoid dioxygenases and abscisic acid accumulation in water stressed vegetative and reproductive tissues of citrus. Plant Sci 172: 85–94 [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Backhaus RA, Camara B. (1998) Induction and control of chromoplast-specific carotenoid genes by oxidative stress. J Biol Chem 273: 30651–30659 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Suire C, Mutterer J, Camara B. (2003) Oxidative remodeling of chromoplast carotenoids: identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in Crocus secondary metabolite biogenesis. Plant Cell 15: 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R, Ducreux LJ, Morris WL, Morris JA, Suttle JC, Ramsay G, Bryan GJ, Hedley PE, Taylor MA. (2010) The metabolic and developmental roles of Carotenoid Cleavage Dioxygenase4 from potato. Plant Physiol 154: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernys JT, Zeevaart JAD. (2000) Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol 124: 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cual AL. (1965) The occurrence of beta-citraurin and of beta-apo-8′-carotenal in the peels of California tangerines and oranges. J Food Sci 30: 13–18 [Google Scholar]
- Cunningham FX, Gantt E. (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 557–583 [DOI] [PubMed] [Google Scholar]
- Cutler AJ, Krochko JE. (1999) Formation and breakdown of ABA. Trends Plant Sci 4: 472–478 [DOI] [PubMed] [Google Scholar]
- Farin D, Ikan R, Gross J. (1983) The carotenoid pigments in the juice and flavedo of a mandarin hybrid (Citrus reticulata) vs Michal during ripening. Phytochemistry 22: 403–408 [Google Scholar]
- Giuliano G, Al-Babili S, von Lintig J. (2003) Carotenoid oxygenases: cleave it or leave it. Trends Plant Sci 8: 145–149 [DOI] [PubMed] [Google Scholar]
- Havaux M. (1998) Carotenoids as membrane stabilizers in chloroplasts. Trends Plant Sci 3: 147–151 [Google Scholar]
- Hofmann K, Stoffel W. (1993) TMbase-A database of membrane spanning protein segments. Biol Chem Hoppe Seyler 374: 166 [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FC, Molnár P, Schwab W. (2009) Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J Exp Bot 60: 3011–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikoma Y, Yano M, Ogawa K, Yoshioka T, Xu ZC, Hisada S, Omura M, Moriguchi T. (1996) Isolation and evaluation of RNA from polysaccharide-rich tissues in fruit for quality by cDNA library construction and RT-PCR. J Jpn Soc Hortic Sci 64: 809–814 [Google Scholar]
- Isaacson T, Ronen G, Zamir D, Hirschberg J. (2002) Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell 14: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M. (2012) Mechanism of carotenoid accumulation in citrus fruit. J Jpn Soc Hortic Sci 81: 219–233 [Google Scholar]
- Kato M, Ikoma Y, Matsumoto H, Sugiura M, Hyodo H, Yano M. (2004) Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol 134: 824–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Matsumoto H, Ikoma Y, Okuda H, Yano M. (2006) The role of carotenoid cleavage dioxygenases in the regulation of carotenoid profiles during maturation in citrus fruit. J Exp Bot 57: 2153–2164 [DOI] [PubMed] [Google Scholar]
- Kita M, Kato M, Ban Y, Honda C, Yaegaki H, Ikoma Y, Moriguchi T. (2007) Carotenoid accumulation in Japanese apricot (Prunus mume Siebold & Zucc.): molecular analysis of carotenogenic gene expression and ethylene regulation. J Agric Food Chem 55: 3414–3420 [DOI] [PubMed] [Google Scholar]
- Kloer DP, Schulz GE. (2006) Structural and biological aspects of carotenoid cleavage. Cell Mol Life Sci 63: 2291–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinsky NI, Landrum JT, Bone RA. (2003) Biologic mechanisms of the protective role of lutein and zeaxanthin in the eye. Annu Rev Nutr 23: 171–201 [DOI] [PubMed] [Google Scholar]
- Ledford HK, Niyogi KK. (2005) Singlet oxygen and photo-oxidative stress management in plants and algae. Plant Cell Environ 28: 1037–1045 [Google Scholar]
- Ma G, Zhang L, Kato M, Yamawaki K, Kiriiwa Y, Yahata M, Ikoma Y, Matsumoto H. (2012) Effect of blue and red LED light irradiation on β-cryptoxanthin accumulation in the flavedo of citrus fruits. J Agric Food Chem 60: 197–201 [DOI] [PubMed] [Google Scholar]
- Mein JR, Dolnikowski GG, Ernst H, Russell RM, Wang XD. (2011) Enzymatic formation of apo-carotenoids from the xanthophyll carotenoids lutein, zeaxanthin and β-cryptoxanthin by ferret carotene-9′,10′-monooxygenase. Arch Biochem Biophys 506: 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa N, Shimada H. (1997) Metabolic engineering for the production of carotenoids in non-carotenogenic bacteria and yeasts. J Biotechnol 59: 169–181 [DOI] [PubMed] [Google Scholar]
- Oberholster R, Cowan AK, Molnár P, Tóth G. (2001) Biochemical basis of color as an aesthetic quality in Citrus sinensis. J Agric Food Chem 49: 303–307 [DOI] [PubMed] [Google Scholar]
- Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K. (2006) Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol 142: 1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ. (2002) Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos G, Naranjo MA, Rodrigo MJ, Alós E, Zacarías L, Cercós M, Talón M. (2010) Identification of a GCC transcription factor responding to fruit colour change events in citrus through the transcriptomic analyses of two mutants. BMC Plant Biol 10: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo MJ, Marcos JF, Zacarías L. (2004) Biochemical and molecular analysis of carotenoid biosynthesis in flavedo of orange (Citrus sinensis L.) during fruit development and maturation. J Agric Food Chem 52: 6724–6731 [DOI] [PubMed] [Google Scholar]
- Rodrigo MJ, Zacarías L. (2007) Effect of postharvest ethylene treatment on carotenoid accumulation and the expression of carotenoid biosynthetic genes in the flavedo of orange (Citrus sinensis L. Osbeck) fruit. Postharvest Biol Technol 43: 14–22 [Google Scholar]
- Rubio A, Rambla JL, Santaella M, Gómez MD, Orzaez D, Granell A, Gómez-Gómez L. (2008) Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativus are both involved in beta-ionone release. J Biol Chem 283: 24816–24825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryle MJ, Hausinger RP. (2002) Non-heme iron oxygenases. Curr Opin Chem Biol 6: 193–201 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR. (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JAD, McCarty DR. (1997) Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA 94: 12235–12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MH, Floss DS, Strack D. (2010) Apocarotenoids: hormones, mycorrhizal metabolites and aroma volatiles. Planta 232: 1–17 [DOI] [PubMed] [Google Scholar]
- Xu J, Liu BZ, Liu X, Gao HJ, Deng XX. (2011) Carotenoids synthesized in citrus callus of different genotypes. Acta Physiol Plant 33: 745–753 [Google Scholar]
- Ytterberg AJ, Peltier JB, van Wijk KJ. (2006) Protein profiling of plastoglobules in chloroplasts and chromoplasts: a surprising site for differential accumulation of metabolic enzymes. Plant Physiol 140: 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ma G, Kato M, Yamawaki K, Takagi T, Kiriiwa Y, Ikoma Y, Matsumoto H, Yoshioka T, Nesumi H. (2012a) Regulation of carotenoid accumulation and the expression of carotenoid metabolic genes in citrus juice sacs in vitro. J Exp Bot 63: 871–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ma G, Shirai Y, Kato M, Yamawaki K, Ikoma Y, Matsumoto H. (2012b) Expression and functional analysis of two lycopene β-cyclases from citrus fruits. Planta 236: 1315–1325 [DOI] [PubMed] [Google Scholar]