A plant-specific chloroplast endopeptidase is identified that functions in protection of the photosynthetic apparatus from photo-oxidative stress.
Abstract
Excess light can have a negative impact on photosynthesis; thus, plants have evolved many different ways to adapt to different light conditions to both optimize energy use and avoid damage caused by excess light. Analysis of the Arabidopsis (Arabidopsis thaliana) mutant snowy cotyledon4 (sco4) revealed a mutation in a chloroplast-targeted protein that shares limited homology with CaaX-type endopeptidases. The SCO4 protein possesses an important function in photosynthesis and development, with point mutations rendering the seedlings and adult plants susceptible to photooxidative stress. The sco4 mutation impairs the acclimation of chloroplasts and their photosystems to excess light, evidenced in a reduction in photosystem I function, decreased linear electron transfer, yet increased nonphotochemical quenching. SCO4 is localized to the chloroplasts, which suggests the existence of an unreported type of protein modification within this organelle. Phylogenetic and yeast complementation analyses of SCO4-like proteins reveal that SCO4 is a member of an unknown group of higher plant-specific proteinases quite distinct from the well-described CaaX-type endopeptidases RAS Converting Enzyme1 (RCE1) and zinc metallopeptidase STE24 and lacks canonical CaaX activity. Therefore, we hypothesize that SCO4 is a novel endopeptidase required for critical protein modifications within chloroplasts, influencing the function of proteins involved in photosynthesis required for tolerance to excess light.
Plants have to acclimate to marked changes in light intensity and quality. Shading effects by neighboring and larger plants as well as short- and long-term variations in illumination intensities during a day or season cause highly variable growth light conditions. Consequently, a strong requirement of acclimation in photosynthetic activity is necessary to establish autotrophy and to limit damage (Kanervo et al., 2005; Dietzel et al., 2008; Pesaresi et al., 2010). This is especially true in young seedlings during the rapid biogenesis of photosystems. For example, lutein- and violaxanthin-deficient lut2aba1 plants have bleached cotyledons and seedlings are often nonviable, yet their mature leaves are green, albeit compromised in different aspects of photosynthesis (Pogson et al., 1998), as opposed to some other carotenoid transgenics (Rissler and Pogson, 2001). A class of mutations that impact chloroplast biogenesis in seedlings was identified by screening for chlorosis during early seedling development. Although referred to as snowy cotyledon (sco; Albrecht et al., 2006, 2008, 2010), several of the SCO genes are essential for plant viability (Albrecht et al., 2006) and others impact photosynthesis and/or photorespiration in mature leaves (Albrecht et al., 2010). This is somewhat similar to the aforementioned lut2aba mutation, although to date, sco mutants have been identified to be defective in novel proteins involved in assembly, targeting, and biogenesis rather than in the core components of photosystems.
In addition to factors required for assembly, recent advances have identified important regulators in photosynthetic acclimation such as the kinases and phosphatases or redox sensors (Bellafiore et al., 2005; Bonardi et al., 2005; Tikkanen et al., 2006; Pesaresi et al., 2009; Dangoor et al., 2012; Samol et al., 2012). It could be shown that Arabidopsis (Arabidopsis thaliana) possesses separate acclimation responses to low-light (LL) and high-light (HL) conditions (Bailey et al., 2004). LL, such as growth in the shade, requires a different composition of proteins for an adequate activity of the photosynthetic apparatus than HL. It has been shown that the concerted action of the kinase STN7 and the phosphatase PPH1TAP38 regulates the adaptation of the photosynthetic apparatus to changes of light quality and quantity (Pribil et al., 2010; Shapiguzov et al., 2010). The phosphorylation/dephosphorylation of LHC II by these proteins results in a remodeling of the thylakoid membrane system and thus also in the reorganization of the electron transfer chain called state transitions (Pribil et al., 2010; Shapiguzov et al., 2010). In addition, remodeling of PSII supercomplexes occurs in short-term acclimations preceding and paralleling state transitions (Dietzel et al., 2011). Long-term acclimation to light quality gradients results in adjustment of the stoichiometry of photosystem reaction centers, improving the photochemical efficiency of the photosynthetic apparatus (Chow et al., 1990; Allen and Pfannschmidt, 2000; Hogewoning et al., 2012). HL acclimation responses involve mainly photoprotection mechanisms such as nonphotochemical quenching (NPQ), which includes a restructuring of the light-harvesting antenna and the dissipation of excess light energy by heat (Kargul and Barber, 2008; Li et al., 2009).
Higher light conditions also result in the release of reactive oxygen species, whose production can be limited or scavenged by, for example, carotenoids (singlet oxygen; Cazzonelli et al., 2009). Most of these components are produced from the plastidic 2-C-methyl-d-erythitol4 phosphate pathway. In addition, it has been shown that in higher plants, the chloroplastic 2-C-methyl-d-erythitol4 phosphate pathway provides geranylgeranyl required for protein isoprenylation, even for cytoplasmic prenylated proteins (Gerber et al., 2009).
Protein isoprenylation, a lipid modification with either a 15-carbon farnesyl group or a 20-carbon geranylgeranyl group, has been demonstrated to be involved in a variety of different processes, such as cell cycle control, signal transduction, cytoskeletal organization, and intracellular vesicle transport (Crowell, 2000). The first step of prenylation in the CaaX-type isoprenylation reaction is the attachment of the prenyl group to a Cys at the fourth-last position of the protein either by a protein farnesyl transferase (FT) or a protein geranylgeranyl transferase (Crowell, 2000; Huizinga et al., 2008). In the second step, the last three amino acids are proteolytically removed by a CaaX-type endopeptidase, with the prenyl-Cys being methylated by an iso-Cys-methyltransferase (Zhang and Casey, 1996). For Arabidopsis, the functions of just two classical type I CaaX-type endopeptidases, STE24 and RCE1/FACE, have been described so far; both of them are located to the cytosolic side of the endomembrane system (Bracha-Drori et al., 2004, 2008). No information is available for loss-of-function alleles of RCE1 and STE24 in Arabidopsis. In mice, the loss of RCE1 is embryo lethal and STE24 mutations result in bone breaking and reduced bone volumes (Bergo et al., 2002a, 2002b).
To date, no indication of protein prenylation occurring in the plastids has been described. However, in spinach chloroplasts, farnesylated proteins have been detected using radioactively labeled farnesyl with a protein FT activity (Parmryd et al., 1997). Here, we describe a sco mutant, sco4, that results in photooxidative bleaching at moderate to higher light intensities and demonstrate that the gene encodes a chloroplast-localized proteinase that is related to CaaX-type proteinases and is required for chloroplast function and photosynthesis.
RESULTS
The Mutation in sco4 Impairs the Acclimation of Chloroplasts to Moderate and Higher Light Intensities
To understand more about the biogenesis and function of chloroplasts, a mutant screen was performed on ethyl methylsulfonate-mutagenized seeds of Arabidopsis (ecotype Landsberg erecta [Ler]) for plants exhibiting pale-green cotyledons in young seedlings. In this screen, the sco4 mutant was isolated, and it exhibits a different phenotype compared with the other sco mutants (Albrecht et al., 2006, 2008, 2010), since the chlorosis is only apparent under moderate to higher light regimes. That is, if seeds were germinated under continuous very low light (vLL) of 20 μmol m−2 s−1, moderate low light (mLL) of 90 μmol m−2 s−1, and moderate high light (mHL) of 230 μmol m−2 s−1, then the sco phenotype was more obvious under mLL and most clear under mHL (Fig. 1A). Mutant seedlings grown under vLL were almost indistinguishable from the wild-type seedlings (Fig. 1A). This was verified by quantification of the chlorophyll content of cotyledons from 14-d-old seedlings with sco4 seedlings under vLL having pigment levels comparable to Ler. In contrast, under mHL conditions, the chlorophyll content in the mutant decreased to around one-third of wild-type levels (Supplemental Fig. S1A).
Figure 1.
Light-dependent phenotype of sco4. A, Phenotype of 8-d-old seedlings grown under continuous light in the different light conditions after light shifts. B, Quantification of the greening process of seedlings of the wild type (squares) and sco4 (diamonds) grown under continuous 20 μmol m−2 s−1 (μE), continuous 90 μmol m−2 s−1, or continuous 230 μmol m−2 s−1.
The question arises if this phenotype is inducible or even reversible. Thus, seedlings were grown under vLL or LL conditions for 3 and 5 d, respectively, and then transferred to mHL. Conversely, seedlings germinated under mHL for the same period were transferred to LL. The seedling phenotype was monitored until day 8 and quantified until the true leaves emerged. Transferring the seedlings to mHL conditions resulted in marked bleaching of sco4 cotyledons and some chlorosis in the Ler seedlings (Fig. 1A). A Scanalyzer (Lemnatec) was used to quantify the greenness per sample per treatment over time. Using this method, the rate of greening of sco4 seedlings was found to be comparable to wild-type seedlings under vLL, whereas under mLL and mHL conditions, the bleaching effect of increasing light intensities on the mutant seedlings was apparent (Fig. 1B). Transfer of the seedlings to mHL after 3 d resulted in almost no greening of the mutant seedlings (Supplemental Fig. S1, C and F), and even transferring the seedlings after 5 d of vLL, the mutant seedlings showed reduced greening (Supplemental Fig. S1, D and G). Transfer of 3-d mHL-grown seedlings to vLL resulted in slight accumulation of chlorophyll in sco4 seedlings (Supplemental Fig. S1J). This was not the case for seedlings transferred after 5 d of mHL (Supplemental Fig. S1K). Thus, the phenotype in sco4 is inducible by increasing light intensity but is not reversible.
The 77 K Fluorescence Emission Spectra in mHL-Grown sco4 Mutants
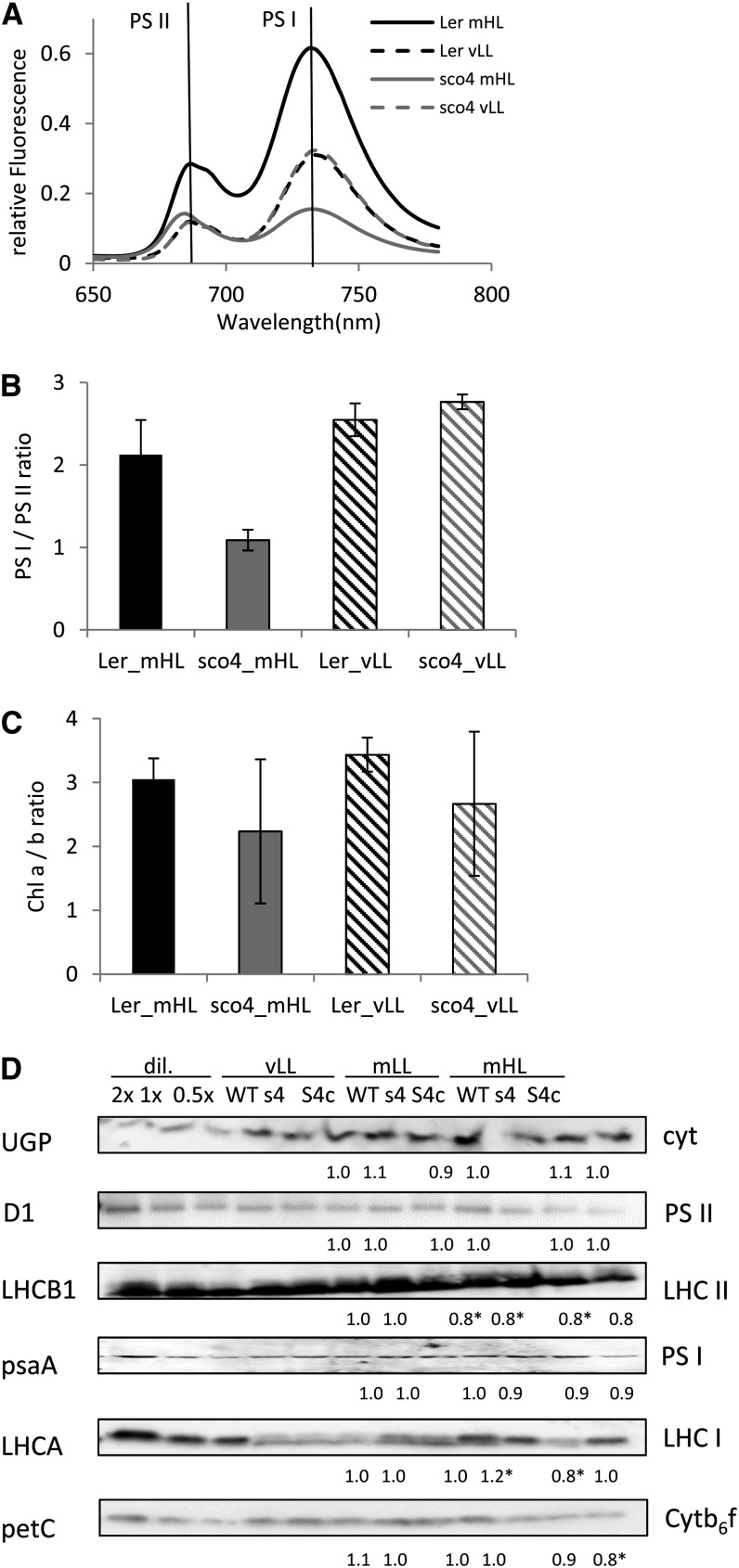
The observed bleaching phenotype of the sco4 cotyledons under increasing light conditions suggests a loss of functional photosystems. To test this, cotyledons from seedlings grown under the aforementioned different light conditions were subjected to 77 K low-temperature fluorescence measurements in order to monitor the chlorophyll fluorescence emitted by functional PSI and PSII. With an excitation wavelength of 440 nm, a semiquantitative measurement of the two photosystems was conducted, with the photosystems emitting fluorescence maxima at either 685 nm for PSII or 733 nm for PSI, using fluorescein as an internal standard for better quantification and comparison. For this, frozen and ground material of the same fresh weight was suspended in a buffer, and the analysis was performed in biological triplicates. Under vLL, sco4 displays an emission spectrum of similar intensity to wild-type plants, suggesting similar fluorescence properties of the two photosystems in sco4 and the wild type (Fig. 2A). At mHL, both peaks of PSI and PSII of sco4 are at decreased levels compared with Ler grown under the same conditions (Fig. 2A). In particular, Ler grown at mHL had elevated peaks at both 685 and 733 nm, probably due to an abundance of chlorophyll on a fresh weight basis, allowing enhanced excitation of both photosystems. A closer examination of the ratio of the 733-nm peak to the 685-nm peak reveals that under vLL, both genotypes are similar. In contrast, under mHL, the ratio of emission from PSI to that from PSII is approximately 1 in the sco4 mutant compared with approximately 2 in the wild type (Fig. 2B). This suggests that the emission from PSII relative to that from PSI may be increased in sco4 mHL plants. The ratio between chlorophyll a and chlorophyll b is highly variable in the sco4 samples, with a tendency to be lower in the mutant compared with the wild type (Fig. 2C). Furthermore, a slight shift from 687 nm in the wild type to 684 nm in the sco4 mutant under mHL could be observed. This might indicate that the transfer of excitation energy from the antenna is impaired.
Figure 2.
Quantification of both photosystems. A, A 77 K fluorescence analysis on cotyledons of 7-d-old seedlings grown under mHL (230 μmol m−2 s−1) or vLL (20 μmol m−2 s−1) compared with an internal fluorescence standard and based on fresh weight. B, Ratio of PSI to PSII in the 77 K fluorescence analysis. C, Ratio of chlorophyll a to b in 7-d-old seedlings grown under mHL and vLL. D, Western analyses on total protein extracted from 7-d-old seedlings grown under vLL, mLL (90 μmol m−2 s−1), or mHL. Numbers below indicate the intensity compared with the wild type (WT) of the same light condition as an average of two independent analyses. Cytb6f, Cytochrome b6f; s4, sco4; S4c, sco4 complemented with wild-type SCO4; cyt, cytosol; dil., dilution series.
Interestingly, when performing a quantitative western analysis with protein-specific antibodies for D1 (PSII), LHCB1 (LHC II), psaA (PSI), LHCA (LHC I), petC (Rieske FeS cytochrome b6f complex), and the cytosolic protein UDP-Glc pyrophosphorylase1, the differences in the PSI- and PSII-associated light-harvesting complexes LHC I and LHC II (Fig. 2D) are not as dramatic as implied by using the 77 K fluorescence measurement. A slight overexpression of SCO4 in the complemented line, though, resulted in a statistically significant reduction of the PetC protein in the cytochrome b6f complex. The observed reductions in LHCB and LHCA proteins under mHL are significant in the sco4 mutant, whereas all other analyzed proteins of the photosynthetic complexes seem to be unchanged in the mutant line. This might be because the fluorescence intensities are compared on the basis of equal fresh weight, whereas the western analysis of proteins is based on equal total protein content.
sco4 Is Not Affected in Its Acclimation to PSI or PSII Light Conditions
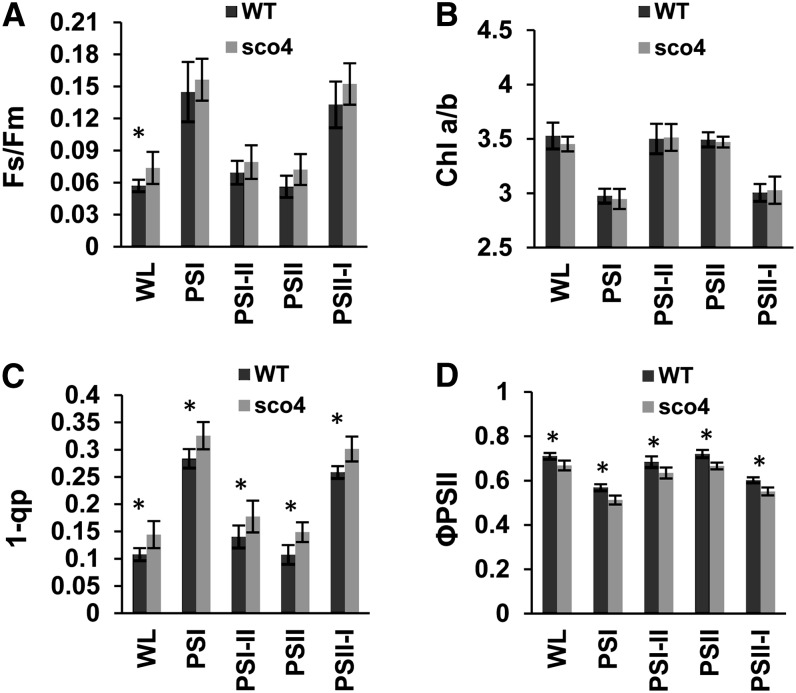
The observed effects in the 77 K fluorescence spectra prompted us to investigate if sco4 is disturbed in its ability to adjust photosystem stoichiometry. To this end, seedlings were grown on Murashige and Skoog medium and acclimated to light sources preferentially exciting either PSI or PSII and then compared with growth under white light. Under such conditions, a long-term response (LTR) is induced, in which photosystem stoichiometry is adjusted in favor of the rate-limiting one. In addition, parallel samples were shifted after acclimation from one light source to the respective other one in order to follow the reversibility of the response as described (Wagner et al., 2008). Photosynthetic parameters were determined to monitor the acclimation capacity of sco4 and Ler. The value of Fs/Fm represents the fraction of the light absorbed by the PSII antenna that is lost by constitutive dissipation, in a light-independent manner, by fluorescence and heat emission (Klughammer and Schreiber, 2008). Growth in light sources that preferentially excite either PSI or PSII typically induces a LTR, which results in a higher PSII/PSI ratio in PSI light and a lower one in PSII light. The measurement of the Fs/Fm value uses a red light source as actinic light, which efficiently excites PSII. Under these experimental conditions, PSI light-acclimated plants absorb more light than they can use and dissipate this as fluorescence, which is significantly higher in the steady state than that from PSII light-acclimated plants. A deficiency in performing a LTR, however, is indicated by the loss of significant changes in Fs/Fm observed in the stn7 mutant of Arabidopsis (Bonardi et al., 2005). In general, Fs/Fm values of wild-type seedlings acclimated to PSI light were relatively high, whereas PSII light-acclimated seedlings displayed significantly lower values, as described earlier. The sco4 mutant displayed wild-type-like changes typical for a functional LTR but exhibited a slight increase in the Fs/Fm values under all acclimation conditions (Fig. 3A). The value of 1 − qP (for photochemical quenching), representing a measure for the reduction state of the primary electron-accepting plastoquinone of PSII and the plastoquinone pool, was also increased in the mutant, whereas the ΦPSII, representing the effective quantum yield, was lower (Fig. 3, C and D). The chlorophyll a/b ratio of sco4 and the changes in it were comparable to the wild type (Fig. 3B). These data imply that the mutant is capable of performing LTR, indicating that the basic redox-controlled acclimation machinery is not disturbed. However, the higher Fs/Fm and 1 − qP values suggest some limitation in the efficiency of linear electron transport in the mutant.
Figure 3.
Determination of the LTR in Arabidopsis wild-type Col (WT) and the mutant sco4 in 7-d-old seedlings. The photosynthetic parameters Fs/Fm (A), chlorophyll (Chl) a/b ratio (B), 1 − qP (C), and ΦPSII (D) were determined. Acclimation conditions are given at the bottom. All values are means from six independent measurements ± sd. Values significantly different between the wild type and sco4 are marked with asterisks (Student’s t test for independent samples; P ≤ 0.05). WL, Normal white light.
The sco4 Mutant Is Not Able to Acclimate Photosynthetic Electron Flow to Increasing Light Intensities
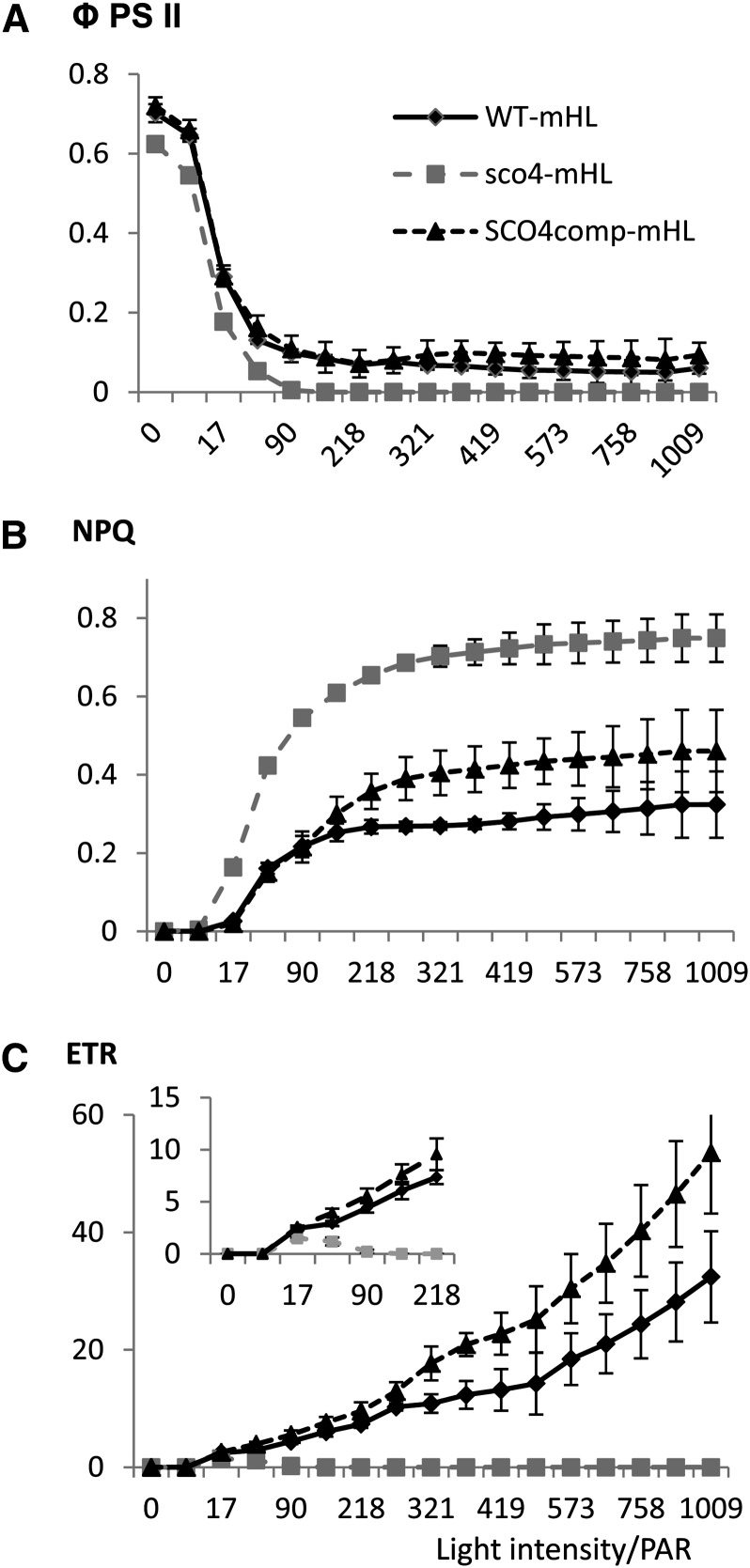
In order to investigate this photosynthetic limitation in more detail, we monitored photosynthesis under different light intensity conditions. Seedlings grown under mHL were dark adapted for 30 min before being transferred to an imaging pulse-amplitude-modulation (Walz; http://www.walz.com/) for photosynthetic measurements with actinic light flashes and a 20-s recovery phase under increasing light intensities (light kinetics from 0 to 1,000 μmol m−2 s−1). Specifically, parameters such as ΦPSII, NPQ, and electron transfer rate (ETR) were considered. When the seedlings were grown under mHL conditions with increasing light intensities, the ΦPSII values dropped for all the lines analyzed (Fig. 4A), whereas the values for both NPQ and ETR increased for the wild type (Fig. 4, B and C). Interestingly, the NPQ value for the sco4 mutant was double that for the wild type (Fig. 4B), while the ETR value dropped to 0 for sco4, even under LL (Fig. 4, C and inset). Thus, in sco4 seedlings, the quick adaptation of the electron transfer between PSII and PSI under quickly increasing light intensities seems to be perturbed when the mutant seedlings have been grown at higher light intensities, although the mechanism is still not understood.
Figure 4.
Determination of photosynthetic parameters in 7-d-old seedlings grown under mHL (230 μmol m−2 s−1). A, ΦPSII. B, NPQ. C, ETR from PSII to PSI. The inset displays the light intensity region up to 218 photosynthetically active radiation (PAR). WT, Wild type.
Increasing Growth Light Intensities Result in Increasing Loss of Photosystem Function in sco4
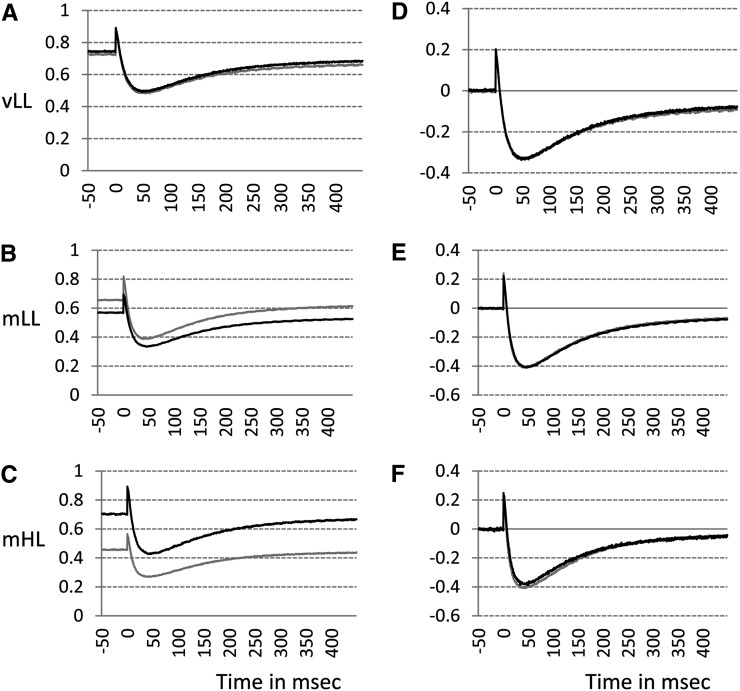
In response to continuous basal far-red light (723 nm), most of the primary electron donor in PSI (P700) is oxidized, indicated by the initial baseline in Figure 5A. The remainder can be momentarily photooxidized by a saturating, single-turnover flash (the spike in Fig. 5A). Subsequently, electrons arrive at P700+ from PSII, tending to rereduce P700+ while continuous far-red light brings the P700+ concentration back to the steady-state level, the initial baseline (Fig. 5A). The maximum magnitude of flash-induced photooxidation of P700 (P700+max) in the presence of continuous background far-red light indicates the content of photooxidizable PSI complex. A gradual decrease of P700+max (normalized to fresh weight) could be seen when the seedlings were grown under increasing light intensities (Fig. 5, A–C), consistent with the small reduction of PSI contents measured by western blotting with psaA antibodies and the substantial disappearance of the 733-nm peak of 77 K fluorescence emission spectra for plants grown under mHL conditions.
Figure 5.
Determination of P700 photooxidation in PSI in 7-d-old seedlings grown under vLL (20 μmol m−2 s−1; A and D), mLL (90 μmol m−2 s−1; B and E), or mHL (230 μmol m−2 s−1; C and F). A to C show the P700+ measurements per 1-cm2 cotyledon, and D to F show the results normalized to the peak height of maximum P700 oxidation. Black lines represent the wild type, and gray lines represent sco4.
It is instructive to normalize the curves in Figure 5, A to C, to P700+max. After normalization of the curves, the “P700 kinetics area” (the area bound by each curve and the horizontal line corresponding to the steady-state baseline) is directly proportional to the functional PSII content (Kou et al., 2012). Since the signals have been normalized to P700+max, the P700 kinetics area is a measure of the ratio of PSII to PSI on an arbitrary scale. The stoichiometry of the two functional photosystems seems to be undisturbed in sco4, as evidenced by little or no difference in the P700 kinetics area between the wild type and sco4 under all growth conditions (Fig. 5, D–F).
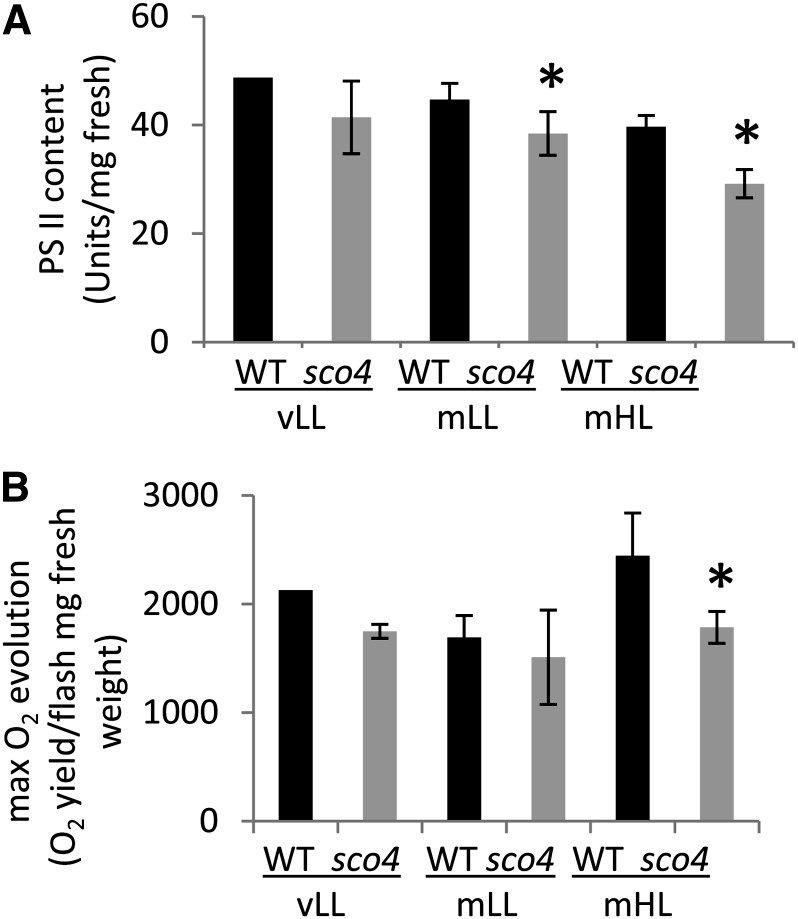
The content of functional PSII per unit of fresh weight was significantly reduced by one-quarter in mHL-grown sco4 mutants (Fig. 6A), consistent with the LHCB1 protein content analyzed in the western analysis (Fig. 2D). The light- and CO2-saturated rate of oxygen evolution, an indication of photosynthetic capacity, also dropped for the mutant to three-quarters of that for the wild type under mHL conditions, but no significant differences were observed under mLL and vLL conditions. This observation is consistent with the decrease in cytochrome b6f content (Fig. 2D). The data on both photosystems, PSI and PSII, indicate that the sco4 mutation affects both photosystems in their content and thus also in their function.
Figure 6.
Determination of PSII content (A) and maximum oxygen evolution rate (B) in seedlings grown under the different light intensities. Asterisks indicate significant differences (Student’s t test; P < 0.05). WT, Wild type.
sco4 Does Overaccumulate Hydrogen Peroxide under Higher Light Intensities
During photosynthesis, upon excitation of the photosystems, electrons are transferred from PSII via the cytochrome b6f/plastoquinone complex to PSI and beyond. This electron transfer can result in the production of superoxide at PSI, which is converted to hydrogen peroxide (H2O2) and effectively scavenged under LL. Increasing light intensities, however, result in the overproduction of these reactive oxygen species. Thus, the plants have to develop a protection system to avoid damage caused by this reactive oxygen species. Since we observed in the sco4 mutant a reduction in the capability to cope with increasing light intensity, resulting in a drop of the ETR, we hypothesized that this might cause increased H2O2 levels. Diaminobenzidine (DAB) staining can be used to detect the accumulation of H2O2 visualized by the accumulation of a brown precipitate. Seedlings grown for 14 d under either vLL or mHL were stained with DAB, using the overexpressor of the thylakoidal ascorbate peroxidase, a scavenger of H2O2 in the plastids (Murgia et al., 2004), as a negative control. As expected, no detectable staining could be observed in the thylakoidal ascorbate peroxidase-overexpressing line under both light conditions. Also, under vLL, no accumulation of H2O2 could be observed in the wild type, whereas under mHL, an accumulation of H2O2 could be detected (Supplemental Fig. S2). In contrast, in the sco4 mutant, already under LL conditions, cotyledons showed a brownish color that increased in the cotyledons and true leaves under mHL, despite a decline in LTR. Thus, an overaccumulation of H2O2 could be observed in sco4 that correlates with the susceptibility to bleaching of the cotyledons under increasing light intensities.
The Bleached True Leaf Phenotype Is Also Observable in sco4 under Specific Conditions
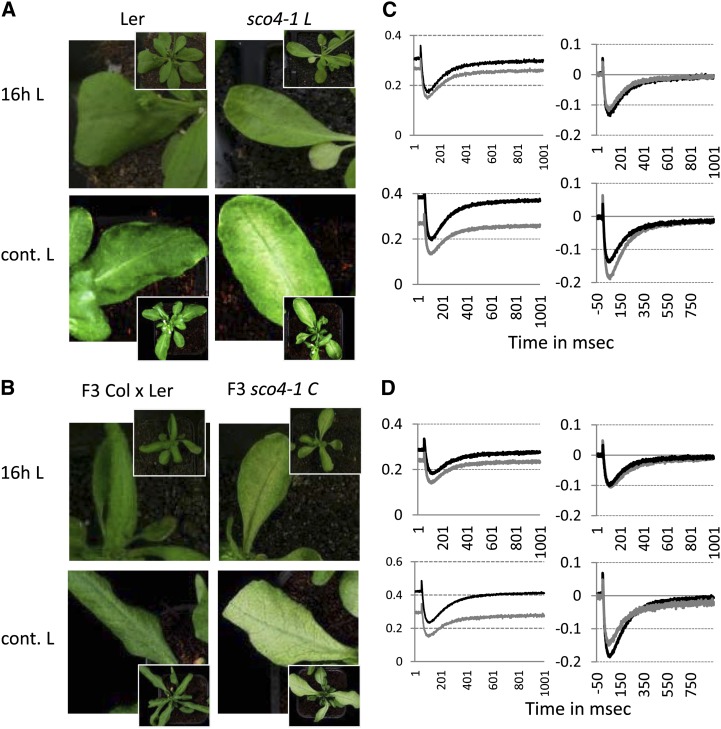
The analyses described for sco4 were performed on the mutant line in Ler under long-day conditions (16 h of light/8 h of dark) and were restricted to the cotyledons (Fig. 7A). Interestingly, when we grew this mutant line under continuous light, photobleaching of the true leaves was observed. Furthermore, when sco4 was crossed into the Columbia (Col) ecotype, the bleaching phenotype for sco4 was also apparent in true leaves under long days and even more enhanced under continuous light (Fig. 7B). A P700+max analysis was performed on the true leaves of long-day- and continuous-light-grown sco4 mutant plants in either background and compared with its corresponding wild type (Fig. 7, C and D). As in cotyledons under continuous light, the P700+max decreases in the mutant compared with the wild type. After normalization of the curves to P700+max, to observe the P700 kinetics area and to measure the ratio of PSII to PSI on an arbitrary scale, under continuous light the P700 kinetics area is reduced for sco4 compared with the wild type, which is not observed under 16 h of light. Thus, the photobleaching phenotype is not only restricted to cotyledons but is light dependent in all green tissues.
Figure 7.
Ecotype-dependent photobleaching of true leaves in sco4. A and B, Photobleaching phenotype in sco4 under 16 h of light (16h L; top row) and under continuous light (cont. L; bottom row) in the Ler (A) and the Col (B) backgrounds. Insets depict the whole plants from which the leaves were magnified. C and D, Determination of P700 photooxidation in PSI in adult plants in the Ler background (C) or once crossed into Col (D). Left panels show the P700+max measurements per 1-cm2 leaf, and right panels show the results normalized to P700+max. Top panels represent plants grown under 16-h light conditions, and bottom panels represent plants grown under continuous light. sco4 was crossed into Col, and homozygous plants were selected for the mutant as well as for the wild type and confirmed in the F3 generation. Black lines represent the wild type, and gray lines represent sco4.
Identification of SCO4
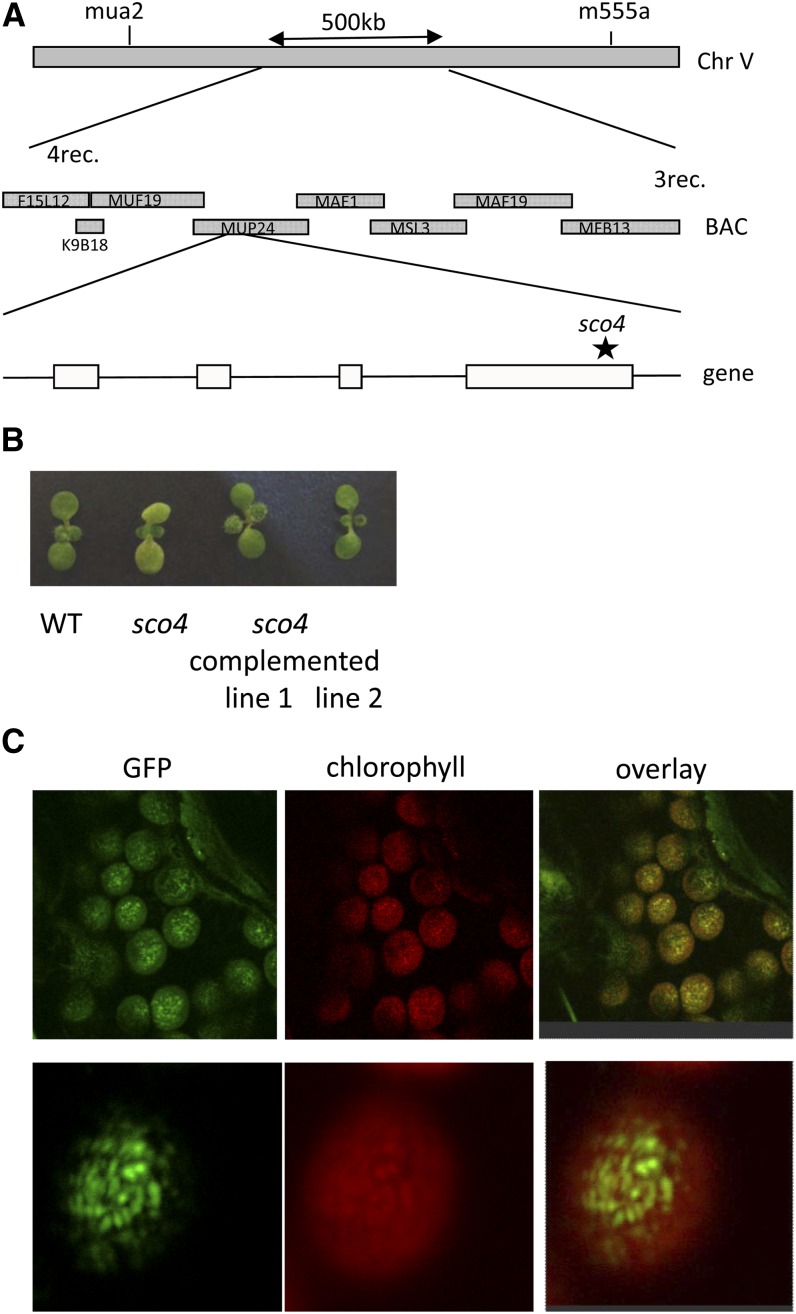
A map-based cloning approach was used to identify the sco4 mutation. Homozygous mutants from the F2 population of a crossing of sco4 with Col were screened with single-nucleotide polymorphism markers. The location of the sco4 mutation could be narrowed down to a 500-kb region at the bottom arm of chromosome 5 (Fig. 8A). Because of the pale-green phenotype, putative candidates predicted to encode proteins located to the chloroplasts were sequenced. A mutation was identified in gene At5g60750, which encodes a proteinase related to CaaX-type prenyl endopeptidases. Several membrane prediction programs are predicting four (http://bp.nuap.nagoya-u.ac.jp) to seven (http://aramemnon.botanik.uni-koeln.de) membrane-spanning domains for SCO4 (Supplemental Fig. S3). The sco4 mutation causes a nucleotide change from guanine to adenosine, which results in an amino acid change from the highly conserved Ala to Thr (A289T) in the conserved and catalytic region between motif 1 and motif 2, characteristic for type II CaaX prenyl proteinases (Supplemental Fig. S4; Pei and Grishin, 2001).
Figure 8.
Identification of sco4. A, Mapping of the sco4 mutation. The asterisk indicates the location of the point mutation in sco4. BAC, Bacterial artificial chromosome. B, Phenotype of sco4 and the complemented sco4 lines compared with the wild type (WT). C, Localization of the SCO4 protein using GFP fused to the C terminus within the chloroplasts. Left, GFP signal; middle, autofluorescence of chlorophyll; right, overlay of GFP and chlorophyll autofluorescence. Top row, whole cell; bottom row, magnification of one chloroplast.
The wild-type complementary DNA (cDNA) of SCO4 was cloned into a plant transformation vector, pCAMBIA1302, sequenced, and subsequently transformed into homozygous sco4 plants. Transformation with the SCO4 gene was able to largely complement the sco4 mutant seedlings recovering the pale-cotyledon phenotype of the mutant (Fig. 8B). Chlorophyll levels in the complemented lines were comparable to that of wild-type seedlings (Supplemental Fig. S5A). Transcript levels of SCO4 in the complemented lines were similar or only slightly increased when compared with wild-type seedlings or adult plants (Supplemental Fig. S5, B and C). Complemented lines were also subjected to the photosynthetic measurements using the light kinetics in the imaging pulse amplitude modulation. Introduction of wild-type SCO4 into sco4 resulted in reduced NPQ levels compared with the mutant, although still increased compared with wild-type seedlings (Fig. 4B), and a rescue of the ETR even higher than observed in wild-type seedlings (Fig. 4C) when grown under mHL. Thus, sco4 was complemented by At5g60750 cDNA, but given that the complementation used the cDNA, without any potential regulatory information and the nonnative 35S promoter, it is perhaps not surprising that the extent of complementation varied for some traits.
The SCO4 Protein Is Localized to the Chloroplasts
The identified mutation in sco4 affects a gene sharing limited homology with CaaX-type endopeptidases. Using the wild-type SCO4 fused to GFP, we could localize the SCO4 protein within the chloroplasts (Fig. 8D). Interestingly, SCO4 seemed not to be localized homogenously within the chloroplasts but rather accumulated in more dot-like structures. Magnification to the size of the chloroplast and comparison with the autofluorescence of chlorophyll suggests an overlap of the SCO4:GFP signals with the increased autofluorescence of the pigments in the stacked thylakoids, the grana. The chloroplast localization was confirmed by immunoblotting of SCO4:GFP using a GFP antiserum, but fractionation of the thylakoids and stroma was not definitive enough to confirm location within the chloroplast, which suggested bands in both thylakoid and stromal fractions (Supplemental Fig. S6). Further analyses would be required to determine its location within the plastid.
SCO4 Belongs to a Highly Conserved Group of Uncharacterized Chloroplast Endopeptidases in Higher Plants
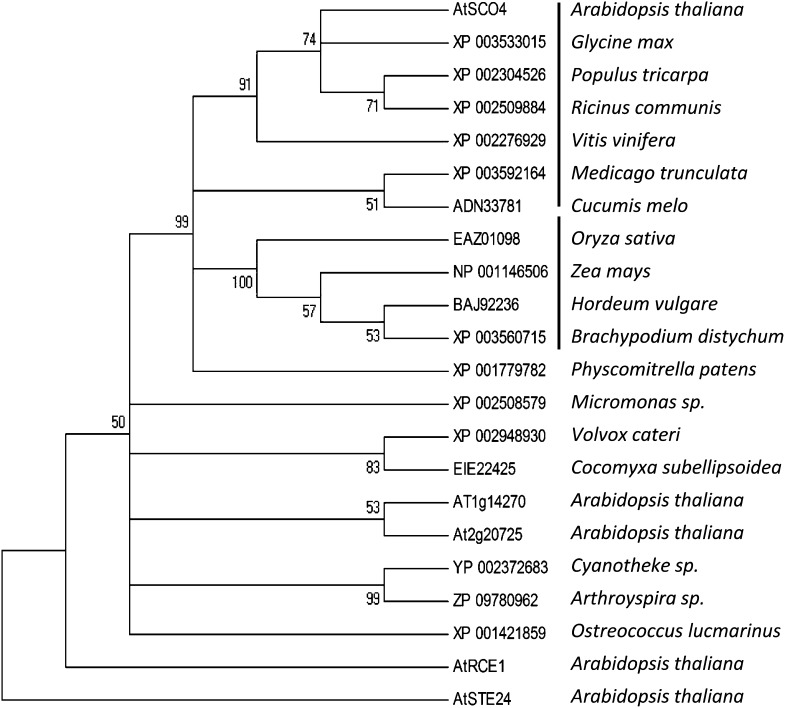
Using the amino acid sequence of SCO4 and comparing it with other proteins in Arabidopsis and other organisms revealed that mostly green organisms such as algae, mosses, and higher plants had similar proteins. All these proteins are indicated to be weakly similar to the CaaX-type endopeptidase RCE1. A cluster analysis using ClustalW-based MEGA4 analysis to create a phylogenetic tree, using STE24, another CaaX-type endopeptidase, as an outgroup, revealed that SCO4 clusters with other chloroplast-targeted proteinases identified in dicotyledonous and monocotyledonous plants but differs from RCE1 (Fig. 9; Supplemental Fig. S7). Indeed, aligning the SCO4-like proteinases from dicotyledonous plants, it appears that the function of SCO4 proteins in chloroplasts is highly conserved, as, except for the chloroplast transit peptide, the C-terminal domains of all identified endopeptidases demonstrate a highly identical amino acid sequence (Supplemental Fig. S8). Thus, SCO4 is a novel plant-specific chloroplast endoproteinase that is related to but distinct from CaaX-type endoproteinases.
In Arabidopsis the most similar proteins are encoded by At1g14270, At2g20725, and At2g36305, the latter encoding the AtFACE2/RCE1 gene, for which the CaaX-type endopeptidase function has already been demonstrated (Cadiñanos et al., 2003). The only similarity between these proteins can be found in the three known motifs conserved in CaaX-type endopeptidases, with FACE2/RCE1 being the most different from SCO4 compared with the other two proteins (Supplemental Fig. S4). Comparison of FACE/RCE1 proteins in the different kingdoms has demonstrated highly conserved amino acids that are required for FACE/RCE1 protein function, such as Glu-164, His-198, His-251, and Cys-254 present in Arabidopsis. The two His residues as well as the Cys are considered to function in metal chelation (Pei and Grishin, 2001). Except for Cys-254, the other three amino acids are also present in SCO4, whereas other conserved amino acids in the FACE/RCE1 proteins are not identified in SCO4 (Supplemental Fig. S4).
SCO4 was not able to facilitate the “A factor” processing in yeast (Saccharomyces cerevisiae) with a deletion in both the rce1 and ste24 genes, both of which encode CaaX-type endopeptidases (Supplemental Fig. S9). Using other yeast strains with deletion of RCE1 and AFC1, both of which are involved in CaaX-type processing, confirmed that SCO4 indeed is not able to process the A factor (Supplemental Fig. S10, A and B). Another assay is the activation of the RAS peptide, which requires CaaX-type proteolysis of RCE1 (Boyartchuk et al., 1997). Yeast not able to modify the RAS peptide are temperature sensitive, so they were impaired in growth at increased temperatures. Transformation of yeast lacking the RCE1 protein, thus being temperature sensitive, with SCO4 results in a partial rescue of the temperature sensitivity at 37°C, which is higher than used in other publications (Supplemental Fig. S11, A and B). Thus, SCO4 appears to possess some capacity for protease activity in yeast, but this is limited, as complementation was only observed in one of the two canonical assays for CaaX proteases. Thus, although SCO4 has limited similarities with the CaaX-type endopeptidases, SCO4 is not able to rescue the A factor or completely rescue RAS factor maturation in yeast and, as such, does not have a typical CaaX-type endopeptidase function.
Chloroplast Proteins with a CaaX Motif
Although SCO4 seems not to have a typical CaaX-type endopeptidase activity, we were interested in determining if proteins in the chloroplasts are present, which might be putative targets for SCO4 in planta if SCO4 has a prenylation-dependent endopeptidase function that has not yet been described. Since normal prenylation occurs at a Cys at the fourth-last position at the C terminus, a genome-wide Arabidopsis predicted proteome analysis was performed to identify proteins with this attribute. A total of 775 proteins in Arabidopsis could be identified that matched this criterion. Using ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) and SUBA (http://suba.plantenergy.uwa.edu.au/), we identified 85 proteins that have either been shown to be targeted or predicted to be targeted to the chloroplast (Supplemental Table S1). Using proteome data sets kindly provided by Klass van Wijk, we analyzed the 85 proteins for peptides obtained in different proteomic analyses. The goal was to identify any modifications to the C terminus consistent with prenylation or CaaX-type modifications, However, the C terminus data were unchanged, inclusive, or not the expected modification if targeted by a CaaX protease (Supplemental Table S2). Modification of the CaaX motif is performed by geranylgeranyl transferase (GGT1 and GGT2), or FT, which require either Leu or Ile for geranylgeranylation or for farnesylation of Met, Ala, Gln, Ser, or Cys. Proteins containing these amino acids in the last position were further analyzed for putative prenylation by either of the enzymes using PrePS (http://mendel.imp.ac.at/sat/PrePS/; Supplemental Table S1). In total, only five of 85 proteins may be modified by GGT or FT, according to this prediction program. The remaining 80 proteins do not contain the CaaX motif in the strict manner to be modified by the known GGT1, GGT2, or FT.
Out of 85 proteins identified above with a putative, although not optimal, CaaX motif, 13 proteins were verified as chloroplast localized using http://ppdb.tc.cornell.edu/ (Friso et al., 2004; Supplemental Table S3). Three of these proteins were identified to be localized to the thylakoids, whereas most of the others are of unspecified localization. However, none of the four published mutants, including NDF5 (a component of the NDH complex involved in cyclic electron flow), of the 13 putative SCO4 targets demonstrates a sco4-like phenotype (Huizinga et al., 2008; Ishida et al., 2009; Comparot-Moss et al., 2010).
Prenylation requires a GGT or a FT. Although several GGTs are localized in the chloroplasts (Supplemental Table S4), none of them seems to be a protein GGT; rather, they seem to have other substrates. A BLAST analysis was performed using a known RAB-type GGT or a protein FT. This approach revealed no GGT or FT with predicted chloroplast localization. Several novel proteins with a GGT or FT motif have been identified in our analysis for which the enzyme activity still has to be verified, but none of them seems to have a GGT1, GGT2, or FT function per se. Therefore, we assume that either protein prenylation is not occurring in chloroplasts or the organelles have developed a different system for this type of protein modification that requires a different set of enzymes than known for eukaryotic cells.
DISCUSSION
SCO4 Is a Novel Plant-Specific Chloroplast Endopeptidase
SCO4 is a protein whose homologs are present in higher plants, according to the data sets produced by GreenCut (Grossman et al., 2010), and, by our analysis, are distantly related to but do not have the function of the canonical CaaX-type endopeptidases, RCE1 and STE24 (Bracha et al., 2002; Bracha-Drori et al., 2008) that are found throughout eukaryotes, both plant and animal (Fig. 9). In the known motifs from endopeptidases, the amino acids required for chelating or binding of metal ions such as zinc (Pei and Grishin, 2001) are still present in SCO4, suggesting that SCO4 is a metalloprotease localized to the chloroplast membranes. RCE1 and STE24 belong to the type I proteases, which are zinc dependent and involved in the modification of CaaX motif-containing prenylated proteins, whereas SCO4 belongs to the type II proteases, for which neither a specific metal ion nor a function has been described (Pei and Grishin, 2001). Interestingly, SCO4 belongs to a chloroplast-specific family of endopeptidases restricted to higher plants, which suggest an evolution of this endopeptidase family away from the CaaX-type endopeptidases such as RCE1 and STE24 to a novel yet unknown function within the chloroplasts. This also explains why SCO4 was not able in our experiments to complement yeast impaired in typical CaaX-type endopeptidases.
Figure 9.
Phyllogenetic tree of CaaX-type endoproteinases. Clustering of CaaX-type endoproteinases into different clusters is shown (MEGA phyllogenetic analysis, bootstrap, neighbor joining). Note the different clustering into dicotyledonous (first clade) and monocotyledonous (second clade) chloroplast SCO4-like proteins, which differ from those in algae and cyanobacteria as well as from AtRCE1 and AtSTE24. The list of proteins from the different species used in this analysis is provided in Supplemental Figure S6.
CaaX-type endopeptidases are involved in protein prenylation, which is a lipid modification at the C terminus of the modified proteins. No indication for prenylation in chloroplasts could be found, since (1) no chloroplast protein GGT or FT could be identified, and (2) although several proteins seem to have a C-terminal CaaX-like motif, very few seem really to be direct target for GGT or FT. Furthermore, SCO4 is only able to partially complement CaaX-type protease-deficient yeast, which, while suggesting that SCO4 may possess limited CaaX proteolytic activity, would require confirmation in planta with different approaches.
Intramembrane metalloproteases form a large and diverse family with more than 5,800 members, which are present in all kingdoms (Pei et al., 2011). For the metalloprotease CaaX-type endopeptidases, the modified proteins are targeted to the membranes. Although no prenylated proteins are known in chloroplasts, the use of radiolabeled farnesyl in spinach (Spinacia oleracea) to visualize prenylated chloroplast proteins indicated the presence of this protein modification and that most of the prenylated proteins in chloroplasts were localized to the larger membrane complexes, particularly pigmented bands, which suggests the colocalization of such modified proteins with PSI and PSII (Parmryd et al., 1997). In sco4, the most apparent phenotypes observed are photobleaching in germinating seedlings and mature leaves, together with impaired photosynthetic reactions under increasing light conditions. Only one chloroplast-localized protein has been described that contains a CaaX motif, ISOPENTENYL TRANSFERASE3, an enzyme of the cytokinin biosynthesis pathway. However, only its unfarnesylated form is chloroplastic, whereas after farnesylation, this protein is localized to the cytoplasm (Galichet et al., 2008). Furthermore, no chloroplast protein GGT or FT could be identified to be encoded in the whole Arabidopsis genome, although GGT and FT are present in the chloroplasts, such as GGPS1 and GGPS3 (Supplemental Table S3; Okada et al., 2000). Although not for proteins, prenylation of other chloroplast substrates, such as plastoquinol9 and phylloquinone, is well described within the chloroplasts (Schultz et al., 1981; Kruk et al., 2003).
We bioinformatically screened for chloroplast proteins that might be modified by prenylation, either farnesylation or geranylgeranylation, and therefore might be potential targets of SCO4. Only 85 proteins with predicted chloroplast localization were identified to have a Cys at the required fourth-last position of the amino acid sequence, but no modifications were detected: this largely reflected incomplete data sets but may be significant, especially as SCO4 is not a typical CaaX-type protease. Although we were not able to prove a CaaX proteinase-specific function for SCO4 using RCE1- and STE24-specific prenylation assays in yeast, the partial complementation is consistent with an ability of SCO4 for protein modification, but the nature of that activity is unknown at this stage. Likewise, whether SCO4 targets one of more proteins is unknown. In this context, it is worth noting that the phenotype is more severe in a Col background having the same point mutation than in Ler, which may be consistent with multiple loci contributing to sco4 photobleaching. It may also reflect different photoprotective capacities in different ecotypes (Jung and Niyogi, 2009) and may not be directly related to SCO4 targets.
sco4 Affects Photosynthetic Reactions under Increased Light Conditions
The most observable phenotype in sco4 is the bleaching of cotyledons and true leaves under increased and/or altered light regimes. We have analyzed different photosynthetic parameters to try to explain the observed phenotype. Under mHL conditions (230 μmol m−2 s−1), the content of photooxidizable P700 decreased by about 35% on a fresh weight basis when compared with the wild type (Fig. 5C). The ratio of PSII and PSI reaction centers did not appear to differ between them, judging by the similarity of the P700 kinetics area bounded by the steady-state horizontal line and the flash-induced P700 redox kinetics curve (Fig. 5F). Therefore, it appears that the contents of both photosystems decrease to a similar extent, as confirmed by a 27% decrease in PSII content in sco4 under mHL conditions (Fig. 6A). With the depletion of both photosystems, we postulate that the density of the photosystem complexes in the thylakoid membrane is much smaller, with the consequence that, on average, PSII and PSI complexes are farther apart.
A greater average separation between PSII and PSI would restrict the spillover of excitation energy from PSII to PSI. This may explain the low ratio of chlorophyll fluorescence emission from PSI to that of PSII (Fig. 2B). With restricted spillover in the sco4 mutant, together with a slight decrease in the cytochrome b6f complex, the primary electron-accepting plastoquinone of PSII would be more readily reduced at a given light intensity (i.e. 1 − qP is higher in the sco4 mutant [Fig. 3C]). Since ΦPSII is the product of qP and the photochemical yield of PSII in the light-acclimated state, a lower qP means a lower ΦPSII in the sco4 mutant, which fell to zero at light intensities above 90 μmol m−2 s−1 (Fig. 4A). Similarly, a lower ΦPSII leads to a lower ETR (Fig. 4). In the sco4 mutant, the decreased capacity to use energy from reducing equivalents for metabolic reactions is compensated by the increase in dissipating heat via NPQ.
CONCLUSION
In this study, we investigated a gene encoding a novel plant-specific metalloendopeptidase, SCO4, which has limited similarities to CaaX-type endopeptidases, although it does not have a CaaX-related function. Although the presence of prenylated proteins in larger complexes within chloroplasts has been demonstrated (Parmryd et al., 1997), our computational analyses could not confirm any proteins within chloroplasts that might be modified by prenylation. Also, SCO4 was not able to completely complement yeast impaired in the typical CaaX endopeptidases RCE1 and STE24. Compromised SCO4 protein function mainly affects the acclimation of photosynthesis to increased light intensities. In mHL-grown seedlings, this resulted in increased NPQ and H2O2 generation but decreased oxygen evolution, impaired ETR under changing light conditions, and a reduction in the content of both photosystems. Identification of the targets of SCO4 and their functions within chloroplasts will help to understand the critical functions of SCO4 and its targets.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Identification of the Gene
A mutagenesis on 75,000 seeds of Arabidopsis (Arabidopsis thaliana) ecotype Ler was performed with ethyl methylsulfonate, and the M2 generation was screened for seedlings exhibiting pale cotyledons and green true leaves (Albrecht et al., 2006). The sco4 mutation was identified by map-based cloning using 1,000 homozygous F2 plants of a crossing with the Col ecotype. For complementation analysis of sco4–1, the full-length cDNA of the identified SCO4 gene (At5g60750) was cloned into the pCAMBIA1302 vector under the control of the cauliflower mosaic virus 35S promoter (http://www.cambia.org/daisy/cambia/585.html), control sequenced, and subsequently transformed into sco4 plants using Agrobacterium tumefaciens-mediated floral dipping (Albrecht et al., 2006). Transformed plants were selected with the corresponding herbicide hygromycin. For most of the experiments, seedlings were grown sterile on Murashige and Skoog medium (Sigma) without additional Suc supplementation. For this, seeds were surface sterilized with 70% ethanol and washed with sterile water. Plates were stratified at 4°C for 24 h before being transferred to the corresponding light conditions. Plants were grown under long-day conditions (16 h of light/8 h of dark) at 21°C and 90 μmol m−2 s−1 except when indicated otherwise. For LL conditions, the plates were covered with filter paper to reduce the light intensity to 20 μmol m−2 s−1. HL treatment was performed with plates transferred to 230 μmol m−2 s−1.
Chloroplast Analyses
Chlorophyll measurement of seedlings was performed as described (Albrecht et al., 2008) using 14-d-old seedlings grown under HL. The 77 K low-temperature fluorescence measurements were performed as described on seedlings grown for 7 d under the corresponding light conditions (Kim et al., 2005). For this, seedlings were harvested and frozen in liquid nitrogen. A dilution series of the internal standard fluorescein was used to identify the proper concentration for the 77 K measurements, and 0.01 μm fluorescein was added to the probe and used as an internal standard in the subsequent measurements (Kim et al., 2005). The quantification has been performed using the fluorescein peak at 480 nm for internal standardization and equal fresh weight of the samples.
Scanalyser Analyses
Seedlings were grown under continuous light in the different light conditions and analyzed every day at the same time for 5 d until the emergence of true leaves using the Scanalyser (www.lemnatec.com). Half of the plate was sown with Ler (wild-type) seedlings and the other half with sco4 seedlings. Darkness versus paleness was analyzed for all seedlings together for one genotype per plate. Analysis was performed in three independent series with three plates for each growth and transfer condition for each series.
Protein Analysis
Total protein was extracted from 7-d-old seedlings as described (Albrecht et al., 2006). Western analysis was performed using 15 μg of protein, separated on a SDS gel, semidry blotted on a polyvinylidene difluoride membrane, and subsequently hybridized with monospecific polyclonal antibodies (Agrisera) and horseradish peroxidase-linked ECL Rabbit IgG (Quantum Scientific). Fluorescence was detected using the ECL Prime western-blotting detection reagent (Amersham).
BLASTP in the National Center for Biotechnology Information (blast.ncbi.nlm.nih.gov/Blast.cgi) was used to identify SCO4-like proteins in other plant species. The identified protein sequences were subsequently used for ClustalW analyses (http://www.ebi.ac.uk/Tools/msa/clustalw2/), and the outcome of this analysis was used in MEGA4 for creating a phylogenetic analysis using bootstrap and neighbor joining as parameters.
Photosynthesis Analyses
We tested whether the defects in the sco4 mutant had an effect on the photosynthetic LTR to light quality gradients. The plants were grown on Murashige and Skoog medium in petri dishes and acclimated to PSI or PSII light or they were acclimated to one light source followed by a shift to the other one in order to follow the reversibility of the response. Subsequently, photosynthetic LTR parameters Fs/Fm, 1 − qP, and ΦPSII were monitored with a FluorCam 700 MF device (Photon Systems Instruments) essentially as described (Wagner et al., 2008). Directly after the fluorescence measurements, the plant material was used for chlorophyll determination (Porra et al., 1989), and chlorophyll a/b ratios were calculated accordingly. Mini pulse amplitude modulation analyses were used to determine the effect of the sco4 mutation compared with the wild type on photosynthetic parameters under different light intensities.
P700 Measurements
Redox changes of P700 were observed with a dual-wavelength (820/870 nm) unit (ED-P700DW) attached to a phase amplitude modulation fluorimeter (Walz) and used in the reflectance mode (Chow and Hope, 2004). To obtain redox changes of P700 due to a flash superimposed on continuous far-red light, a steady state was sought by illumination with far-red light from a light-emitting diode (approximately 12 µmol photons m−2 s−1) for about 10 s. Then, a single-turnover xenon flash (XST 103 xenon flash; Walz) was applied, triggered by a pulse/delay generator (model 565; Berkeley Nucleonics). Flashes were given at 0.2 Hz, and four consecutive signals were averaged (time constant = 95 μs). Measurements were normalized to the fresh weight of 7-d-old cotyledons or 1-cm2 true leaves and performed in triplicate.
The “dip” in the oxidation level of P700 induced by the single-turnover flash reflects the number of electrons that arrive from PSII per flash (Losciale et al., 2008); therefore, the integrated transient flow of electrons from PSII that arrive at P700+ after a flash is essentially given by the area between the dipping curve and the horizontal line corresponding to the steady state. This P700 kinetics area (being normalized to the total photooxidizable P700) can indicate the ratio of PSII to PSI reaction centers on an arbitrary scale (Kou et al., 2012).
Functional PSII Content and Maximum Oxygen Evolution Rate
The content of functional PSII complexes was determined from the oxygen yield per single-turnover saturating flash on leaf segments in 1% CO2 in an oxygen electrode chamber (Hansatech), according to Chow et al. (1989, 1991). Repetitive single-turnover xenon flashes were applied at 10 Hz. A slight heating artifact was taken into account. The estimation of the content of functional PSII centers is based on the assumption that for every four flashes, four electrons are transferred through each functional PSII, resulting in the evolution of one oxygen molecule. The functional PSII content was expressed on the basis of leaf mass unit of cotyledons from 7-d-old seedlings.
Maximum oxygen evolution at HL (1,000 μmol photons m−2 s−1) was determined in 1% CO2 in air using an oxygen electrode (Hansatech). The slope of the electrode voltage over time in the postillumination dark period was taken to be the baseline associated with mitochondrial respiration and any instrumental drift, and the gross rate of oxygen evolution was determined relative to it. The maximum oxygen evolution rate was expressed based on leaf mass unit of cotyledons from 7-d-old seedlings. Measurements were performed in triplicate with plants grown under vLL, LL, and mHL conditions.
DAB Staining
The accumulation of H2O2 was visualized using DAB staining. For this, 14-d-old seedlings were incubated in a 0.4% DAB and 0.4 NiCl2 buffered in 0.5 m Tris at pH 7.4, and subsequently destained using 70% ethanol.
Yeast Complementation Assays
For yeast halo analyses using the yeast A factor processing as parameter, the SCO4 cDNA was cloned into the pJR1138 vector and, after sequence confirmation, was transformed into the yeast strains JRY6659 (Mat a Δste24Δrce1), JRY6961 (Mat α Δrce1), and JRY 6962 (Mat α Δ ste24) according to protocol (Bracha-Drori et al., 2008; Tanz et al., 2012). The halo test was performed as described (Bracha-Drori et al., 2004, 2008). Another assay to verify CaaX-type proteolysis in yeast is the complementation of temperature-sensitive yeast strains impaired in the maturation of the RAS factor (Boyartchuk et al., 1997). These strains were provided by Jasper Rine and were transformed with the same vectors used for the former assay, and the positively transformed yeast were plated on yeast peptone dextrose for monitoring growth at room temperature, 30°C, and 37°C, as described (Boyartchuk et al., 1997).
Sequence data from this article can be found in the TAIR data libraries under accession number A5g60750.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantification of the greening process of seedlings.
Supplemental Figure S2. Visualization of H2O2 in seedlings.
Supplemental Figure S3. Prediction of transmembrane-spanning domains of AtSCO4.
Supplemental Figure S4. Alignment of proteins of Arabidopsis exhibiting similar CaaX-type proteinase features as SCO4.
Supplemental Figure S5. Complementation of sco4.
Supplemental Figure S6. Protein detection in fractionated chloroplasts.
Supplemental Figure S7. List of CaaX-type endoproteinases from the different species used in the ClustalW and subsequent phyllogenetic analysis.
Supplemental Figure S8. The SCO4 protein is highly conserved within chloroplast CaaX-type proteinases located in chloroplasts in dicotyledonous plants.
Supplemental Figure S9. Yeast complementation assays with the SCO4 protein with yeast halo assay.
Supplemental Figure S10. Yeast complementation assays with the SCO4 protein with yeast halo assay in AFC1 deletion mutants.
Supplemental Figure S11. Yeast complementation assays with the SCO4 protein for temperature sensitivity.
Supplemental Table S1. List of proteins containing a putative CaaX motif predicted to be localized to the chloroplasts.
Supplemental Table S2. Proteomic analysis of data sets provided by Klass van Wijk for peptides containing a modified C terminus.
Supplemental Table S3. List of identified chloroplast proteins with a putative CaaX motif present in the Plant Proteome Database.
Supplemental Table S4. BLAST analysis for proteins in plant organelles that might be geranylgeranyl pyrophosphate synthases or farnesylase transferases.
Acknowledgments
We thank Shaul Yalowski (Tel Aviv University) for providing us with the yeast strains for the yeast halo assay and Jasper Rine (University of California, Berkeley) for the yeast strains required for the analysis of RAS activation. Some of the primary antibodies for the detection of chloroplast proteins were provided by Susanne von Cammerer (Australian National University). We thank Klaas van Wijk (Cornell University) for bioinformatic analyses of chloroplast proteome data. We also thank Diep Ganguly (Australian National University) for his help.
Glossary
- NPQ
nonphotochemical quenching
- Ler
Landsberg erecta
- vLL
very low light
- mLL
moderate low light
- mHL
moderate high light
- LTR
long-term response
- Fs/Fm
fraction of light absorbed by the PSII antenna that is lost by constitutive dissipation, in a light-independent manner, by fluorescence and heat emission
- ΦPSII
effective quantum yield
- 1 − qP
reduction state of primary electron-accepting plastoquinone of PSII and the plastoquinone pool
- ETR
electron transfer rate
- DAB
diaminobenzidine
- H2O2
hydrogen peroxide
- Col
Columbia
- cDNA
complementary DNA
- FT
farnesyl transferase
- GGT
geranylgeranyl transferase
- P700
primary electron donor in PSI
- P700+max
maximum magnitude of flash-induced photooxidation of P700
- LL
low light
- HL
high light
Footnotes
This work was supported by the Australian Research Council (grant no. DP1093827 to W.S.C.) and by the Australian Research Council Centre of Excellence in Plant Energy Biology (grant no. CE0561495).
The online version of this article contains Web-only data.
References
- Albrecht V, Ingenfeld A, Apel K. (2006) Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: the impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol Biol 60: 507–518 [DOI] [PubMed] [Google Scholar]
- Albrecht V, Ingenfeld A, Apel K. (2008) Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Mol Biol 66: 599–608 [DOI] [PubMed] [Google Scholar]
- Albrecht V, Simková K, Carrie C, Delannoy E, Giraud E, Whelan J, Small ID, Apel K, Badger MR, Pogson BJ. (2010) The cytoskeleton and the peroxisomal-targeted SNOWY COTYLEDON3 protein are required for chloroplast development in Arabidopsis. Plant Cell 22: 3423–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF, Pfannschmidt T. (2000) Balancing the two photosystems: photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philos Trans R Soc Lond B Biol Sci 355: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Horton P, Walters RG. (2004) Acclimation of Arabidopsis thaliana to the light environment: the relationship between photosynthetic function and chloroplast composition. Planta 218: 793–802 [DOI] [PubMed] [Google Scholar]
- Bellafiore S, Barneche F, Peltier G, Rochaix JD. (2005) State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Bergo MO, Ambroziak P, Gregory C, George A, Otto JC, Kim E, Nagase H, Casey PJ, Balmain A, Young SG. (2002a) Absence of the CAAX endoprotease Rce1: effects on cell growth and transformation. Mol Cell Biol 22: 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo MO, Gavino B, Ross J, Schmidt WK, Hong C, Kendall LV, Mohr A, Meta M, Genant H, Jiang Y, et al. (2002b) Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc Natl Acad Sci USA 99: 13049–13054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D. (2005) Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437: 1179–1182 [DOI] [PubMed] [Google Scholar]
- Boyartchuk VL, Ashby MN, Rine J. (1997) Modulation of Ras and A-factor function by carboxyl-terminal proteolysis. Science 275: 1796–1800 [DOI] [PubMed] [Google Scholar]
- Bracha K, Lavy M, Yalovsky S. (2002) The Arabidopsis AtSTE24 is a CAAX protease with broad substrate specificity. J Biol Chem 277: 29856–29864 [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N. (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Lubetzky TC, Yalovsky S. (2008) Functional analysis of Arabidopsis postprenylation CaaX processing enzymes and their function in subcellular protein targeting. Plant Physiol 148: 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadiñanos J, Varela I, Mandel DA, Schmidt WK, Díaz-Perales A, López-Otín C, Freije JM. (2003) AtFACE-2, a functional prenylated protein protease from Arabidopsis thaliana related to mammalian Ras-converting enzymes. J Biol Chem 278: 42091–42097 [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, Finnegan EJ, Turnbull C, Pogson BJ. (2009) Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21: 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow WS, Hope AB. (2004) Electron fluxes through photosystem I in cucumber leaf discs probed by far-red light. Photosynth Res 81: 77–89 [DOI] [PubMed] [Google Scholar]
- Chow WS, Hope AB, Anderson JM. (1989) Oxygen per flash from leaf disks quantifies photosystem II. Biochim Biophys Acta 973: 105–108 [Google Scholar]
- Chow WS, Hope AB, Anderson JM. (1991) Further studies on quantifying photosystem II in vivo by flash-induced oxygen yield from leaf discs. Aust J Plant Physiol 18: 397–410 [Google Scholar]
- Chow WS, Melis A, Anderson JM. (1990) Adjustments of photosystem stoichiometry in chloroplasts improve the quantum efficiency of photosynthesis. Proc Natl Acad Sci USA 87: 7502–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comparot-Moss S, Kötting O, Stettler M, Edner C, Graf A, Weise SE, Streb S, Lue WL, MacLean D, Mahlow S, et al. (2010) A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves. Plant Physiol 152: 685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell DN. (2000) Functional implications of protein isoprenylation in plants. Prog Lipid Res 39: 393–408 [DOI] [PubMed] [Google Scholar]
- Dangoor I, Peled-Zehavi H, Wittenberg G, Danon A. (2012) A chloroplast light-regulated oxidative sensor for moderate light intensity in Arabidopsis. Plant Cell 24: 1894–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel L, Bräutigam K, Pfannschmidt T. (2008) Photosynthetic acclimation: state transitions and adjustment of photosystem stoichiometry. Functional relationships between short-term and long-term light quality acclimation in plants. FEBS J 275: 1080–1088 [DOI] [PubMed] [Google Scholar]
- Dietzel L, Bräutigam K, Steiner S, Schüffler K, Lepetit B, Grimm B, Schöttler MA, Pfannschmidt T. (2011) Photosystem II supercomplex remodeling serves as an entry mechanism for state transitions in Arabidopsis. Plant Cell 23: 2964–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ. (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16: 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galichet A, Hoyerová K, Kamínek M, Gruissem W. (2008) Farnesylation directs AtIPT3 subcellular localization and modulates cytokinin biosynthesis in Arabidopsis. Plant Physiol 146: 1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber E, Hemmerlin A, Hartmann M, Heintz D, Hartmann MA, Mutterer J, Rodríguez-Concepción M, Boronat A, Van Dorsselaer A, Rohmer M, et al (2009) The plastidial 2-C-methyl-d-erythritol 4-phosphate pathway provides the isoprenyl moiety for protein geranylgeranylation in tobacco BY-2 cells. Plant Cell 21: 285–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AR, Karpowicz SJ, Heinnickel M, Dewez D, Hamel B, Dent R, Niyogi KK, Johnson X, Alric J, Wollman FA, et al (2010) Phylogenomic analysis of the Chlamydomonas genome unmasks proteins potentially involved in photosynthetic function and regulation. Photosynth Res 106: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogewoning SW, Wientjes E, Douwstra P, Trouwborst G, van Ieperen W, Croce R, Harbinson J. (2012) Photosynthetic quantum yield dynamics: from photosystems to leaves. Plant Cell 24: 1921–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizinga DH, Omosegbon O, Omery B, Crowell DN. (2008) Isoprenylcysteine methylation and demethylation regulate abscisic acid signaling in Arabidopsis. Plant Cell 20: 2714–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Takabayashi A, Ishikawa N, Hano Y, Endo T, Sato F. (2009) A novel nuclear-encoded protein, NDH-dependent cyclic electron flow 5, is essential for the accumulation of chloroplast NAD(P)H dehydrogenase complexes. Plant Cell Physiol 50: 383–393 [DOI] [PubMed] [Google Scholar]
- Jung HS, Niyogi KK. (2009) Quantitative genetic analysis of thermal dissipation in Arabidopsis. Plant Physiol 150: 977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanervo E, Suorsa M, Aro EM. (2005) Functional flexibility and acclimation of the thylakoid membrane. Photochem Photobiol Sci 4: 1072–1080 [DOI] [PubMed] [Google Scholar]
- Kargul J, Barber J. (2008) Photosynthetic acclimation: structural reorganisation of light harvesting antenna. Role of redox-dependent phosphorylation of major and minor chlorophyll a/b binding proteins. FEBS J 275: 1056–1068 [DOI] [PubMed] [Google Scholar]
- Kim EH, Chow WS, Horton P, Anderson JM. (2005) Entropy-assisted stacking of thylakoid membranes. Biochim Biophys Acta 1708: 187–195 [DOI] [PubMed] [Google Scholar]
- Klughammer C, Schreiber U. (2008) Complementary PS II quantum yields calculated from simple fluorescence parameters measured by PAM fluorometry and the saturation pulse method. Pancreas 1: 27–35 [Google Scholar]
- Kou J, Oguchi R, Fan DY, Chow WS. (2012) The time course of photoinactivation of photosystem II in leaves revisited. Photosynth Res 113: 157–164 [DOI] [PubMed] [Google Scholar]
- Kruk J, Jemioła-Rzemińska M, Strzałka K. (2003) Cytochrome c is reduced mainly by plastoquinol and not by superoxide in thylakoid membranes at low and medium light intensities: its specific interaction with thylakoid membrane lipids. Biochem J 375: 215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wakao S, Fischer BB, Niyogi KK. (2009) Sensing and responding to excess light. Annu Rev Plant Biol 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Losciale P, Oguchi R, Hendrickson L, Hope AB, Corelli-Grappadelli L, Chow WS. (2008) A rapid, whole-tissue determination of the functional fraction of PSII after photoinhibition of leaves based on flash-induced P700 redox kinetics. Physiol Plant 132: 23–32 [DOI] [PubMed] [Google Scholar]
- Murgia I, Tarantino D, Vannini C, Bracale M, Carravieri S, Soave C. (2004) Arabidopsis thaliana plants overexpressing thylakoidal ascorbate peroxidase show increased resistance to paraquat-induced photooxidative stress and to nitric oxide-induced cell death. Plant J 38: 940–953 [DOI] [PubMed] [Google Scholar]
- Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y. (2000) Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol 122: 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmryd I, Shipton CA, Swiezewska E, Dallner G, Andersson B. (1997) Chloroplastic prenylated proteins. FEBS Lett 414: 527–531 [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. (2001) Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem Sci 26: 275–277 [DOI] [PubMed] [Google Scholar]
- Pei J, Mitchell DA, Dixon JE, Grishin NV. (2011) Expansion of type II CAAX proteases reveals evolutionary origin of γ-secretase subunit APH-1. J Mol Biol 410: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Hertle A, Pribil M, Kleine T, Wagner R, Strissel H, Ihnatowicz A, Bonardi V, Scharfenberg M, Schneider A, et al (2009) Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21: 2402–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Hertle A, Pribi M, Schneider A, Kleine T, Leister D. (2010) Optimizing photosynthesis under fluctuating light: the role of the Arabidopsis STN7 kinase. Plant Signal Behav 5: 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Niyogi KK, Björkman O, DellaPenna D. (1998) Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc Natl Acad Sci USA 95: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll-a and chlorophyll-b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Pribil M, Pesaresi P, Hertle A, Barbato R, Leister D. (2010) Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol 8: e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissler HM, Pogson BJ. (2001) Antisense inhibition of the beta-carotene hydroxylase enzyme in Arabidopsis and the implications for carotenoid accumulation, photoprotection and antenna assembly. Photosynth Res 67: 127–137 [DOI] [PubMed] [Google Scholar]
- Samol I, Shapiguzov A, Ingelsson B, Fucile G, Crevecoeur M, Vener AV, Rochaix JD, Goldschmidt-Clermont M. (2012) Identification of a photosystem II phosphatase involved in light acclimation in Arabidopsis Plant Cell 24: 2596–2609Schultz G, Ellerbrock BH, Soll J. (1981) Site of prenylation reaction in synthesis of phylloquinone (vitamin K1) by spinach chloroplasts. Eur J Biochem 117: 329–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiguzov A, Ingelsson B, Samol I, Andres C, Kessler F, Rochaix JD, Vener AV, Goldschmidt-Clermont M. (2010) The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis. Proc Natl Acad Sci USA 107: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz SK, Kilian J, Johnsson C, Apel K, Small I, Harter K, Wanke D, Pogson B, Albrecht V. (2012) The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LCHB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings. Plant J 69: 743–754 [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Piippo M, Suorsa M, Sirpiö S, Mulo P, Vainonen J, Vener AV, Allahverdiyeva Y, Aro EM. (2006) State transitions revisited: a buffering system for dynamic low light acclimation of Arabidopsis. Plant Mol Biol 62: 779–793 [DOI] [PubMed] [Google Scholar]
- Wagner R, Dietzel L, Bräutigam K, Fischer W, Pfannschmidt T. (2008) The long-term response to fluctuating light quality is an important and distinct light acclimation mechanism that supports survival of Arabidopsis thaliana under low light conditions. Planta 228: 573–587 [DOI] [PubMed] [Google Scholar]
- Zhang FL, Casey PJ. (1996) Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem 65: 241–269 [DOI] [PubMed] [Google Scholar]