A single-stranded DNA-binding protein represses expression and delays leaf senescence in a developmental stage-dependent manner in Arabidopsis.
Abstract
Leaf senescence in plants involves both positive and negative transcriptional regulation. In this work, we show evidence for the single-stranded DNA-binding protein WHIRLY1 (WHY1) that functions as an upstream suppressor of WRKY53 in a developmental stage-dependent manner during leaf senescence in Arabidopsis (Arabidopsis thaliana). The why1 mutant displayed an early-senescence phenotype. In this background, the expression levels of both WRKY53 and the senescence-associated protease gene SAG12 increased. WHY1 bound to the sequence region that contains an elicitor response element motif-like sequence, GNNNAAATT, plus an AT-rich telomeric repeat-like sequence in the WRKY53 promoter in in vivo and in vitro mutagenesis assays as well as in a chromatin immunoprecipitation assay. This binding to the promoter of WRKY53 was regulated in a developmental stage-dependent manner, as verified by chromatin immunoprecipitation-polymerase chain reaction assay. This direct interaction was further determined by a transient expression assay in which WHY1 repressed β-GLUCURONIDASE gene expression driven by the WRKY53 promoter. Genetic analysis of double mutant transgenic plants revealed that WHY1 overexpression in the wrky53 mutant (oeWHY1wrky53) had no effect on the stay-green phenotype of the wrky53 mutant, while a WHY1 knockout mutant in the wrky53 mutant background (why1wrky53) generated subtle change in the leaf yellow/green phenotype. These results suggest that WHY1 was an upstream regulator of WRKY53 during leaf senescence.
Senescence is a highly regulated and energy-consuming process (Guo et al., 2004). Early expression profiling and transcriptome analyses have revealed that a big portion of specific transcription factors are reprogrammed during leaf senescence (Chen et al., 2002; Buchanan-Wollaston et al., 2003, 2005; Guo et al., 2004; Zentgraf et al., 2004; Balazadeh et al., 2008; Breeze et al., 2011). These transcription factors are characterized in the protein families NAC, WRKY, MYB, C2H2 zinc finger, bZIP, and AP2/EREBP. Among them, the NAC (Balazadeh et al., 2010, 2011) and WRKY (Miao et al., 2004; Ülker et al., 2007; Zentgraf et al., 2010; Zhou et al., 2011) family members have been proven to play central roles in controlling leaf senescence in Arabidopsis (Arabidopsis thaliana). Besides transcription factors, many other proteins with diverse functions, such as the activation domain (AD) protein (Miao et al., 2008), the epithiospecifying protein ESR/ESP (Miao and Zentgraf, 2007), the SUVH2 histone methyltransferase (Ay et al., 2009), and the WHIRLY2 (WHY2) protein (Maréchal et al., 2008), have also been enumerated to participate in the regulation of plant senescence and cell death processes.
The WHY proteins were first identified as transcription factors in mediating elicitor-induced gene expression of the pathogenesis-related gene PR-10a in potato (Solanum tuberosum; Després et al., 1995). StWHY1 forms a homotetramer with high preference for single-stranded DNA (ssDNA) binding. It has been proposed that this protein might bind to melted promoter regions to modulate transcriptional activities. The interaction of StWHY1 with the PR-10a promoter was mapped to a region termed the elicitor response element (ERE) for elicitor-induced gene expression (Després et al., 1995; Desveaux et al., 2000, 2002). The Arabidopsis genome encodes three WHY proteins: AtWHY1 and AtWHY3 contain plastid-targeting signal, whereas AtWHY2 localizes to mitochondria (Krause et al., 2005). Similar to StWHY1, AtWHY1 has been proven to have a relationship with disease resistance (Desveaux et al., 2004, 2005). A number of potential WHY1 target genes were proposed based on the occurrence of ERE sequences in their promoters, but experimental data are lacking. Until now, only the PR1 gene has been proven to be down-regulated as a result of a TILLING mutation in WHY1 in Arabidopsis (Desveaux et al., 2004, 2005). Besides the activity as a transcriptional activator, AtWHY1 was also identified in a fraction of telomere-binding proteins, and its knockout mutant appeared to have a shorter telomere (Yoo et al., 2007). This result led the authors to suggest a role for WHY1 in telomerase inhibition (Yoo et al., 2007). In addition, WHY1 also functions as a repressor in the salicylic acid-mediated pathogen-responsive pathway by binding to the GAGAAATT motif of the kinesin AtKP1 promoter (Xiong et al., 2009). The above studies established the nucleus functions of WHY1 even though it contains plastid-targeting signal sequences. However, the dual localization of the native protein in two compartments of the same cell has so far only been confirmed for the barley (Hordeum vulgare) protein HvWHY1, which was detected in the nucleus and plastids of leaf material by immunohistochemical methods (Grabowski et al., 2008). Using bimolecular fluorescence complementation assays, it has been shown that HvWHY1 formed homooligomers in the nucleus (Grabowski et al., 2008).
Although the organellar isoforms of WHY proteins seem to be associated with the organellar DNA, they might not function as organellar transcription factors. In fact, it was proposed that plastidic WHY1 in maize (Zea mays) and barley both have functions in RNA processing and that the plastidic WHY proteins in Arabidopsis have a function as antirecombination proteins that could be involved in maintaining plastome DNA stability (Maréchal et al., 2008, 2009; Prikryl et al., 2008; Cappadocia et al., 2010; Maréchal and Brisson, 2010; Melonek et al., 2010).
Here, we present evidence that WHY1 acts as an upstream suppressor of WRKY53 in a developmental stage-dependent manner during leaf senescence in Arabidopsis. Our results are based on the phenotypic analysis of WHY1 transfer DNA (T-DNA) insertion mutants, expression profiling of senescence-associated genes, and a physical interaction between WHY1 and a GNNNAAATT motif plus an AT-rich telomeric repeat-like sequence in the WRKY53 promoter in vitro and in a chromatin immunoprecipitation (ChIP) assay. In addition, genetic analysis of the double mutant why1wrky53 and oeWHY1wrky53 transgenic plants indicates that WHY1 is an upstream regulator of WRKY53. Finally, ChIP-PCR analysis of WRKY53 promoter activity, at different developmental stages, in leaves of the why1 mutant and the function-restored transgenic plant PWHY1-HA demonstrates that the nuclear isoform of the WHY1 protein developmentally controls the expression of WRKY53.
RESULTS
Mutation of WHY1 Causes a Severe Early-Senescence Phenotype in Rosette Leaves of Arabidopsis
In order to determine the relationship of WHY1 with senescence, we collected and generated transgenic plants: two independent homozygous lines carrying a T-DNA insertion in the first exon of WHY1 (why1-1 and why1-2), two WHY1 antisense lines (awhy1-1 and awhy1-6), two independent lines overexpressing hemagglutinin (HA)-tagged WHY1 (oeWHY1-HA5 and oeWHY1-HA6 [oe5 and oe6 for short]), and one functional complementation line (PWHY1-HA) that harbored HA-tagged WHY1 coding sequence in the why1-1 background under the control of its own promoter (Pwhy1:WHY1-HA/why1-1). All of them were subjected to northern-blot and quantitative reverse transcription (qRT)-PCR analyses. The results are listed in Supplemental Figure S1. Both mutants showed a single weak transcript of the same size as in wild-type plants that were obviously caused by cross hybridization to the closely related WHY3 gene, since this band was abolished in the why1why3 double mutant (ko1/3). Two WHY1 antisense lines showed reduced transcript levels. Enhanced transcript levels were observed in all WHY1-overexpressing lines.
After confirming the alteration of WHY1 expression in the mutants and the different transgenic plants, the leaf senescence progression of these plants was systematically analyzed. First, the photochemical efficiency of PSII (Fv/Fm) value was compared as a marker for senescence in rosette leaves of mutant, transgenic, and wild-type plants. The Fv/Fm value typically decreases to below 0.8 when senescence begins (Humbeck et al., 1996; Oh et al., 1996). The plants were monitored once a week for several weeks, starting from 5-week-old plants. Leaf 7 was labeled for subsequent measurement of Fv/Fm value and chlorophyll content. Individual rosette leaves of 9-week-old plants showed the same pattern of differential onset of senescence. However, the Fv/Fm values in the two WHY1 mutants decreased to below 0.5 in leaf 7, whereas a value of 0.7 was still maintained in the wild type and the WHY1 functional complementation line. None of the two WHY1-overexpressing lines dropped below 0.8 in leaf 7 (Fig. 1A). To juxtapose the chlorophyll content, the highest value in the oe5 line was set as 100%. In the wild type and the complementation line, chlorophyll contents declined to 80%, while in why1 mutants and the two antisense lines, the values dropped sharply to 43% (Fig. 1B).
Figure 1.
Senescence phenotype of wild-type and WHY1 mutant plants. A and B, Fv/Fm (A) and chlorophyll content (B) of leaf 7 in 9-week-old plants. Means and se of five independent measurements are shown. Different letters indicate significant differences at P ≤ 0.05 based on Student’s t test. C, Leaf senescent phenotypes of 10-week-old plants. Numbers above the images indicate leaf age, with leaf 1 being the oldest. D, Senescent leaf fraction from 12 plants at week 10. Means and se from two independent experiments with six plants each are shown. why1-1 and why1-2 are two WHY1 T-DNA insertion mutant lines; awhy1-1 and awhy1-6 are two WHY1 antisense lines; oe5 and oe6 are two WHY1 overexpression lines; PWHY1 is a complementary line harboring WHY1 coding sequence driven by its own promoter in the why1-1 background (why1-1 Pwhy1:WHY1-HA). WT, Wild-type plants.
A visual comparison of all rosette leaves from a representative plant of each mutant and transgenic line after 10 weeks of growth confirmed the above results (Fig. 1C). Each leaf of a rosette was marked with color-coded threads after its emergence, as described previously (Hinderhofer and Zentgraf, 2001), and numbered progressively. Leaves 1 to 6 from 10-week-old wild-type plants showed a senescent phenotype with severe yellowing. At the same stage, the two independent WHY1 mutants showed more severe yellowing up to leaf 10, confirming a stronger early-senescence phenotype, whereas all leaves of the two independent overexpressing lines still appeared green (Fig. 1C). The why1-1 mutant exhibiting the strongest yellowing phenotype was complementally restored by WHY1 under the control of its own promoter, ensuring that the mutant phenotype was really due to the T-DNA insertion in the WHY1 gene (Fig. 1). Furthermore, a statistical analysis of 12 plants was performed by categorizing the leaves into four groups according to leaf-yellowing degrees (green, green/yellow, full yellow, and brown/dry; Fig. 1D). Using this categorization, phenotype analysis was done with multiple individuals and, in addition, was analyzed with different generations of the mutants (Supplemental Table S1; Supplemental Fig. S2). Although a few leaves of why1-1 and why1-2 plants seemed to be pale green, these pale-green leaves consequently occurred from old leaves to young leaves during development. This phenomenon is most likely a senescent phenotype. However, the why1why3 double mutant (ko1/3) line did not show any senescent phenotype, but it was reported that 5% variegation plants of the why1why3 line have the pale-green phenotype during seedling development (Maréchal et al., 2009; Cappadocia et al., 2010; Maréchal and Brisson, 2010).
The Expression of Senescence-Related Genes Is Altered in why1 Mutants
To get an overview of whether the expression levels of senescence-related genes are affected by the loss of WHY1 expression, a semiquantitative reverse transcription-PCR screen was performed in 7- and 9-week-old why1 mutant and wild-type plants. This screen included all three WHY genes, part of the target genes of WHY proposed by Desveaux et al. (2005), and several key genes known to be involved in the regulation of senescence in Arabidopsis (Miao et al., 2004). This pilot study revealed that the transcript levels of both WHY paralogs (WHY2 and WHY3) are increased to some extent in the absence of WHY1. A strong increase was observed for SAG12 (He and Gan, 2002), and a somewhat weaker up-regulation of WRKY53 (Hinderhofer and Zentgraf, 2001) and WRKY33 (Zheng et al., 2006) was seen. The PR1 (Uknes et al., 1992; Desveaux et al., 2004; Xiong et al., 2009) and AD1 (Miao et al., 2008) genes were found to be clearly down-regulated, while the expression of more than 30 other genes was not affected in the why1 mutant (Supplemental Table S2).
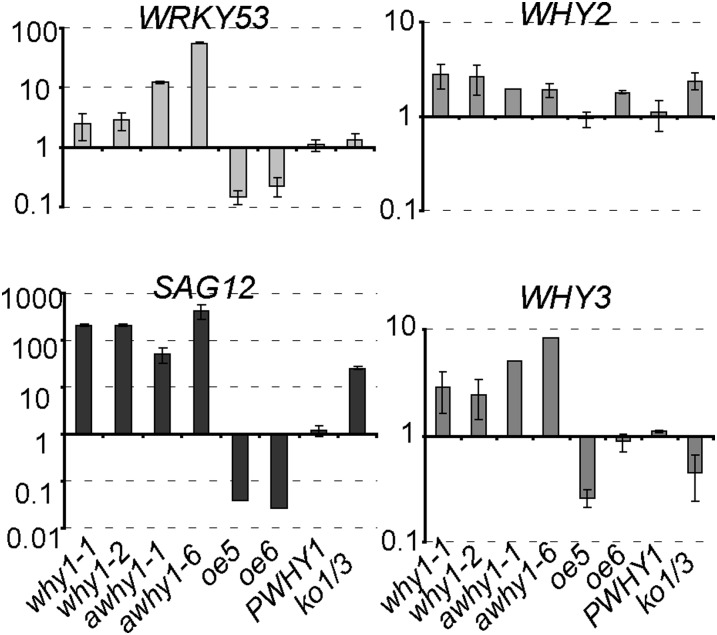
To further confirm the results, a qRT-PCR analysis was performed with the same why1 mutants and the other aforementioned transgenic plants. The complementation line came out as the wild type. The mRNAs were isolated from the rosette leaves of 8-week-old plants grown under 16 h of illumination. The results confirmed that WRKY53, SAG12, WHY2, and WHY3 were up-regulated or down-regulated to various degrees in plants of different background (Fig. 2). All of the why1 mutants and antisense lines showed an average increase in transcript levels of 12- to 56-fold for WRKY53, 2.8-fold for WHY2, and 5- to 8-fold for WHY3. For SAG12, its expression was drastically enhanced at 56- to 427-fold (Fig. 2). In contrast, the WHY1 overexpression lines showed 440- and 600-fold increased transcript levels of WHY1, but in this background, the average transcript levels declined 5- and 10-fold for WRKY53, 250- and 300-fold for SAG12, and rendered no significant difference for WHY2 and WHY3. The why1why3 double mutant (ko1/3) showed a 3-fold reduced transcript level of WHY3 and a 45-fold increase for SAG12 but no significant difference for WRKY53. It should be noted that SAG12 is one of the target genes directly regulated by WRKY53 (Miao et al., 2004), but WHY3 was not involved in the WRKY53-mediated pathway.
Figure 2.
qRT-PCR analysis of gene expression in the WHY1 mutant and transgenic lines. why1-1 and why1-2 are two WHY1 T-DNA insertion lines; awhy1-1 and awhy1-6 are two WHY1 antisense lines; oe5 and oe6 are two lines overexpressing WHY1; PWHY1 is a complementary line harboring WHY1 coding sequence driven by its own promoter in the why1-1 background (why1-1Pwhy1:WHY1-HA); ko1/3 is a double knockout why1why3 line. Samples were taken from 8-week-old plants. ACTIN2 was used as an internal standard. The y axes indicate fold changes of mutants with respect to wild-type delta threshold cycle (CT) normalized to 1. Means and se from three technical replicates of two biological replicates are shown.
The WHY1 Protein Binds to the Promoter of WRKY53
The DNA-binding motifs recognized by WHY1 have been well characterized by others (Després et al., 1995; Desveaux et al., 2000, 2002; Yoo et al., 2007; Xiong et al., 2009). We used a ChIP assay to screen downstream targets of WHY1. To achieve this, a transgenic line (ID-WHY1-HA) expressing human influenza HA-tagged WHY1 driven by an estradiol-inducible promoter was constructed. Plants treated with estradiol for 2 h led to protein expression at approximately the same level as in the 7-week-old Pwhy1:WHY1-HA plants (Supplemental Fig. S3). According to the frequency and variation of DNA fragments, 30 candidate genes were selected after sequencing analysis (Supplemental Table S4). They all contained the ERE motif or AT-rich telomeric repeat-like sequence or both in their promoter regions. Among them, six WRKY genes (WRKY53, WRKY33, WRKY67, WRKY66, WRKY71, and WRKY9) were selected for further analysis.
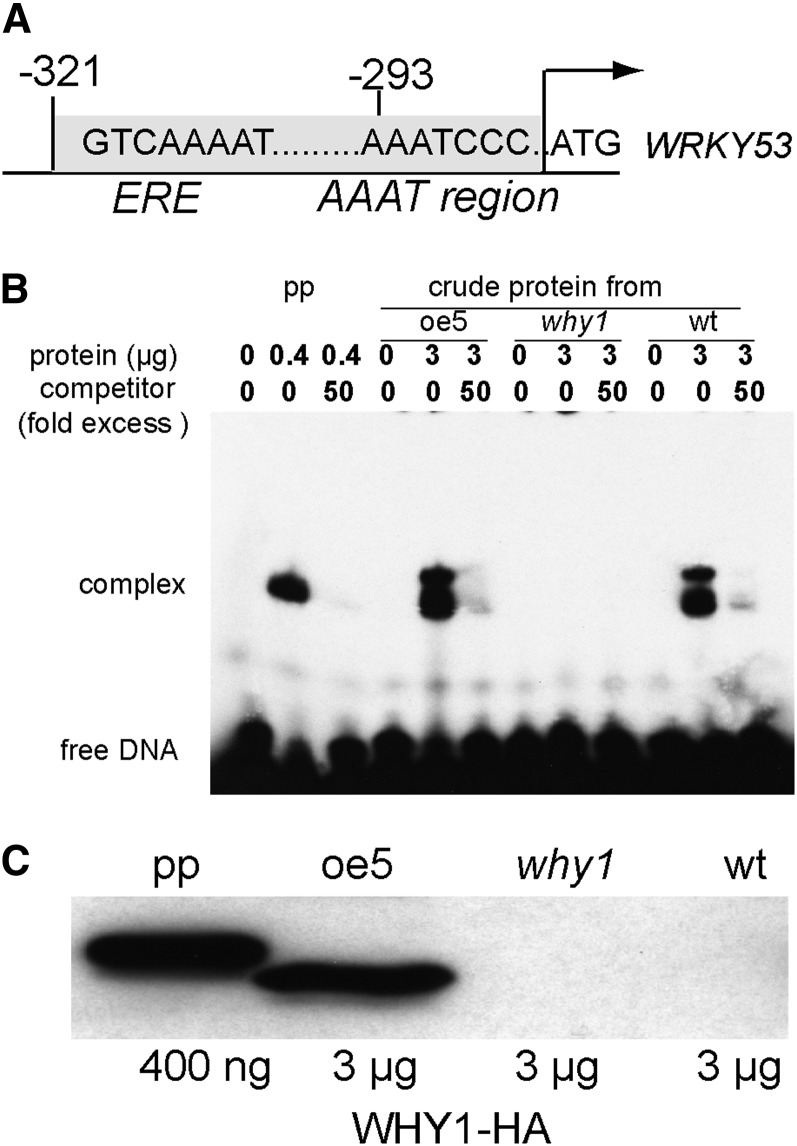
The promoter of the WRKY53 gene contains a sequence stretch with high similarity to both the ERE motif and the reverse telomere-like sequence AAATCCC or “AAAT region” (Desveaux et al., 2000; Yoo et al., 2007; Fig. 3A) that implies a direct interaction between WHY1 and the promoter of WRKY53. Therefore, we conducted an electrophoretic mobility shift assay (EMSA) using ssDNA of the WRKY53 promoter region (−321 to −287 upstream of ATG; Fig. 3A) with either purified recombinant 6xHis-tagged WHY1-HA protein expressed in Escherichia coli (Fig. 3B, PP) or nuclear proteins extracted from 6-week-old Arabidopsis plants overexpressing HA-tagged WHY1 under the control of a 35S promoter (Fig. 3B, line oe5). The ssDNA of the WRKY53 promoter formed a complex with bacterial purified recombinant WHY1 protein or plant nuclear WHY1-HA protein that was blocked by adding 50-fold amounts of unlabeled oligonucleotides as competitors (Fig. 3B). This interaction with ssDNA was specific to WHY1 protein, since no band shifts were observed with the nuclear proteins extracted from why1-1, although double bands appeared for extracts from both wild-type and WHY1-HA-overexpressing plants but only a single band appeared for the recombinant WHY1 proteins (Fig. 3B). Equal input of the overexpressed WHY1 protein in the EMSA was confirmed by western blotting with an antibody specific for the HA tag. In addition, as seen on the western blot, the smaller size of the nuclear WHY1 proteins compared with the full-length WHY1 expressed in E. coli (Fig. 3C) suggested that this nuclear WHY1 resembled the cleaved plastid WHY1 protein (Grabowski et al., 2008; Isemer et al., 2012). Furthermore, WHY1 formed a complex only with the sense DNA strand but not with the antisense DNA strand (Fig. 4B).
Figure 3.
WHY1 protein binds to the WRKY53 promoter. A, Schematic depiction of the ERE (GTCAAAAT) and AAATCCC sequence motifs found in the promoter region of WRKY53. The region from −321 to −287 nucleotides upstream of the ATG codon of WRKY53 is used for EMSA. B, EMSA performed with purified protein (pp) from E. coli extracts containing recombinant 6xHis-WHY1-HA or crude extract protein from the transgenic plants oeWHY1-HA5 (oe5), the why1-1 mutant (why1), and wild-type plants (wt). Unlabeled oligonucleotides were used in excess amounts (50-fold) as competitors. C, Western blot showing the relative amounts of WHY1 protein for a loading control. An antibody against the HA tag was used to detect HA-tagged WHY1 proteins.
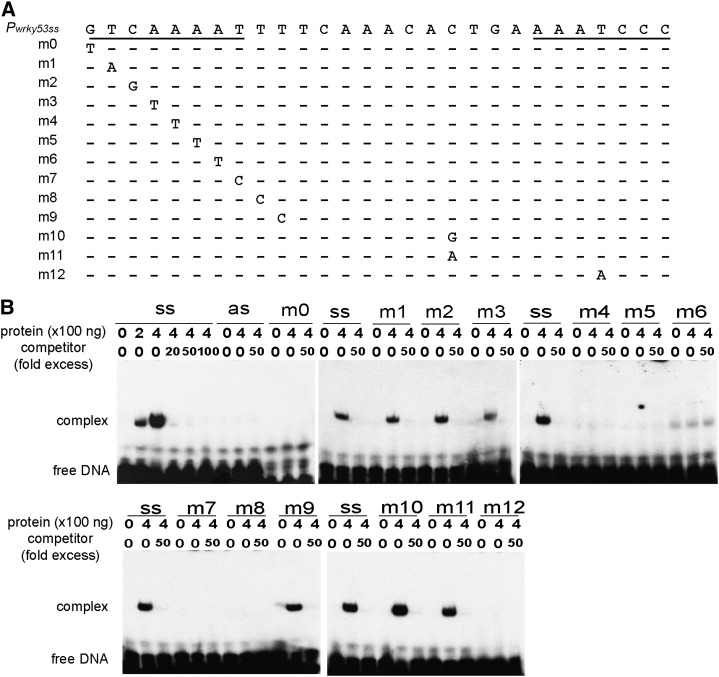
Figure 4.
Analysis of the WHY1-binding motif in the WRKY53 promoter region. A, A series of single-nucleotide exchanges (m0–m12) were introduced into the putative binding site of WHY1. B, EMSA showing the interaction of the purified recombinant WHY1 protein with the sense strand (ss), antisense strand (as), or mutated fragments m0 to m12. The sense strand binding reaction was run on all gels for a reference of complex size. Unlabeled oligonucleotides were used in excess amounts as competitors.
Single-nucleotide mutagenesis in the promoter region of WRKY53 showed that the binding with WHY1 was abolished when nucleotides of the ERE motif were mutated in several positions (Fig. 4). In the AAAT telomere-like sequence region, downstream of the ERE motif, single-nucleotide exchanges also abolished the binding of WHY1, while mutations in the region between the two motifs had no effect (Fig. 4B). Taken together, these results demonstrate a direct and specific binding of WHY1 to the single-stranded sequence GNNNAAATT within the ERE-like motif of the WRKY53 promoter and also addressed the importance of the downstream AAAT motif for the interaction.
WHY1 Acts as a Direct Repressor of WRKY53 Gene Expression
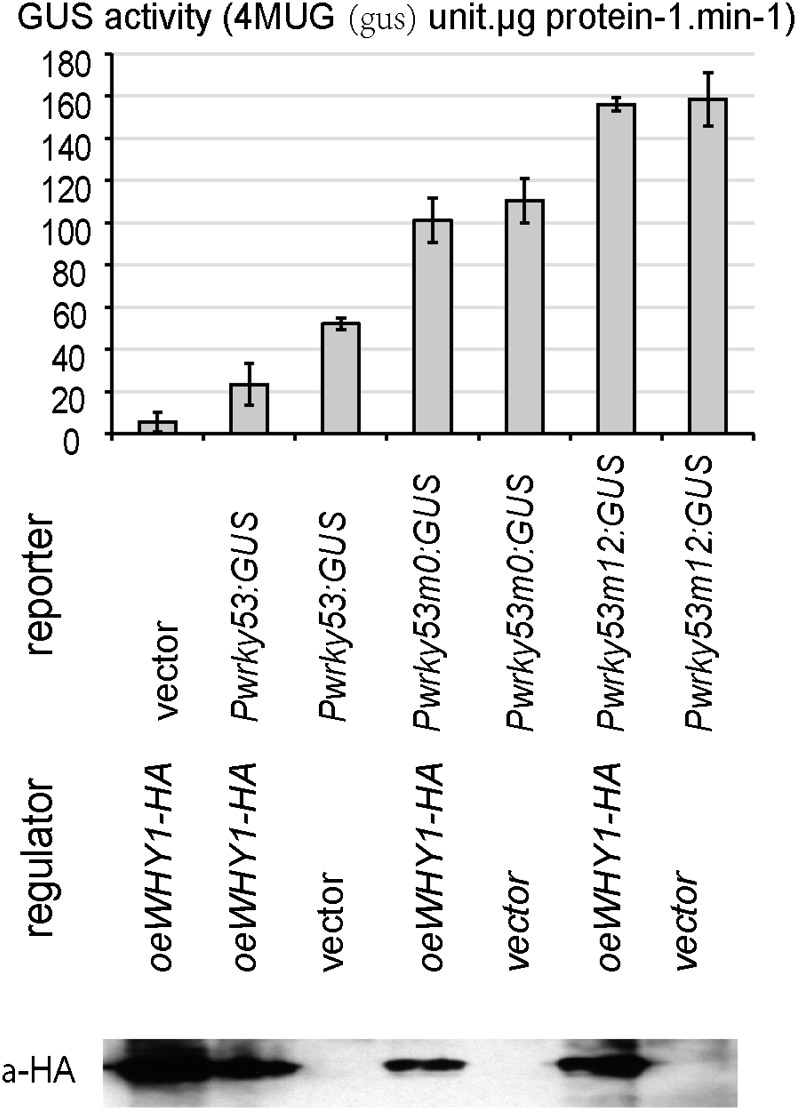
In order to investigate whether the binding of WHY1 to the WRKY53 promoter has an impact on WRKY53 expression in vivo, a construct giving rise to the accumulation of WHY1-HA was used to coexpress with a construct harboring the GUS/uidA reporter gene driven by the WRKY53 promoter in tobacco (Nicotiana tabacum) leaves. Proteins of the overexpressed WHY1 in the transfected tobacco tissues were checked by immunodetection with an antibody specific for the HA tag (Fig. 5). The resultant fluorimetric GUS activity indicated that the WRKY53 promoter effectively drove the expression of the reporter gene, which in turn was repressed by the accumulation of WHY1 (Fig. 5, oeWHY1-HA/Pwrky53:GUS). When constructs with mutation m0 (ERE motif) or m12 (AAAT region) in the WRKY53 promoter were used instead of the wild-type promoter, the measured GUS activity increased significantly, independent of the presence or absence of the introduced WHY1 (Fig. 5). This was probably due to the activities of the endogenous tobacco WHY1, which repressed the native, unaltered WRKY53 promoter activity to a certain degree but had no inhibitory effect on the mutated versions. Together with EMSA, these results suggest that both ERE and the AAAT motif in the WRKY53 promoter are indeed involved in direct interaction with WHY1, and both motifs are necessary to achieve efficient transcriptional repression of WRKY53.
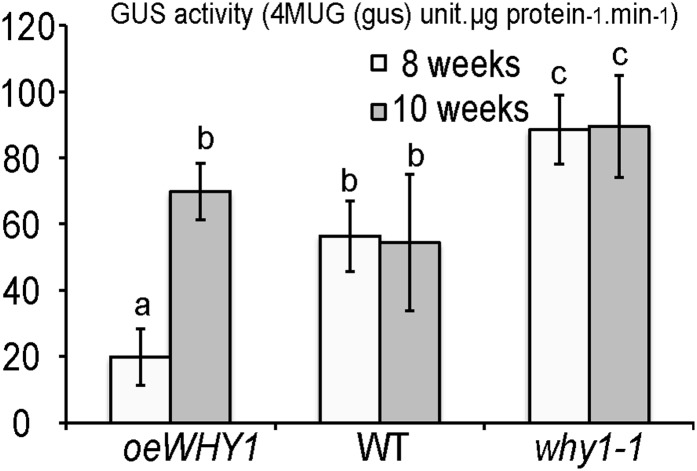
Figure 5.
Transient GUS reporter gene expression in transformed tobacco leaf. Tobacco leaves were cotransformed with a combination of regulator and reporter plasmids as indicated. Means and se of 12 independent measurements (three biological replicates with four technical replicates each) of GUS enzyme activities are indicated. An antibody against the HA tag was used to detect HA-tagged WHY1 proteins. oeWHY1-HA is overexpressing WHY1-HA; Pwrky53:GUS is GUS driven by the WRKY53 promoter; Pwrky53m0:GUS is GUS driven by the m0-mutated WRKY53 promoter; Pwrky53m12:GUS is GUS driven by the m12-mutated WRKY53 promoter; 4MUG is 4-Methylumbelliferyl-β-D-glucuronide.
WHY1 Could Not Change the Delayed-Senescence Phenotype of the wrky53 Mutant
A delayed-senescence phenotype was previously reported in the WRKY53 T-DNA insertion mutant (Miao et al., 2004; Miao and Zentgraf, 2007). We took advantage of the opposite phenotype in WHY1 mutants or overexpression by crossing the two homozygous plants separately with the wrky53 mutant line. PCR screening identified why1wrky53 double mutant and oeWHY1wrky53 plants, thus enabling us to analyze the genetic interaction between WRKY53 and WHY1. As shown in Figure 6, the oeWHY1wrky53 plants kept the same stay-green phenotype as strong as the wrky53 mutant, and both the Fv/Fm value and the chlorophyll content of leaf 7 in 9-week-old plants were similar in these two lines (Fig. 6, A and B). Visual analysis of leaf senescence and a statistical analysis of six 10-week-old plants further showed no phenotypic difference between them (Fig. 6, C and D). This result supported the assumption that WHY1 was an upstream regulator of the WRKY53-mediated plant senescence pathway. However, the phenotype in the why1wrky53 double mutant showed an intermediate phenotype between wild-type and wrky53 plants and did not exhibit a conclusively delayed senescence phenotype as strong as the wrky53 mutant. This might point to an inhibitory activity of WHY1 on genes downstream of WRKY53 or the existence of other targets of WHY1 that regulate senescence.
Figure 6.
Senescence phenotype of the why1-1wrky53 and oeWHY1wrky53 double mutants. A and B, Fv/Fm (A) and chlorophyll content (B) of leaf 7 in 9-week-old plants. Means and se of five independent measurements are shown. Different letters indicate significant differences at P ≤ 0.05 based on Student’s t test. C, Senescent leaf fraction from 12 plants at 10 weeks old. Means and se from two independent experiments with six plants each are indicated. D, Leaf phenotypes of 10-week-old plants. Numbers above the images indicate leaf age, with 1 being the oldest. WT, Wild type.
Repression of WRKY53 Expression by WHY1 Is Developmental Stage Dependent
Next, we introduced reporter constructs harboring the GUS/uidA reporter gene driven by the WRKY53 promoter into wild-type, why1 mutant, or WHY1 overexpression line plants to follow the temporal regulation of WHY1 on WRKY53 promoter activity during leaf senescence. In the why1 mutant, elevated GUS activity over that of the wild type appeared in 8- and 10-week-old plants, which means a loss of regulatory control of WHY1 on the promoter activity of WRKY53, whereas a significant decline of GUS activity was observed in 8-week-old plants of the WHY1 overexpression line (Fig. 7). However, in 10-week-old plants, the difference in GUS activity between the overexpressing line and the wild type was negligible (Fig. 7). This suggested that WHY1 repression on WRKY53 promoter activity mainly occurred at an early stage of senescence. It should be noted that, in the overexpression line, WHY1 was constantly driven by the 35S promoter; therefore, one could speculate that only the binding activity of WHY1 per se might be regulated by the stage of leaf development. On the contrary, in wild-type plants, WHY1 protein level might already be controlled by leaf development.
Figure 7.
GUS expression analysis driven by the WRKY53 promoter in different WHY1 transgenic plants. GUS fluorometric quantitative activity was measured from leaves of 8- or 10-week-old transgenic plants (oeWHY1-HA, why1-1, and the wild type [WT]) stably transformed with the Pwrky53:GUS construct. Means and se of nine independent measurements are shown. The same letters mean no significant difference at P ≤ 0.05 based on Student’s t test. 4MUG, 4-Methylumbelliferyl-β-D-glucuronide.
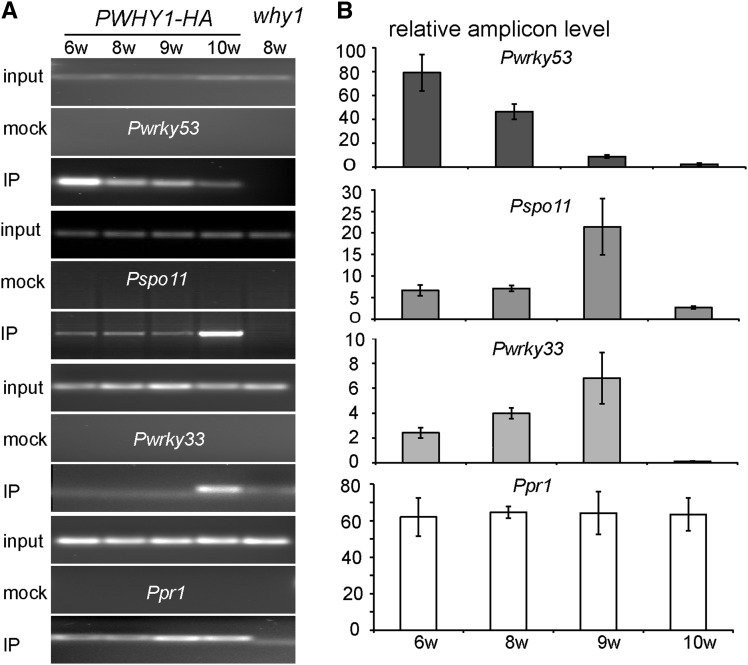
To further confirm this developmental stage dependence of WRKY53 repression by WHY1, ChIP-PCR was performed with a transgenic line harboring Pwhy1:WHY1-HA (PWHY1-HA) in the why1-1 background at different developmental stages. Under the normal (16-h) illumination condition, WHY1-HA protein could be detected at week 6, declined at week 8, and afterward increased again up to a peak at week 10 (Supplemental Fig. S3); a similar WHY1 expression profile was seen by qRT-PCR at this illumination condition but not in plants with 10 h of illumination, under which leaf senescence was postponed and extended (Supplemental Fig. S4). Thus, we used leaves from 6-, 8-, 9-, and 10-week-old PWHY1-HA plants under 10 and 16 h of illumination for the isolation of chromatins, and leaves of the why1-1 line were used as a parental control. PCR amplification of immunoprecipitated DNA, unimmunoprecipitated DNA, and input of PWHY1-HA or why1-1 under 10 h of the illumination condition was first performed by using semiquantitative PCR to check pull-down efficiency (Fig. 8A). According to the ChIP-sequencing results (Supplemental Table S3) and based on previous reports that WHY1 could bind to the telomere sequence in the promoter of PR1 and that WHY1 had functions in the DNA repair of double-strand breaks (DSB; Desveaux et al., 2004; Yoo et al., 2007; Maréchal et al., 2009; Maréchal and Brisson, 2010), the PR1 promoter was selected as a positive candidate control. As such, WRKY53, WRKY33, and SPO11 were detected in each DNA sample (Fig. 8A). The quantitative PCR amplicons of the promoter region of the PR1 gene were constantly amplified during developmental stages under 16 h of illumination, but those of the WRKY53 gene were strongly amplified at the 6-week-old stage and then subsequently decreased until they disappeared at the 10-week-old stage. In contrast, the promoter region of WRKY33 and SPO11 was abundantly amplified at the 9-week-old stage (Fig. 8B). This adverse effect likely suggested that WHY1 binding to WRKY33 and SPO11 promoters might have functions in the late developmental stage or might be involved in DSB-related DNA damage repair and cell death events.
Figure 8.
ChIP assays were performed using chromatin fragments from leaves of different developmental stages of the PWHY1-HA line and chromatin from the parental line why1 as a control. A, PCR amplification of the promoter region of the WRKY53 gene (Pwrky53), the WRKY33 gene (Pwrky33), and the PR gene (Ppr1) from input chromatin (input), nonimmunoprecipitation (mock), and immunoprecipitated DNA (IP). B, Quantitative PCR of the promoter regions of WRKY53, WRKY33, SPO11, and PR1 genes from immunoprecipitated DNA at different developmental stages (6, 8, 9, and 10 weeks old) under 16 h of illumination. The relative amplicon level was first normalized to the corresponding ACTIN amplicon, followed by a second normalization to the why1 line. Means and se from six independent experiments (two biological replicates and three technique replicates) are shown.
DISCUSSION
WHY proteins have been previously assigned roles in pathogen response, cell death regulation (Desveaux et al., 2000; Maréchal et al., 2008), and telomere length regulation in the nucleus (Yoo et al., 2007). This study revealed that WHY1 proteins also directly regulate WRKY53 expression and leaf senescence in a developmental stage-dependent manner. Since WRKY53 regulates several genes important for plant aging and leaf senescence (Miao et al., 2004), the specific repression of WRKY53 by WHY1 suggests that WHY1 is a key upstream regulator controlling senescence in Arabidopsis.
WHY1 functions as a transcriptional activator of PR genes in the pathogen response pathway (Desveaux et al., 2000; Maréchal et al., 2008). Several PR genes were suggested to be putative targets of WHY1 based on the presence of an ERE motif in their promoters (Desveaux et al., 2004, 2005). However, under our experimental conditions, most of their transcript level did not show any obvious alteration in WHY1 mutants or overexpression plants compared with the wild-type plants (Supplemental Table S2). Our ChIP-sequencing analyses showed that WHY1 could bind to the promoters of at least 30 downstream target genes, including WRKY53 and other WRKY members. More than 1.5-fold increase or decrease of the transcript level of these genes in why1 mutants was detected by qRT-PCR (Supplemental Table S3). Experiments with the promoter single-nucleotide mutation further proved that the nucleotides GNNNAAATT of the ERE-like motif and AAAT of the AT-rich region (Yoo et al., 2007) in target genes were crucial for the DNA-binding activity of WHY1, consistent with the analysis of WHY1 protein structure data. In the crystal structure of the WHY1 protein, the DNA ends of the WHY1 downstream promoter sequence are located in close proximity between adjacent 24-mers, thus raising the possibility that two pathways exist for the GNNNAAATT and AAAT-rich ssDNA in the crystal: one goes to the adjacent promoter of the same tetramer and the other goes to an adjacent 24-mers (Cappadocia et al., 2012). Similarly, the binding affinity of WHY1 is sensitive to changes in the AT-rich sequence downstream of the ERE motif (Fig. 3). These results support the idea that transcriptional regulation by WHY1 can be more complex than originally assumed and can be governed by a combination of DNA sequence motifs and different interacting factors. Therefore, both the ERE motif and the AT-rich telomeric repeat-like sequence in the promoter region of WRKY53 are equally important for WHY1 binding and regulation.
The recent scene of regulation of WRKY53 is intricate. At the protein level, WRKY53 may be modified by ubiquitination by the E3 ligase UPL5, or phosphorylated by MEKK1, or antagonistically interacted with an ESR/ESP protein (Miao et al., 2007; Miao and Zentgraf, 2007, 2010). These events represent a network of fine-turning the function of WRKY53 during leaf senescence (Zentgraf et al., 2010). At the transcriptional level, WRKY53 may bind to its own promoter, a self-repression mechanism, contrasting to the action of AD protein (Miao et al., 2008). Previously reported interactions between proteins and the WRKY53 promoter involved the double-stranded DNA. Binding of WHY1 protein to ssDNA of the promoter sequence of WRKY53 may hint at a new mechanism for WRKY53-dependent leaf senescence regulation (Figs. 1 and 8).
Interestingly, WHY1 also regulates the transcription of other WRKY family members and other downstream target genes, including the topoisomerase gene SPO11 that is essential for the DSB-induced initiation of meiotic recombination (Keeney and Neale, 2006; Hartung et al., 2007; Supplemental Table S3). WHY1 binding to ssDNA in plastids had a function in the DNA repair of detrimental lesions and prevents the spurious annealing of resected DNA molecules with other regions in the plastid genome (Maréchal et al., 2009; Cappadocia et al., 2010, 2012). However, whether nuclear WHY1 bound to the ssDNA of the WRKY53 promoter has the same DSB-related function is still unclear, since DSB in the nuclear genome were observed so far only during meiosis (Keeney and Neale, 2006) or under UV radiation and oxidation stress. Therefore, a link between DSB repair and WRKY53-mediated leaf senescence has not yet been established. In contrast, WHY1 might be involved in the preinitiation complex formation of the transcription of WRKY53. During the preinitiation complex formation, unwound ssDNA is introduced to the active RNA polymerase II site. In addition, several different histone modifications are deposited on nucleosomes at the promoter, including acetylation, H3K4 trimethylation, phosphorylation, and H2B monoubiquitylation. It is known that overexpression of one of the histone methyltransferases (SUVH2) in Arabidopsis leads to a repression of WRKY53, and the H3K4 histone modification of the WRKY53 promoter is developmental stage dependent (Ay et al., 2009). However, the DNA methylation of the WRKY53 promoter exhibits no difference during development (Zentgraf et al., 2010).
Another interesting feature of WHY1 is that it can bind to telomere sequences and regulate telomere length homeostasis (Yoo et al., 2007). With the identification of a telomere repeats-like sequence (TTTAGGG) in the noncoding region of the WRKY53 promoter (Fig. 5A), an intriguing link might be expected. Although the telomere length during leaf senescence progression in Arabidopsis has no significant differences (Zentgraf, 1995) and shorter telomeres (approximately 250–500 bp per generation) were tolerated for at least five to six generations in telomerase-deficient mutants, finally leading to instability of the genome and abnormal phenotypes in the following five generations, the late generation plants remained metabolically active and showed no signs of early senescence or apoptosis but, surprisingly, exhibited extended life spans (Riha et al., 2001). In mammals, telomere shortening is a characteristic of cellular aging. Telomeres and subtelomeric regions are enriched in epigenetic marks (histone methylation), and telomere shortening to a critical length affects the epigenetic status of telomeres and subtelomeres (Blasco, 2007). Therefore, binding of WHY1 to Arabidopsis telomeres not only shortens the telomeres (Yoo et al., 2007) but may also change the epigenetic status of telomeres, resulting in heterochromatin reorganization. Thus, it is likely that the binding of WHY1 to the telomere-like sequences in the WRKY53 promoter might also lead to a change in the epigenetic status of the WRKY53 promoter (Ay et al., 2009), a possible new regulatory pathway in addition to the direct transcriptional control.
WHY1 is located in both the nucleus and the plastids (Grabowski et al., 2008). In our experiments, WHY1 was necessary and sufficient to restore the phenotype of the why1 mutant, and only WHY1 in the nucleus had a suppressing effect on the activity of the WRKY53 promoter at the early stage of senescence (Fig. 1). The redundant gene WHY3 is partially involved in these processes (Fig. 2); however, the exact functions of WHY3 are elusive in this regard. It is reported that double knockout of WHY1 and WHY3 (why1why3) results in plants with variegated green/white/yellow leaves, suggesting a protective function of WHY1 and WHY3 against microhomology-mediated break-induced replication events in chloroplasts (Maréchal et al., 2009; Cappadocia et al., 2010; Maréchal and Brisson, 2010). Therefore, WHY proteins might have different functions depending on intracellular localization and developmental stage.
Overall, we have provided clear evidence showing the involvement of WHY1 proteins in the regulation of Arabidopsis leaf senescence through its repression of the senescence-activating WRKY53 transcription factor.
MATERIALS AND METHODS
Plant Material
Plants of Arabidopsis (Arabidopsis thaliana) ecotype Columbia were grown in a growth chamber at 22°C with 10 h of illumination under low-light conditions (70 µmol s−1 m−2). Under these conditions, plants developed flowers within 11 to 12 weeks, and mature seeds could be harvested after 14 to 15 weeks. Colored threads were used to label entire rosette leaves after their emergence, as described previously (Hinderhofer and Zentgraf, 2001). Plants grown at 22°C with 16 h of illumination were used for qRT-PCR.
T-DNA insertion lines Salk_023713 (why1-1) and Salk_147680 (why1-2) for WHY1 were provided by the European Arabidopsis Stock Centre. These mutant lines were confirmed by PCR with the primers suggested by the T-DNA Express tool of the SALK Institute and by northern blot (Supplemental Fig. S1).
Overexpression and Induction Expression of WHY1, PWHY1-HA, and Antisense WHY1 Constructs
The full-length coding sequence of WHY1 and the antisense sequence of WHY1 without plastid transit peptide sequence were amplified by PCR using the complementary DNA (cDNA) U10139, cloned into pENTR/TOPO Gateway vector, and sequenced to verify PCR products. The HA tag was included in the sequence of the reverse primer. The 790-bp promoter of WHY1 was amplified from genomic DNA, cloned into pGEM-T Easy vector, and sequenced to verify the PCR product. The promoter was digested with NotI, subcloned to the upstream region of the WHY1 coding sequence in pENTR/TOPO vector, and verified by sequencing. After transfer of the WHY1 construct to the binary destination vectors pB2GW7.0 (Karimi et al., 2002) and pMDC7 (Curtis and Grossniklaus, 2003) and of the Pwhy1:WHY1-HA cassette to the modified binary destination vector pBGWL7 (Karimi et al., 2002), in which the LUC coding sequence was deleted, the constructed vectors were transformed into why1-1 mutant, wrky53 mutant, and wild-type plants using Agrobacterium tumefaciens-mediated vacuum infiltration (Miao et al., 2004) to produce the transgenic plants oeWHY1-HA, Pwhy1:WHY1-HA (PWHY1-HA), antisense-WHY1 (aWHY1), and oeWHY1-HAwrky53. The transgenic seedlings were selected by spraying with 0.1% (w/v) glufosinate ammonium (Basta; Bayer Crop Science).
Transient Infiltration of Tobacco Leaves
The 1.2-kb WRKY53 promoter fused to the GUS reporter cassette (Miao et al., 2007) was subcloned in the pCB308kan binary vector (Xiang et al., 1999). Mutated WRKY53 promoters Pwrky53m0 and Pwrky53m12 were assembled by site-directed mutagenesis according to the manufacturer’s instructions (Stratagene). These constructs were transiently coexpressed in leaves of wild-type tobacco (Nicotiana tabacum) plants with a plasmid allowing overexpression of the full-length WHY1 gene tagged with a HA epitope using A. tumefaciens infiltration as described previously in Arabidopsis (Miao and Zentgraf, 2007). Protein expression in the infiltrated areas was examined after 24 to 48 h of incubation in a growth chamber. Total protein content was measured using Bradford reagent, and immunodetection of WHY1-HA was done with an antibody directly against the HA tag (Roche Diagnostics). GUS enzyme activity was determined using the 4MUG fluorometric quantitative assay as described by Jefferson et al. (1987).
Expression of a Pwrky53-GUS Reporter Gene Construct in WHY1 Overexpression and Mutant Plants
The 1.2-kb WRKY53 promoter fused with the GUS reporter plasmid was transformed by vacuum infiltration to the WHY1 overexpression line (oeWHY1-HA5, oe5), why1-1 mutant, and wild-type plants. Transgenic seedlings were selected by growing on kanamycin-containing medium and spraying with 0.1% (w/v) Basta. GUS fluorometric quantitative measurement was done on transgenic T2 plants after 8 and 10 weeks of growth.
Measurements of Chlorophyll Fluorescence and Chlorophyll Content
Chlorophyll fluorescence was measured noninvasively using an Imaging-PAM-M series (Heinz Walz) after a 15-min dark incubation of leaf 7 from 9-week-old plants, and three regions were measured per leaf. As a measure of the photochemical efficiency of PSII, the Fv/Fm value was calculated. For the monitoring of whole plant senescence, the average Fv/Fm of all rosette leaves from five individual plants was calculated. For leaf senescence, the same leaf 7 from five different plants was measured and used to determine Fv/Fm. Chlorophylls were extracted from whole rosettes with hot methanol in the presence of magnesium hydroxide carbonate. Chlorophyll concentrations were determined according to Lichtenthaler (1987).
mRNA Preparation and qRT-PCR Analysis
mRNA was extracted from Arabidopsis entire rosettes using the Chemagic mRNA Direct Kit (Chemagen). First-strand cDNA was synthesized using the qScript cDNA SuperMix Kit (Quanta Biosciences). Gene-specific primers were developed for the genes SAG12, WRKY53, WHY1, WHY2, and WHY3 as well as for ACTIN2 as an internal control. Primer pairs are shown in Supplemental Table S4. The expression of the genes was analyzed using the qPCR SYBR Green Kit (Quanta Biosciences) according to the manufacturer’s protocol. Data analysis was accomplished using the iCycler (Bio-Rad). To calculate PCR efficiencies, four different cDNA dilutions were applied. To determine the relative expression rate, data were normalized to the expression level in wild-type or in 3-week-old plants, these basic points were set to 1, and the relative expression formula described by Pfaffl (2001) was used. mRNA template and no-template controls were performed to exclude the amplification of unspecific products. Additionally, the end-time PCR products were separated on agarose gels to ensure the amplification of a single specific product. Each value represents three technical replicates of two biological replicates (six measurements altogether).
Production of Recombinant WHY1 Protein
The WHY1 sequence was cloned into the Gateway protein expression vector pDEST17 containing a 6xHis tag for purification (Invitrogen). The recombinant protein was expressed in Escherichia coli BL21-S1, purified by nickel-nitrilotriacetic acid agarose affinity chromatography, and dialyzed according to the manufacturer’s protocol.
EMSA
EMSA was performed essentially as described by Miao et al. (2004). A total of 400 ng of purified recombinant WHY1 protein from bacteria or 3 µg of crude nuclear extracts from different Arabidopsis lines was incubated with 0.02 pmol of a [γ-32P]ATP-labeled ssDNA fragment in a total volume of 20 µL. The reaction products were analyzed on 5% (w/v) polyacrylamide gels under nondenaturing conditions. To show the specificity of the DNA-protein interaction, a 20-, 50-, or 100-fold excess of the unlabeled DNA fragment was added.
ChIP Assay
ChIP assays were performed using the method described by Ay et al. (2009) with some modifications. A total of 1.5 g of leaf tissue from entire rosettes of 7-week-old plants harboring an estradiol induced promoter-WHY1-HA transgene was treated with 20 μm estradiol for 1, 2, 4, 6, 8, or 16 h or not treated. In contrast, the why1 mutant function-restored transgenic line PWHY1-HA was grown on soil under 10 or 16 h of illumination, and 1.5 g of leaf tissue from entire rosettes of 6-, 8-, 9-, and 10-week-old plants was continually dissected. The wild type and the why1-1 line were grown under the same conditions. The dissected leaves were cross linked with 1% formaldehyde by vacuum infiltration for 15 min and then quenched with Gly (0.135 m). Nuclei were extracted, and the chromatin contained was resuspended in 1% SDS nuclear lysis buffer with the addition of complete protease inhibitor cocktail tablets (Roche Diagnostics). Samples were then sonicated to chromatin fragments of about 300 to 500 bp (determined for each experiment). Chromatin was immunoprecipitated overnight with antibodies against the HA tag at 4°C or was processed with no antibody as a control (mock precipitation). The immunoprecipitated DNA was isolated by the Nucleospin Extract II kit (Macherey-Nagel), an A tailing was added, then it was cloned to pGEM-T Easy vector and sequenced. Using real-time PCR, the enrichment of the immunoprecipitated DNAs was determined. Data analysis was adapted to published work (Fode et al., 2008), where the amplicon level in the why1 mutant was calibrated to 1 and the relative fold enrichment in the PWHY1-HA line was calculated upon the why1 mutant. For estimation of the percentage input, a standard curve using the following dilutions of the input DNA (DNA from chromatin without immunoprecipitation) was generated for each amplicon: 0.12%, 0.06%, 0.03%, 0.015%, 0.0075%, and 0.00375%. ACTIN served as an internal reference for the first normalization, and the 2−ΔΔCT method was used for data analysis. First, the ΔCT value for each sample is determined by calculating the difference between the CT value of the target gene and the CT value of the endogenous reference gene. This is determined for each unknown sample as well as for the calibrator sample: ΔCT (sample) = CT target gene − CT ACTIN, ΔCT (why1) = CT target gene − CT ACTIN. Next, the ΔΔCT value for each sample is determined by subtracting the ΔCT value of the calibrator from the ΔCT value of the sample: ΔΔCT = ΔCT (sample) − ΔCT (why1). If the PCR efficiencies of the target gene and the endogenous reference gene are comparable, the normalized level of target gene expression is calculated by using this formula: Normalized target gene expression level in sample = 2−ΔΔCT. Each ChIP assay was performed in two biological replicates and three technique replicates. For each gene, specific PCR primer sets were designed (Supplemental Table S4), and fragments of approximately 100 to 300 bp were amplified. The end-time PCR products were separated on 1% (w/v) agarose gels to ensure the amplification of a single specific product.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_101308 for WHY1 (At1g14410), NM_105795 for WHY2 (At1g71260), NM_126329 for WHY3 (At2g02740), NM_118512 for WRKY53 (At4g23810), NM_123957 for SAG12 (At5g45890), NM_129404 for WRKY33 (At2g38470), NM_127025 for PR1 (At2g14610), and NM_105072 for SPO11 (At1g63990).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic representation of the two WHY1 T-DNA insertion lines, the overexpression lines, and the complementation line.
Supplemental Figure S2. Senescence phenotype analysis of whole plants (whole rosettes upside down) from different WHY1 transgenic plants at 8 weeks old under 16 h of illumination.
Supplemental Figure S3. Immunodetection of WHY1-HA protein in the estradiol-inducible promoter driving WHY1-HA after 20 µm estradiol treatment for 0, 1, 2, 4, 6, 8, and 16 h and in the PWHY1-HA line during developmental stages from 6 to 11 weeks.
Supplemental Figure S4. qRT-PCR analyses of gene expression during the development of wild-type plants under 10 and 16 h of illumination.
Supplemental Table S1. Phenotype analyses of progeny of different transgenic plants of the WHY1 gene.
Supplemental Table S2. List of 30 other nonaffected genes by the why1 mutant.
Supplemental Table S3. List of downstream candidate genes of WHY1 identified by ChIP.
Supplemental Table S4. Primer sequences used for semiquantitative reverse transcription-PCR, qRT-PCR, and quantitative PCR.
Acknowledgments
We thank Prof. Karin Krupinska (University of Kiel) and Prof. Ulrike Zentgraf (University of Tuebingen) for providing laboratory facilities for experiments and helpful discussions, Prof. Karin Krupinska (University of Kiel) for providing the oeWHY1 lines, and Prof. Normand Brisson (University of Montreal) for providing the why1why3 double mutants. We thank Ms. Rena Isemer (University of Kiel) for helping with phenotype analyses and Ms. Ulrike Voigt (University of Kiel) for crossing the why1 mutant with the wrky53 mutant.
Glossary
- ssDNA
single-stranded DNA
- ERE
elicitor response element
- ChIP
chromatin immunoprecipitation
- T-DNA
transfer DNA
- HA
hemagglutinin
- qRT
quantitative reverse transcription
- Fv/Fm
photochemical efficiency of PSII
- EMSA
electrophoretic mobility shift assay
- DSB
double-strand breaks
- cDNA
complementary DNA
Footnotes
This work was supported by the German Research Foundation (grant no. MI1392/1–1), the National Science Foundation of China, and the Deutsche Akademische Austausch Dienst program (Programme des Projektbezogenen Personenaustauschs China).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ay N, Irmler K, Fischer A, Uhlemann R, Reuter G, Humbeck K. (2009) Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J 58: 333–346 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B. (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4: 346–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B. (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol (Stuttg) (Suppl 1) 10: 63–75 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B. (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62: 250–264 [DOI] [PubMed] [Google Scholar]
- Blasco MA. (2007) The epigenetic regulation of mammalian telomeres. Nat Rev Genet 8: 299–309 [DOI] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D. (2003) The molecular analysis of leaf senescence: a genomics approach. Plant Biotechnol J 1: 3–22 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Cappadocia L, Maréchal A, Parent JS, Lepage E, Sygusch J, Brisson N. (2010) Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 22: 1849–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappadocia L, Parent JS, Zampini E, Lepage E, Sygusch J, Brisson N. (2012) A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage. Nucleic Acids Res 40: 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, et al. (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C, Subramaniam R, Matton DP, Brisson N. (1995) The activation of the potato PR-10a gene requires the phosphorylation of the nuclear factor PBF-1. Plant Cell 7: 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Allard J, Brisson N, Sygusch J. (2002) A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat Struct Biol 9: 512–517 [DOI] [PubMed] [Google Scholar]
- Desveaux D, Després C, Joyeux A, Subramaniam R, Brisson N. (2000) PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12: 1477–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Maréchal A, Brisson N. (2005) Whirly transcription factors: defense gene regulation and beyond. Trends Plant Sci 10: 95–102 [DOI] [PubMed] [Google Scholar]
- Desveaux D, Subramaniam R, Després C, Mess JN, Lévesque C, Fobert PR, Dangl JL, Brisson N. (2004) A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev Cell 6: 229–240 [DOI] [PubMed] [Google Scholar]
- Fode B, Siemsen T, Thurow C, Weigel R, Gatz C. (2008) The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 20: 3122–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski E, Miao Y, Mulisch M, Krupinska K. (2008) Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol 147: 1800–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cai Z, Gan S. (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27: 521–549 [Google Scholar]
- Hartung F, Wurz-Wildersinn R, Fuchs J, Schubert I, Suer S, Puchta H. (2007) The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19: 3090–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Gan S. (2002) A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 14: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderhofer K, Zentgraf U. (2001) Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213: 469–473 [DOI] [PubMed] [Google Scholar]
- Humbeck K, Quast S, Krupinska K. (1996) Functional and molecular changes in the photosynthetic apparatus during senescence of flag leaves from field-grown barley plants. Plant Cell Environ 19: 337–344 [Google Scholar]
- Isemer R, Mulisch M, Schäfer A, Kirchner S, Koop HU, Krupinska K. (2012) Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett 586: 85–88 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Keeney S, Neale MJ. (2006) Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans 34: 523–525 [DOI] [PubMed] [Google Scholar]
- Krause K, Kilbienski I, Mulisch M, Rödiger A, Schäfer A, Krupinska K. (2005) DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett 579: 3707–3712 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–383 [Google Scholar]
- Maréchal A, Brisson N. (2010) Recombination and the maintenance of plant organelle genome stability. New Phytol 186: 299–317 [DOI] [PubMed] [Google Scholar]
- Maréchal A, Parent JS, Sabar M, Véronneau-Lafortune F, Abou-Rached C, Brisson N. (2008) Overexpression of mtDNA-associated AtWhy2 compromises mitochondrial function. BMC Plant Biol 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Parent JS, Véronneau-Lafortune F, Joyeux A, Lang BF, Brisson N. (2009) Whirly proteins maintain plastid genome stability in Arabidopsis. Proc Natl Acad Sci USA 106: 14693–14698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melonek J, Mulisch M, Schmitz-Linneweber C, Grabowski E, Hensel G, Krupinska K. (2010) Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta 232: 471–481 [DOI] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U. (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Miao Y, Laun TM, Smykowski A, Zentgraf U. (2007) Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol Biol 65: 63–76 [DOI] [PubMed] [Google Scholar]
- Miao Y, Smykowski A, Zentgraf U. (2008) A novel upstream regulator of WRKY53 transcription during leaf senescence in Arabidopsis thaliana. Plant Biol (Stuttg) (Suppl 1) 10: 110–120 [DOI] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U. (2007) The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U. (2010) A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J 63: 179–188 [DOI] [PubMed] [Google Scholar]
- Oh SA, Lee SY, Chung IK, Lee CH, Nam HG. (1996) A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol Biol 30: 739–754 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. (2008) A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res 36: 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Griffing LR, Shippen DE. (2001) Living with genome instability: plant responses to telomere dysfunction. Science 291: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B, Shahid Mukhtar M, Somssich IE. (2007) The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226: 125–137 [DOI] [PubMed] [Google Scholar]
- Xiang CB, Han P, Lutziger I, Wang K, Oliver DJ. (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40: 711–717 [DOI] [PubMed] [Google Scholar]
- Xiong JY, Lai CX, Qu Z, Yang XY, Qin XH, Liu GQ. (2009) Recruitment of AtWHY1 and AtWHY3 by a distal element upstream of the kinesin gene AtKP1 to mediate transcriptional repression. Plant Mol Biol 71: 437–449 [DOI] [PubMed] [Google Scholar]
- Yoo HH, Kwon C, Lee MM, Chung IK. (2007) Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J 49: 442–451 [DOI] [PubMed] [Google Scholar]
- Zentgraf U. (1995) Telomere-binding proteins of Arabidopsis thaliana. Plant Mol Biol 27: 467–475 [DOI] [PubMed] [Google Scholar]
- Zentgraf U, Jobst J, Kolb D, Rentsch D. (2004) Senescence-related gene expression profiles of rosette leaves of Arabidopsis thaliana: leaf age versus plant age. Plant Biol (Stuttg) 6: 178–183 [DOI] [PubMed] [Google Scholar]
- Zentgraf U, Laun T, Miao Y. (2010) The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur J Cell Biol 89: 133–137 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T. (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]
- Zhou X, Jiang Y, Yu D. (2011) WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol Cells 31: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]