Arabidopsis tetraspanins show conserved cellular and molecular features with functional relevance in the context of intercellular interactions, with specificity that might be determined by the composition of tetraspanin complexes and their binding partners at the cell surface of specific cell types.
Abstract
Tetraspanins are evolutionary conserved transmembrane proteins present in all multicellular organisms. In animals, they are known to act as central organizers of membrane complexes and thought to facilitate diverse biological processes, such as cell proliferation, movement, adhesion, and fusion. The genome of Arabidopsis (Arabidopsis thaliana) encodes 17 members of the tetraspanin family; however, little is known about their functions in plant development. Here, we analyzed their phylogeny, protein topology, and domain structure and surveyed their expression and localization patterns in reproductive tissues. We show that, despite their low sequence identity with metazoan tetraspanins, plant tetraspanins display the typical structural topology and most signature features of tetraspanins in other multicellular organisms. Arabidopsis tetraspanins are expressed in diverse tissue domains or cell types in reproductive tissues, and some accumulate at the highest levels in response to pollination in the transmitting tract and stigma, male and female gametophytes and gametes. Arabidopsis tetraspanins are preferentially targeted to the plasma membrane, and they variously associate with specialized membrane domains, in a polarized fashion, to intercellular contacts or plasmodesmata. A membrane-based yeast (Saccharomyces cerevisiae) two-hybrid system established that tetraspanins can physically interact, forming homo- and heterodimer complexes. These results, together with a likely genetic redundancy, suggest that, similar to their metazoan counterparts, plant tetraspanins might be involved in facilitating intercellular communication, whose functions might be determined by the composition of tetraspanin complexes and their binding partners at the cell surface of specific cell types.
In all multicellular organisms, cell-to-cell communication is fundamental in diverse biological events such as morphogenesis, differentiation, development, and reproduction. In plants, the reproductive process is highly dependent on a successful communication between the male gametophyte (pollen grain and pollen tube) and different female tissues or cells. However, we still lack information of potential mediators of these intercellular signaling interactions. Upon pollination, a primary species-specific recognition event occurs between a quiescent pollen grain and the receptive surface of the female stigmatic papilla, initiating a series of cellular events by which a pollen grain fully hydrates and regains metabolic activity. The pollen grain then germinates, producing a tip-growing cytoplasmic extension, the pollen tube, which grows within the female reproductive tract, transporting both nonmotile male gametes (sperm cells; for review, see Boavida et al., 2005). During this journey, pollen tubes acquire the competence to perceive localized female external cues that will be decoded and converted into changes of growth and direction (Palanivelu and Preuss, 2006), guiding each pollen tube to an available and unfertilized ovule. Finally in the embryo sac, the pollen tube enters a receptive synergid and bursts (Huck et al., 2003), delivering the two sperm cells into the close vicinity of the female gametes (Hamamura et al., 2011). Each sperm cell fuses with the egg or the central cell (double fertilization), initiating a new developmental program leading to the formation of a diploid embryo and the triploid nourishing endosperm, respectively.
The transition to a multicellular existence involved major genomic innovations that led to the emergence of novel signal and adhesion proteins fundamental for the organization of stable cellular interactions. Although multicellularity evolved independently and at multiple times in different lineages, genomic comparisons showed that basic elements conserved in all major multicellular lineages were already present in their unicellular ancestors (Rokas, 2008). From a functional perspective, some of these key elements could have been part of a primitive signal mechanism that was coopted to meet the need for effective intercellular communication in multicellular organisms. There are several examples of common signal elements functioning in animals and plants. For instance, important key players in cell-cell communication in animal central nervous systems, such as nitric oxide, d-Ser, γ-amino butyric acid, and Glu-like receptors (Traynelis et al., 2010), are known to perform crucial functions in plant reproduction by controlling signal events required for pollen tube growth and guidance (Palanivelu et al., 2003; Prado et al., 2004; Michard et al., 2011).
Using bioinformatics and a survey of reproductive tissue-related microarray data sets, we identified several members of the tetraspanin gene family for which the transcriptional profiles in reproductive cells (gametophytes and gametes; Honys and Twell, 2004; Pina et al., 2005; Borges et al., 2008; Wuest et al., 2010) or regulation upon pollination (Qin et al., 2009; Boavida et al., 2011) were consistent with a function in the reproductive process. Tetraspanins are a well-known evolutionary conserved gene family present in all major eukaryotic lineages and were associated with the appearance of multicellularity (Huang et al., 2005). As eukaryotes diverged, tetraspanins may have been lost or become divergent in the distinct lineages, as they are present in protozoan amoeba, fungi, and plants, but are absent from the unicellular chlorophyte algae Chlamydomonas reinhardtii and Volvox carteri. In metazoans, tetraspanins act as components or mediators of different biological processes such as cell adhesion, proliferation, migration, and fusion (Rubinstein, 2011). Some well-known examples include CD9, an oocyte-expressed tetraspanin essential in oocyte-sperm fusion in mice (Le Naour et al., 2000; Jégou et al., 2011) and the fungal tetraspanin PSL1, required for appressorial binding and penetration during infection of host plants (Veneault-Fourrey et al., 2006). Tetraspanins are known to act as membrane organizers, interacting with other tetraspanins and recruiting binding partners, such as key adhesion molecules and signaling receptors, which assemble in macromolecular membrane microdomains termed tetraspanin-enriched microdomain complexes or “tetraspanin webs” (Hemler, 2005). Despite their established importance in diverse biological processes and organisms, their function in plants is still poorly understood. Evidence for a possible involvement of plant tetraspanins in intercellular signaling comes from genetic interactions of mutants defective for TETRASPANIN1/TORNADO2 (TET1/TRN2/EKEKO) with mutants for TRN1, encoding a leucine-rich repeat protein and affecting leaf and root patterning (Cnops et al., 2000, 2006; Olmos et al., 2003), and with WINDHOSE1 (WIH1) and WIH2 in female gametophyte development (Lieber et al., 2011). tet1/trn2/ekeko mutants exhibit severe pleiotropic developmental defects affecting cell positioning/differentiation and overproliferation, phenotypes with similarities to the known cellular functions of animal tetraspanins (Rubinstein, 2011).
To better understand if plant tetraspanins maintained related functions in intercellular signaling, we combined phylogenetic analysis and predictions of protein structure with a detailed description of their expression patterns in reproductive tissues, as well as yeast (Saccharomyces cerevisiae) two-hybrid assays and phenotypic analysis of Arabidopsis (Arabidopsis thaliana) mutants. Arabidopsis tetraspanins show a distinct and often overlapping expression at the cell surface of specific reproductive cell types and tissue domains, and this expression is responsive to pollination. Comparable to tetraspanins in metazoans, Arabidopsis tetraspanins can assemble in homo- and heterodimers when expressed in yeast. Our results indicate that despite a large evolutionary distance, plant tetraspanins conserved cellular and molecular features that are functionally relevant in the context of intercellular interactions, providing a basis for the functional analysis of tetraspanins in diverse plant development processes, such as the reproductive process.
RESULTS AND DISCUSSION
Gene Structure and Phylogeny
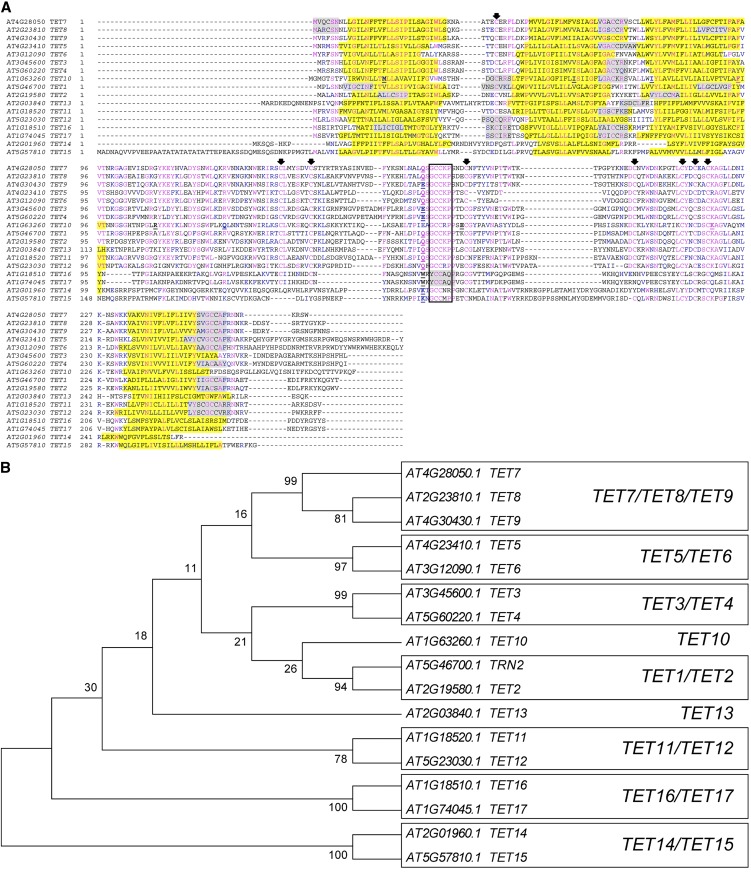
The Arabidopsis genome has 17 tetraspanin (TET) members, identified here as TET1 to TET17. The gene structure of most Arabidopsis TETs is characterized by a single intron (approximately 1 kb), with a conserved intron/exon junction (Fig. 1A; Supplemental Data File S1). TET2, TET5, and TET6 contain an additional short intron in either the 5′ or 3′ of the coding sequence (Supplemental Data File S1), while TET10 has 10 introns. Plant and animal tetraspanins appear to share a common ancestor that predates the divergence of the Opisthokont lineage, as the conserved intron position of plant tetraspanins appears to be the most ancient intron present in animal tetraspanins (Garcia-España and DeSalle, 2009). Despite their common ancestry, plant tetraspanins are quite dissimilar at the amino acid level from their metazoan counterparts, making it impossible to identify direct orthologs. In addition, they lack several highly conserved residues in the Transmembrane Domain2 (TM2)-Intracellular Loop (ICL)-TM3 of the so-called “tetraspanin signature” defined in metazoan tetraspanins (Seigneuret et al., 2001; Kovalenko et al., 2005). To evaluate the similarity among Arabidopsis tetraspanins, the predicted amino acid sequences of the 17 TETs were aligned using ClustalW (Fig. 1A). Pairwise comparisons showed that Arabidopsis TETs share a relatively low amino acid identity, on average 30% identity and 51% similarity (Supplemental Table S1), comparable to values usually found for tetraspanins within a species in other organisms (Seigneuret et al., 2001). An unrooted tree constructed with the neighbor-joining method resolved Arabidopsis tetraspanins into nine paralogous clades, six of which comprise member pairs; one contains three members, while the other two represent singletons (Fig. 1B). The tree structure is supported by the higher values of amino acid identity/similarity of members within the same clade (Supplemental Table S1). We then retrieved putative ortholog sequences from genomes in Phytozome (http://www.phytozome.net/) to evaluate the conservation in domain structure of plant tetraspanins with other metazoan tetraspanins and their evolutionary and functional diversification. We selected 122 amino acid sequences belonging to 13 plant species representative of each divergent evolutionary branch and generated a neighbor-joining phylogenetic tree rooted to Dictyostelium discoideum (social amoeba) as an outgroup (Supplemental Fig. S1). We excluded incomplete fragments and sequences that did not conform to the major tetraspanin structural features (Fig. 2). A previous phylogenetic analysis (Wang et al., 2012) using 115 genes representing 11 plant species grouped plant tetraspanins into seven functional groups. Despite the low bootstrap support for basal branches in both trees, the overall tree structure is consistent with the plant phylogenetic affinities, although the topology and organization of orthologous clades is different. For example, our analyses support TET13, TET14-TET15, and TET16-TET17 as distinct orthologous clades, instead of a single clade (Wang et al., 2012). This discrepancy likely resulted from the methodology or sequence alignment rather than from the number of sequences or species used for tree construction, as the same type of group associations were obtained when establishing paralogous genes in Arabidopsis (Fig. 1B). The phylogenetic analysis points to a common ancestor for orthologs within the same clade, suggesting that diversification of plant tetraspanins occurred before the monocot/dicot split. However, the TET1 and TET2 paralogous members appear to have diversified independently, which is consistent with their different gene structures (Supplemental Data File S1). In fact, TET1 is the only Arabidopsis tetraspanin member to date with an identified developmental phenotype (Cnops et al., 2000, 2006; Olmos et al., 2003; Lieber et al., 2011), supporting their functional divergence. Only two orthologous clades include representatives in the lycophyte Selaginella moellendorffii (TET13 and TET10) and the bryophyte Physcomitrella patens (TET10), suggesting that these clades could be the most ancient. In agreement with this hypothesis, Selaginella and Physcomitrella representative sequences within the TET10 and TET13 clades share the same gene structure with their direct orthologs in monocot and dicot species (10 introns and 1 intron, respectively). Other clades are represented predominantly by dicot sequences (TET13, TET11-TET12, TET14-TET15, and TET16-TET17). While this could be due to incompleteness of available sequences not included in the alignment, to gene copies that were not yet identified in thoroughly sampled genomes of dicot and monocot species, or to gene loss in other plant lineages, it is also possible that they arose from independent gene duplications within dicot branches. Taken together, the phylogenetic relationships are consistent with plant evolutionary trends, showing that plant tetraspanins share a common origin, pointing to TET13 and TET10 as founding members. Independent expansion by gene duplications or reduction by gene loss in some plant lineages likely resulted in the functional diversification of plant tetraspanins with the appearance of dicot-specific clades. Thus, paralogous genes are expected to share similar or complementary functions and, if coexpressed, to have redundant biological functions.
Figure 1.
Phylogenetic analysis of Arabidopsis tetraspanins. A, Amino acid sequence alignment of Arabidopsis tetraspanins. Identical residues are highlighted in blue and conserved residues in magenta. The transmembrane domains are shaded in yellow and predicted palmitoylation sites in gray. Intron positions are indicated by an underlined bold letter. Conserved Cys residues are indicated by arrows. Unfilled black box indicates the GCCK/RP motif conserved among all Viridiplantae tetraspanins. B, Unrooted neighbor-joining tree of Arabidopsis tetraspanins. The evolutionary distances were computed using the Dayhoff matrix-based method using MEGA5. The bootstrap consensus tree was inferred from 1,000 replicates. Numbers above branches indicate bootstrap percentage values for a particular branch. Boxes represent clades supported by bootstrap values above 50%. [See online article for color version of this figure.]
Figure 2.
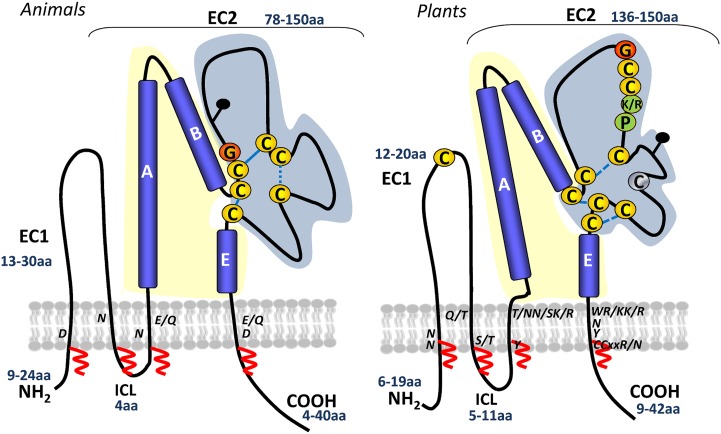
Schematic representation of animal and plant tetraspanin topologies. A generic topology for animal tetraspanins is shown on the left (adapted from Hemler, 2005). A predicted plant tetraspanin topology is shown on the right, inferred from protein structure predictions and by comparison with the known topology of animal tetraspanins. Numbers in blue indicate the range of amino acids in the small extracellular loop (EC1), large extracellular domain (EC2), ICL, and C-terminal tail. Yellow and blue shading represent the variable and conserved domains of EC2, respectively. Conserved cysteines and the plant GCCK/RP motif in EC2 are marked. Cysteines in yellow are 100% conserved, and those in gray are 90% conserved. Conserved polar residues in the TM are indicated. Predicted disulfide bridges are shown with dashed blue lines. Potential palmitoylation sites in the transmembrane domains are indicated with red zigzag lines. Black pins indicate predicted glycosylation sites. [See online article for color version of this figure.]
Protein Structure
The functional versatility of metazoan tetraspanins results from a fold-forming structural core domain maintained by a few highly conserved residues that are interspersed with freely variable regions thought to allow substantial diversification in the specificity of interacting proteins (Stipp et al., 2003). We performed a detailed prediction analysis of tetraspanin protein sequences to disclose conserved regions or motifs that might be critical for protein function or structure. The sequence alignment shows that plant tetraspanins preserved the core architecture of metazoan tetraspanins, defined by N- and C-terminal short cytoplasmic tails and four TMs, delineating three main domains with relatively well-conserved sizes, the ICL, EC1 (small extracellular domain), and EC2 (large extracellular domain; Fig. 2). Secondary structure predictions indicate that, similar to metazoan tetraspanins, the EC2 domain of Arabidopsis tetraspanins is organized in two distinct subdomains, one with a conserved topology consisting of helixes A, B, and E and a second variable subdomain, where short sequence stretches are interspersed by conserved cysteines located at defined distances from the TM domains (Figs. 1A and 2). In metazoans, the EC2 variable region is known to be functionally important, mediating interactions with specific binding partners, while the more conserved region is involved in homodimerization (Yáñez-Mó et al., 2001). In plants, the EC2 variable region contains a conserved plant-specific “GCCK/RP” motif, which differs in position and sequence from the “CCG” motif present in animal tetraspanins. In the unicellular amoeba D. discoideum, TspanE contains the typical “CCG” motif found in metazoan tetraspanins, while the other four Tspans show a modified “CCK/Y/C” motif, which is more similar to some variants present in the land plant lineage. The plant “GCCK/RP” motif is also present in the moss P. patens and in the lycophyte S. moellendorffii TETs, suggesting that this motif arose early in the land plant lineage, either representing an innovation (DeSalle et al., 2010) or resulting from modification or reorganization of an ancestral motif before the divergence of land plants. TET16/TET17, instead of the “GCCK/RP” motif, has a variant “YCCAQ,” while TET14/TET15 has “GCCM/NR/P.” Plant tetraspanins contain a conserved Cys residue in EC1 and up to nine cysteines in EC2, instead of the typical four to six cysteines of metazoan tetraspanins. The cysteines in EC2 are predicted to be involved in disulfide binding, contributing to maintain the typical structural folding of tetraspanins (Fig. 2). However, TET14 and TET15 lack the conserved Cys in EC1 and two or three cysteines in EC2. As in animal TETs, the cysteines juxtaposed to TMs in plant TETs are predicted to be palmitoylated (Figs. 1A and 2), and potential N-glycosylation sites were found in the EC2 variable region of TET1 to TET4, TET8, TET10, TET13, and TET14 but not in TET5 to TET7, TET9, TET11, TET12, and TET15 to TET17; only TET3 and TET4 have a predicted glycosylation site in EC1. In metazoans, such posttranslational modifications are necessary for tetraspanin-tetraspanin interactions (Charrin et al., 2002) or tetraspanin interactions with other binding partners (Baldwin et al., 2008); disruption of these modifications also affected subcellular localization or proper tetraspanin folding and function (Yang et al., 2002). Despite the differences in amino acid sequence, the nature of most residues indicated as important by structural and mutational analysis of the mammalian proteins (Stipp et al., 2003; Kovalenko et al., 2005) were also functionally important in plants (Cnops et al., 2006; Figs. 1A and 2). Specifically, these include some conserved polar residues in the TMs (Fig. 2) and charged residues in the C-terminal cytoplasmic tail (Fig. 1A), as well as conserved and distinctive residues/motifs, such as the Cys residues in EC2 and adjacent to the TMs, and the Gly residue of the plant-specific “GCCK/RP” motif. Moreover, several conserved hydrophobic residues, namely the Gly residues in TM1 and TM2, highly conserved in Dictyostelium spp. and metazoan tetraspanins, are also found in plant tetraspanins. Thus, amino acid conservation in these regions might be important for protein function.
Despite the significant divergence in primary sequences with metazoan tetraspanins, plant TETs conserved structural features by preserving motifs and residues crucial for TET homo- and heterodimerization and for interactions with specific binding partners.
Arabidopsis Tetraspanins Preferentially Localize at the Plasma Membrane
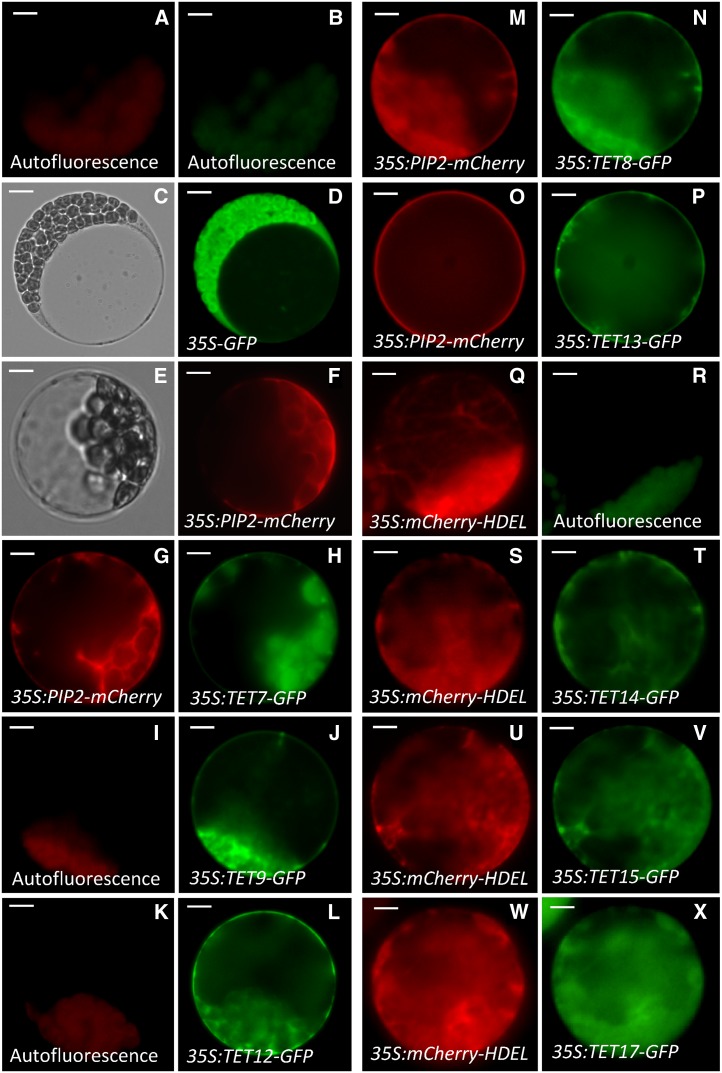
The subcellular targeting predictions for Arabidopsis tetraspanins suggested plasma membrane localization, with the exception of TET15, for which a stretch of several amino acids generated an unusual and longer cytoplasmic N terminus. The subcellular localization prediction for TET15 was uncertain, with higher scores for mitochondria. In mammals, tetraspanins are generally found associated with the plasma membrane, though in some cases they accumulate in intracellular compartments, such as late endosomes and lysosomes, and in specialized vesicles as exosomes (Pols and Klumperman, 2009). To investigate the subcellular localization of Arabidopsis tetraspanins, we generated GFP C-terminal protein fusions for some tetraspanins (TET7 to TET9, TET11, TET13 to TET15, and TET17) under the control of the constitutive Cauliflower mosaic virus 35S (CaMV-35S) promoter. TETs’ localization was confirmed by coexpression with fluorescent markers targeted to specific subcellular compartments (Fig. 3). TET7 to TET9, TET12, and TET13 appeared to be plasma membrane localized (Fig. 3, g–p). To confirm this localization, we cotransformed protoplasts with a known plasma membrane marker, the mCherry-tagged Plasma membrane Intrinsic Protein (PIP2; Nelson et al., 2007), confirming their colocalization at the plasma membrane (Fig. 3, g and h and m–p). However, in TET14, TET15, and TET17, the subcellular localization was distinct from that of the remaining TETs and accumulated in cytoplasmic organelles. To identify their precise localization, we cotransformed protoplasts of TET14-, TET15-, and TET17-GFP stable lines with an endoplasmic reticulum (ER)-tagged mCherry fusion (HDEL, ER retention signal; Nelson et al., 2007), confirming their potential association with the ER (Fig. 3, Q–X).
Figure 3.
Representative images of subcellular localization of selected Arabidopsis tetraspanins using GFP translational fusions driven by a constitutive promoter (CaMV-35S). Chloroplast background autofluorescence in the GFP (green) and RFP (red) emission channel (a and b) in nontransformed mesophyll protoplasts. Representative DIC and fluorescent images of mesophyll protoplasts transformed with empty destination vector (35S-GFP) and an intrinsic plasma membrane protein (35S:PIP2-mCherry) as controls for cytoplasmic (c and d) and membrane (e and f) localization, respectively. Chloroplast autofluorescence (i and k) and GFP fluorescence of protoplasts transformed with 35S:TET9-GFP (j) and 35S:TET12-GFP (l), respectively. Cotransformation of 35S:PIP2-mCherry with 35S:TET7-GFP (g and h), 35S:TET8-GFP (m and n), and 35S:TET13-GFP (o and p), respectively. ER-tagged mCherry (35S:mCherry-HDEL) control for ER localization (q) and chloroplast autofluorescence (r). Cotransformation of 35S:mCherry-HDEL with 35S:TET14-GFP (s and t), 35S:TET15-GFP u and v), and 35S:TET17-GFP (w and x), respectively. Bars = 5 μm (a, b, e–x), 10 μm (c and d). [See online article for color version of this figure.]
In mammals, the localization of tetraspanins is often associated with specific plasma membrane subdomains, namely with intercellular contacts, such as gap junctions or immune synapses. These membrane subdomains are thought to represent tetraspanin-enriched microdomains that result from TET associations with cell type-specific binding partners (Yáñez-Mó et al., 2001). In plants, plasmodesmata are considered to have analogous functions to gap junctions, and this type of localization in membrane microdomains seems plausible, as TET3 was recently isolated as a plasmodesmata-associated protein and its localization confirmed in mesophyll cells (Fernandez-Calvino et al., 2011).
Our results support that plant tetraspanins preferentially associate with the plasma membrane or ER when overexpressed in vivo. However, their localization in membrane microdomains might depend on interactions with cell type-specific binding partners.
Arabidopsis Tetraspanins Show Unique But Overlapping Expression Patterns in Reproductive Cells or Tissue Domains
Our phylogenetic analysis (Fig. 1B) supported distinct tetraspanin functional groups, and these might reflect specific tissue expression patterns. Transcriptomic data from vegetative and reproductive tissues/cells also supported this hypothesis (Supplemental Fig. S2). In addition, the enriched expression of some members in reproductive tissues/cells and their developmental regulation upon pollination suggested potential functions in the reproductive process. For instance, TET7, TET8, TET11 to TET13, and TET15 were expressed in pollen (Honys and Twell, 2004; Pina et al., 2005), TET7, TET8, TET11, and TET12 in sperm cells (Borges et al., 2008), and TET7 to TET9 in female gametophytic cells (Wuest et al., 2010). In addition, some members were up-regulated during in vitro (TET7, TET8, TET11, TET13, and TET16; Wang et al., 2008), semi in vivo (TET4, TET7, TET8, TET11 to TET13, and TET16; Qin et al., 2009), and during in vivo (TET3, TET12, and TET13; Boavida et al., 2011) pollen tube growth (Supplemental Fig. S2).
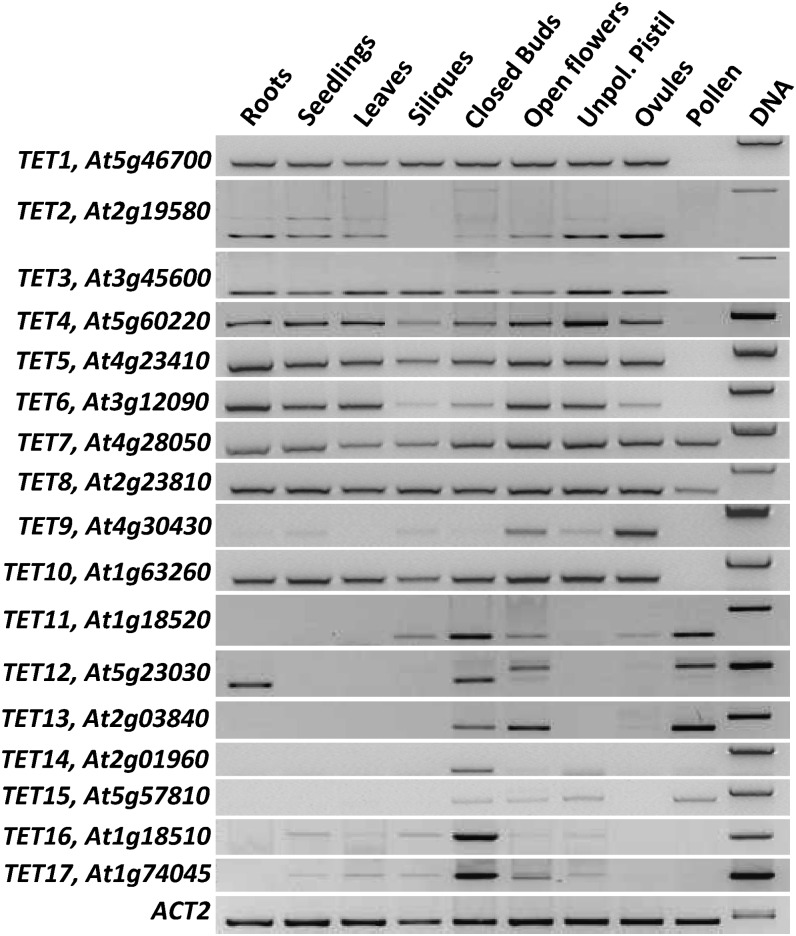
To evaluate the extent of TETs’ expression in vegetative and reproductive tissues, we first analyzed their steady-state transcript levels by reverse transcription (RT)-PCR (Fig. 4). While for some tetraspanins (TET1–TET8 and TET10), the expression was almost ubiquitous, other members (TET9 and TET11–TET17) had more limited patterns of expression (Fig. 4), suggesting some type of cell or tissue specificity. For two specific tetraspanins (TET2 and TET12), we detected two transcripts of distinct sizes and abundance. In both cases, the smaller transcripts corresponded to the annotated and correctly spliced sense transcript, while the longer transcripts could have resulted from intron retention. In fact, the TET2 larger transcript conforms to an annotated cis-natural antisense transcript (cis-nat; At2g19582), whose expression was restricted to vegetative tissues and absent from ovules and open flowers (Fig. 4). We confirmed by strand-specific RT-PCR in pollen that the TET12 longer transcript also represents a cis-nat transcript (Supplemental Fig. S3) exclusively expressed in pollen and open flowers (Fig. 4). Though the function of antisense transcripts is still poorly understood, the presence of antisense transcripts is usually associated with regulatory functions, as recently shown for a cis-nat functionally relevant in the context of male gametes and in the success of double fertilization (Ron et al., 2010).
Figure 4.
RT-PCR expression analysis of Arabidopsis tetraspanins. Locus name and gene identifier numbers are shown on the left. ACT2 was used as internal control.
To gain further insights into their expression patterns and possible tissue/cell specificity, we generated C-terminal NLS3xeGFP (for localization signal with 3 copies of eGFP) transcriptional and GFP translational fusions for each tetraspanin, under the control of their endogenous promoters. Given TETs predominant expression in reproductive tissues, we focused our expression analysis during the reproductive process (before/after pollination and fertilization), surveying potential changes in the expression patterns that could be pollination dependent, as the transcriptomic data suggested (Supplemental Fig. S2). When relevant, pollen expression was monitored in early stages of development or after in vitro or semi-in vivo pollen germination.
TETs expression was associated with particular reproductive cell types or tissue domains that could partially overlap or be distinctive among the TET members (Figs. 5–7). In general, the GFP expression patterns were consistent for both transcriptional and translational fusions of a given tetraspanin member. However, for TET7 and TET8, the translational activity of the endogenous promoter was slightly different from the transcriptional fusions (see below). We failed to recover transgenic plants for the translational fusion of TET2. For clarity in the following text, transcriptional fusions will be referred to in italic case and translational fusions in roman case.
Figure 5.
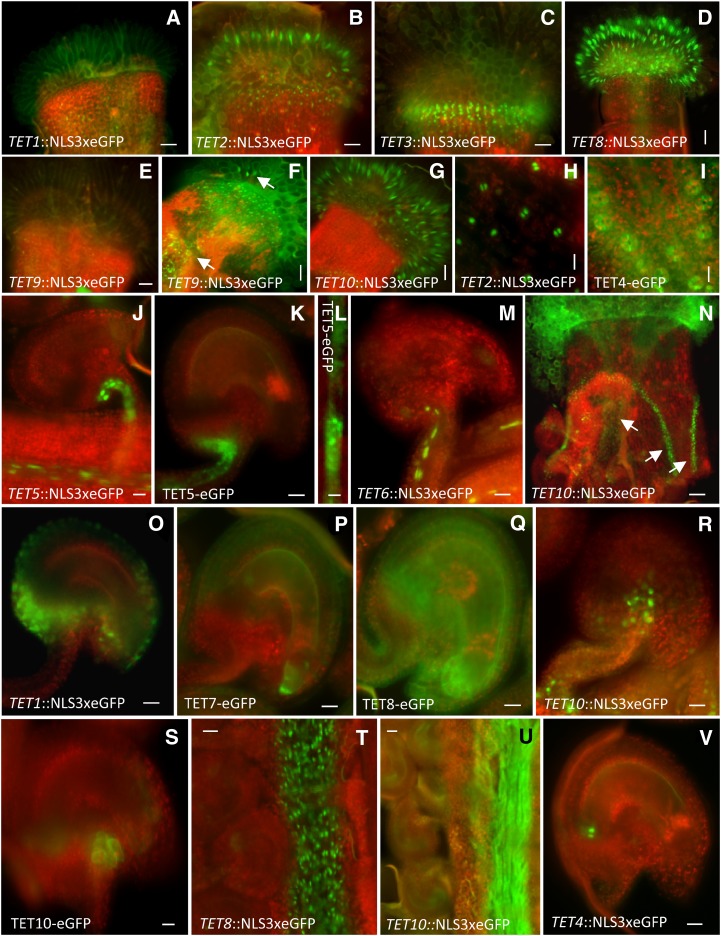
Representative images of Arabidopsis tetraspanin expression patterns in diploid reproductive tissues. Transcriptional GFP fusions of various tetraspanins expressed in stigmatic carpel and papilla (a–g). TET9 expression in stigmatic papilla before (e) and after pollination (f). Arrows in (f) point to TET9 expression in transmitting tissue and papilla. Localization of TET2 transcriptional fusion (h) and expression of TET4 translational fusion (i) in carpel stomata. Localization of TET5 and TET6 in vascular tissues (j–m). Magnification of a vascular strand showing punctuated deposition (l). TET10 transcriptional fusion expressed in transmitting tissue (after pollination) and valve margins of the carpel (n). Transcriptional and translational fusions of various tetraspanins in ovule tissues after pollination (o–s). TET8 transcriptional fusion (t) and TET10 translational fusion (u) in the transmitting tract after pollination. Localization of transcriptional fusion of TET4 in ovules (v). Green fluorescent refers to GFP, and red fluorescent signal represents tissue autofluorescence. Bars = 50 μm (a–g, t, and u), 100 μm nN), 25 μm (j, k, m, o–s, and v), and 10 μm (h, i, and l).
Figure 7.
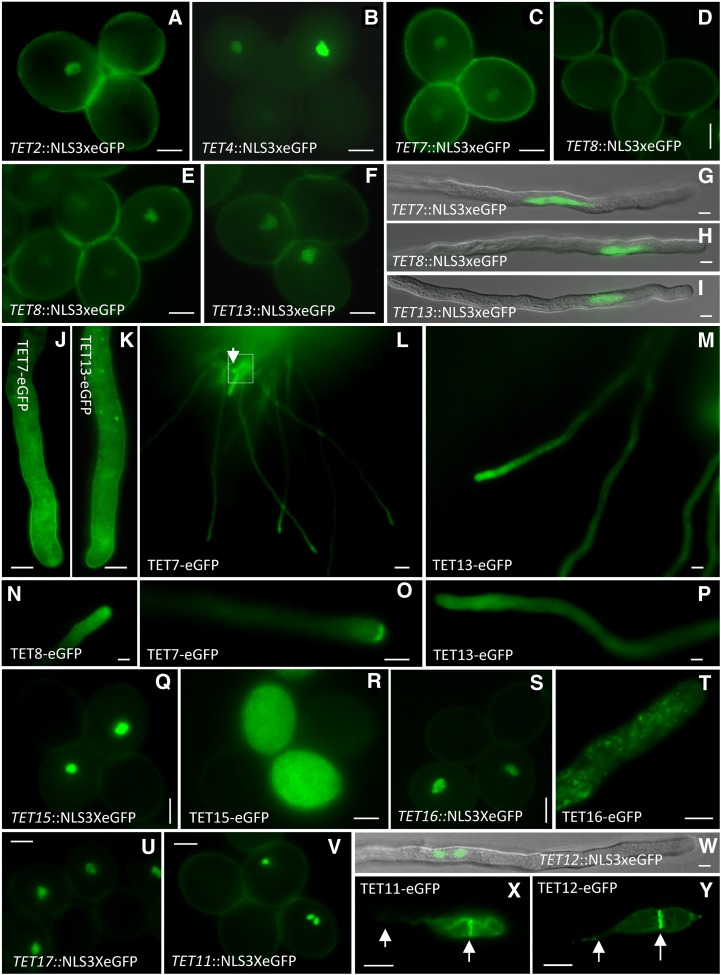
Representative images of Arabidopsis tetraspanins expression patterns in male gametophyte and gametes. Expression in the vegetative nucleus of transcriptional fusions of TET2 (a) and TET4 (b). TET7 transcriptional fusion expressed in the vegetative nucleus of mature pollen (c). TET8 transcriptional fusion in fresh dehiscent mature pollen (d) and activation after 4 h of hydration (e). TET13 expression in mature pollen (f). TET7, TET8, and TET13 transcriptional fusion expressed in the vegetative nucleus of in vitro-germinated pollen tubes (g–i). TET7 and TET13 expression in semi-in vitro-germinated pollen tubes showing plasma membrane localization and in cytoplasmic organelles (j and k). TET7, TET8, and TET13 semi-in vivo pollen germinations showing preferential accumulation at the apical dome (l–p). Arrow (l) points to pollen tube showing TET7 accumulation at the pollen tube tip (o). Transcriptional and translational fusions of TET15 (q and r) and TET16 (s) in mature pollen and germinated in vitro (t). TET17 transcriptional fusion in mature pollen (u). Transcriptional fusions of TET11 (mature pollen; v) and TET12 (pollen tube; w) in sperm cells. TET11 (x) and TET12 (y) expression in sperm cells within a pollen tube. Arrows point to the plasma membrane of the sperm cell tail and to the tetraspanin-enriched microdomain at the interface of the sperm cells. Bars = 10 μm (a–k, n–t, and u–w), 15 μm (m), 50 μm (l), and 5 μm (r and s).
Expression in Diploid Reproductive Tissues
The expression patterns of TETs in reproductive diploid tissues were diverse and point to a potential involvement in different cellular processes, from basic to more direct functions in the reproductive process. For instance, in the carpel, TET1 (Fig. 5a), TET2 (Fig. 5b), TET3 (Fig. 5c), and TET9 (Supplemental Fig. S4) were expressed at the base of the stigma. However, TET2 expression also extended to the receptive stigmatic papilla (Fig. 5B), where it is apparently coexpressed with other tetraspanins (TET8–TET10; Fig. 5, d–g). Interestingly, while TET8 (Fig. 5d) and TET10 (Fig. 5g) expression in stigmatic papilla was detected before pollination, TET9 expression was induced in response to pollination (Fig. 5, e and f; Supplemental Fig. S4).
TET2 and TET4 expression were detected in carpel guard cells (Fig. 5, h and i). The TET4 protein fusion localized at the plasma membrane and was visible at the interface of the guard cells (Fig. 5i), suggesting that coexpression of TET2/TET4 might define specific functions in these cells. TET5 and TET6 were expressed in vascular tissues in carpels and ovules (Fig. 5, j–m). The stronger expression of TET5 allowed detection of a distinctive punctate pattern at the membrane (Fig. 5l), which would be consistent with the known abundance of plasmodesmata in vascular companion cells (van Bel, 1996). This potential association of TET5 with plasmodesmata is not exclusive among TETs, as TET3 was identified as a plasmodesmata-associated protein (Fernandez-Calvino et al., 2011). Also, in carpel walls, TET10 had a characteristic expression in the valve margins flanking the central carpel replum (Fig. 5n). This characteristic expression pattern is known for several transcription factors involved in valve margin development and silique dehiscence, such as SHATTERPROOF1 (SHP1), SHP2, and INDEHISCENT (Liljegren et al., 2004). The tet1/trn2 mutant was reported to have defects in carpel fusion (Olmos et al., 2003), suggesting that TET1 and TET10 could be expressed in similar tissue domains in early stages of carpel development.
Several tetraspanins were expressed in ovule integuments. After pollination, TET1 was restricted to the ovule outer integument (Fig. 5o), while TET7 (Fig. 5p) and TET8 (Fig. 5q) were expressed in both integuments. However, the transcriptional fusions of TET7 and TET8 were weakly detected in ovules before pollination, though TET8 expression appeared to be induced by pollination (Supplemental Fig. S4). TET10 was detected in both integuments just before pollination (Supplemental Fig. S4). In early stages, TET1 is known to act in a signaling pathway controlled by WUSCHEL and involving WIH1 and WIH2 in the nucellus, promoting megaspore mother cell formation and megasporogenesis (Lieber et al., 2011). Thus, TET1 expression is developmentally regulated, acquiring different tissue localizations during ovule development. The similarity of expression patterns with other TETs members (TET7, TET8, and TET10) suggests that some TETs might share complementary or similar functions in specific ovule domains.
TETs’ expression also seem to be responsive to pollination in other tissues with more direct functions in the reproductive process, such as those associated with pollen tube growth or guidance. For example, before pollination, TET9 and TET10 have a wider expression in ovules (Supplemental Fig. S4) that becomes restricted to specific tissue domains or cells upon pollination (Figs. 5r and 6, d and e). After pollination, TET9 is detected in a cluster of cells in the upper part of the funiculus (Fig. 6, d and e) and in a few cells at the ovule micropyle flanking the egg apparatus in the embryo sac (Fig. 6e), while TET10 becomes restricted to 10 to 12 cells in the upper part and at the bottom of the funiculus (Fig. 5, r and s). The more restricted expression of TET9/TET10 in the upper part of the funiculus is consistent with a possible role in pollen tube guidance, as this cellular domain could represent a localized site for production of female signals promoting exit from the transmitting tissue and directional growth into the ovule micropyle (Hulskamp et al., 1995; Shimizu and Okada, 2000). Interestingly, TETs expression in the transmitting tract is also pollination dependent. Before pollination, TET8 is expressed in the transmitting tract (Fig. 5t), and this expression persists during pollen tube growth, while TET9 and TET10 expression was induced upon pollination (Fig. 5, f and u; Supplemental Fig. S4). Curiously, the TET8 translational fusion was weakly detected in transmitting tissue and stigma before pollination but became reasonably strong upon pollination (Supplemental Fig. S4).
Figure 6.
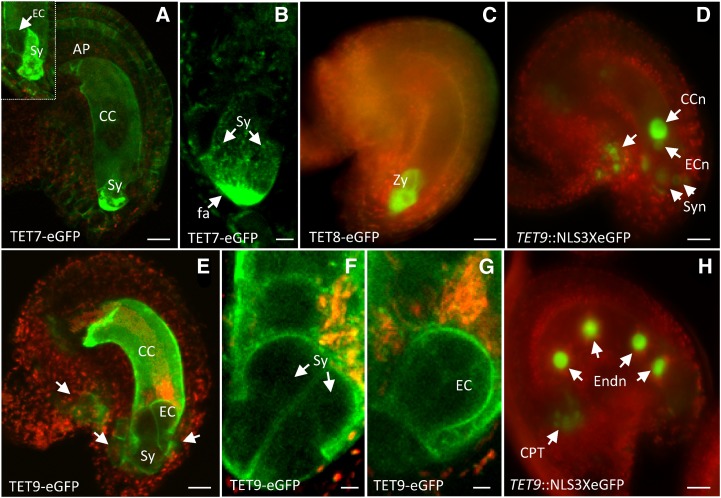
Representative images of Arabidopsis tetraspanin expression patterns in female gametophyte and gametes. Expression of TET7 translational fusion in the plasma membrane of central cell (CC), antipodals (AP), and accumulation in the filiform apparatus (fa) of synergids (Sy). Inset shows an optical section pointing to egg cell lacking TET7 expression (a). Optical section of filiform apparatus showing TET7 granules distributing in a gradient within synergids (b). Representative image of TET8 translational fusion in atypical zygotes (Zy; c). TET9 expression before fertilization (d), showing egg cell nucleus (ECn), central cell nucleus (CCn), and synergid nuclei (Syn); expression of TET9 translational fusion in ovule and female gametophytic cells. Arrows point to individual cells in the micropyle and flanking the egg cell apparatus (e). Detail of TET9 translational fusion in the membrane of both synergids (f) and egg cell (EC; g). TET9 transcriptional fusion after fertilization (h) showing expression in endosperm nuclei (Endn) and chalazal proliferating tissue (CPT; arrows). Green fluorescent signal refers to GFP, and red fluorescent signal represents tissue autofluorescence. Bars = 20 μm (a and c–f), 10 μm (b), and 5 μm (g and h).
TET4 was detected in a pair of cells facing the chalazal region of the ovule, outside the embryo sac and above the vascular tissue (Fig. 5v). This TET4-restricted localization broadened in fertilized ovules, similar to the expression observed in TET8 (Supplemental Fig. S4) and the weaker expression intensity in TET9 (Fig. 6h). This expression domain might define the chalazal proliferating tissue, a cluster of densely cytoplasmic cells that lies close to the site of unloading from the vascular tissue, thought to be related to the transfer of reserve materials, such as nutrients, enzymes, and hormones, to the embryo sac or endosperm (Debeaujon et al., 2003). After fertilization, TET7 and TET8 were not detected in ovules, while TET9 and TET10 were associated with seed integuments (Supplemental Fig. S4).
Expression in Female Gametophyte and Gametes
Transcriptomic data supported the expression of several TET members in gametophytes and gametes (Borges et al., 2008; Wuest et al., 2010). Although TET7, TET8, and TET9 transcripts were detected in female gametophytic cells (Wuest et al., 2010), we failed to detect TET7 and TET8 transcriptional activity in female gametophytic cells (Supplemental Fig. S4). The translational fusion of TET7 was detected at the plasma membrane of synergids, the central cell, and antipodals (Figs. 5e and 6a) but was apparently absent from the egg cell (Fig. 6a, inset). The differences observed between the TET7 and TET8 transcriptional and translational fusions might be explained by a weak promoter activity. Interestingly, TET7 showed a distinctive accumulation at the filiform apparatus, a specialized membrane domain of synergids. Despite the strong GFP signal, a more detailed observation of the filiform apparatus revealed that TET7 accumulated in intracellular organelles that appeared to distribute over a gradient within the synergids (Fig. 6, a and b). This distinctive and polarized TET7 accumulation in the filiform apparatus is similar to that first described for FERONIA, a receptor-like kinase involved in pollen tube reception (Escobar-Restrepo et al., 2007). It would be interesting to know whether the apparent coexpression of TET7/FERONIA reflects a functional molecular interaction in synergid cells. We failed to detect any TET8 transcriptional activity in unfertilized female gametophytic cells (Fig. 5p; Supplemental Fig. S4), despite screening several hundred ovules to survey embryo sacs before and after fertilization. Though of rare occurrence in wild-type pistils (1 d after pollination), some ovules can contain arrested zygotes. In TET8-GFP transgenic plants, we observed few ovules containing arrested zygotes with strong GFP accumulation in the cytoplasm (Fig. 6c). The atypical zygotes expressing GFP presented the distinctive inversion in polarity observed after fertilization (Faure et al., 2002) but were clearly arrested in development, as they failed to elongate and develop (Fig. 6c). No expression was observed in normal unfertilized or developing fertilized ovules. Contrasting with TET7 and TET8, TET9 transcriptional activity was strongly detected in the central cell, while it was relatively weaker in the egg cell, synergids, and antipodals (Fig. 6d). Consistently, TET9 was detected in the plasma membrane of egg and central cells (Fig. 6, e and g), with a slightly weaker expression in synergid cells (Fig. 6, e and f). TET9 expression was uniformly distributed in the plasma membrane of synergids and did not show the polarization in the filiform apparatus seen for TET7. Upon fertilization, TET9 was first internalized and accumulated in the cytoplasm of the central cell and egg cell and eventually disappeared; however, the transcriptional fusion was still detected in the endosperm after several nuclear divisions (Fig. 6h; Supplemental Fig. S4). Our results suggest that three tetraspanins (TET7–TET9) are predominantly expressed in female gametophytic cells, with distinct levels of expression and cell type specificity; both TET7 and TET9 expression appeared to overlap in the central cell and synergids but not in synergids, while the stable expression of TET9 in the egg cell and the possibility of a transient induction of TET8 in the egg suggests that their potential coexpression might be functionally relevant just before or after fertilization.
Expression in Male Gametophyte and Gametes
In general, the expression of TET members in mature pollen (Fig. 7) or during pollen development (TET7, TET8, and TET13–TET17) were consistent with the transcriptomic data (Honys and Twell, 2004; Pina et al., 2005). The transcriptomic data also suggested that expression of some TETs could be up-regulated during in vitro pollen tube growth (TET7, TET8, and TET13) and promoted by interaction with female tissues (TET7, TET8, TET13, and TET16; Supplemental Fig. S2). While TET2 and TET4 were detected in mature pollen (Fig. 7, a and b), neither the microarray data nor the RT-PCR supported this expression, which might be explained by their low expression levels. TET7, TET8, and TET13 transcripts were detected in mature pollen (Fig. 4), but only the transcriptional fusion of TET7 (Fig. 7c) and TET13 (Fig. 7f) were detectable at this stage with a relative weak intensity, while we failed to detect TET8 (Fig. 7d). TET8-GFP signal was only detectable 3 to 4 h after hydration (Fig. 7, d and e), which would be consistent with induction after pollen metabolic activation, likely delayed by a slow GFP maturation time (Chiu et al., 1996). Meanwhile, this potential TET8 induction upon pollen hydration led us to reevaluate the occasional expression detected in zygotes: though TET8 transcript could be present in the egg cell before fertilization, TET8 translation might be induced in the egg cell in a transient manner just before or upon fertilization. This hypothesis might explain our failure to consistently detect GFP expression in egg cells or normal developing zygotes, as this transient expression would cause a rapid protein turnover preventing detection of the GFP signal, unless a failed fertilization would allow for protein accumulation (Fig. 6c).
To better assess if TET7, TET8, and TET13 were regulated during pollen germination and if changes in the expression patterns were dependent on interaction with female tissues, we examined in vitro and semi-in vivo pollen tube growth (Fig. 7, g–P). TET7, TET8, and TET13 expression was strongly induced during in vitro pollen tube growth, as shown by the robust accumulation of nuclear-localized GFP associated with the vegetative nucleus (Fig. 7, g–i). The translational fusions confirmed this induction and showed that TET7/TET13 expression was associated with the subapical and apical pollen tube plasma membrane and with small cytoplasmic granules (Fig. 7, j–k). In semi-in vivo conditions, TET7, TET8, and TET13 accumulated preferentially at the apical domain of pollen tubes (Fig. 7, l–p). Interestingly, we observed that about 10% of the TET7 pollen tubes grown through female tissues showed an enriched deposition at the pollen tube tip (Fig. 7, l and o). While seen only in some pollen tubes, this pattern was consistent and verified only in semi-in vivo conditions. We were not able to correlate this pattern with pollen tubes exiting the pistil (Fig. 7l) with proximity to an ovule or with growth arrest. While comparisons of the transcriptional profiles of mature pollen and in vitro and semi-in vivo pollen tube growth supported a possibility of de novo transcription in mature pollen or during pollen tube growth (Supplemental Fig. S2), we presented here in vivo validation for the occurrence of such cellular events.
The results obtained for TET14 to TET17 were not fully consistent with the RT-PCR results. The transcriptional activity was preferentially detected in early stages of pollen development (unicellular and bicellular stages), though TET15 to TET17 were also detected in mature pollen (Fig. 7, q–u). However, we failed to detect expression of the TET17 translational fusion at any pollen developmental stage. In addition, the subcellular localization of TET15 and TET16 clearly differed from the remaining pollen-expressed TETs, with accumulation in abundant cytoplasmic organelles (Fig. 7, r and t), similar to the subcellular localization found in mesophyll protoplasts (Fig. 3).
The sperm cell transcriptome (Borges et al., 2008) predicted the enriched expression of specific tetraspanin members in sperm cells (TET7, TET8, TET11, and TET12). While we failed to detect any transcriptional or translational activity for TET7 and TET8 in sperm cells, TET11 and TET12 showed a strict sperm cell-specific activity (Fig. 7, v and w). Interestingly, the TET11/TET12 translational GFP fusions revealed a distinctive protein accumulation in a membrane subdomain at the interface contact between sperm cells, which also delineates the membrane projection that extends from one sperm cell to the vegetative nucleus (Fig. 7, x and y). This distinctive expression clearly defined the male germ unit structure (McCue et al., 2011). However, the cellular resolution is currently limited, as we lacked a visual marker for the pollen vegetative endomembrane (McCue et al., 2011). Nevertheless, this characteristic localization appears to define a sperm-specific tetraspanin-enriched microdomain, as the ectopic overexpression of TET9, a female gametophyte-enriched tetraspanin (Fig. 6, e–g), in sperm cells led to a similar polarized localization (Sprunck et al., 2012). While the biological function of TET11/TET12 in this microdomain is unknown, in view of known tetraspanin functions (Mazurov et al., 2006; Barreiro et al., 2008; Evans, 2012), it is plausible that TET11/TET12 could be involved in facilitating cellular adhesion or intercellular communication. The localization at the interface of sperm cells would be consistent with a function in maintaining the physical stability of the male germ unit during pollen tube growth, until the sperm cell pair is delivered inside the embryo sac. It could also be important to establish cellular contacts that facilitate an effective intercellular communication, allowing synchronization of cellular activities (analogous to the function of cytomitic channels linking pollen mother cells; Mursalimov and Deineko, 2011) required for proper sperm cell differentiation, distribution of cellular components to be delivered to female gametes (Bayer et al., 2009), or transport of silencing information in light of a possible directional transport of small interfering RNAs from the vegetative nucleus to sperm cells (Slotkin et al., 2009).
Overall, the expression analysis revealed that distinct Arabidopsis tetraspanins are enriched in specific reproductive cell types of tissue domains, sometimes with overlapping expression patterns, and that the expression often exhibits a pollination-dependent regulation (Supplemental Table S2). The results also support that paralogous members share more similar expression patterns, which might reflect related biological functions through the occurrence of cellular interactions in specific cell types.
Arabidopsis Tetraspanins Are Likely Genetically Redundant
Coexpression and colocalization of TET members with complementary or similar functions might result in functional redundancy. To investigate this possibility, we obtained and analyzed several available transfer DNA (T-DNA) insertion lines for different tetraspanin members (Supplemental Data File S1). Homozygous plants were recovered, and T-DNA insertion locations confirmed and correlated with alterations in gene expression by RT-PCR (Supplemental Data File S1). We identified homozygous lines in which the T-DNA insertions resulted in overexpression (TET7 and TET11) or absence of a full-length transcript that likely rendered the protein nonfunctional (TET1, TET3, TET5, TET7, TET8, TET11, TET13, TET14, TET16, and TET17). All but one of the insertion lines produced no noticeable phenotypes, suggesting that knockouts of higher order are required to overcome genetic redundancy between homologous genes. The exception was tet1/trn2 knockout mutant, whose phenotypes were consistent with those previously reported (Olmos et al., 2003; Cnops et al., 2006). No null mutant is currently available for TET2, but the existence of a phenotype in the TET1 knockout line supports functional divergence within the TET1/TET2 clade (Supplemental Fig. S1). Our results also suggest that functional redundancy could exist among nonparalogous tetraspanin members. For example, for TET10 and TET13 (both singleton members), no phenotype was found in the single knockout line, suggesting that other TET members expressed in the same cell types could function redundantly. Four insertion lines had gametophytic-associated phenotypes linked to nonrelated secondary T-DNA insertions (see description in Supplemental Data File S1).
Arabidopsis Tetraspanins Homo- and Heterodimerize When Expressed in Yeast
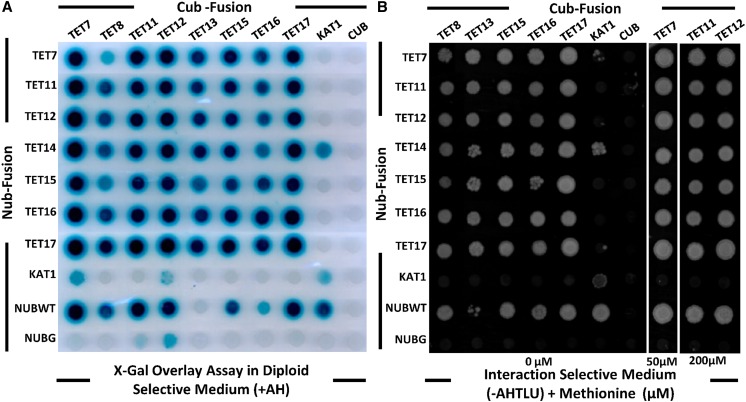
In other organisms, tetraspanins associate in multimolecular membrane complexes, whose functions are dependent on patterns of coexpression and the presence of specific cell type interaction partners (Levy and Shoham, 2005a). Tetraspanins’ heterogeneity in cellular composition and the dynamics of regulation is thought to provide the flexibility needed to participate in many different biological functions (Yunta and Lazo, 2003). Our findings fully support this view for Arabidopsis tetraspanins, where distinct members could physically interact in specific cellular contexts and contribute to the apparent functional redundancy. To validate this hypothesis, we performed combinatory interactions using a yeast membrane mating-based split-ubiquitin system (mbSUS; Obrdlik et al., 2004; Grefen et al., 2009) with tetraspanins preferentially expressed in reproductive tissues (i.e. TET7–TET17; Fig. 8). TETs’ full-length complementary DNAs (cDNAs) were fused to the C-terminal ubiquitin (Cub-protein A-LexA-VP16) fragment or used to generate N-terminal fusions with the N-terminal ubiquitin (Nub) fragment. After mating, the presence of both plasmids (diploids) and binary interactions were assayed by growth on appropriate media (Fig. 8, A and B). No growth was detected for the TET9, TET10, and TET14 Cub fusions with wild-type Nub, so these clones were excluded from the interaction assays. X-Gal assays (Fig. 8A) showed a slight autoactivation for TET7, TET11, and TET12 Cub fusions, which was repressed by addition of different Met concentrations to the interaction-selective medium (Fig. 8B). With these conditions and for all other Cub fusions, no growth was observed when TETs were coexpressed with the mutant N-terminal half of ubiquitin (NubG) empty vector (Fig. 8B). All possible tetraspanin combinations were tested, and growth was induced on interaction-selective medium (Fig. 8B). Binary interactions for TET7, TET11, TET12, TET15, and TET17 were particularly strong, with yeast growth detected after 2 d, while for the remaining constructs, growth was only detected after 5 d. The results from the interaction assays confirmed that TETs could physically interact in vivo and that TET-TET combinations (homo- and heterodimers) could be diverse. We also showed that TET interactions were tetraspanin specific, as all but one TET fusions did not yield positive interactions with KAT1, a potassium channel included as control for interaction specificity (a positive KAT1 interaction was detected for TET14). TET-TET interactions are thought to occur laterally through transmembrane domains (Levy and Shoham, 2005b), and no evidence exists to support tetraspanin interactions across membranes of different cells. Tetraspanin oligomer complexes should assemble in a cell type-specific manner, e.g. TET11 and TET12 colocalize in sperm cells, and thus TET11/TET12 complexes should perform specific functions in male gametes. These results also reinforce the hypothesis that functional redundancy may result from the diversity of TET-TET interactions, where the recruitment of cell type-specific binding partners could determine distinct biological functions.
Figure 8.
Arabidopsis tetraspanins physically interact when expressed in yeast. Yeast THY.AP4 and THY.AP5 clones expressing full-length tetraspanins were mated and grown in SC medium (–TLU; Trp, Leu, and uracil) for diploid selection (A) or interaction-selective medium (–AHTLU; adenine, His, Trp, Leu, and uracil) supplemented with different concentrations of Met (B). NubWT was used as a positive control, and NubG was used as a negative control. The potassium channel KAT1 was used as an interaction specificity control. [See online article for color version of this figure.]
CONCLUSION
We showed that plant tetraspanins maintain many of the functional and structural features present in other organisms. Despite their wide distribution in diverse tissues, Arabidopsis tetraspanins have unique expression patterns that in general are confined to specific tissue domains or cell types where expression can overlap. In addition to their predominant expression in the plasma membrane, tetraspanins can localize in membrane microdomains in a cell type-dependent manner. In addition, we showed that plant tetraspanins, similar to those in other organisms, physically interact with other tetraspanin members, supporting the hypothesis that plant tetraspanins assemble in multimeric complexes in plant cells. The diversity of expression patterns in different cell types and their interacting partners will determine the composition of individual complexes bestowing the functional flexibility known for tetraspanins and contributing to maintaining their cellular robustness.
The unique patterns of expression of particular tetraspanin members in reproductive tissues and their regulation in response to pollination makes us envision their biological relevance in diverse cellular events during the reproductive process, but also in more basic but vital processes in plant development. Tetraspanins also constitute excellent reporter markers for particular cell types and for addressing the function of specific tissue domains in vegetative or reproductive tissues. Thus, this study provides basic knowledge about this important gene family in plants that we think will be instrumental for future functional studies of tetraspanin interactions in diverse plant developmental processes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
T-DNA insertion lines were obtained from the Arabidopsis Stock Center (Arabidopsis Biological Resource Center [ABRC]), the Nottingham Arabidopsis Stock Centre, or the Institut National de la Recherche Agronomique and are listed in Supplemental Data File S1. Seeds were surface sterilized and plated on basal Murashige and Skoog medium supplemented with Arabvitamins (1 mg L–1 thiamine, 0.5 mg L–1 pyridoxine, 0.5 mg L–1 nicotinic acid, and 0.1 mg L–1 myoinositol), 0.5 g L–1 MES (Sigma), 1% (w/v) Suc (Fisher Scientific), and 0.8% (w/v) agar (Fisher Scientific), adjusted to pH 5.7. Seeds were stratified for 3 d at 4°C in the dark and grown under long-day conditions (16 h light at 21°C) for 14 d before transfer to soil. Transgenic plants and mutant lines were preselected on Murashige and Skoog medium containing the appropriate antibiotic or herbicide. Resistant seedlings were transferred to soil and grown in a greenhouse with 16-h light, day/night temperatures of 22°C/18°C, and approximately 50% relative humidity. The quartet 1-2 (qrt1-2; (Copenhaver et al., 2000) mutant in ecotype Columbia background was used for stable transformations (Clough, 2005). Ten to 20 resistant plants of each transformation were genotyped by PCR (see list of primers used in Supplemental Table S3) and phenotyped, and the expression patterns were analyzed. One or two lines were chosen for further analysis based on the presence of single insertions and stable GFP expression.
Bioinformatics Analysis
The amino acid sequences of Arabidopsis (Arabidopsis thaliana) tetraspanins and putative orthologs in other species were retrieved from The Arabidopsis Information Resource (http://www.arabidopsis.org) and from plant genomes in Phytozome (http://www.phytozome.net), respectively. The amino acid sequences were aligned and used for the determination of the phylogenetic relationships using the default parameters for ClustalW in MEGA5 software (Tamura et al., 2011). Only sequences containing the full or partial protein length that included key features of the tetraspanin signature were used in the final alignment. For the sake of clarity in the phylogenetic tree, we only considered founder gene family members of each divergent angiosperm evolutionary branch. The following species were surveyed: Selaginella moellendorffii, Physcomitrella patens, Brachypodium distachyon, Oryza sativa, Zea mays, Sorghum bicolor, Vitis vinifera, Carica papaya, Arabidopsis lyrata, Arabidopsis, Glycine max, Medicago truncatula, and Populus trichocarpa, with Dictyostelium discoideum as an outgroup. The best alignment was used to derive a phylogenetic tree constructed by the Dayhoff method using the neighbor-joining algorithm implemented in MEGA5 (Tamura et al., 2011). Bootstrap analysis with 1,000 replicates was performed to test the significance of nodes. Homologous groups were defined based on a bootstrap value greater than 50%. Secondary structure was predicted with PsiPred (http://bioinf.cs.ucl.ac.uk/psipred). Transmembrane domains were predicted using TMpred (http://www.ch.embnet.org/software/TMPRED_form.html). The disulfide bond topology of tetraspanins was predicted using DiANNA 1.1 (Ferré and Clote, 2006). Palmitoylation sites were predicted using Palmitoylation CSS-Palm 2.0 (Ren et al., 2008) and N-glycosylation sites using the NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc). The amino acid similarity and identity matrix was generated using the Matrix Global Alignment Tool (http://iubio.bio.indiana.edu/soft/molbio/evolve).
RT-PCR Expression Analysis
Total RNA was extracted from open flowers, closed flower buds, roots, seedlings, rosette leaves, siliques, unpollinated pistils, unfertilized ovules, and pollen of wild-type Columbia plants using the RNeasy Plant mini kit (Qiagen) with on-column DNase treatment. Two micrograms of total RNA was reverse transcribed with SuperScript III (Invitrogen) according to the manufacturer’s instructions using oligo(dT) primers in a 20-µL reaction. PCR reactions were performed using 30 cycles if not otherwise indicated. Actin2 (ACT2, AT3G18780) was used as internal control. The primers were designed to flank intron sequences. Primer sequences and description of their use can be found in Supplemental Table S3 and Supplemental Data File S1, respectively.
Localization Assays
All PCR amplifications performed from genomic DNA or cDNA used Phusion Taq polymerase (Finnzymes), according to the manufacturer’s instructions (Supplemental Table S3). All clones were constructed using Gateway technology (Invitrogen). PCR products were directionally cloned into PentrSD/D/Topo or via a BP reaction into pDONR-207 (Invitrogen) and verified by sequencing. For subcellular localization assays, the predicted coding sequence of tetraspanins was fused in frame with GFP driven by the CaMV-35S promoter using the Gateway destination vector pB7FWG2 (Karimi et al., 2002). These constructs were transiently expressed in Arabidopsis protoplasts via polyethylene glycol-mediated transformation (Yoo et al., 2007) or used to generate stable transgenic lines. Transformed protoplasts were held in the dark at 22°C for 12 h and then imaged. The 35S:PIP2-mCherry plasmid (ABRC clone CD3-1007) and ER-tagged mCherry (HDEL, ER retention signal; ABRC clone CD3-959) were used as positive controls for plasma membrane and ER localization in protoplast transient assays or used for cotransformation analysis in stable transgenic lines.
Transcriptional and Translational GFP Fusions
For transcriptional fusions, the native tetraspanin promoters were amplified from genomic DNA by PCR (Supplemental Table S3) to generate Gateway entry clones that were then recombined into the Gateway-compatible version (Zheng et al., 2011) of the pGreenII-based vector NLS3xeGFPnopaline synthase terminator (Takada and Jürgens, 2007). These constructs were introduced into Agrobacterium tumefaciens strain GV3101 harboring the pGreenII helper plasmid pSOUP and transformed in Arabidopsis qrt1-2 Columbia plants using the floral dipping method (Clough, 2005). For C-terminal translational fusions with eGFP, endogenous promoters and the predicted coding sequence lacking the STOP codon were amplified from genomic DNA (Supplemental Table S3), directionally cloned into the entry vector, and sequenced. The destination vector pB7FWG2 (Karimi et al., 2002) was digested with SacI and SpeI to remove the CaMV-35S promoter and then blunted and religated to generate pB7FWG2*. The binary vector for expression of C-terminal eGFP fusions was then generated via an LR reaction using the corresponding entry vector. Constructs were introduced into A. tumefaciens strain GV3101 and used to transform Arabidopsis qrt1-2 Columbia plants.
Yeast mbSUS
cDNA clones in Gateway-compatible entry vectors were acquired from the ABRC stock center, or, when not available, annotated open reading frames were amplified from cDNA of Arabidopsis open flowers or pollen (Supplemental Table S4) and cloned into a Gateway entry vector as described previously. These clones were then used to perform LR recombination reactions with pNX32-DEST for mutant N-terminal half of ubiquitin (NubG) fusions or with pMETYC-DEST for C-terminal Cub fusions. Expression of Cub fusions was regulated by a Met-repressive promoter. The destination clones were transformed into the yeast (Saccharomyces cerevisiae) haploid strains THY.AP5 (MATα) for NubG fusions and THY.AP4 (MATα) for Cub fusions using the lithium acetate method (Gietz and Woods, 2002). Cells harboring the Cub plasmids were grown on synthetic complete (SC) medium supplemented with Trp, His, adenine, and uracil, and cells with the NubG plasmid were grown on SC medium supplemented with Leu, adenine, and His. Diploid cells were selected on SC medium supplemented with adenine and His, while selection for protein-protein interactions was performed on SC medium alone or supplemented with Met when increased interaction stringency was required. Clones expressing the Cub fusions were mated with either the soluble wild-type Nub clone or soluble NubG clone alone as controls for false negatives and false positives, respectively. Only Cub fusions showing no growth when mated with a strain containing soluble NubG (i.e. a Cub fusion that did not autoactivate the reporters) and growth for soluble wild-type Nub (indicating protein expression) were used for interaction tests. The potassium channel KAT1 (Obrdlik et al., 2004) was used as an interaction specificity control. Yeast mating and the β-galactosidase activity of cells was performed as described in Grefen et al. (2009).
In Vivo Expression Analysis and Microscopy
Arabidopsis in vitro pollen germinations were performed as described in Boavida and McCormick (2007). Semi-in vivo assays were performed according to Palanivelu and Preuss (2006). Imaging was performed with a Zeiss Axiophot 2 microscope using fluorescence or DIC optics. Images were captured with Axiovision 4.3 software using an AxioCamMR camera. Confocal imaging was performed using a Zeiss LSM510 META confocal laser-scanning microscope. Excitation was performed using a HeNe laser set to 543 nm, and emission was detected with a 560-nm long-path barrier filter. Image analysis was performed using ImageJ (Abramoff et al., 2004).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of plant tetraspanins.
Supplemental Figure S2. Tetraspanin expression from microarrays during pollen development and in vivo, in vitro, and semi-in vivo pollen tube growth.
Supplemental Figure S3. Confirmation of TET12 antisense transcript by strand-specific RT-PCR in pollen.
Supplemental Figure S4. Arabidopsis tetraspanins whose expression is altered by pollination.
Supplemental Table S1. Tetraspanin amino acid identity and similarity matrix.
Supplemental Table S2. Summary of TETs expression patterns in transcriptional and translational fusions.
Supplemental Table S3. Primer sequences (listed 5′ to 3′) and description of their use.
Supplemental Table S4. cDNA clones and primers used for the mbSUS.
Supplemental Data File S1. Analysis of T-DNA insertion lines.
Acknowledgments
We thank Christopher Greffen for providing the mbSUS “DEST” Gateway-compatible vectors and Mily Ron for providing the Gateway-compatible vector pGreenII-NLS3xeGFP and for technical advice.
Glossary
- CaMV-35S
Cauliflower mosaic virus 35S
- RT
reverse transcription
- T-DNA
transfer DNA
- cDNA
complementary DNA
- ABRC
Arabidopsis Biological Resource Center
- mbSUS
mating-based split-ubiquitin system
- SC
synthetic complete
References
- Abramoff MD, Magalhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Baldwin G, Novitskaya V, Sadej R, Pochec E, Litynska A, Hartmann C, Williams J, Ashman L, Eble JA, Berditchevski F. (2008) Tetraspanin CD151 regulates glycosylation of α3β1 integrin. J Biol Chem 283: 35445–35454 [DOI] [PubMed] [Google Scholar]
- Barreiro O, Zamai M, Yáñez-Mó M, Tejera E, López-Romero P, Monk PN, Gratton E, Caiolfa VR, Sánchez-Madrid F. (2008) Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J Cell Biol 183: 527–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer M, Nawy T, Giglione C, Galli M, Meinnel T, Lukowitz W. (2009) Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488 [DOI] [PubMed] [Google Scholar]
- Boavida LC, Borges F, Becker JD, Feijó JA. (2011) Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiol 155: 2066–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boavida LC, McCormick S. (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J 52: 570–582 [DOI] [PubMed] [Google Scholar]
- Boavida LC, Vieira AM, Becker JD, Feijó JA. (2005) Gametophyte interaction and sexual reproduction: how plants make a zygote. Int J Dev Biol 49: 615–632 [DOI] [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD. (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol 148: 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin S, Manié S, Oualid M, Billard M, Boucheix C, Rubinstein E. (2002) Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett 516: 139–144 [DOI] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Clough SJ. (2005) Floral dip: agrobacterium-mediated germ line transformation. Methods Mol Biol 286: 91–102 [DOI] [PubMed] [Google Scholar]
- Cnops G, Neyt P, Raes J, Petrarulo M, Nelissen H, Malenica N, Luschnig C, Tietz O, Ditengou F, Palme K, et al. (2006) The TORNADO1 and TORNADO2 genes function in several patterning processes during early leaf development in Arabidopsis thaliana. Plant Cell 18: 852–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnops G, Wang X, Linstead P, Van Montagu M, Van Lijsebettens M, Dolan L. (2000) Tornado1 and tornado2 are required for the specification of radial and circumferential pattern in the Arabidopsis root. Development 127: 3385–3394 [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Keith KC, Preuss D. (2000) Tetrad analysis in higher plants. A budding technology. Plant Physiol 124: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L. (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSalle R, Mares R, Garcia-España A. (2010) Evolution of cysteine patterns in the large extracellular loop of tetraspanins from animals, fungi, plants and single-celled eukaryotes. Mol Phylogenet Evol 56: 486–491 [DOI] [PubMed] [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U. (2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660 [DOI] [PubMed] [Google Scholar]
- Evans JP. (2012) Sperm-egg interaction. Annu Rev Physiol 74: 477–502 [DOI] [PubMed] [Google Scholar]
- Faure JE, Rotman N, Fortuné P, Dumas C. (2002) Fertilization in Arabidopsis thaliana wild type: developmental stages and time course. Plant J 30: 481–488 [DOI] [PubMed] [Google Scholar]
- Fernandez-Calvino L, Faulkner C, Walshaw J, Saalbach G, Bayer E, Benitez-Alfonso Y, Maule A. (2011) Arabidopsis plasmodesmal proteome. PLoS ONE 6: e18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrè F, Clote P. (2006) DiANNA 1.1: an extension of the DiANNA web server for ternary cysteine classification. Nucleic Acids Res 34: W182–W185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-España A, DeSalle R. (2009) Intron sliding in tetraspanins. Commun Integr Biol 2: 394–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Grefen C, Obrdlik P, Harter K. (2009) The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS). Methods Mol Biol 479: 217–233 [DOI] [PubMed] [Google Scholar]
- Hamamura Y, Saito C, Awai C, Kurihara D, Miyawaki A, Nakagawa T, Kanaoka MM, Sasaki N, Nakano A, Berger F, et al. (2011) Live-cell imaging reveals the dynamics of two sperm cells during double fertilization in Arabidopsis thaliana. Curr Biol 21: 497–502 [DOI] [PubMed] [Google Scholar]
- Hemler ME. (2005) Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 6: 801–811 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D. (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Yuan S, Dong M, Su J, Yu C, Shen Y, Xie X, Yu Y, Yu X, Chen S, et al. (2005) The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics 86: 674–684 [DOI] [PubMed] [Google Scholar]
- Huck N, Moore JM, Federer M, Grossniklaus U. (2003) The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130: 2149–2159 [DOI] [PubMed] [Google Scholar]
- Hulskamp M, Schneitz K, Pruitt RE. (1995) Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell 7: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jégou A, Ziyyat A, Barraud-Lange V, Perez E, Wolf JP, Pincet F, Gourier C. (2011) CD9 tetraspanin generates fusion competent sites on the egg membrane for mammalian fertilization. Proc Natl Acad Sci USA 108: 10946–10951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kovalenko OV, Metcalf DG, DeGrado WF, Hemler ME. (2005) Structural organization and interactions of transmembrane domains in tetraspanin proteins. BMC Struct Biol 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. (2000) Severely reduced female fertility in CD9-deficient mice. Science 287: 319–321 [DOI] [PubMed] [Google Scholar]
- Levy S, Shoham T. (2005a) The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol 5: 136–148 [DOI] [PubMed] [Google Scholar]
- Levy S, Shoham T. (2005b) Protein-protein interactions in the tetraspanin web. Physiology (Bethesda) 20: 218–224 [DOI] [PubMed] [Google Scholar]
- Lieber D, Lora J, Schrempp S, Lenhard M, Laux T. (2011) Arabidopsis WIH1 and WIH2 genes act in the transition from somatic to reproductive cell fate. Curr Biol 21: 1009–1017 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AH, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF. (2004) Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116: 843–853 [DOI] [PubMed] [Google Scholar]
- Mazurov D, Heidecker G, Derse D. (2006) HTLV-1 Gag protein associates with CD82 tetraspanin microdomains at the plasma membrane. Virology 346: 194–204 [DOI] [PubMed] [Google Scholar]
- McCue AD, Cresti M, Feijó JA, Slotkin RK. (2011) Cytoplasmic connection of sperm cells to the pollen vegetative cell nucleus: potential roles of the male germ unit revisited. J Exp Bot 62: 1621–1631 [DOI] [PubMed] [Google Scholar]
- Michard E, Lima PT, Borges F, Silva AC, Portes MT, Carvalho JE, Gilliham M, Liu LH, Obermeyer G, Feijó JA. (2011) Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil d-serine. Science 332: 434–437 [DOI] [PubMed] [Google Scholar]
- Mursalimov SR, Deineko EV. (2011) An ultrastructural study of cytomixis in tobacco pollen mother cells. Protoplasma 248: 717–724 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, et al. (2004) K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci USA 101: 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos E, Reiss B, Dekker K. (2003) The ekeko mutant demonstrates a role for tetraspanin-like protein in plant development. Biochem Biophys Res Commun 310: 1054–1061 [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Brass L, Edlund AF, Preuss D. (2003) Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114: 47–59 [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Preuss D. (2006) Distinct short-range ovule signals attract or repel Arabidopsis thaliana pollen tubes in vitro. BMC Plant Biol 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijó JA, Becker JD. (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols MS, Klumperman J. (2009) Trafficking and function of the tetraspanin CD63. Exp Cell Res 315: 1584–1592 [DOI] [PubMed] [Google Scholar]
- Prado AM, Porterfield DM, Feijó JA. (2004) Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 131: 2707–2714 [DOI] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R. (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. (2008) CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel 21: 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A. (2008) The molecular origins of multicellular transitions. Curr Opin Genet Dev 18: 472–478 [DOI] [PubMed] [Google Scholar]
- Ron M, Alandete Saez M, Eshed Williams L, Fletcher JC, McCormick S. (2010) Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev 24: 1010–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein E. (2011) The complexity of tetraspanins. Biochem Soc Trans 39: 501–505 [DOI] [PubMed] [Google Scholar]
- Seigneuret M, Delaguillaumie A, Lagaudrière-Gesbert C, Conjeaud H. (2001) Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J Biol Chem 276: 40055–40064 [DOI] [PubMed] [Google Scholar]
- Shimizu KK, Okada K. (2000) Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127: 4511–4518 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzić M, Becker JD, Feijó JA, Martienssen RA. (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, Dresselhaus T. (2012) Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 338: 1093–1097 [DOI] [PubMed] [Google Scholar]
- Stipp CS, Kolesnikova TV, Hemler ME. (2003) Functional domains in tetraspanin proteins. Trends Biochem Sci 28: 106–112 [DOI] [PubMed] [Google Scholar]
- Takada S, Jürgens G. (2007) Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development 134: 1141–1150 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. (2010) Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62: 405–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel AJ. (1996) Interaction between sieve element and companion cell and the consequences for photoassimilate distribution. Two structural hardware frames with associated physiological software packages in dicotyledons? J Exp Bot 47: 1129–1140 [DOI] [PubMed] [Google Scholar]
- Veneault-Fourrey C, Lambou K, Lebrun MH. (2006) Fungal Pls1 tetraspanins as key factors of penetration into host plants: a role in re-establishing polarized growth in the appressorium? FEMS Microbiol Lett 256: 179–184 [DOI] [PubMed] [Google Scholar]
- Wang F, Vandepoele K, Van Lijsebettens M. (2012) Tetraspanin genes in plants. Plant Sci 190: 9–15 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH. (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol 148: 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenführer J, von Mering C, Grossniklaus U. (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Yáñez-Mó M, Tejedor R, Rousselle P, Sánchez -Madrid F. (2001) Tetraspanins in intercellular adhesion of polarized epithelial cells: spatial and functional relationship to integrins and cadherins. J Cell Sci 114: 577–587 [DOI] [PubMed] [Google Scholar]
- Yang X, Claas C, Kraeft SK, Chen LB, Wang Z, Kreidberg JA, Hemler ME. (2002) Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol Biol Cell 13: 767–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yunta M, Lazo PA. (2003) Tetraspanin proteins as organisers of membrane microdomains and signalling complexes. Cell Signal 15: 559–564 [DOI] [PubMed] [Google Scholar]
- Zheng B, Chen X, McCormick S. (2011) The anaphase-promoting complex is a dual integrator that regulates both microRNA-mediated transcriptional regulation of cyclin B1 and degradation of cyclin B1 during Arabidopsis male gametophyte development. Plant Cell 23: 1033–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]