An Arabidopsis family of phospholipases, probed using reverse genetics for involvement in cell wall-based defense to nonhost powdery mildew, identifies one isoform as a component in the defense reaction.
Abstract
Plants have evolved a complex array of defensive responses against pathogenic microorganisms. Recognition of microbes initiates signaling cascades that activate plant defenses. The membrane lipid phosphatidic acid, produced by phospholipase D (PLD), has been shown to take part in both abiotic and biotic stress signaling. In this study, the involvement of PLD in the interaction between Arabidopsis (Arabidopsis thaliana) and the barley powdery mildew fungus Blumeria graminis f. sp. hordei (Bgh) was investigated. This nonadapted pathogen is normally resisted by a cell wall-based defense, which stops the fungal hyphae from penetrating the epidermal cell wall. Chemical inhibition of phosphatidic acid production by PLD increased the penetration rate of Bgh spores on wild-type leaves. The analysis of transfer DNA knockout lines for all Arabidopsis PLD genes revealed that PLDδ is involved in penetration resistance against Bgh, and chemical inhibition of PLDs in plants mutated in PLDδ indicated that this isoform alone is involved in Bgh resistance. In addition, we confirmed the involvement of PLDδ in penetration resistance against another nonadapted pea powdery mildew fungus, Erysiphe pisi. A green fluorescent protein fusion of PLDδ localized to the plasma membrane at the Bgh attack site, where it surrounded the cell wall reinforcement. Furthermore, in the pldδ mutant, transcriptional up-regulation of early microbe-associated molecular pattern response genes was delayed after chitin stimulation. In conclusion, we propose that PLD is involved in defense signaling in nonhost resistance against powdery mildew fungi and put PLDδ forward as the main isoform participating in this process.
A wide range of potentially pathogenic microbes are found in the environment of plants. Nevertheless, plants are able to resist the great majority of microbes they encounter and are susceptible to only a small number of specifically adapted pathogens. This capacity hinges primarily on a broad-range form of immunity called “nonhost resistance” (NHR), which is defined as the immunity displayed by an entire plant species against all genetic variants of a pathogen (Thordal-Christensen, 2003). NHR against pathogens of distantly related host species is believed to be due to basal defense activated after the recognition of microbe-associated molecular patterns (MAMPs; Schulze-Lefert and Panstruga, 2011). While adapted pathogens have evolved effector proteins that target distinct plant mechanisms to, for instance, suppress plant defenses (Jones and Dangl, 2006), the effectors of nonadapted pathogens are not able to sufficiently suppress plant defenses. In this way, NHR is provided by multiple genes, which makes it effective and genetically robust (Schulze-Lefert and Panstruga, 2011). This form of resistance has thus gained particular interest for its agroeconomic potential.
A well-studied model system for this type of NHR is the interaction between Arabidopsis (Arabidopsis thaliana) and nonadapted powdery mildew fungi (Micali et al., 2008). After germination on a leaf, fungal spores develop a specialized hyphal structure, called the “appressorium,” which attaches to the epidermal cell wall. Subsequently, the fungus attempts to penetrate the cell from the attachment site. Here, plants are thought to recognize fungal MAMPs and induce localized defense responses at the site of interaction. Fungal penetration is usually stopped by the formation of a multilayered cell wall reinforcement known as a papilla. The papilla is rich in callose and other antimicrobial substances, which provides both a physical and a chemical barrier against the invading pathogen (Hardham et al., 2007; Hückelhoven, 2007). When powdery mildew fungi manage to evade this defense, they form a haustorium, a feeding structure that invades the host cell. Successfully penetrating spores are stopped either by a second level of defense, confinement of the haustorium within a callose encasement, or a third level of defense, a hypersensitive cell death response to stop further fungal development (Lipka et al., 2008). Thus, NHR against nonadapted powdery mildews and other biotrophic fungi can be conceptually divided into pre- and postinvasion defenses.
It is well established that cell wall-based pathogen recognition in plants is mediated by transmembrane pattern recognition receptors (PRRs) that recognize highly conserved MAMPs. Examples of fungal MAMPs are xylanase and chitin (Boller and Felix, 2009; Monaghan and Zipfel, 2012). The corresponding PRRs, ETHYLENE INDUCING-XYLANASE2 in tomato (Solanum lycopersicum; Ron and Avni, 2004), CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) in Arabidopsis (Miya et al., 2007), and CHITIN ELICITOR-BINDING PROTEIN in rice (Oryza sativa; Shimizu et al., 2010), have been identified. Notably, the chitin receptor CERK1 has been found to be essential for resistance against a powdery mildew fungus in Arabidopsis (Wan et al., 2008). Several molecular and physiological changes are initially brought about by MAMP perception. These include fluxes of various ions including Ca2+ over the plasma membrane, protein phosphorylation, generation of reactive oxygen species and nitric oxide, activation of mitogen-activated protein kinase, Ca2+-dependent protein kinase cascades, and, finally, transcriptional changes in defense genes (Hückelhoven, 2007; Schwessinger and Zipfel, 2008; Boller and Felix, 2009). These early responses to MAMP detection are well studied and known to depend on one another (Ogasawara et al., 2008; Ma et al., 2009). However, thorough insight into the regulation and the genetic basis of these events remains fragmented.
Three major components of preinvasion defense were identified in forward genetic screens for Arabidopsis mutants with enhanced penetration frequencies of the nonadapted powdery mildew fungus Blumeria graminis f. sp. hordei (Bgh; Collins et al., 2003; Lipka et al., 2005; Stein et al., 2006). These correspond to the PENETRATION1 (PEN1), PEN2, and PEN3 genes, which encode a plasma membrane-localized syntaxin, a glycoside hydrolase, and a pleiotropic drug resistance ATP-binding cassette transporter, respectively. The current model predicts the existence of a PEN1-defined pathway where this syntaxin is involved in polar secretion of material at sites of attempted fungal ingress, together with an ADP ribosylation factor, GTPase, and the corresponding GTP exchange factor, GNOM (Böhlenius et al., 2010; Nielsen et al., 2012). In a second pathway, PEN2 is required for the production of an antifungal glucosinolate product, secreted into the apoplast by PEN3 (Bednarek et al., 2009). The three PEN proteins accumulate at the incipient fungal entry site: PEN1 in exosomes in the papilla body (Meyer et al., 2009; Nielsen et al., 2012), PEN2 in peroxisomes in the cytoplasmic area surrounding the papilla (Lipka et al., 2005), and PEN3 at the plasma membrane (Stein et al., 2006).
Several lipids and lipid metabolites have been shown to function in the signal transduction pathway leading to the activation of plant defense responses (Laxalt and Munnik, 2002; Kachroo and Kachroo, 2009). A prominent role has been postulated for phosphatidic acid (PA), whose levels increase within minutes in plant cells recognizing different MAMPs (van der Luit et al., 2000) or upon the recognition of pathogen effector proteins (de Jong et al., 2004; Andersson et al., 2006; Kirik and Mudgett, 2009). PA has been implicated in the modulation of mitogen-activated protein kinase activity, Ca2+ influx from the apoplast, and oxidative burst during abiotic and biotic stresses (Testerink and Munnik, 2011). In plants, PA can be formed via two main pathways: the direct hydrolysis of phospholipids by phospholipase D (PLD) or the combined action of phospholipase C and diacylglycerol kinase (Wang, 2004). Both pathways are involved in the early signaling events of plant-pathogen interactions and contribute to the activation of plant defenses. Specifically, PLD activity has been found to be required for the hypersensitive response induced by the recognition of bacterial effector proteins (Andersson et al., 2006; Kirik and Mudgett, 2009).
The 12 different PLD isoforms encoded in the Arabidopsis genome are classified into six groups (α, β, γ, δ, ε, and ζ) based on sequence similarity and in vitro activity. Two HxKxxxxD (HKD) motifs that interact with each other are essential for lipase activity in all eukaryotic PLD isoforms (Bargmann and Munnik, 2006). Although an increase in PLD activity upon treatment with various MAMPs has been reported for different cell culture systems (van der Luit et al., 2000; Yamaguchi et al., 2005; Suzuki et al., 2007), genetic evidence for the involvement of specific PLD isoforms in pathogen defense so far is missing. Here, we reveal a role for PLD activity in Arabidopsis cell wall-based defense against powdery mildew fungi and genetically identify PLDδ as the responsible PLD isoform. We also report that this PLD is involved in MAMP signaling at a more general level.
RESULTS
Inhibition of PLD-Generated PA Causes Increased Bgh Penetration in Arabidopsis
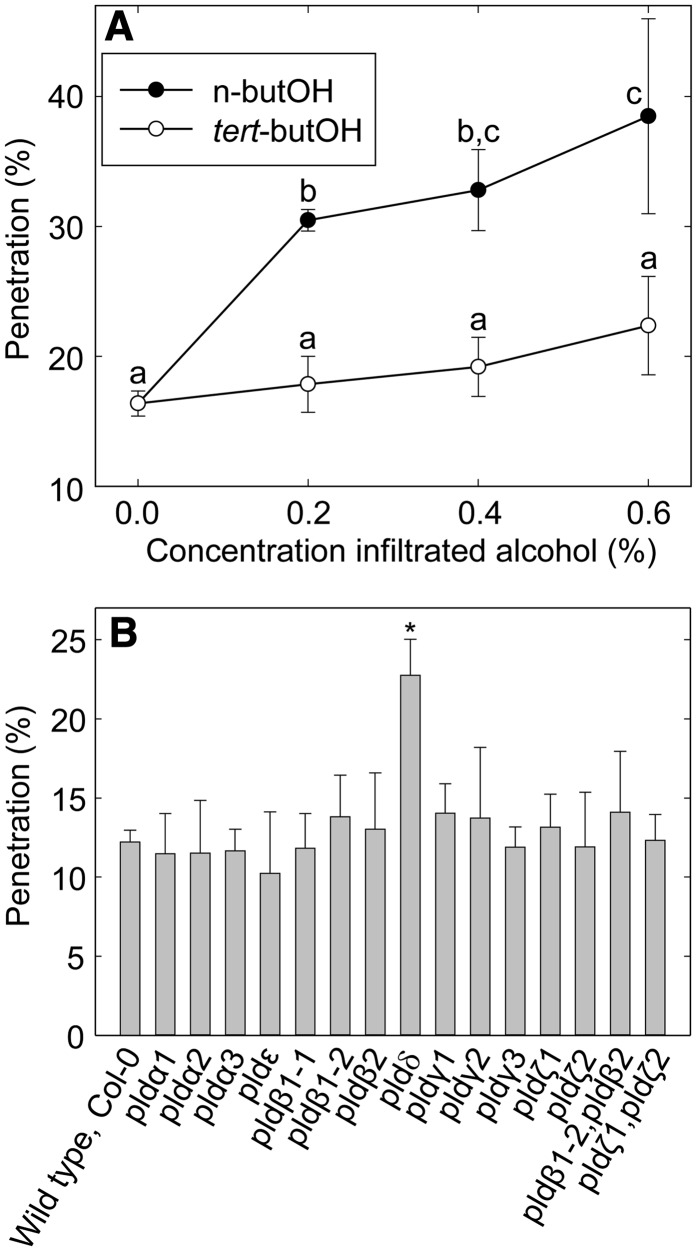
The development of Bgh spores on Arabidopsis leaves is synchronous, and penetration attempts occur in a narrow time frame around 12 h post inoculation (hpi; Assaad et al., 2004). It is thus possible to score and compare fungal penetration on different plants at a certain time point. To test a possible contribution of PLD-generated PA to plant resistance against Bgh penetration, we exploited the transphosphatidylation activity of PLD, which uses primary alcohols as substrates to form an artificial phosphatidyl alcohol. The preferential formation of this compound impairs PA production (Munnik et al., 1995). Thus, we employed n-butanol to inhibit PA formation by PLD. Leaves were infiltrated with different n- or tert-butanol concentrations 3 h before inoculation, and penetration was scored at 3 d post inoculation (dpi). Increasing n-butanol concentrations caused progressively higher Bgh penetration rates, from 16% (sd 1.0) for a water control to 38% (sd 7.5) for leaves treated with 0.6% n-butanol (Fig. 1A). In contrast, the penetration rate for leaves infiltrated with tert-butanol, an alcohol with no inhibitory effects on PA formation, was not significantly different compared with that of the water control. These results thus indicate the involvement of PLD and PA in cell wall-based defense against Bgh. We next attempted to measure PLD activity and PA accumulation during the defense response. To this end, leaf explants were labeled with 33PO4 overnight before inoculation with Bgh spores. However, no increase in PA label could be measured at 4, 8, 12, and 24 hpi (Supplemental Fig. S1).
Figure 1.
PLDδ-generated PA contributes to Arabidopsis penetration resistance against Bgh spores. A, Bgh penetration rate (means ± sd; n = 3) at 3 dpi on Arabidopsis leaves infiltrated with different n-butanol and tert-butanol concentrations. Lowercase letters indicate significantly different samples as determined by multiple comparison using one-way ANOVA (P < 0.02), with 6 degrees of freedom. B, Screening of single and double Arabidopsis PLD T-DNA lines for Bgh penetration rate (means ± sd; n = 4). Three-week-old plants were inoculated with Bgh and observed at 2 dpi. The asterisk indicates a significant difference for pldδ compared with the wild type (Col-0) and all other mutant lines as determined by multiple comparison using one-way ANOVA (P < 0.01), with 15 degrees of freedom. Three independent experiments were performed with similar results.
PLDδ Is the Only PLD Isoform Involved in Penetration Resistance against Bgh
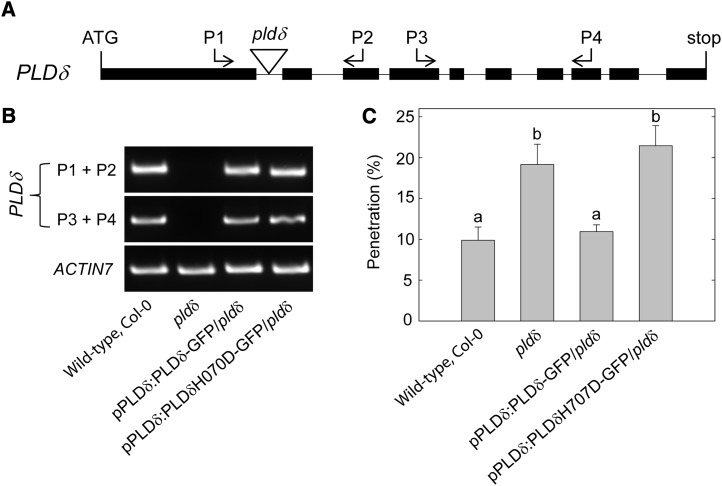
We set out to identify which, if any, of the 12 known Arabidopsis PLD isoforms contributes to penetration resistance against Bgh. For this purpose, we identified at least one transfer DNA (T-DNA) insertion line for each PLD gene. The insertion site of each T-DNA line was verified by sequencing (Supplemental Table S1; Supplemental Fig. S2). Homozygous plants for T-DNA insertion lines not previously published were tested by reverse transcription (RT)-PCR for the presence of the corresponding PLD transcript using primers flanking the insertion site (Supplemental Fig. S3; Supplemental Table S2). Absence of transcripts confirmed that the T-DNA lines indeed are knockout (KO) mutants of the PLD genes. Absence of transcript and/or protein in the pldα1, pldα3, pldζ1, and pldζ2 mutants was described in previous publications (Zhang et al., 2004; Li et al., 2006; Hong et al., 2008). The different PLD single KO lines and two PLD double KO lines (pldβ1-2 pldβ2 and pldζ1 pldζ2) were scored for Bgh penetration resistance by counting interaction events in leaves stained by trypan blue 2 d after inoculation with Bgh spores. Among all, only the PLDδ KO line (pldδ) displayed a significantly higher Bgh penetration rate compared with the wild type (Fig. 1B). The penetration rate in this mutant was approximately doubled compared with the wild type, representing an increase from 12% (sd 0.8) penetration in the wild type to over 23% (sd 2.3) in pldδ. None of the pld mutants investigated was affected in the cell death response to successfully penetrating spores (Supplemental Fig. S4). Furthermore, none of the lines displayed any visible growth or other phenotype under our cultivation regime (Supplemental Fig. S5).
Absence of the PLDδ-specific transcript in pldδ was verified with a primer pair annealing downstream of the insertion site in addition to the T-DNA flanking pair (Fig. 2, A and B). To confirm that the penetration phenotype of pldδ is caused by the specific loss of the PLDδ protein, the mutant was complemented with PLDδ coding sequence fused to GFP under the transcriptional regulation of a 1.4-kb PLDδ promoter region. This restored wild-type levels of Bgh penetration (Fig. 2C). In addition, pldδ plants were transformed with a mutated version of the pPLDδ:PLDδ-GFP transgene, which carried a point mutation turning a His into an Asp residue (H707D) in one of the catalytically active HKD sites of PLDδ. This His-to-Asp mutation was previously shown to abolish the enzymatic activity of a PLD isoform in cabbage (Brassica oleracea; Lerchner et al., 2006). Transgenic pldδ plants expressing the pPLDδ:PLDδH707D-GFP construct displayed Bgh penetration levels similar to the untransformed pldδ (Fig. 2, B and C), further supporting that PLDδ activity is essential for normal penetration resistance.
Figure 2.
PLDδ enzymatic activity is required for Arabidopsis penetration resistance against Bgh spores. A, Schematic representation of PLDδ gene structure and the T-DNA insertion site of the pldδ mutant. Black bars and lines indicate exons and introns, respectively. Proportions are not represented in scale. B, Semiquantitative RT-PCR analysis of the wild type, the pldδ mutant, the PLDδ-GFP fusion line complementing pldδ, and the mutated PLDδ-GFP fusion line with pldδ background (for description, see text). The relative positions and orientations of the primers employed for the amplification of PLDδ cDNA are indicated in A. Amplification of ACTIN7 cDNA was used as an internal control. C, Bgh penetration rate (means ± sd; n = 4) at 2 dpi on leaves of the wild type, the pldδ mutant, the PLDδ-GFP fusion line complementing pldδ, and the mutated PLDδ-GFP fusion line with pldδ background. Lowercase letters indicate significant differences between genotypes as determined by multiple comparison using one-way ANOVA (P < 0.01), with 3 degrees of freedom. Two independent experiments were performed with similar results.
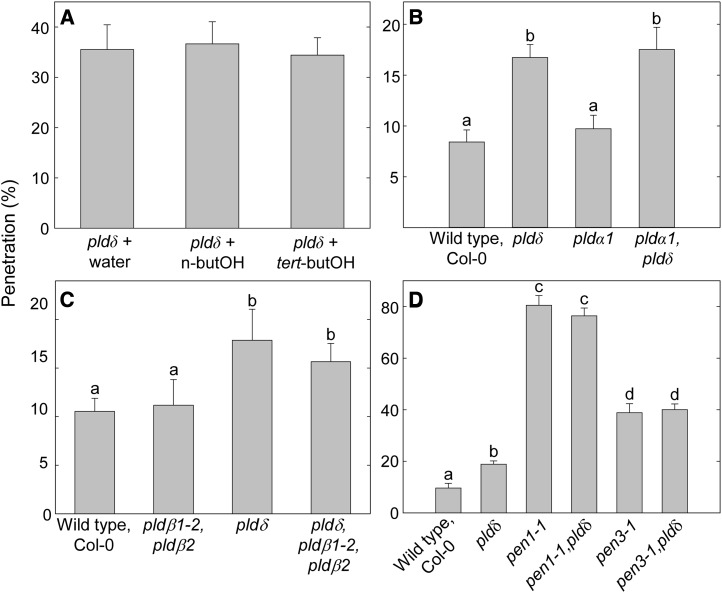
To verify that PLDδ is the only PLD isoform that plays a role in penetration resistance against Bgh, the pldδ mutant was treated with n-butanol before inoculation with Bgh. This treatment caused no further increase in Bgh penetration rate (Fig. 3A). The transcripts of PLDα1 and PLDδ are the most abundant among the PLD genes in green tissues (Li et al., 2006), while PLDβ1 and PLDβ2 are the two isoforms most closely related to PLDδ (Qin and Wang, 2002). Bgh penetration was scored in pldα1 pldδ double and pldβ1-2 pldβ2 pldδ triple mutants, but none of these displayed higher levels than the pldδ single mutant (Fig. 3, B and C). This, together with the n-butanol treatment of pldδ, verified that PLDδ is the sole PLD isoform involved in penetration resistance. Double mutants combining the pldδ mutation with the pen1-1 or the pen3-1 mutation did not display higher Bgh penetration frequencies than the respective parent pen mutants (Fig. 3D).
Figure 3.
PLDδ is the only PLD isoform contributing to penetration resistance against Bgh. A, Bgh penetration rate of pldδ leaves infiltrated with water and 0.6% n-butanol or tert-butanol. No significant difference in penetration was observed for pldδ + n-butanol, indicating that no other PLD isoform contributes to penetration resistance. B and C, Bgh penetration rate at 2 dpi on leaves of different 3-week-old PLD mutant plants. KO mutations of PLDα1 (B) or PLDβ1 and PLDβ2 (C) in the pldδ background do not cause a further increase in Bgh penetration rate. D, PLDδ KO in the pen1 or pen3 mutant background does not result in any further increase of Bgh penetration frequencies. Lowercase letters indicate significant differences between genotypes (P < 0.05) as determined by multiple comparison using one-way ANOVA, with 3 (A–C) or 5 (D) degrees of freedom. Means ± sd are shown (n = 4). Three independent experiments were performed with similar results.
PLDδ Contributes to Penetration Resistance against the Nonhost Pea Powdery Mildew
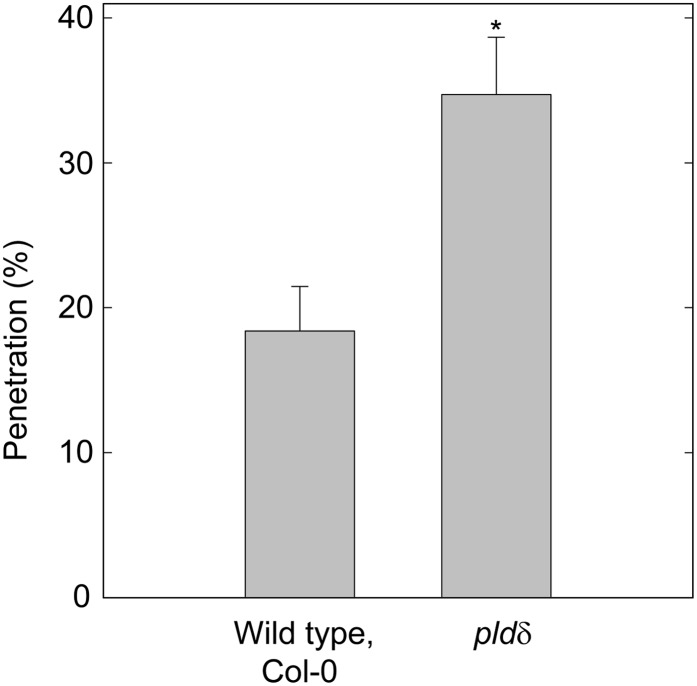
To investigate the conservation of the involvement of PLDδ in penetration resistance against powdery mildew fungi, we used the Arabidopsis nonhost fungal pathogen Erysiphe pisi (Ep), which is an adapted powdery mildew fungus on pea (Pisum sativum) plants. The Ep penetration rate on wild-type Arabidopsis leaves was higher than that of Bgh, suggesting a higher level of adaptation to Arabidopsis. The pldδ mutant displayed a significantly higher penetration rate compared with the wild type for Ep (Fig. 4). The penetration success rate increased from 18% (sd 3.1) of the germinated spores on wild-type leaves to 35% (sd 4.0) on the pldδ mutant. This clearly demonstrates that the PLDδ gene is involved in penetration resistance against this fungus as well.
Figure 4.
PLDδ contributes to penetration resistance against the nonhost powdery mildew fungus Ep. Scoring of penetration events was carried out at 2 dpi on wild-type and pldδ leaves. Means ± sd are represented (n = 4; Student’s t test, P < 0.01). Three independent experiments were performed with similar results.
PLDδ in Chitin-Induced MAMP-Response Gene Activation
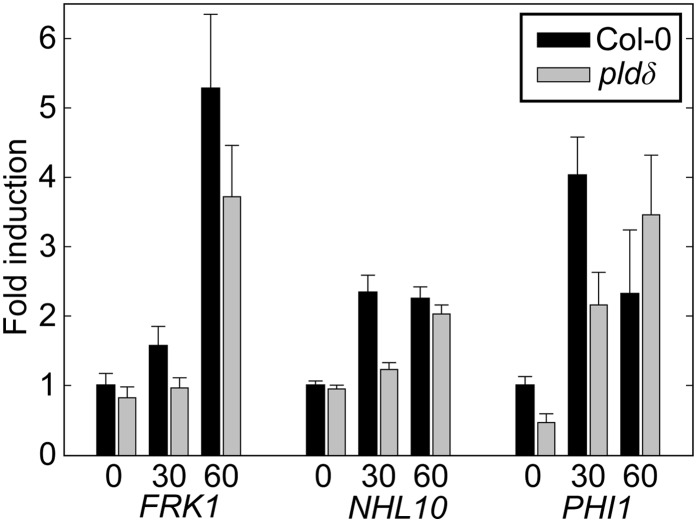
Since KO of the PLDδ gene caused decreased penetration resistance against two different nonhost powdery mildew fungi, and since the gene appears not specifically linked to either PEN1- or PEN2/PEN3-defined penetration resistance pathways, we hypothesized that the activity of the PLDδ isoform might have a more general role in plant pathogen defense. In particular, it seems reasonable to suggest that the gene might be involved in signaling after recognition of MAMPs. It would be interesting to test the response of the pldδ mutant to MAMPs, such as chitin, since a chitin receptor mutant has been previously found to have reduced powdery mildew resistance (Wan et al., 2008). For this purpose, wild-type and pldδ mutant seedlings were treated with a chitin extract, and transcriptional changes of the early MAMP-induced genes, FLG22-INDUCED RECEPTOR-LIKE KINASE1 (FRK1), NDR1/HIN1-LIKE10 (NHL10) and PHOSPHATE-INDUCIBLE1 (PHI-1; Boudsocq et al., 2010; Kwaaitaal et al., 2011), were evaluated by quantitative RT-PCR (Fig. 5). The transcript levels of all three genes rapidly increased following treatment with chitin in both wild-type and mutant plants. PHI1 and NHL10 both reached a peak of transcript accumulation 30 min after treatment, whereas FRK1 transcript increased throughout the experiment. However, the pldδ mutant showed a trend for an approximately 50% reduction in transcript accumulation after 30 min for PHI1 and NHL10. For all three genes, a trend for a delayed chitin-induced accumulation of transcripts was observed. Observations for NHL10 were consistent for all biological replicates. Results for PHI-1 and FRK1 demonstrated more variability between experiments and, in some instances, revealed a lower amount of transcript at the zero time point in the pldδ mutant. These observations suggest that PLDδ is needed for rapid and timely activation of CERK1-controlled defense signaling cascades.
Figure 5.
Chitin-induced defense transcript accumulation is delayed in the pldδ mutant. Col-0 and pldδ seedlings were grown in liquid culture on a microtiter plate. Ten-day-old seedlings were treated with chitin extract, and 0, 30, and 60 min later, the seedlings were harvested. Total RNA of approximately 20 seedlings was extracted per sample for transcript analysis. Transcript levels of the early MAMP-activated defense genes NHL10, FRK1, and PHI-1 were determined by quantitative PCR. Relative expression levels compared with Col-0 at the zero time control are presented. The data were normalized using At4g26410 as a reference gene. The average of three technical replicates is shown, and error bars show se. At least three independent experiments were performed with similar results.
PLDδ Is Localized to the Plant Plasma Membrane at Bgh Attack Sites
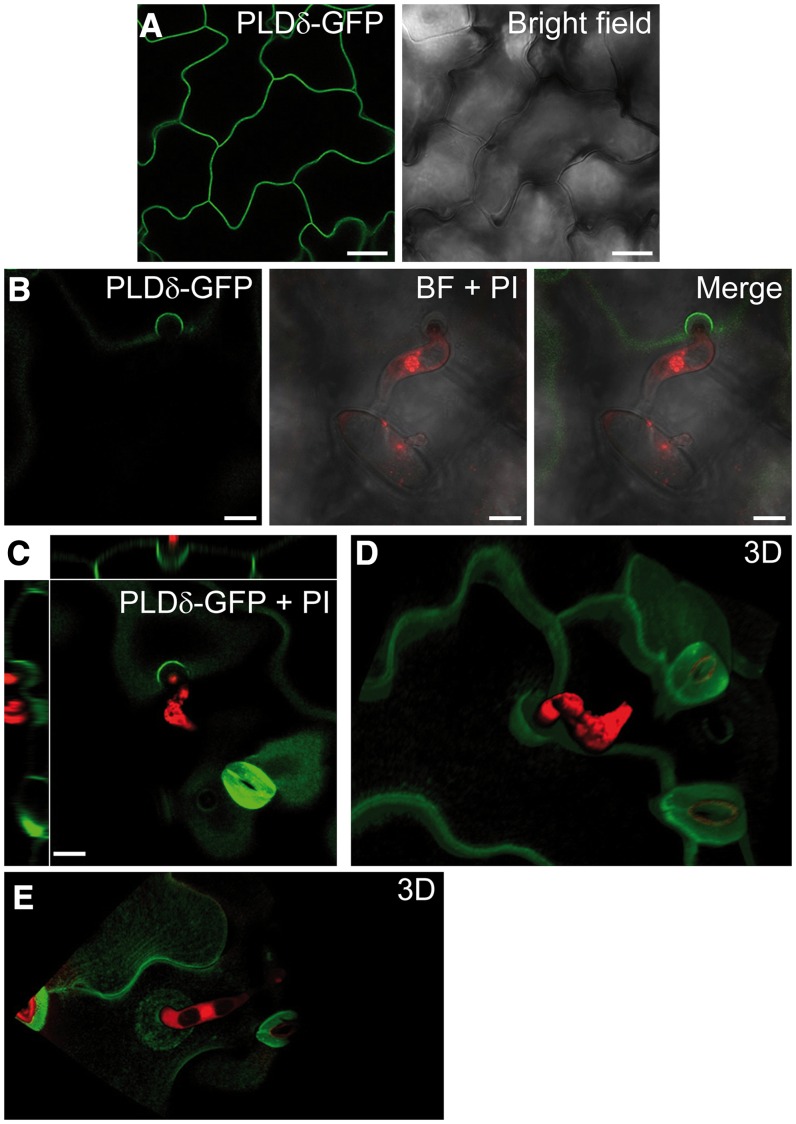
The complementation of pldδ phenotype by the expression of the pPLDδ:PLDδ-GFP transgene indicates that the PLDδ-GFP translational fusion is functional. Although the fluorescent signals of the pPLDδ:PLDδ-GFP and pPLDδ:PLDδH707D-GFP lines were clearly visible in leaves observed with an epifluorescence microscope (Supplemental Fig. S6), they were not sufficiently high for confocal imaging. Thus, we placed the coding sequence for the PLDδ-GFP fusion under the transcriptional regulation of the 35S promoter and transformed this transgene into wild-type plants. The p35S:PLDδ-GFP line proved to be suitable for confocal microscopy, and PLDδ-GFP clearly localized to the plasma membrane of leaf epidermal cells (Fig. 6A). Twenty-four hours after Bgh inoculation, PLDδ-GFP was visible in the plasma membrane in general and at the regions of fungal attack where the appressoria attempted penetration. Here, PLDδ-GFP fluorescence labeled the rim of the forming papilla (Fig. 6B). Z-stack observations and three-dimensional visualization of several fungal attack sites displayed an accumulation of PLDδ-GFP at the plasma membrane, which forms a cup-like shape around the papilla (Fig. 6, C and D; Supplemental Movie S1). Occasionally, the accumulation of PLDδ-GFP around Bgh interaction sites extended to the surrounding membrane in the shape of a compact disk (Fig. 6E). Three-dimensional reconstruction of these entry sites showed a small plasma membrane invagination corresponding to a small papilla (Supplemental Movie S2). Nontransformed wild-type leaf epidermal cells exhibited low autofluorescence in the GFP channel before (Supplemental Fig. S6A) and 24 h after (Supplemental Fig. S6B) inoculation with Bgh spores. The autofluorescence was mainly associated with stomatal guard cell walls and did not coincide with PLDδ-GFP signal.
Figure 6.
Bgh induces the accumulation of PLDδ at the plasma membrane around the site of fungal attack. A, Plasma membrane localization of PLDδ-GFP in leaf epidermal cells of noninoculated p35S::PLDδ-GFP plants. B, Plasma membrane accumulation of PLDδ-GFP at the site of attempted Bgh penetration. Fungal structures are stained with propidium iodide (PI). BF, Bright field. C, Image depicts xy plane of PLDδ-GFP accumulation at the Bgh attack site and side views of it along the z axis. D and E, Three-dimensional (3D) reconstruction of the Bgh attack site and PLDδ-GFP localization around it (Supplemental Movies S1 and S2). Images were taken at 24 hpi with Bgh. Two independent experiments were performed with similar results. Bars = 25 µm (A and B) and 10 µm (C).
DISCUSSION
Effective resistance against pathogenic microbes depends on the timely deployment of the appropriate defense responses. A functional signaling pathway ensures that pathogen recognition is promptly communicated to the cellular machinery responsible for the execution of defense. Our results clearly indicate that the enzymatic activity of PLDδ contributes to the accomplishment of penetration resistance against nonadapted powdery mildew fungi in Arabidopsis. This is supported by the observation that the penetration resistance was decreased by the PLD inhibitor n-butanol and by KO of the PLDδ gene. Finally, introduction of a construct encoding a catalytically inactive PLDδ did not complement the loss of the PLDδ gene, whereas a functional copy did complement the mutant phenotype. The involvement of PLDδ in cell wall-based defense apparently extends to at least two different nonadapted powdery mildew fungi. However, we failed to detect an increase in PA levels after inoculation of wild-type Arabidopsis plants with Bgh. It seems likely that the limited number of leaf epidermal cells interacting with Bgh spores is not sufficient to provoke a detectable increase in PA against the background of the whole leaf tissue. The PA signal might also be more restricted in time than what we addressed. Nevertheless, we demonstrated that chemical inhibition of PA production via PLD enhanced the Bgh penetration rate and provided clear genetic evidence that specific enzymatic activity of PLDδ is required for the full penetration resistance to nonhost powdery mildew fungi in Arabidopsis.
PLDδ has previously been implicated in the freezing tolerance of Arabidopsis. Loss of or overexpression of PLDδ resulted in reduced and increased PA levels, respectively, but did not alter cold-regulated gene expression (Li et al., 2004). Thus, PLDδ seems to control unique PA-dependent cellular signaling events in response to cold stress. In this study, we cannot determine, based on the penetration resistance phenotype alone, where mechanistically in the defense pathways PLDδ acts. However, we tentatively propose that PLD activity is involved in upstream early signaling events rather than in the execution phase of penetration resistance. Mutations in PEN1, PEN2, and PEN3 result in a severalfold increase in the Bgh penetration rate (Collins et al., 2003; Lipka et al., 2005; Stein et al., 2006). Based on their cellular and molecular functions, the three PEN proteins are regarded as main components in the execution of preinvasion resistance. The effect of the pldδ mutation on Bgh penetration resistance was clearly much lower than that caused by the loss of any of the PEN genes. In addition, introduction of the pldδ mutation into the pen1 and pen3 backgrounds did not cause additional effects on the frequency of Bgh penetration in any of the combinations. Thus, it appears that PLDδ cannot be directly linked to either the PEN1 or the PEN2/PEN3 pathway. This leaves room for several different interpretations. However, as the forms of NHR studied here are considered to be triggered by fungal MAMPs (Schulze-Lefert and Panstruga, 2011), and as we observe that chitin-triggered defense gene transcript accumulation is delayed in the pldδ mutant, we propose a role for PLDδ early in MAMP-triggered penetration resistance. Furthermore, this is in agreement with the documented role of a chitin receptor in powdery mildew resistance (Wan et al., 2008).
Mutants in PEN1 (Assaad et al., 2004) and its ortholog ROR2 in barley (Hordeum vulgare; Böhlenius et al., 2010) both exhibit delayed callose deposition, which correlates with the increased susceptibility of these mutants. This illustrates the importance of a timely activation of defense to repel invading pathogens. Therefore, delayed activation of defense downstream of MAMP-induced activation of PRR in pldδ might have a significant impact on the success of a single cell in defending itself against pathogen invasion. The pldδ mutation and n-butanol treatment cause only a partial loss of resistance, likely reflecting a large degree of redundancy in the signaling pathway. Such a signaling function of PLDδ in preinvasion defense is in agreement with the role of PA as a second messenger in cellular responses to different biotic stimuli (de Jong et al., 2004; Andersson et al., 2006; Kirik and Mudgett, 2009). An alternative interpretation to the involvement of PLDδ in penetration resistance might be that the PLD activity is involved in controlling the polar secretion of antimicrobial compounds by, for instance, aiding in vesicle fusion to the plasma membrane (Jenkins and Frohman, 2005). This model, however, is less easily reconcilable with the effect of the pldδ mutation on MAMP-triggered transcriptional activation of defense transcripts. Alternatively, PLDδ might be involved in regulating the amount and targeting of PRRs to the plasma membrane. Finally, a combination of the three might be a possibility. However, the most consistent model with the data presented in this and previous studies seems to be that PA is generated as a second messenger following MAMP recognition and that PA acts as one part of an otherwise partly redundant signaling network including Ca2+ and kinases.
Gene expression studies of the whole Arabidopsis genome in response to Bgh inoculation did not reveal any change in PLDδ transcription during NHR (Zimmerli et al., 2004). Considering previous studies demonstrating the induction of PA production via PLD within minutes after the elicitation of tomato cells with fungal MAMPs (van der Luit et al., 2000; Raho et al., 2011), it is likely that PLDδ is activated at a posttranslational level upon the recognition of a Bgh penetration attempt. Further support for this notion is the fact that the PLDδ gene together with PLDα1 is the most highly expressed PLD isoform in leaves (Li et al., 2006). Notably, none of the other PLD isoforms appeared to be involved in preinvasion defense, although they were all expressed in wild-type leaves. This likely reflects the differences among the 12 PLD isoforms in terms of activation requirements, subcellular localization, and substrate preferences (Li et al., 2009). The PLD isoforms were shown to be differentially involved in the plant response to specific stress conditions (Zhang et al., 2003, 2004; Li et al., 2006; Hong et al., 2008). The expression profile, the subcellular localization, and the biochemical properties of PLDδ together make it a candidate PLD isoform responsive to the stimulus of fungal penetration attempts. The plasma membrane localization shown here for a PLDδ-GFP fusion protein confirmed previous reports based on immunochemical assays (Wang and Wang, 2001). PRRs perceive fungal MAMPs at the cell surface and trigger downstream signaling cascades (Monaghan and Zipfel, 2012). Hence, PLDδ activation might be an early event at the plasma membrane after MAMP recognition, and the subsequently produced PA could stimulate the activation of plant defenses.
This is also supported by the effect of the pldδ mutation on transcriptional changes of early MAMP-activated defense genes. The observation that PHI-1 and NHL10 exhibit an altered chitin-triggered transcriptional up-regulation in the pldδ mutant links PLDδ action to CERK1-activated Ca2+ signaling (Boudsocq et al., 2010). In parallel, PLDδ activation might contribute to the complex lipid rearrangements occurring at the plasma membrane, which lead to the formation of a lipid raft-like microdomain at sites of fungal penetration attempt (Bhat et al., 2005). In this sense, the presence of PLDδ at fungal interaction sites shown here is significant and further supports the role of PLDδ in penetration resistance. Interestingly, PLD was also shown to cluster in regions of the plasma membrane adjacent to bacterial attack sites in rice leaves resistant to Xanthomonas oryzae pv oryzae (Young et al., 1996). In vitro PLDδ activity is stimulated by oleic acid and phosphatidylinositol bisphosphate and requires high micromolar to low millimolar Ca2+ concentrations (Wang and Wang, 2001; Qin and Wang, 2002). The influx of Ca2+ from the extracellular space is a well-established event occurring after the recognition of MAMPs and effector proteins (Ma and Berkowitz, 2007). An increase in cytosolic Ca2+ levels was shown to trigger a PLD-dependent accumulation of PA after the recognition of two bacterial effectors in Arabidopsis (Andersson et al., 2006). Stimulation of PLDδ activity by oleic acid was found to be a surface-dilution effect, as Triton X-100 was also shown to stimulate this activity (Qin et al., 2002). However, the levels of free unsaturated fatty acids, including oleate, in plant cells have been shown to increase after treatment with a fungal elicitor (Kirsch et al., 1997).
Recently, PLDδ has been implicated in a PA-dependent growth response to the root endophytic fungus Piriformospora indica (Camehl et al., 2011). This observation, together with our data, raises the question of whether PLDδ is part of a common signaling mechanism communicating the occurrence of a biotic stimulus at the cell surface. PLDδ activity is also stimulated by hydrogen peroxide (Zhang et al., 2003), and this activation was recently shown to require the association of PLDδ to cytosolic glyceraldehyde-3-phosphate dehydrogenases (Guo et al., 2012). Hydrogen peroxide production is a well-known event occurring early in plant-pathogen interactions and in response to MAMP perception (Torres et al., 2006). It thus seems plausible that a stimulation of PLDδ activity might depend on rapid hydrogen peroxide production during a penetration attempt of a nonhost powdery mildew fungus. Details of how PLDδ is activated, as well as the function and nature of the targets of the produced PA, remain to be determined.
To conclude, we herein demonstrate that the enzymatic activity of PLDδ and thus the membrane lipid PA contributes to cell wall-based defense against nonadapted powdery mildew fungi in Arabidopsis. We further propose that PA produced by this PLD isoform is part of a signaling network activated by the recognition of fungal MAMPs.
MATERIALS AND METHODS
Plant Growth Conditions and Mutant Isolation
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) lines were grown under short-day conditions (8-h day and 16-h night, 180 µmol m−2 s−1) at 22°C and 60% relative humidity. For analysis of chitin-induced transcript accumulation, Col-0 and pldδ mutant seeds were sterilized and grown on 1× Murashige and Skoog medium for 11 to 12 d with 16 h of light (at 21°C) and 8 h of dark (at 19°C) as described (Kwaaitaal et al., 2011). SALK (Alonso et al., 2003) and GABI-Kat (Kleinboelting et al., 2012) T-DNA lines were obtained from the Nottingham Arabidopsis Stock Centre (Scholl et al., 2000). Identifier numbers and gene-specific primers used for the isolation of homozygous lines are listed in Supplemental Table S1. The LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′) and GK_8409 (5′-ATATTGACCATCATACTCATTGC-3′) oligonucleotides were used as T-DNA-specific primers for genotyping the SALK and GABI-Kat lines, respectively. The pldζ1-1 pldζ2-1 double mutant line was kindly provided by Prof. Xuemin Wang.
Semiquantitative RT-PCR
Total RNA was extracted from 100 mg of leaf tissue using the GeneJET RNA purification kit (Thermo Scientific) and treated with DNase I (Thermo Scientific) to remove traces of genomic DNA. RT of mRNA was achieved with the RevertAid Premium First Strand cDNA Synthesis kit (Thermo Scientific) using oligo(dT)18. Subsequent PCR amplification of PLD-specific transcripts was performed with the primers listed in Supplemental Table S2. Amplification of the ACTIN7 gene (At5g09810) was used as a control.
Fungal Inoculation and Scoring of Fungal Penetration
The nonhost powdery mildew fungi Blumeria graminis f. sp. hordei (isolate DH14) and Erysiphe pisi (isolate CO-01) were propagated on barley (Hordeum vulgare) and pea (Pisum sativum) plants, respectively. Inoculations were carried out as described previously (Zhang et al., 2008). Leaves were stained with trypan blue as described (Koch and Slusarenko, 1990). Successful penetration and cell death were determined by the presence of a fungal haustorium and trypan blue uptake, respectively. At least 50 interaction sites of two leaves from three or four independent plants were counted for each treatment/mutant line. The average frequency of penetrations from two leaves from each plant was used for multiple comparison with one-way ANOVA with Fisher’s lsd using the software package GraphPad Prism 6.0 (Graphpad Software) or paired Student’s t test using Excel 2010. To study the effect of Bgh inoculation on phospholipid metabolism, leaf explants were labeled with 33PO4 overnight before Bgh inoculations. The lipids were extracted and separated by thin-layer chromatography as described previously (Andersson et al., 2006), and lipid radiolabel was quantified after liquid scintillation of PA-containing regions of the thin-layer plates.
Butanol Infiltration
Leaves were infiltrated with alcohol solutions in deionized water using a 1-mL syringe without needle from their abaxial side using gentle pressure. Attention was paid to ensure complete diffusion of the solution to the whole leaf lamina. Plants were subsequently returned to the growth chamber for 3 h before fungal inoculation to allow absorption of the solution and to let the leaves dry.
MAMP Treatment and Quantitative RT-PCR
Experiments were performed as described by Kwaaitaal et al. (2011) with the following modifications. Total seedling RNA was isolated using TRIzol (www.lifetechnologies.com/) according to the protocol provided with the reagent. Subsequently, 2 µg of total RNA was used to generate complementary DNA (cDNA) using the Advantage RT-for-PCR Kit (www.clontech.com) in a total volume of 20 µL. Finally, 80 µL of double distilled water was added to the reaction mixture. A volume of 2.4 µL of cDNA was used in the quantitative PCRs, with final primer concentrations of 0.5 mm of the forward and reverse primers for the reference gene At4g26410, FRK1, and NHL10 and 0.125 mm of the forward and reverse primers for PHI-1 (Boudsocq et al., 2010). Primer sequences are listed in Supplemental Table S3. The volume was adjusted to 6 µL, and 6 µL of 2× SYBR Green Jumpstart Taq ReadyMix (www.sigma.com) was added. The quantitative PCR was run in a Stratagene Mx3000P (Amersham Biosciences, GE Healthcare) thermal cycler, and the results were analyzed using Stratagene MxPro Mx3005P software (Amersham Biosciences, GE Healthcare). The cycling program used was as follows: 10 min at 95°C, then 40 cycles of 30 s at 95°C, 30 s at 62°C, and 25 s at 72°C. The melting curves of the PCR products were recorded between 55°C and 95°C.
Generation of the PLDδ-GFP Transgene and Site-Directed Mutagenesis of PLDδ
The complete coding sequence of the PLDδ gene excluding the final stop codon was amplified from wild-type leaf cDNA (generated as described above) using the PLDδ_cds primers (Supplemental Table S4). The obtained product was inserted into the pENTR/D-TOPO vector through directional TOPO cloning (Invitrogen) and subsequently verified by sequencing. To obtain a PLDδ 3′ terminal fusion to GFP, the PLDδ coding sequence was transferred using Gateway technology (Invitrogen) to the pB7FWG2 vector (Karimi et al., 2005), resulting in the p35S:PLDδ-GFP T-DNA construct. For generation of the pPLDδ:PLDδ-GFP transgene, the pB7FWG2 vector was modified prior to recombination with the PLDδ coding sequence as follows. A 1.4-kb PLDδ untranslated promoter region upstream of the first ATG was amplified from wild-type genomic DNA using the pPLDδ primers (Supplemental Table S4). The amplified fragment was used to replace the p35S region in the pB7FWG2 vector (Karimi et al., 2005) through digestion with HindIII and SpeI endonucleases and subsequent ligation. The modified vector was verified by sequencing. The obtained T-DNA plasmids carrying the p35S:PLDδ-GFP and pPLDδ:PLDδ-GFP transgenes were introduced into Agrobacterium tumefaciens strains GV3101 and C58C1, respectively. Transformation of wild-type and pldδ plants was achieved through the floral dip method (Clough and Bent, 1998). The pENTR/D-TOPO vector containing the PLDδ coding sequence was employed to obtain the PLDδΗ707D mutant open reading frame (ORF). The C-to-G mutation at position 2,152 of the PLDδ ORF was introduced by amplifying the whole plasmid by PCR (26 cycles) with the phosphorylated PLDδΗ707D ORF primers (Supplemental Table S4). The original template plasmid was digested with DpnI endonuclease (acting on methylated DNA), and the PCR product was subsequently circularized by ligation. The mutant PLDδΗ707D ORF was verified by sequencing and subsequently transferred to the pB7FWG2 vector (modified as described above) in order to generate the pPLDδ:PLDδH707D-GFP transgene. The obtained plasmid was introduced into A. tumefaciens strain C58C1 and transformed into pldδ plants.
Fluorescence and Confocal Microscopy
Transgenic PLDδ-GFP lines were scored for GFP fluorescence in leaves using a 20× objective mounted on a Zeiss Axio Scope.A1 epifluorescence microscope. Localization of PLDδ-GFP was investigated with a Zeiss LSM 700 confocal laser scanning microscope using a 40× water-immersion objective (numerical aperture, 1.1). For visualization of fungal spores, inoculated leaves were mounted in a solution of 2.5% mannitol/0.1% (v/v) Silwet/0.02 mg mL−1 propidium iodide. Images were analyzed with Volocity (Perkin-Elmer) and ImageJ software.
Sequence data of the genes used in this article can be found in the GenBank/EMBL data libraries. Gene accession numbers are listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantification of radiolabeled PA after Bgh inoculation.
Supplemental Figure S2. Schematic representation of the structure of the PLD genes and the T-DNA insertion sites.
Supplemental Figure S3. Semiquantitative RT-PCR analysis of PLD T-DNA lines.
Supplemental Figure S4. No postpenetration cell death phenotype was observed in pld mutants.
Supplemental Figure S5. Growth phenotypes of the PLD T-DNA lines used in this study.
Supplemental Figure S6. Expression of pPLDδ::PLDδ-GFP and pPLDδ::PLDδH707D-GFP transgenes in the pldδ mutant background.
Supplemental Figure S7. Control image of the autofluorescence of noninoculated and Bgh-inoculated leaves.
Supplemental Table S1. T-DNA mutant alleles of Arabidopsis PLD genes and gene-specific primers used for genotyping.
Supplemental Table S2. Primers used for semiquantitative RT-PCR analysis of PLD T-DNA mutant lines.
Supplemental Table S3. Primers used for quantitative RT-PCR analysis of MAMP-responsive genes.
Supplemental Table S4. Primers used for the molecular cloning of PLDδ transgenes.
Supplemental Movie S1. Three-dimensional reconstruction of the Bgh attack site and PLDδ-GFP localization around it at 24 hpi of p35S::PLDδ-GFP plants.
Supplemental Movie S2. PLDδ-GFP localization around a small papilla at 24 hpi of p35S::PLDδ-GFP plants.
Acknowledgments
We are very grateful to the Centre for Cellular Imaging (Sahlgrenska Academy, Gothenburg University) for the use of confocal microscopes and helpful assistance, to Associate Prof. Johan Uddling (Department of Biology and Environmental Sciences, University of Gothenburg) for advice on statistical testing and the interpretation thereof, and to Prof. Xuemin Wang (University of Missouri) for providing the pldζ1-1 pldζ2-1 double mutant.
Glossary
- NHR
nonhost resistance
- MAMP
microbe-associated molecular pattern
- PRR
pattern recognition receptor
- Bgh
Blumeria graminis f. sp. hordei
- PA
phosphatidic acid
- PLD
phospholipase D
- hpi
hours post inoculation
- dpi
days post inoculation
- T-DNA
transfer DNA
- RT
reverse transcription
- KO
knockout
- Ep
Erysiphe pisi
- Col-0
Columbia
- cDNA
complementary DNA
- ORF
open reading frame
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Kourtchenko O, Dangl JL, Mackey D, Ellerström M. (2006) Phospholipase-dependent signalling during the AvrRpm1- and AvrRpt2-induced disease resistance responses in Arabidopsis thaliana. Plant J 47: 947–959 [DOI] [PubMed] [Google Scholar]
- Assaad FF, Qiu J-L, Youngs H, Ehrhardt D, Zimmerli L, Kalde M, Wanner G, Peck SC, Edwards H, Ramonell K, et al. (2004) The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell 15: 5118–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann BOR, Munnik T. (2006) The role of phospholipase D in plant stress responses. Curr Opin Plant Biol 9: 515–522 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R. (2005) Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci USA 102: 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhlenius H, Mørch SM, Godfrey D, Nielsen ME, Thordal-Christensen H. (2010) The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell 22: 3831–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camehl I, Drzewiecki C, Vadassery J, Shahollari B, Sherameti I, Forzani C, Munnik T, Hirt H, Oelmüller R. (2011) The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis. PLoS Pathog 7: e1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu J-L, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SCet al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- de Jong CF, Laxalt AM, Bargmann BOR, de Wit PJGM, Joosten MHAJ, Munnik T. (2004) Phosphatidic acid accumulation is an early response in the Cf-4/Avr4 interaction. Plant J 39: 1–12 [DOI] [PubMed] [Google Scholar]
- Guo L, Devaiah SP, Narasimhan R, Pan X, Zhang Y, Zhang W, Wang X. (2012) Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 24: 2200–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardham AR, Jones DA, Takemoto D. (2007) Cytoskeleton and cell wall function in penetration resistance. Curr Opin Plant Biol 10: 342–348 [DOI] [PubMed] [Google Scholar]
- Hong Y, Pan X, Welti R, Wang X. (2008) Phospholipase Dα3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20: 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R. (2007) Cell wall-associated mechanisms of disease resistance and susceptibility. Annu Rev Phytopathol 45: 101–127 [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Frohman MA. (2005) Phospholipase D: a lipid centric review. Cell Mol Life Sci 62: 2305–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Kachroo P. (2009) Fatty acid-derived signals in plant defense. Annu Rev Phytopathol 47: 153–176 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kirik A, Mudgett MB. (2009) SOBER1 phospholipase activity suppresses phosphatidic acid accumulation and plant immunity in response to bacterial effector AvrBsT. Proc Natl Acad Sci USA 106: 20532–20537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch C, Takamiya-Wik M, Reinold S, Hahlbrock K, Somssich IE. (1997) Rapid, transient, and highly localized induction of plastidial ω-3 fatty acid desaturase mRNA at fungal infection sites in Petroselinum crispum. Proc Natl Acad Sci USA 94: 2079–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B. (2012) GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch E, Slusarenko A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaaitaal M, Huisman R, Maintz J, Reinstädler A, Panstruga R. (2011) Ionotropic glutamate receptor (iGluR)-like channels mediate MAMP-induced calcium influx in Arabidopsis thaliana. Biochem J 440: 355–365 [DOI] [PubMed] [Google Scholar]
- Laxalt AM, Munnik T. (2002) Phospholipid signalling in plant defence. Curr Opin Plant Biol 5: 332–338 [DOI] [PubMed] [Google Scholar]
- Lerchner A, Mansfeld J, Kuppe K, Ulbrich-Hofmann R. (2006) Probing conserved amino acids in phospholipase D (Brassica oleracea var. capitata) for their importance in hydrolysis and transphosphatidylation activity. Protein Eng Des Sel 19: 443–452 [DOI] [PubMed] [Google Scholar]
- Li M, Hong Y, Wang X. (2009) Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim Biophys Acta 1791: 927–935 [DOI] [PubMed] [Google Scholar]
- Li M, Qin C, Welti R, Wang X. (2006) Double knockouts of phospholipases Dζ1 and Dζ2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol 140: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li M, Zhang W, Welti R, Wang X. (2004) The plasma membrane-bound phospholipase Ddelta enhances freezing tolerance in Arabidopsis thaliana. Nat Biotechnol 22: 427–433 [DOI] [PubMed] [Google Scholar]
- Lipka U, Fuchs R, Lipka V. (2008) Arabidopsis non-host resistance to powdery mildews. Curr Opin Plant Biol 11: 404–411 [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Ma W, Berkowitz GA. (2007) The grateful dead: calcium and cell death in plant innate immunity. Cell Microbiol 9: 2571–2585 [DOI] [PubMed] [Google Scholar]
- Ma W, Qi Z, Smigel A, Walker RK, Verma R, Berkowitz GA. (2009) Ca2+, cAMP, and transduction of non-self perception during plant immune responses. Proc Natl Acad Sci USA 106: 20995–21000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Pajonk S, Micali C, O’Connell R, Schulze-Lefert P. (2009) Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J 57: 986–999 [DOI] [PubMed] [Google Scholar]
- Micali C, Göllner K, Humphry M, Consonni C, Panstruga R. (2008) The powdery mildew disease of Arabidopsis: a paradigm for the interaction between plants and biotrophic fungi. The Arabidopsis Book 6: e0115, doi/10.1199/tab.0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C. (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15: 349–357 [DOI] [PubMed] [Google Scholar]
- Munnik T, Arisz SA, De Vrije T, Musgrave A. (1995) G protein activation stimulates phospholipase D signaling in plants. Plant Cell 7: 2197–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen ME, Feechan A, Böhlenius H, Ueda T, Thordal-Christensen H. (2012) Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc Natl Acad Sci USA 109: 11443–11448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara Y, Kaya H, Hiraoka G, Yumoto F, Kimura S, Kadota Y, Hishinuma H, Senzaki E, Yamagoe S, Nagata K, et al. (2008) Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J Biol Chem 283: 8885–8892 [DOI] [PubMed] [Google Scholar]
- Qin C, Wang C, Wang X. (2002) Kinetic analysis of Arabidopsis phospholipase Dδ: substrate preference and mechanism of activation by Ca2+ and phosphatidylinositol 4,5-biphosphate. J Biol Chem 277: 49685–49690 [DOI] [PubMed] [Google Scholar]
- Qin C, Wang X. (2002) The Arabidopsis phospholipase D family: characterization of a calcium-independent and phosphatidylcholine-selective PLDζ1 with distinct regulatory domains. Plant Physiol 128: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raho N, Ramirez L, Lanteri ML, Gonorazky G, Lamattina L, ten Have A, Laxalt AM. (2011) Phosphatidic acid production in chitosan-elicited tomato cells, via both phospholipase D and phospholipase C/diacylglycerol kinase, requires nitric oxide. J Plant Physiol 168: 534–539 [DOI] [PubMed] [Google Scholar]
- Ron M, Avni A. (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH. (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P, Panstruga R. (2011) A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci 16: 117–125 [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Zipfel C. (2008) News from the frontline: recent insights into PAMP-triggered immunity in plants. Curr Opin Plant Biol 11: 389–395 [DOI] [PubMed] [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku Het al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou B-H, Molina A, Schulze-Lefert P, Lipka V, Somerville S. (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Yano A, Nishiuchi T, Nakano T, Kodama H, Yamaguchi K, Shinshi H. (2007) Comprehensive analysis of early response genes to two different microbial elicitors in tobacco cells. Plant Sci 173: 291–301 [Google Scholar]
- Testerink C, Munnik T. (2011) Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J Exp Bot 62: 2349–2361 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H. (2003) Fresh insights into processes of nonhost resistance. Curr Opin Plant Biol 6: 351–357 [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL. (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Luit AH, Piatti T, van Doorn A, Musgrave A, Felix G, Boller T, Munnik T. (2000) Elicitation of suspension-cultured tomato cells triggers the formation of phosphatidic acid and diacylglycerol pyrophosphate. Plant Physiol 123: 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang X. (2001) A novel phospholipase D of Arabidopsis that is activated by oleic acid and associated with the plasma membrane. Plant Physiol 127: 1102–1112 [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2004) Lipid signaling. Curr Opin Plant Biol 7: 329–336 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Minami E, Ueki J, Shibuya N. (2005) Elicitor-induced activation of phospholipases plays an important role for the induction of defense responses in suspension-cultured rice cells. Plant Cell Physiol 46: 579–587 [DOI] [PubMed] [Google Scholar]
- Young SA, Wang X, Leach JE. (1996) Changes in the plasma membrane distribution of rice phospholipase D during resistant interactions with Xanthomonas oryzae pv oryzae. Plant Cell 8: 1079–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X. (2004) Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wang C, Qin C, Wood T, Olafsdottir G, Welti R, Wang X. (2003) The oleate-stimulated phospholipase D, PLDδ, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15: 2285–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lenk A, Andersson MX, Gjetting T, Pedersen C, Nielsen ME, Newman M-A, Hou B-H, Somerville SC, Thordal-Christensen H. (2008) A lesion-mimic syntaxin double mutant in Arabidopsis reveals novel complexity of pathogen defense signaling. Mol Plant 1: 510–527 [DOI] [PubMed] [Google Scholar]
- Zimmerli L, Stein M, Lipka V, Schulze-Lefert P, Somerville S. (2004) Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J 40: 633–646 [DOI] [PubMed] [Google Scholar]