Selective elimination of uniparental chromosomes occurred in both embryo and endosperm during the formation of haploid in maize.
Abstract
Producing maternal haploids via a male inducer can greatly accelerate maize (Zea mays) breeding process. However, the mechanism underlying haploid formation remains unclear. In this study, we constructed two inducer lines containing cytogenetic marker B chromosome or alien centromeric histone H3 variant-yellow fluorescent protein vector to investigate the mechanism. The two inducer lines as the pollinators were crossed with a hybrid ZhengDan958. B chromosomes were detected in F1 haploids at a low frequency, which was direct evidence to support the occurrence of selective chromosome elimination during haploid formation. We found that most of the inducer chromosomes were eliminated in haploid embryonic cells during the first week after pollination. The gradual elimination of chromosomes was also detected in the endosperm of defective kernels, although it occurred only in some endosperm cells as late as 15 d after pollination. We also performed a genome-wide identification of single nucleotide polymorphism markers in the inducers, noninducer inbred lines, and 42 derived haploids using a 50K single nucleotide polymorphism array. We found that an approximately 44-Mb heterozygous fragment from the male parent was detected in a single haploid, which further supported the occurrence of paternal introgression. Our results suggest that selective elimination of uniparental chromosomes leads to the formation of haploid and possible defective kernels in maize as well, which is accompanied with unusual paternal introgression in haploid cells.
Doubled haploid (DH) technology is widely used in maize (Zea mays) breeding. Although the life cycle of most sexually reproducing plant species alternates between a diploid sporophytic phase and a highly reduced gametophytic haploid phase, it is possible to obtain haploid plants containing the same number of chromosomes in their somatic cells as do the normal gametes of the species (Dunwell, 2010). Haploids can be obtained by in vitro or in vivo approaches. Anther and microspore culture are the most commonly used in vitro approaches; however, many species and genotypes are recalcitrant to these processes (Forster et al., 2007). Interspecies cross, which is an in vivo approach, induces haploids via chromosome elimination (Forster et al., 2007). For example, the cross between Hordeum vulgare and Hordeum bulbosum produces haploids in barley (Kasha and Kao, 1970; Sanei et al., 2011); maize and pearl millet (Pennisetum glaucum) are used as pollinators to produce haploids in wheat (Triticum aestivum; Laurie and Bennett, 1988; Gernand et al., 2005). There are two approaches for in vivo haploid induction in maize. One is using an ig mutant to generate both maternal and paternal haploids (Kermicle, 1969; Evans, 2007), and the other is using Stock6-derived inducers to produce maternal haploids only. The application of Stock6-derived inducers has become the foundation for modern DH technology (Prigge and Melchinger, 2012; Xu et al., 2013). The haploid induction rate (HIR) of inducer line Stock6 is 1% to 2% (Coe, 1959), which is 10 to 20 times higher than the spontaneous HIR in maize. The haploid-inducing capacity of inducers can be improved by selection (Sarkar et al., 1972). New inducers with improved HIRs have been developed, such as WS14 (2%–5% induction rate; Lashermes et al., 1988), RWS (Röber et al., 2005), and CAU5 (Xu et al., 2013).
The first haploid inducer line Stock6 was discovered 50 years ago, and DH technology based on in vivo induction of maternal haploids has been widely used in maize breeding (Geiger, 2009). The mechanism underlying haploid formation remains unclear. Two hypotheses have been proposed for the mechanism. The first hypothesis is that one of the two sperm cells fails to fuse with an egg cell, but instead triggers haploid embryogenesis. In the second hypothesis, the two sperm cells fuse with an egg cell and a central cell, and the chromosomes from the inducer degenerate and are eliminated stepwise in the primordial cells during subsequent cell divisions.
Evidence supporting the first hypothesis was reported by Bylich and Chalyk (1996). They found that 6.3% of the pollen grains of the ZMS haploid inducer line have two sperms showing different morphology. Thus, they proposed that the morphological defects of one of the sperms interfere in the sperm’s function, causing single fertilization. Another abnormality of inducer lines was described by Chalyk et al. (2003). They found that the frequency of aneuploid microsporocytes is much higher in haploid inducers than that in normal maize.
Wedzony et al. (2002) studied the ovaries of inducer line RWS during the first 20 d after self-pollination. They found that about 10% of the embryos contain micronuclei with various sizes in the cytoplasm of every cell of shoot primordium. Their result supports the second hypothesis, which is selective chromosome elimination. Moreover, Fischer (2004) speculated that maternal haploids might possess small fractions of inducer genome. Consistently, previous studies (Zhang et al., 2008; Li et al., 2009) demonstrated morphological and molecular evidence for paternal DNA introgression in haploids, indicating the mechanism of chromosome elimination.
Robust evidence to support either hypothesis is still missing due to lack of cytogenetic makers to trace chromosomes from inducers. In this study, we used B chromosomes and centromeric histone H3 variant (CENH3)-yellow fluorescent protein (YFP) as cytogenetic markers to discover direct evidence supporting selective chromosome elimination in both embryo and endosperm during haploid formation. In addition, we performed a genome-wide identification of single nucleotide polymorphism (SNP) markers in inducers, noninducer inbred lines, and 42 haploids using a 50K SNP array and detected unusual DNA introgression from inducer lines during haploid formation.
RESULTS
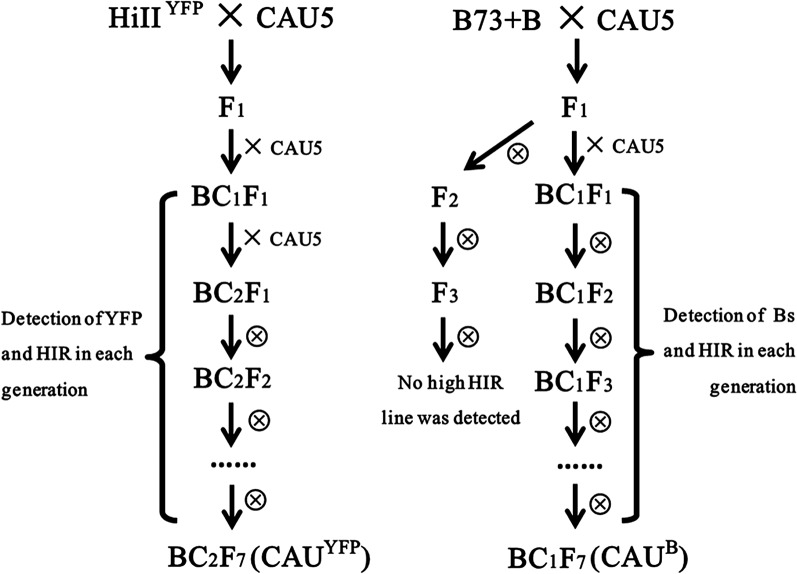
The Development of the New Inducer Lines CAUYFP and CAUB
To develop cytogenetic makers to trace chromosomes from inducers, we intended to add CENH3-YFP or B chromosomes into inducer lines. A new inducer line with CENH3-YFP was generated from the cross of HiIIYFP × CAU5 (Fig. 1). In each generation from BC1F1 to BC2F7, YFP signals and HIR (based on R1-nj color marker) were determined in each plant, and only the plants containing both HIR and YFP signals were maintained. A BC2F7 line with a HIR of 11.26% (Table I) and strong CENH3-YFP signals (Fig. 2B; Supplemental Fig. S1) was named as CAUYFP and used in the study.
Figure 1.
Experimental flow chart for developing and identifying haploid induction lines containing B chromosome and CENH3-YFP. The CAU5 with a high HIR of 8.79% was the recurrent parent, while HiIIYFP and B73+B were used as the donor parents, respectively. YFP signals or Bs were detected in the high HIR lines in each generation.
Table I. The HIR of the developed inducer lines.
| Name | Haploid | Total | HIR |
|---|---|---|---|
| % | |||
| CAU5 | 210 | 2179 | 8.79 |
| CAUYFP | 216 | 1703 | 11.26 |
| CAUB | 172 | 2375 | 6.75 |
Figure 2.
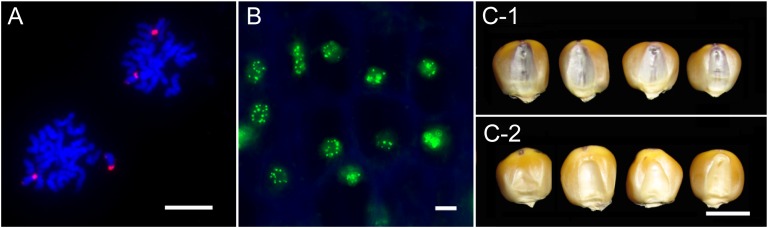
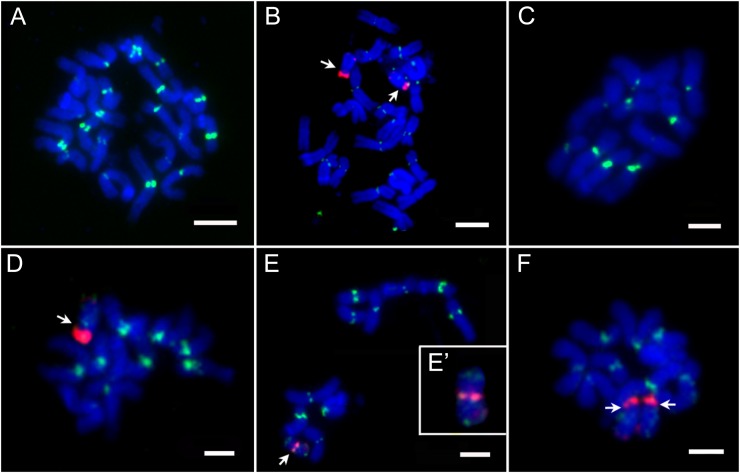
Identification of YFP signals and B chromosomes in the haploid induction lines. A, Metaphase chromosomes were hybridized with ZmBs probe (red) from the CAUB line. Bar = 10 μm. B, YFP signals from the CAUYFP line were observed directly under epifluorescence microscope. Bar = 10 μm. Diploid (C-1) and Haploid (C-2) was offspring from ZD958×CAUB. Bar = 1 cm.
An inducer line with a B chromosome was also generated. We used different breeding strategy to maintain a high HIR level and accumulate as many B chromosomes as possible in the B chromosome-containing inducers (Fig. 1). The inducer line CAU5 was crossed with B73+B and then followed by one generation of backcrossing and self-pollination for six generations. The B chromosomes were detected (Fig. 2A) in each generation of lines with a high HIR, and a final line with a HIR of 6.75% (Table I) was developed in BC1F7 and named as CAUB.
These two new inducer lines were used as male parents in cross with an elite commercial hybrid ZhengDan958 (ZD958) to produce F1 offspring carrying B chromosomes (Fig. 2A) or YFP (Fig. 2B). The F1 offspring were used for analyzing chromosome behavior.
Haploid Contained B Chromosomes from the Paternal Genome (Inducer)
Our fluorescence in situ hybridization (FISH) results exhibited an unambiguous separation of B and normal A chromosomes. B-specific repeats (ZmBs) are usually located in the following two regions: intense hybridization at the cytologically defined centromere and a minor site in the distal region of the long arm. Diploid and haploid kernels derived from ZD958×CAUB were screened for B chromosomes by FISH assay (Fig. 3; Table II). We found that most of the haploids contained the 10 normal A chromosomes (Fig. 3C). Surprisingly, some of the haploids contained B chromosome(s). We isolated four B chromosome-containing haploids from 148 haploids in two locations during 2010 and 2011 (Table II). These four B chromosome-containing haploids were not identical. Two of the haploids contained only one normal B chromosome (Fig. 3D). The third haploid had two normal B chromosomes (Fig. 3F). The fourth haploid had a truncated B chromosome, in which strong ZmBs FISH signals appeared in the intermediate region instead of in the terminal region and weak FISH signals were located on both ends of the chromosome arms (Fig. 3, E and E′). The fact that B chromosome can only come from the male parent CAUB indicates that the maternal and the paternal genomes are once fused together and then A chromosomes from the paternal genome are eliminated. Conversely, a few B chromosomes escape the elimination.
Figure 3.
Karyotyping of the somatic chromosomes of progeny from the cross of ZD958 × CAUB probed with CentC (green) and ZmBs (red). A, Diploid without B chromosome. B, Diploid with two B chromosomes. C, Haploid without B chromosome. D, Haploid with one B chromosome. E and E’, Haploid with one abnormal B chromosome. F, Haploid with two B chromosomes. Arrows in B and D to and F denote the B chromosome(s). Bars = 10 μm.
Table II. The B chromosome distribution in progeny from ZD958×CAUB in two locations for 2 years.
| Year | Diploid |
Haploid |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Bsa | 0B | Total | B fre.b | 1B | 2Bs | 0B | Total | B fre.b | |
| 2010 | 16 | 35 | 51 | 52.6 | 1 | 0 | 29 | 30 | 2.7 |
| 2011 | 55 | 29 | 84 | 2 | 1 | 115 | 118 | ||
Bs represents one to three B chromosomes.
The B chromosome frequency in progeny from ZD958×CAUB for 2 years in two locations; B fre., the plants with B chromosomes/total plants × 100%.
Chromosome Elimination within the First Week after Pollination Led to Haploid Formation
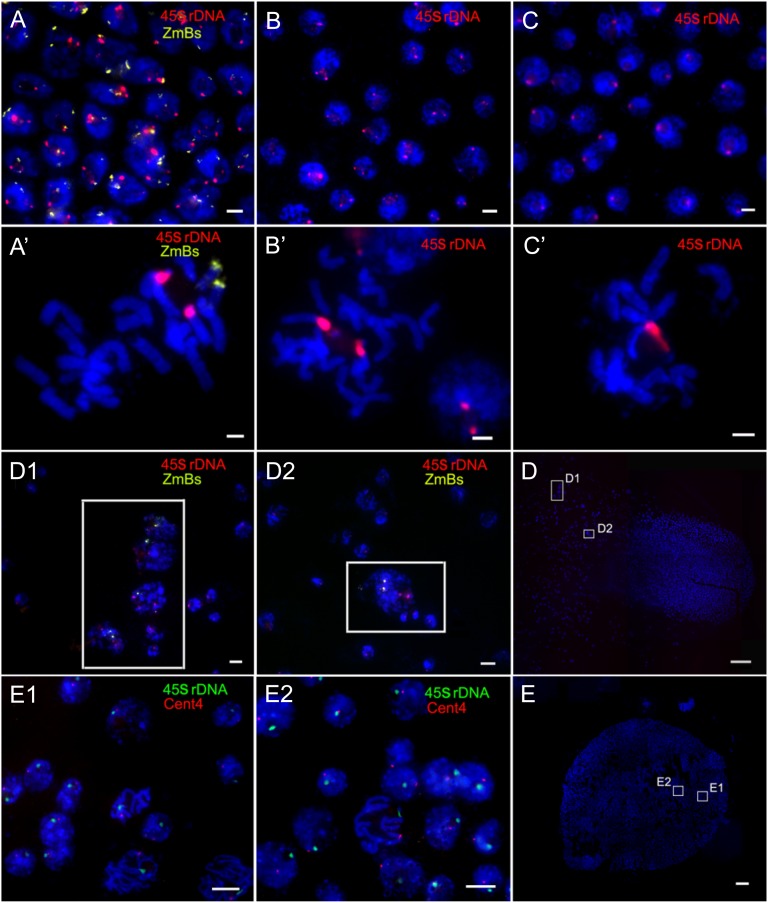
In order to determine the approximate time window of haploid formation, we used the 45S ribosomal (r)DNA and ZmBs as FISH probes to determine the ploidy of embryos and ovaries from F1 ears fixed 7 d after pollination (DAP). Only one copy of the 45S signals in one cell generally indicates haploid (diploid if two), while in some cases, it is possible that there could be only a loss of chromosome 6 (aneuploidy). Out of the 275 independently observed immature embryos, 263 had two 45S rDNA FISH signals with (Fig. 4, A and A′) or without B chromosomes (Fig. 4, B and B′), indicating that they are diploid. Eleven out of the rest 12 embryos had only one 45S rDNA FISH signal (Fig. 4, C and C′) in every cell, which confirmed that they were haploids at 7 DAP. However, one embryo showed mixoploid (Fig. 4, D1 and D2). FISH results revealed that in this embryo, 2853 (99%) out of 2880 cells were haploid without B chromosome, while the remaining 27 (1%) cells were diploid cells exhibiting two 45S rDNA and one or two ZmBs FISH signal. More interestingly, these diploid cells were located either on the edge or on the suspensor of the embryo (Fig. 4D), indicating that the process of chromosome elimination is not completed in this 7-DAP embryo. Our results showed that there were only 12 haploids, including the mixoploid one, detected in the 275 immature embryos. Although haploid-inducing frequency detected in the immature embryos (4.36%) was lower than that in the mature seeds (6.75%), the values were not significantly different between the two groups (P = 0.06; Table I). This result might due to the fact that immature haploid embryos are smaller than diploid embryos, which makes it difficult to find and dissect immature haploid embryos from ovaries, resulting in underestimation of HIR in immature embryos. In addition, we used 45S rDNA and Cent4 as the FISH probes to identify the ploidy of embryos from F1 ears fixed at 10 DAP. The cells in 127 out of the 133 independently observed immature embryos showed two 45S rDNA and two Cent4 signals, indicating the cells possible are diploid. The assay for 45S rDNA and Cent4 only determines the copy number of chromosomes 6 and 4, respectively, and it is possible that other chromosomes might not be diploid. The cells in the remaining six embryos only exhibited one signal of each cytogenetic marker, suggesting that the cells are haploid (Fig. 4, E, E1, and E2). The HIR was 4.51%. Therefore, our results suggest that the inducer chromosomes are mostly eliminated from primordial cells during the first week after pollination, leading to haploid formation.
Figure 4.
Ploidy determination in the 7- and 10-DAP embryos of ZD958×CAUB by the number of 45S rDNA and/or Cent4 signals. A to D2 are 7 DAP, and E to E2 are 10 DAP. FISH was conducted with 45S rDNA (red) and ZmBs (yellow). A and A’, Diploid with two B chromosomes. B and B’, Diploid without the B chromosome. C and C’, Haploid without the B chromosome. D, A wide-angle view of the mixoploid embryo at 7 DAP. Further enlarged cell clusters with 45S rDNA (red) and ZmBs (yellow) are shown in the inset of D1 and D2. E, A wide-angle view of the embryo at 10 DAP. Further enlarged cell clusters with Cent4 (red) and 45S rDNA (green) are shown in E1 and E2. Bars = 10 μm in A to C, A’ to C’, D1, D2, E1, and E2. Bars = 1 mm in D and E.
CENH3-YFP Signal Was Detected in the Endosperm of All the Kernels from the Cross of ZD958×CAUYFP
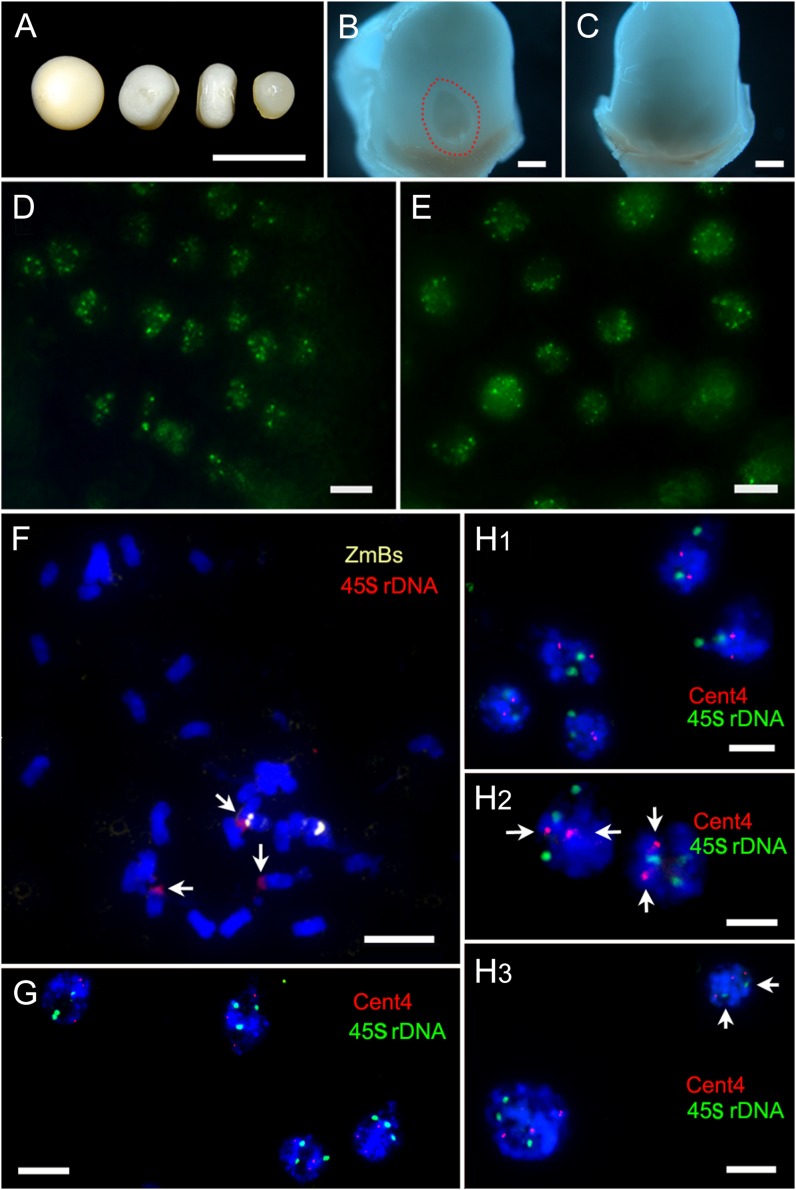
Defective kernels have been found to be closely associated with haploid kernels in maternal haploid induction (Xu et al., 2013). The immature kernels derived from the cross of ZD958×CAUYFP were classified into two different subgroups: the normal kernels (including diploid and haploid) and the defective kernels (Fig. 5, A and C). We found that all the defective kernels were significantly smaller than the normal kernels, and the sizes of defective kernels varied (Fig. 5A).
Figure 5.
Ploidy detection of aborted endosperm at different days after pollination. A to E are 13 DAP, F and G are 5 DAP, and H1 to H3 are 9 to 15 DAP. A, The first one was normal kernel and the others were defective kernels with different sizes. B, Enlarged view of the normal kernel in A with embryo (outlined with red), and the ovary wall was removed. C, Enlarged view of the defective kernel in A without embryo, and the ovary wall was removed. Bars = 500 μm. D, YFP signals in the endosperm of normal kernel. E, YFP signals in the endosperm of defective kernel. F, Spread of aborted endosperm at 5 DAP with three 45S rDNA (red, arrows) and two ZmBs (yellow). G, Interphase nucleus of an aborted endosperm at 5 DAP with three 45S rDNA (red) and three Cent4 (green). H1, Interphase nucleus of an aborted endosperm with two 45S rDNA (green) and two Cent4 (red). H2, An aborted endosperm with three 45S rDNA (green) and two Cent4 (red, arrows). H3, An aborted endosperm showed mixoploid. One cell showed three 45S rDNA (green) and three Cent4 (red); another cell showed two 45S rDNA (green, arrows) and three Cent4 (red). Bars = 10 μm
We monitored YFP signals in endosperm and embryonic cells in different kernel subgroups. Our results showed that YFP signals started to appear at 7 DAP in both the endosperm and the embryo of the normal kernel, and then the proportion of cells displaying YFP signals gradually increased during 9 to 14 DAP in both the endosperm and the embryo (Fig. 5D). At 16 to 20 DAP, the number of cells having YFP signals dropped in the endosperm, which might be associated with the programmed cell death of endosperm (Young and Gallie, 2000). There were only a small number of cells showing YFP signals at 30 DAP in the endosperm; however, the YFP signals in the embryo remained robust. We examined the YFP signals in 210 normal kernels from 13 to 15 DAP (Table III). Based on the status of YFP signals, we classified the normal kernels into two subtypes: The first subtype including 187 kernels out of the 210 observed kernels (89.0%) exhibited YFP signals in both endosperm and embryo (Fig. 5, B and D), and they were diploid. The second subtype showed YFP signals only in endosperm, including 23 kernels out of the 210 observed kernels (11.0%). By chromosome counting, we found that the embryo of the second subtype of normal kernels was haploid. Thus, the HIR calculated based on YFP signal status was 11.0%, which was similar to the HIR calculated according to R1-nj (11.26%). The frequency of haploid formation was stable during the development of immature embryo (13–15 DAP) into mature kernels, indicating that haploid formation occurs at least before 13 DAP.
Table III. Detection of YFP signal in the immature kernels from 13 to 15 DAP derived from the cross of ZD958×CAUYFP.
A majority of the defective kernels (97.75%; Table III) lacked an embryo (Fig. 5C) when they were examined at early stage (13–15 DAP); however, a few defective kernels had a much smaller embryo compared with the embryo of normal seeds. We then examined the YFP signals in the endosperm and the embryo of the defective kernels separately. CENH3-YFP signals were always detectable in all the endosperm of the defective kernels (n = 356; Fig. 5E; Table III). Although only a small number of the defective kernels had an embryo (n = 8), all the embryos exhibited obvious YFP signals (Table III). These results confirm that the endosperms of defective kernels undergo normal fertilization and cell division in the early stage of kernel development.
Mixoploids (or Aneuploids) Were Detected in the Endosperm of the Defective Kernels from the Cross of ZD958×CAUYFP
In order to further determine the ploidy of endosperm in the defective kernels (without embryo), we isolated aborted endosperms and used 45S rDNA and (or) Cent4 as FISH probes to determine the ploidy. As mentioned before, a loss of one copy of the 45S or Cent4 signals in one cell generally indicates a loss of one copy of haploid, while in few cases a cell contains two 45S but one Cen4 (or vise versa), which most likely suggests an aneuploid cell. We screened 42 defective kernels (about 2,520 cells) and determined the number of FISH signals in aborted endosperms from 5 to 15 DAP. To increase the accuracy of FISH signal estimation, we only evaluated the nuclei exhibiting strong fluorescent signals at both emission wavelengths (green and red). Four hundred and seventy out of 497 nuclei (94.6%) from 10 of the 5- to 7-DAP aborted endosperms had three 45S rDNA loci, suggesting that the occurrence frequency of three 45S rDNA loci in defective kernels is not significantly different from that in normal kernels (155 of 163 nuclei had three 45S rDNA loci; 95.1%). This result indicates that the patch of each aborted endosperm is 3n. Our results from chromosome counting assay also supported this conclusion (Fig. 5F). In contrast, during 9 to 15 DAP, the number of FISH signals in 14 out of the remaining 32 aborted endosperms was significantly different from that of normal endosperms. In normal endosperms (3n = 30), approximately 94.9% and approximately 94.5% cells had three 45S rDNA and Cent4 signals, respectively (Fig. 5G). In the 14 aborted endosperms, three types of abnormal FISH signal distribution were detected: (1) cells with two 45S rDNA and two Cent4 loci (Fig. 5H1), (2) cells with three 45S rDNA and two Cent4 loci (Fig. 5H2), and (3) cells with two 45S rDNA and three Cent4 loci (Fig. 5H3). As shown in Supplemental Figure S2, the number of endosperms with three types of abnormal cells was gradually increased from 5 to 7 DAP to 13 to 15 DAP. These results suggest that the ploidy in some endosperm cells changes (mixoploids or aneuploids) during the development of defective kernels from 5 to 15 DAP, and chromosome elimination gradually occurs during this period as well.
To confirm our hypothesis, we performed FISH assay to determine the ploidy of additional eight aborted endosperms. We isolated two patches of cells at different position of the same endosperm (Supplemental Fig. S3A). Two of the eight endosperms showed significantly different ploidy between the paired patches. As highlighted in Supplemental Figure S3, all the cells from one patch showed three 45S rDNA and three Cent4 loci (3n; Supplemental Fig. S3A1), while the cells from the other patch showed two 45S rDNA and two Cent4 loci (2n; Supplemental Fig. S3A2). These results indicate that the process of chromosome elimination in endosperm cells is partial (mosaic) and gradual, and the time window for the occurrence of chromosome elimination in endosperm cells is different from that in embryo (within the first week after pollination).
In summary, the endosperm of defective kernels underwent normal fertilization and cell division in the early stage of development. The occurrence of chromosome elimination in some endosperm cells might result in defective kernels with various sizes. Meanwhile, embryonic development in most defective kernels was arrested and formed embryo-less defective kernels eventually.
No Difference in the Coding Sequence and the Expression of the CENH3 Gene Was Detected among CAU5, ZD958, and B73
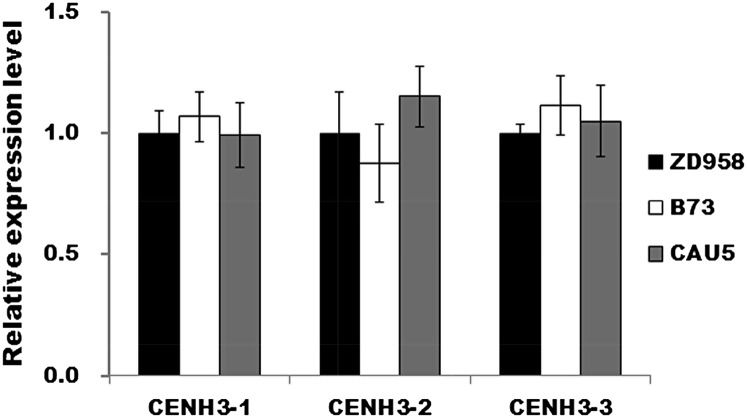
The functional defection of the CENH3 gene has been found to be associated with haploid formation during plant hybridization (Ravi and Chan, 2010; Sanei et al., 2011). To determine whether the CENH3 gene was identical in inducer lines and noninducer lines, we isolated the gene from CAU5, ZD958, and B73 and examined the coding sequence (CDS) and the expression level of the CENH3 gene. No difference in the CENH3 gene sequence was found among the three lines (Supplemental Fig. S4), suggesting that the inducing capability of CAU5 is not due to the variation of CDS of the CENH3 gene. Moreover, we performed quantitative reverse transcription (qRT)-PCR assay to measure the relative expression level of the CENH3 gene in CAU5, ZD958, and B73. The expression level of the CENH3 gene in ZD958 was arbitrarily set to 1.0. The relative expression level of CENH3 in CAU5 and B73 was normalized to the level in ZD958 (Fig. 6). No significant difference in the expression level of CENH3 gene was found among the lines. Therefore, the inducing capability of CAU5 was not derived from the CENH3 gene. This result is consistent with the quantitative trait locus (QTL) mapping results of haploid induction trait (Prigge and Melchinger, 2012), which show that the major QTL for haploid induction trait is located on bin 1.04 from chromosome 1, but the CENH3 gene is located at a region on chromosome 6 where no QTL for haploid induction trait has been detected.
Figure 6.
Relative expression level of CENH3 in ZD958, CAU5, and B73 from different positions within CENH3 CDS. Fourteen-day-old seedlings grown in a greenhouse were sampled for gene expression analysis. The expression level was determined by qRT-PCR. Actin was used as the internal standard.
Inducer DNA Introgression during Haploid Formation Was Detected by the 50K SNP Array
Among all the 50,904 SNP markers, 17,905 makers showed SNP polymorphisms between the parental lines Z58 and CAUHOI, and 17,910 makers exhibited polymorphisms between line Z58 and CAU5. No polymorphisms were identified in six of the haploids derived from the cross of Z58 × CAUHOI, while 332 markers were found to carry polymorphisms in the rest 36 haploids derived from the cross of Z58 × CAU5. Three hundred and twenty-six out of the 332 markers originated from a single haploid plant (B7), and the remaining six markers originated from five other haploids.
When ignoring the haploid B7, the mean frequency of segment introgression in the haploid derived from the cross of Z58 × CAUHOI was zero, and the mean frequency in the haploid derived from the cross of Z58 × CAU5 was 0.0012%. More interestingly, we found that all the 326 makers, which had polymorphisms and originated from haploid B7, showed heterozygous genotype (the genotype of both parental lines Z58 and CAU5). Moreover, 320 out of the 326 markers were located in the region from 105,475,851 to 149,288,994 bp, which was close to the centromere of chromosome 4 (Supplemental Fig. S5). When the SNP markers without polymorphism between the parental lines were considered, the whole region from 105,475,851 to 149,288,994 bp also showed heterozygous genotype in B7.
To further determine whether the haploid B7 had a heterozygous genotype for the 43.8-Mb fragment derived from paternal introgression, we used 84 simple sequence repeat (SSR) markers (75 were newly developed and nine were from the database MaizeGDB) and 32 insertion-deletion polymorphism (IDP) markers (from the database MaizeGDB) from the region of 105,475,851 to 149,288,994 bp to examine the fragment. Among all the 116 markers, three fragment length polymorphic markers were found to have the heterozygote genotype of both parents (Fig. 7). Based on these results, we proposed a model to explain the existence of heterozygous genotype in a haploid (Supplemental Fig. S5). After an inducer sperm fertilized the Z58 egg, a haploid was formed accompanied with a 43.8-Mb fragment paternal introgression, while all other paternal chromosomes were lost. The fragment was integrated into a sister chromatid or attached to a maternal chromosome. Subsequently, this heterozygous chromosome produced two daughter chromosomes, each of which carried a different homozygous genotype (one with the paternal introgression and the other without it) during following cell division, resulting in a heterozygous haploid (Supplemental Fig. S5A). Another possibility is that the DNA fragment might exist independently as a mini chromosome (Supplemental Fig. S5B).
Figure 7.
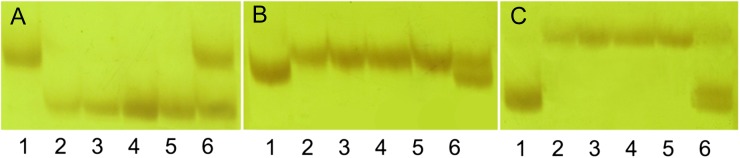
The polymorphisms of SSR markers among haploid plants derived from the cross Z58 × CAU5. The lanes from 1 to 6 were CAU5, Z58, B18, B27, B33, and B7, respectively. B18, B27, B33, and B7 are haploids derived from the parents Z58 and CAU5. A, Polymorphism detected by marker X10. B, Polymorphism detected by marker X35. C, Polymorphism detected by marker umc1317. [See online article for color version of this figure.]
DISCUSSION
The Mechanism Underlying the in Vivo Induction of Maternal Haploid in Maize
The mechanism underlying the haploid induction by inducer lines remains unclear due to the absence of direct evidence. We developed two new inducer lines, CAUB and CAUYFP, which contained B chromosomes and CENH3-YFP, respectively, to pollinate elite hybrid ZD958. The B chromosome(s) from the male parent was observed in a few haploid embryos and haploid plants, indicating that haploid formation undergoes double fertilization. In addition, the embryo at 7 DAP of one haploid was found to contain mixoploid cells, among which the diploid cells had 20 normal A chromosomes plus B chromosomes, and the haploid cells only had 10 normal A chromosomes. Our findings provide direct evidence to support that selective chromosome elimination is involved in the mechanism underlying in vivo haploid induction. Nevertheless, we could not exclude the possibility that other mechanisms might contribute to in vivo haploid induction as well. For instance, one of the two sperm cells might fail to fuse with an egg cell and instead might trigger haploid embryogenesis. Our results showed that most defective kernels did not contain an embryo, potentially because of the absence of fertilization of the egg. Alternatively, the embryos could be aneuploid haploid, which might abort early before detection or a normal embryo might abort because of the defective endosperm.
The time window of haploid formation was determined by FISH analysis on the embryos at 7 and 10 DAP. Except that one of the embryos was mixoploid, all the other embryos developed either diploid or haploid. Micronucleus was not detected in the embryos at both 7 and 10 DAP. The percentage of haploid embryos was 4.36%, 4.51%, and 6.75% in 7-DAP, 10-DAP, and mature seeds, respectively, which was not significantly different. Therefore, the inducer chromosomes were completely eliminated within the first 7 DAP in haploid embryos. It has been reported that about 56.4% of the radicles from the haploid kernels of sweet corn induced by HZI1 are mixoploid (Zhang et al., 2008). However, in our study, we did not find mixoploid in radicles and even in the embryo at 7 to 10 DAP. The discrepancy might result from different materials and methods used in our study. Our results were different from the observation in the studies of wide cross. The paternal chromosomes of wide cross are gradually lost in wheat and pearl millet crosses (Gernand et al., 2005). The percentage of cells containing pearl millet-positive micronuclei reaches the maximum (30%) in the embryos 6 to 8 DAP, while in the embryos between 17 and 23 DAP, micronuclei are only occasionally observed (Gernand et al., 2005). In our study, micronuclei were not found in the embryos at 7 or 10 DAP. Therefore, our results suggested that chromosome elimination during maternal haploid formation in maize might occur at a very early stage of embryonic development (during the first a few cycles of cell division).
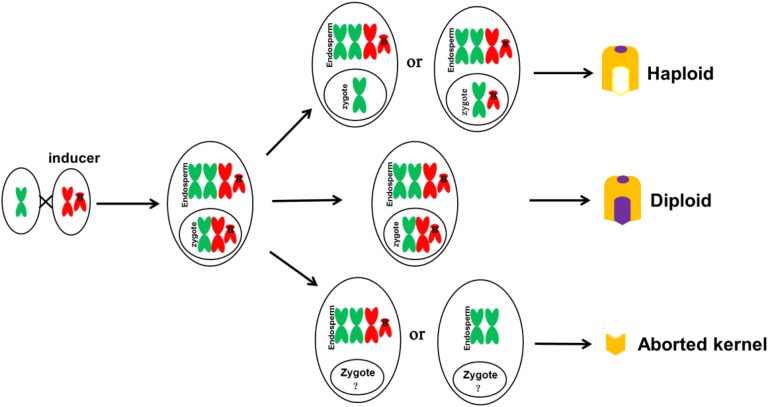
In addition, we performed a genome-wide identification of genotypic difference by using a 50K SNP array to further confirm the occurrence of large paternal fragment introgression. The paternal introgression indicates the occurrence of double fertilization during haploid formation. Our results suggest that an embryo sac could have three possible fates after fertilization (Fig. 8). Most embryo sacs develop into diploid kernels containing B chromosome. A small number of them develop into haploid kernels with (a few) or without B chromosome. The rest of the embryos become defective kernels.
Figure 8.
Schematic illustration of the fate of fertilized embryo sac with B chromosomes (without considering the mechanism of B chromosomes accumulation). Most embryo sacs developed into diploid kernels with B chromosomes. A small number of them developed into haploid kernels with (seldom) or without B chromosome. The rest of embryos became defective kernels. The development of zygotes is complicated, and it is yet to be confirmed whether the embryo will be produced
Several hypotheses have been proposed to explain uniparental chromosome elimination during hybrid embryo development in plants, such as asynchrony in cell cycle (Gupta, 1969), nucleoprotein metabolism (Bennett et al., 1976; Laurie and Bennett, 1989), malfunction of sister chromatids segregation (Ishii et al., 2010), and inactive centromere (Kim et al., 2002; Jin et al., 2004; Mochida et al., 2004). Chromosome elimination has been found to be associated with the formation of multipolar spindles (Subrahmanyam and Kasha, 1973) or nuclear extrusions (Gernand et al., 2005, 2006). The mechanism underlying haploid formation recently has been found to be related with the CENH3 gene. Sanei et al. (2011) found that CENH3 of H. bulbosum is inactive and/or CENH3 of H. vulgare cannot be successfully incorporated into the male parent in the classic barley interspecies cross (H. vulgare × H. bulbosum), which causes the failure of kinetochore assembly and leads to female haploid formation. In addition, chromosomes of the parent containing null CENH3 are eliminated, leading to haploids in Arabidopsis (Arabidopsis thaliana; Ravi and Chan, 2010). In our study, we found that the maize inducer lines produced haploids when the inducer lines were selfed with female and male gametophytes carrying identical CENH3 genes, suggesting that the CENH3 gene did not contribute to haploid-inducing capacity directly. This finding is consistent with the study on QTL mapping analysis (Prigge and Melchinger, 2012). However, CENH3 still might be associated with chromosome elimination during haploid formation in maize. In this study, we did not detect any differences in the CDSs and mRNA expression level between the inducer and noninducer lines, while the splice variants, translation, modification, and other regulatory level of CENH3 might be different. It has been shown that CENH3 acts epigenetically in plants (Han et al., 2009; Ravi and Chan, 2010; Sanei et al., 2011).
Haploid Formation with Rare Inducer Fragment Introgression
Our results from the genome-wide SNP array demonstrated that few inducer fragments were introgressed during haploid induction. No polymorphism was identified in six of the haploids derived from the cross of Z58 × CAUHOI. Similarly, only six discontinuous SNP markers were found to originate from the inducer in five out of 35 haploids derived from the cross of Z58 × CAU5, which indicated that the introgression frequency of inducer DNA fragments in these haploids was extremely low. In contrast, in our previous study, we showed that the introgression frequency was 1.84% (Li et al., 2009). The discrepancy might due to the following three factors: (1) Different female parents were used in the two studies. The hybrid ZD958 was used in the previous research, while the inbred line Z58 (with more pure genetic background) was used in this study. (2) Different types of markers were used. The PCR-based SSR markers were used in previous study, while the sequencing based SNP markers were used in this study. (3) The number of molecular markers used in the two studies was different. Only 40 markers were used in the previous study, which might cause a high false-positive rate, while 56,100 markers were used in this study. Our SNP array has limitations. For example, the marker coverage is low (the mean distance between adjacent SNP markers is 40.7 kb) and the markers are not evenly distributed. Meanwhile, the genetic similarity of the genotypes is also a limitation; only approximately 17,900 SNP makers exhibited polymorphisms between two parents in our study. Thus, we might miss some small inducer segments in our detection. In addition, we removed those SNPs with three or more heterozygous genotypes, which could possibly underestimate the frequency of paternal introgression. A next-generation sequencing-based approach might be helpful to detect smaller inducer segments and provide more detailed information.
Unexpectedly, a large paternal fragment of approximately 44 Mb (spanning 320 SNP markers) was detected in one haploid (B7). The genotype of this region was heterozygous (Z58/CAU5), indicating that illegitimate recombination or similar unbalanced translocation process might be triggered as a result of chromosome fragmentation during the process of genome elimination after the fusion of egg and sperm cells (Supplemental Fig. S5A). The paternal chromosome is eliminated during the subsequent mitosis, resulting in mosaic haploid daughter cells. Consistently, a truncated B chromosome was found during haploid formation in our study (Fig. 3, E and E′). The other possibility is that the DNA fragment might exist independently as a mini chromosome (Supplemental Fig. S5B). Since the DNA fragment that shows heterozygosity in the haploid maps very close to the centromere of chromosome 4, this 44-Mb DNA fragment along with either complete or truncated centromere repeat sequences can be stably maintained as a mini chromosome if the broken DNA ends are repaired.
Chromosome Elimination Occurred in Both Embryo and Endosperm
After sexual hybridization, some hybrid embryos undergo chromosome elimination to produce haploids with normal endosperm, while some of the other endosperms experience abortion to produce defective kernel, which barely has vitality and fertility (Xu et al., 2013). It is not clear whether chromosome elimination occurring during endosperm development is similar to that during embryonic development. In our study, CENH3-YFP signals were detected in all types of kernels (including defective kernels and haploids) derived from the cross of ZD958 × CAUYFP at 13 to 15 DAP, suggesting that the chromosome of the male parent, which carries YFP signals, is involved in the fertilization.
In addition, we determined the ploidy of the endosperm in defective kernels at 5 to 15 DAP by FISH assay (Fig. 5, F–H). Normal endosperm cells should contain three copies of 45S rDNA or Cent4 signals. Our results showed that some cells of endosperms from 9 to 15 DAP contained only two copies of chromosomes 6 or 4, which indicates that these endosperms are mixoploids or aneuploids. These results support that chromosome elimination occurs in aborted endosperm. It has been shown that the endosperms of defective kernels contain the paternal haplotype of sed1 at the mature stage (Xu et al., 2013). Consistently, we also detected YFP signals in all the aborted endosperms at 15 DAP. In each aborted kernel, YFP signals were detected in only partial cells but not all the cells. Chromosome elimination is thought to be a gradual process and might only occur in some endosperm cells (Xu et al., 2013). Our results suggest that chromosome elimination in embryo is completed at an early stage of embryonic development, while chromosome elimination in endosperm abortion is a slower process, although the two processes could be controlled by the same genes.
B Chromosomes Evaded Chromosome Elimination
We found that a few B chromosomes were able to evade elimination. The percentage of the haploid derived from the cross of ZD958 × CAUB and containing B chromosomes was 2.7%. The percentage of the diploid plants containing B chromosomes and derived from the same cross was 52.6% (Table II), which was significantly higher than that of the haploid plants. Thus, B chromosomes were eliminated in majority of the haploids and survived only in a very small number of the haploid plants. These results indicate that all chromosomes, including B chromosome(s), undergo the process of chromosome elimination. Although no A chromosome aneuploidy in the haploid was detected in our study, we could not exclude the possibility that extra A chromosome could show a similar pattern. However, due to the extreme aneuploidy in the haploid, A chromosome aneuploidy at an early developmental stage could be lethal.
We propose that as soon as a sperm cell and an egg cell fuse together, the fate of chromosomes (including B chromosomes) is decided and then chromosome elimination occurs in a very narrow time window (during the first a few cycles of cell division). If normal A chromosomes or B chromosome(s) were able to survive the first few cell divisions, they would be preserved in the following cell divisions, producing normal diploids or B chromosome-containing haploids.
We speculate that the B chromosome preservation is associated with the intrinsic characteristics of B chromosomes, such as B chromosome accumulation mechanisms (Roman, 1947, 1948; Carlson, 2007; Han et al., 2007), effect of B chromosomes on crossing over in A chromosomes (Hanson, 1969; Robertson, 1984; Carlson, 1994), or the function of B centromere (Jin et al., 2005). In particular, the uniqueness of B heterochromatin involved in the different timing of DNA replication between A and B chromosomes (Pryor et al., 1980) and in the histone modifications associated with both A and B chromosomes might contribute to B chromosome survival (Jin et al., 2008).
MATERIALS AND METHODS
Plant Materials
Maize (Zea mays) haploid inducer line CAU5, B chromosomes containing line B73+B, and the transgenic line HiIIYFP (kindly provided by Dr. James Birchler, University of Missouri-Columbia) carrying a CENH3-YFP fusion expression vector (Jin et al., 2008) were used to develop new inducers, which contained either CENH3-YFP or B chromosomes. A maize commercial hybrid ZD958 was used for haploid production. In addition, the haploids derived from crosses between the elite maize inbred line Z58 and two maize haploid inducer lines CAU5 and CAUHOI (Li et al., 2009) were used for SNP analysis.
Haploid Identification and HIR Determination
Haploid identification was based on the R1-nj color marker (Li et al., 2009). Putative haploids were identified as having a purple endosperm and colorless embryo (indicating a female parent derived embryo), and diploids were identified as containing both a purple endosperm and purple embryo (indicating crossed embryo). HIR was determined as described previously (Li et al., 2009). At least three ears of ZD958 were used as the female tester. To determine the significance of the difference of HIR among inducer lines, we performed a U test at the level of a = 0.05.
Root Tip and Ovary Preparation for FISH Assay
Analyses on root tip chromosomes were performed according to the procedures described previously (Wang et al., 2011). Kernels were germinated at 28°C for 4 d. The root tips were then pretreated in 0.002 mol/L 8-hydroxyquinoline at room temperature for 2 h to accumulate metaphase cells. Then, the root tips were placed directly into Carnoy’s solution (ethanol:glacial acetic = 3:1) for fixation and stored at −20°C until use. The root tips were digested with 2% cellulose and 1% pectolyase at 37°C for 2 h. Squashes were prepared in the same manner.
For the preparation of ovaries, 7- to 10-DAP ovaries were isolated and fixed in ethanol/acetic acid (3:1). Embryos were dissected under a stereomicroscope (Olympus CX41) using fine needles. Immature embryos were slightly digested by 2% cellulose and 1% pectolyase at 37°C for 10 min and squashed in a 50% acetic acid solution. Slides with chromosomes were frozen in liquid nitrogen. After the coverslips were removed, the slides were refixed in Carnoy’s solution. For the preparation of defective kernels, endosperms were isolated from ovaries at 5 to 15 DAP and dissected into patches with scalpel. Slides were processed as described above.
Probes and FISH Assay
The plasmids harboring maize tandem repeat 45S rDNA (Kato et al., 2004), maize centromeric satellite repeat CentC (Ananiev et al., 1998), maize centromere 4-specific sequences Cent4 (Page et al., 2001), and B chromosome specific repeat ZmBs (Alfenito and Birchler, 1993) were purified and labeled with digoxigenin-11-dUTP (Roche), biotin-11-dUTP (Vector Laboratories), or diethylaminocoumarin-5-dUTP (NEN Life Science Products) via nick translation reaction. The digoxigenin- and biotin-labeled probes were detected by antidigoxigenin antibody conjugated with Rhodamin (Roche) and antiavidin antibody conjugated with fluorescein isothiocyanate (Vector Laboratories), respectively. Sequential FISH was conducted according to the protocol published previously (Zhao et al., 2011). Slides were counterstained with 4′,6-diamino-phenylindole (Vector Laboratories).
FISH images were captured digitally using a CCD camera (QImaging; RETGA-SRV FAST 1394) attached to an Olympus BX61 epifluorescence microscope. Chromosome spread analysis was performed using Image-Pro Plus 6.0 software (Media Cybernetics), and image adjustments were performed with Adobe Photoshop CS 3.
YFP Signal Detection in Maize Root Tips and Kernels
For CENH3-YFP signal detection, root tips cut from 4-d-old seedlings were directly placed on the slide and squashed in phosphate-buffered saline buffer (0.13 M NaCl, 0.007 M Na2HPO4, and 0.003 M NaH2PO4, pH 7.4). Imaging was performed using an Olympus BX61 epifluorescence microscope and a QImaging CCD camera. The detection of YFP signals in immature kernels between 3 and 30 DAP was performed according to the same protocol, except that the ears from different pollination days were fixed in 4% paraformaldehyde for 20 min as described previously (Jin et al., 2008). After dissection, endosperm and embryo of kernels were checked separately.
Sequence Analysis and qRT-PCR
To analyze the sequence of the CENH3 gene, total RNA was extracted using an RNA-pure high-purity total rapid extract kit (BioTeke) from 14-d-old seedlings of CAU5, ZD958, and B73. cDNA was synthesized with the reverse transcription (RT) reagent kit (Life Technologies) according to the manufacturer’s protocol. The sequences for CENH3-specific primers, CENH3-FL-F and CENH3-FL-R, for RT-PCR are listed in Supplemental Table S1. The RT-PCR products were cloned into pGEM-T Easy Vector (Promega) according to the manufacturer’s instructions and sequenced at Invitrogen Biotechnology Co. Sequences were analyzed using ClustalX software (Thompson et al., 2002). For qRT-PCR analysis, the CENH3-specific primers were designed to cover different exons: exons 1 to 4 for CENH3-1, exons 3 to 5 for CENH3-2, and exons 4 to 7 for CENH3 (Supplemental Table S1). qRT-PCR was conducted on an ABI 7500 (Life Technologies) with the SYBR RT-PCR kit (Takara). Relative expression level of the CENH3 gene was calculated according to the 2−ΔΔCT (cycle threshold) method (Livak and Schmittgen, 2001) with actin as the internal control.
SNP and SSR Analysis
SNP genotyping screening of two haploid inducers (CAU5 and CAUHOI), one noninducer inbred line (Z58), and 42 haploids (including six haploids derived from the cross of Z58 × CAUHOI and 36 from the cross of Z58 × CAU5) was performed using the Illumina SNP chip MaizeSNP50 (Illumina), which contains 56,110 SNPs. Data were analyzed using the Illumina BeadStudio genotyping software. The quality of SNPs was controlled by removing SNPs that did not meet the following criteria: (1) <10% missing values and (2) no more than three heterozygous genotypes. After this filtering, 50,904 SNPs were retained for further analysis. The chromosomal positions of the SNPs were determined using the B73 reference genome (B73 RefGen_v1). The mean distance between adjacent SNP markers was 40.7 kb. For SSR analysis, PCR-based marker development (primer sequences for each of these markers are provided in Supplemental Table S2) and genotyping were conducted according to the protocol of Yang et al. (2010).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Chromosomes with CENH3-YFP signal (green) in the root tips of CAUYFP.
Supplemental Figure S2. The percentage of endosperms with three types of abnormal cells at 5 to 7 DAP, 9 to 11 DAP, and 13 to 15 DAP by FISH.
Supplemental Figure S3. Determination of ploidy at the different position of the same aborted endosperm by FISH.
Supplemental Figure S4. Sequence alignment of CENH3 CDS from ZD958, CAU5, and B73.
Supplemental Figure S5. Schematic diagram for fragment introgression from an inducer in haploid B7.
Supplemental Table S1. The sequences for the primers used in CENH3 analysis.
Supplemental Table S2. The sequences for the primers used in SNP analysis.
Acknowledgments
We thank Drs. Gui Su and Huihuang Yan (University of Wisconsin-Madison) for their valuable comments.
Glossary
- DH
doubled haploid
- SNP
single nucleotide polymorphism
- YFP
yellow fluorescent protein
- FISH
fluorescence in situ hybridization
- DAP
days after pollination
- CDS
coding sequence
- RT
reverse transcription
- QTL
quantitative trait locus
- SSR
simple sequence repeat
- HIR
haploid induction rate
- qRT
quantitative reverse transcription
References
- Alfenito MR, Birchler JA. (1993) Molecular characterization of a maize B chromosome centric sequence. Genetics 135: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev EV, Phillips RL, Rines HW. (1998) Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc Natl Acad Sci USA 95: 13073–13078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Finch RA, Barclay IR. (1976) The time rate and mechanism of chromosome elimination in Hordeum hybrids. Chromosoma 54: 175–200 [Google Scholar]
- Bylich VG, Chalyk ST. (1996) Existence of pollen grains with a pair of morphologically divergent sperm nuclei as a possible cause of the haploid-inducing capacity in ZMS line. Maize Genet. Coop. News Lett. 70: 33 [Google Scholar]
- Carlson WR. (1994) Crossover effects of B chromosomes may be ‘selfish’. Heredity 72: 636–638 [Google Scholar]
- Carlson WR. (2007) Locating a site on the maize B chromosome that controls preferential fertilization. Genome 50: 578–587 [DOI] [PubMed] [Google Scholar]
- Chalyk S, Baumann A, Daniel G, Eder J. (2003) Aneuploidy as a possible cause of haploid-induction in maize. Maize Genet. Coop. News Lett. 77: 29–30 [Google Scholar]
- Coe EH. (1959) A line of maize with high haploid frequency. Am Nat 93: 381–382 [Google Scholar]
- Dunwell JM. (2010) Haploids in flowering plants: origins and exploitation. Plant Biotechnol J 8: 377–424 [DOI] [PubMed] [Google Scholar]
- Evans MMS. (2007) The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development. Plant Cell 19: 46–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E (2004) Molekulargenetische untersuchungen zum vorkommen paternaler DNA-Übertragung bei der in-vivo-Haploideninduktion bei Mais (Zea mays L.). PhD thesis. University of Hohenheim, Stuttgart, Germany [Google Scholar]
- Forster BP, Heberle-Bors E, Kasha KJ, Touraev A. (2007) The resurgence of haploids in higher plants. Trends Plant Sci 12: 368–375 [DOI] [PubMed] [Google Scholar]
- Geiger HH (2009) Doubled haploids. In JL Bennetzen, S Hake, eds, Handbook of Maize Genetics and Genomics. Springer, Berlin, pp 641–657 [Google Scholar]
- Gernand D, Rutten T, Pickering R, Houben A. (2006) Elimination of chromosomes in Hordeum vulgare x H. bulbosum crosses at mitosis and interphase involves micronucleus formation and progressive heterochromatinization. Cytogenet Genome Res 114: 169–174 [DOI] [PubMed] [Google Scholar]
- Gernand D, Rutten T, Varshney A, Rubtsova M, Prodanovic S, Brüss C, Kumlehn J, Matzk F, Houben A. (2005) Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell 17: 2431–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SB. (1969) Duration of mitotic cycle and regulation of DNA replication in Nicotiana plumbaginifolia and a hybrid derivative of N. tabacum showing chromosome instability. Can J Genet Cytol 11: 133–142 [Google Scholar]
- Han FP, Gao Z, Birchler JA. (2009) Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell 21: 1929–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han FP, Lamb JC, Yu W, Gao Z, Birchler JA. (2007) Centromere function and nondisjunction are independent components of the maize B chromosome accumulation mechanism. Plant Cell 19: 524–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson GP. (1969) B chromosome-stimulated crossing over in maize. Genetics 63: 601–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Ueda T, Tanaka H, Tsujimoto H. (2010) Chromosome elimination by wide hybridization between Triticeae or oat plant and pearl millet: pearl millet chromosome dynamics in hybrid embryo cells. Chromosome Res 18: 821–831 [DOI] [PubMed] [Google Scholar]
- Jin WW, Lamb JC, Vega JM, Dawe RK, Birchler JA, Jiang JM. (2005) Molecular and functional dissection of the maize B chromosome centromere. Plant Cell 17: 1412–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin WW, Lamb JC, Zhang WL, Kolano B, Birchler JA, Jiang JM. (2008) Histone modifications associated with both A and B chromosomes of maize. Chromosome Res 16: 1203–1214 [DOI] [PubMed] [Google Scholar]
- Jin WW, Melo JR, Nagaki K, Talbert PB, Henikoff S, Dawe RK, Jiang JM. (2004) Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell 16: 571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasha KJ, Kao KN. (1970) High frequency haploid production in barley (Hordeum vulgare L.). Nature 225: 874–876 [DOI] [PubMed] [Google Scholar]
- Kato A, Lamb JC, Birchler JA. (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA 101: 13554–13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle JL. (1969) Androgenesis conditioned by a mutation in maize. Science 166: 1422–1424 [DOI] [PubMed] [Google Scholar]
- Kim NS, Armstrong KC, Fedak G, Ho K, Park NI. (2002) A microsatellite sequence from the rice blast fungus (Magnaporthe grisea) distinguishes between the centromeres of Hordeum vulgare and H. bulbosum in hybrid plants. Genome 45: 165–174 [DOI] [PubMed] [Google Scholar]
- Lashermes P, Gaillard A, Beckert M. (1988) Gynogenetic haploid plants analysis for agronomic and enzymatic markers in maize (Zea mays L.). Theor Appl Genet 76: 570–572 [DOI] [PubMed] [Google Scholar]
- Laurie DA, Bennett MD. (1988) The production of haploid wheat plants from wheat × maize crosses. Theor Appl Genet 76: 393–397 [DOI] [PubMed] [Google Scholar]
- Laurie DA, Bennett MD. (1989) The timing of chromosome elimination in hexaploid wheat × maize crosses. Genome 32: 953–961 [Google Scholar]
- Li L, Xu XW, Jin WW, Chen SJ. (2009) Morphological and molecular evidences for DNA introgression in haploid induction via a high oil inducer CAUHOI in maize. Planta 230: 367–376 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mochida K, Tsujimoto H, Sasakuma T. (2004) Confocal analysis of chromosome behavior in wheat x maize zygotes. Genome 47: 199–205 [DOI] [PubMed] [Google Scholar]
- Page BT, Wanous MK, Birchler JA. (2001) Characterization of a maize chromosome 4 centromeric sequence: evidence for an evolutionary relationship with the B chromosome centromere. Genetics 159: 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge V, Melchinger AE. (2012) Production of haploids and doubled haploids in maize. Methods Mol Biol 877: 161–172 [DOI] [PubMed] [Google Scholar]
- Pryor A, Faulkner K, Rhoades MM, Peacock WJ. (1980) Asynchronous replication of heterochromatin in maize. Proc Natl Acad Sci USA 77: 6705–6709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M, Chan SWL. (2010) Haploid plants produced by centromere-mediated genome elimination. Nature 464: 615–618 [DOI] [PubMed] [Google Scholar]
- Röber FK, Gordillo GA, Geiger HH. (2005) In vivo haploid induction in maize: performance of new inducers and significance of doubled haploid lines in hybrid breeding. Maydica 50: 275–283 [Google Scholar]
- Robertson DS. (1984) Different frequency in the recovery of crossover products from male and female gametes of plants hypoploid for B-A translocations in maize. Genetics 107: 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman H. (1947) Mitotic nondisjunction in the case of interchanges involving the B-type chromosomein maize. Genetics 32: 391–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman H. (1948) Directed fertilization in maize. Proc Natl Acad Sci USA 34: 36–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanei M, Pickering R, Kumke K, Nasuda S, Houben A. (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci USA 108: E498–E505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar KR, Panke S, Sachan JKS. (1972) Development of maternal-haploidy-inducer lines in maize. Indian J. Agric. Sci. 42: 781–786 [Google Scholar]
- Subrahmanyam NC, Kasha KJ. (1973) Selective chromosomal elimination during haploid formation in barley following interspecific hybridization. Chromosoma 42: 111–125 [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. In AD Baxevanis, ed, Current Protocols in Bioinformatics, Chapter 2, Unit 2.3. John Wiley and Sons, New York, pp 1–22 [DOI] [PubMed] [Google Scholar]
- Wang GX, He QY, Liu F, Cheng ZK, Talbert PB, Jin WW. (2011) Characterization of CENH3 proteins and centromere-associated DNA sequences in diploid and allotetraploid Brassica species. Chromosoma 120: 353–365 [DOI] [PubMed] [Google Scholar]
- Wedzony M, Röber FK, Geiger HH (2002) Chromosome elimination observed in selfed progenies of maize inducer line RWS. In XVIIth International Congress on Sex Plant Reproduction. Maria Curie-Sklodowska University Press, Lublin, Poland, p 173 [Google Scholar]
- Xu XW, Li L, Dong X, Jin WW, Melchinger AE, Chen SJ. (2013) Gametophytic and zygotic selection leads to segregation distortion through in vivo induction of a maternal haploid in maize. J Exp Bot 64: 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Yin G, Guo Y, Zhang D, Chen S, Xu M. (2010) A major QTL for resistance to Gibberella stalk rot in maize. Theor Appl Genet 121: 673–687 [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR. (2000) Programmed cell death during endosperm development. Plant Mol Biol 44: 283–301 [DOI] [PubMed] [Google Scholar]
- Zhang ZL, Qiu FZ, Liu YZ, Ma KJ, Li ZY, Xu SZ. (2008) Chromosome elimination and in vivo haploid production induced by Stock 6-derived inducer line in maize (Zea mays L.). Plant Cell Rep 27: 1851–1860 [DOI] [PubMed] [Google Scholar]
- Zhao X, Lu JY, Zhang ZH, Hu JJ, Huang SW, Jin WW. (2011) Comparison of the distribution of the repetitive DNA sequences in three variants of Cucumis sativus reveals their phylogenetic relationships. J Genet Genomics 38: 39–45 [DOI] [PubMed] [Google Scholar]