Two different synthases are responsible for the high production of eugenol in the green achene and the ripe receptacle of the strawberry fruit.
Abstract
Eugenol is a volatile that serves as an attractant for pollinators of flowers, acts as a defense compound in various plant tissues, and contributes to the aroma of fruits. Its production in a cultivated species such as strawberry (Fragaria × ananassa), therefore, is important for the viability and quality of the fruit. We have identified and functionally characterized three strawberry complementary DNAs (cDNAs) that encode proteins with high identity to eugenol synthases from several plant species. Based on a sequence comparison with the wild relative Fragaria vesca, two of these cDNAs, FaEGS1a and FaEGS1b, most likely correspond to transcripts derived from allelic gene variants, whereas the third cDNA, FaEGS2, corresponds to a different gene. Using coniferyl acetate as a substrate, FaEGS1a and FaEGS1b catalyze the in vitro formation of eugenol, while FaEGS2 catalyzes the formation of eugenol and also of isoeugenol with a lower catalytic efficiency. The expression of these genes is markedly higher in the fruit than in other tissues of the plant, with FaEGS1a and FaEGS1b mostly expressed in the green achenes, whereas FaEGS2 expression is almost restricted to the red receptacles. These expression patterns correlate with the eugenol content, which is highest in the achene at the green stage and in the receptacle at the red stage. The transient expression of the corresponding cDNAs in strawberry fruit and the subsequent volatile analyses confirm FaEGSs as genuine eugenol synthases in planta. These results provide new insights into the diversity of phenylpropene synthases in plants.
Phenylpropenes such as eugenol and isoeugenol are produced by plants, where they play roles as floral attractants for pollinators and defense compounds against animals and microorganisms (Koeduka et al., 2006; Pasay et al., 2010). Phenylpropenes are also produced by fruit, contributing to its aroma (Jordán et al., 2001; Aubert and Pitrat, 2006; Ortiz-Serrano and Gil, 2010). In strawberry (Fragaria spp.), eugenol production by ripe fruit has been reported (Pyysalo et al., 1979; Zorrilla-Fontanesi et al., 2012), but there is no information about its production along the growth and ripening processes or about the enzymes that synthesize this compound. Interestingly, the production of eugenol in Fragaria species has diminished with domestication. The amount of this volatile is significantly lower in the fruits of the cultivated strawberry (Fragaria × ananassa) compared with the wild diploid Fragaria vesca (Pyysalo et al., 1979). Partial restoration of wild strawberry flavor in cultivated varieties was performed through metabolic engineering by diverting the flavonoid pathway to phenylpropene synthesis (Hoffmann et al., 2011).
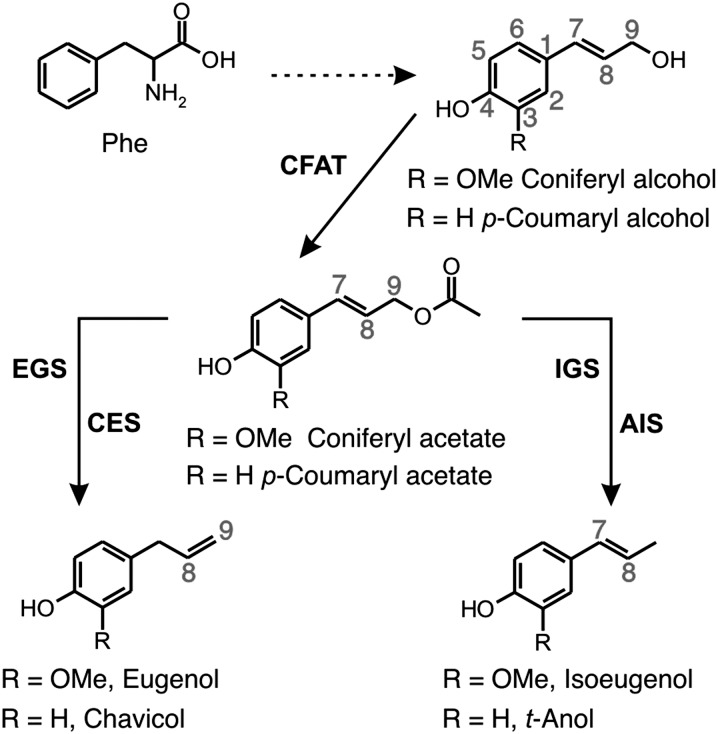
As precursors of phenylpropene production, the monolignol alcohols first undergo acetylation of the C9 hydroxyl group (Dexter et al., 2007) and then reductive cleavage of the acetate moiety to yield the propenyl side group (Koeduka et al., 2006; Fig. 1). Distinct NADPH-dependent enzymes that are involved in the biosynthesis of phenylpropenes have been characterized. The first such enzymes are a eugenol synthase (EGS) from basil (Ocimum basilicum), ObEGS1, and an isoeugenol synthase (IGS) from petunia (Petunia × hybrida), PhIGS1 (Koeduka et al., 2006). These enzymes use coniferyl acetate as a substrate for the in vitro production of eugenol and isoeugenol, respectively. Next, a chavicol/EGS from Larrea tridentata (LtCES1) was characterized and shown to be able to use both coniferyl acetate to form eugenol and p-coumaryl acetate or p-coumaryl coumarate to synthesize chavicol (Vassão et al., 2007). Later, an EGS from petunia (PhEGS1), together with two EGSs and an IGS from Clarkia breweri (CbEGS1, CbEGS2, and CbIGS1), all of which use coniferyl acetate as a substrate, were reported (Koeduka et al., 2008). Recently, a t-anol/IGS from anise (Pimpinella anisum), PaAIS1, was shown to convert p-coumaryl and coniferyl acetates into t-anol and isoeugenol, respectively (Koeduka et al., 2009). All of these enzymes are closely related to a number of NADPH-dependent reductases that act on different phenylpropanoid-derived substrates. Collectively, these enzymes constitute the PIP family, named after the three initially identified members, pinoresinol lariciresinol reductase, isoflavone reductase, and phenylcoumaran benzylic ether reductase, which are involved in the central steps of the biosynthesis of various important bioactive lignans and isoflavonoids (Gang et al., 1999; Min et al., 2003).
Figure 1.
Biochemical reactions leading to the phenylpropenes eugenol, chavicol, isoeugenol, and t-anol from Phe. CFAT, Coniferyl alcohol acyltransferase. The carbon numbering system used in this paper is shown in gray.
The crystal structures of apo-ObEGS1 and holo-ObEGS1 and a ternary complex with the cofactor NADP+ and the competitive inhibitor (7S,8S)-ethyl(7,8-methylene)-dihydroferulate have been resolved by x-ray crystallography (Louie et al., 2007). These structures have led to the proposal of a mechanistic scheme in which the binding of the coniferyl acetate substrate within the active site of EGS leads to the deprotonation of the p-hydroxy group coupled with the expulsion of the acetate ion from the C1 substituent (Louie et al., 2007). The resultant extended quinone-methide intermediate serves as a hydride acceptor at C7 to yield the product eugenol. However, hydride attack at C9 presumably leads to the formation of isoeugenol (Supplemental Fig. S1).
Despite eugenol having been identified in a variety of fruits (Jordán et al., 2001; Aubert and Pitrat, 2006; Ortiz-Serrano and Gil, 2010), including strawberry (Pyysalo et al., 1979; Zorrilla-Fontanesi et al., 2012), no EGS from this species has thus far been characterized. However, previous studies have shown that strawberry bears a functional phenylpropene biosynthetic pathway (Hoffmann et al., 2011). In addition, a recent study of the protein variations occurring between green and red strawberry achenes has demonstrated the existence, in green achenes, of at least one EGS (Aragüez et al., 2013). We have isolated three strawberry complementary DNAs (cDNAs) whose encoded proteins exhibit the highest identity to known EGSs. In addition, we have also assessed the in planta activity of these enzymes by transient expression experiments in strawberry fruit. Here, we provide a detailed biochemical characterization of the encoded proteins as well as fruit tissue- and development-specific expression patterns of the corresponding genes.
RESULTS
Strawberry Genes Encoding Proteins Highly Homologous to EGSs
We conducted a search for cDNAs encoding proteins with sequence similarity to known EGSs in the strawberry EST Database FREST at http://fresa.uco.uma.es/srs71 (Bombarely et al., 2010). Two ESTs with high identity to LtCES1, CbEGS2 and PhEGS1, were identified, and they corresponded to the genes named FaEGS1 and FaEGS2. Using the sequence information from the ESTs, two cDNAs for FaEGS1 (FaEGS1a and FaEGS1b) and one cDNA for FaEGS2 were cloned (GenBank accession numbers KF562264, KF562265, and KF562266, respectively), all of which were predicted to encode full-length proteins of 309 amino acids. The identity, at the nucleotide level, between FaEGS1a and FaEGS1b was 97.8%, whereas it was approximately 69% between these two genes and FaEGS2. BLASTN analyses using the wild relative F. vesca Genome Browser at www.rosaceae.org (Shulaev et al., 2011) identified gene15165 and gene25260 as the orthologs of FaEGS1 and FaEGS2, respectively. These genes were named FvEGS1 and FvEGS2 based on their homology to strawberry genes. An additional gene (gene15166), located in tandem with FvEGS1, was identified as a putative EGS and named FvEGS3. The sequence homology and nucleotide alignment of the FaEGSs with the putative FvEGSs found in the F. vesca genome were in agreement with FaEGS1a and FaEGS1b being possible alleles of the F. vesca ortholog FvEGS1. Nucleotide identity between FaEGS1a,b and FvEGS1 was around 96%, whereas it was only 81% when compared with FvEGS3 (Supplemental Fig. S2). We also identified two F. vesca genes homologous to IGSs, gene14701 and gene14703, named FvIGS1 and FvIGS2 on the basis of their putative function.
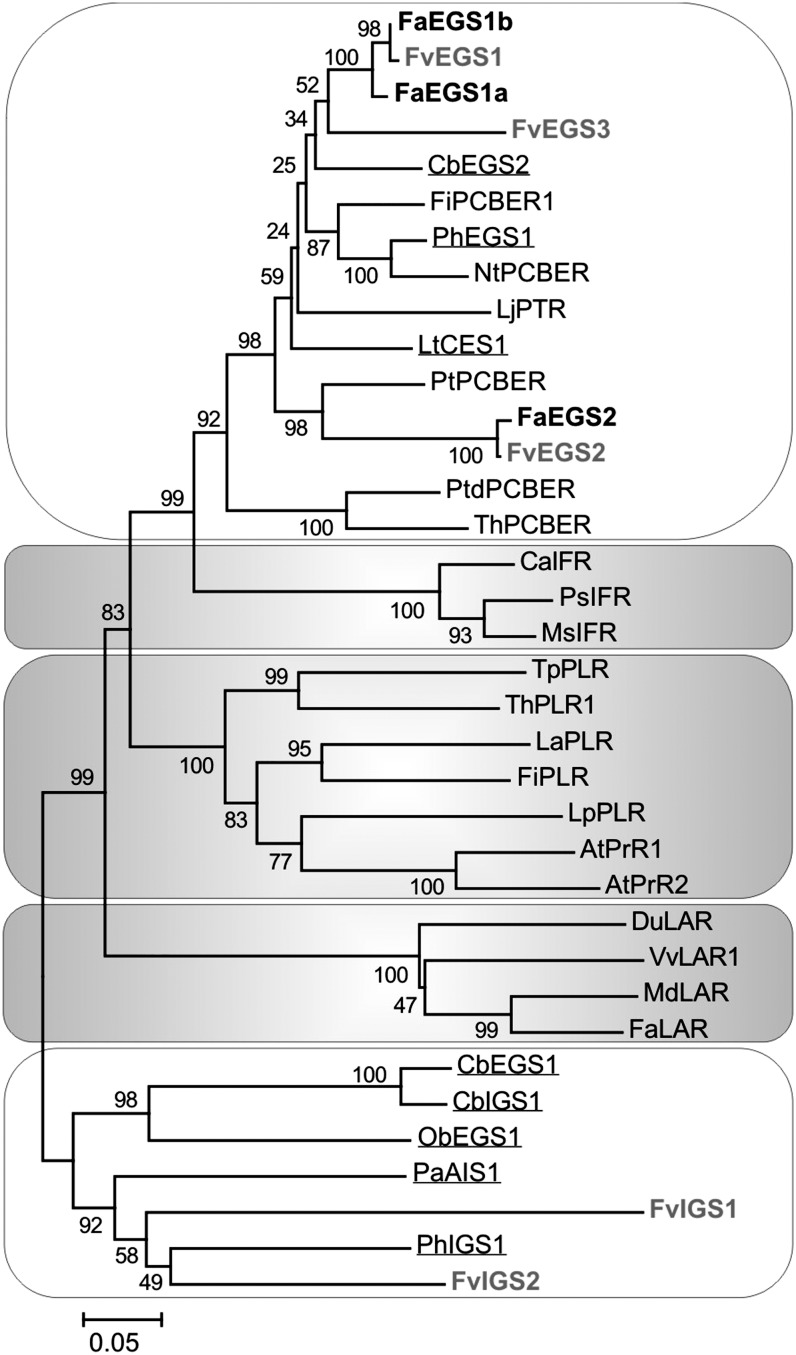
An unrooted phylogenetic tree with the FaEGSs, FvEGSs, FvIGSs, and selected enzymes of the PIP family of NADPH-dependent reductases grouped leucoanthocyanidin reductases, pinoresinol lariciresinol reductases and pinoresinol reductases, and isoflavone reductases into three clades (Fig. 2, gray background). The proteins described to have EGS or IGS activities were grouped in two different clades (white backgrounds), as reported formerly (Koeduka et al., 2008). FaEGSs and FvEGSs were grouped within the same clade as previously characterized enzymes that exhibited EGS activity, such as CbEGS2, PhEGS1 (Koeduka et al., 2008), and LtCES1 (Vassão et al., 2007), together with several enzymes described to have phenylcoumaran benzylic ether reductase and pterocarpan reductase activities (Gang et al., 1999). In a different clade were the enzymes PaAIS1, PhIGS1, CbEGS1, and CbIGS1, jointly with the putative FvIGSs.
Figure 2.
Unrooted phylogenetic tree of selected protein sequences in the PIP family of NADPH-dependent reductases, including FaEGS1a, FaEGS1b, and FaEGS2, as well as putative FvEGSs and FvIGSs. The two clades in which eugenol- and isoeugenol-forming enzymes are grouped are shown with a white background. The different clades of isoflavone reductases (IFRs), pinoresinol lariciresinol reductases (PLRs) together with pinoresinol reductases (PrRs), and leucoanthocyanidin reductases (LARs) are shown with a gray background. FaEGSs (black) and F. vesca putative EGSs and IGSs (gray) are shown in boldface. The proteins with EGS or IGS activity characterized in C. breweri, O. basilicum, P. × hybrida, P. anisum, and L. tridentata are underlined. Sequence analysis was performed by using ClustalW, and the neighbor-joining method was used to generate the tree. The percentages of replicate trees in which the associated proteins clustered together in the bootstrap test (1,000 trials) are shown next to the branches. The evolutionary distances were computed by using the p-distance method (this distance is the proportion, p, of amino acid sites at which the two sequences to be compared are different), and the scale is in units of the number of amino acid differences per site. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011). The accession numbers corresponding to the sequence data used in this study are listed in Supplemental Table S1. Cb, Clarkia breweri; Fi, Forsythia × intermedia; Nt, Nicotiana tabacum; Lj, Lotus japonicus; Lt, Larrea tridentata; Pt, Populus trichocarpa; Ptd, Pinus taeda; Th, Tsuga heterophylla; Ca, Cicer arietinum; Ps, Pisum sativum; Ms, Medicago sativa; Tp, Thuja plicata; La, Linum album; Lp, Linum perenne; At, Arabidopsis thaliana; Du, Desmodium uncinatum; Vv, Vitis vinifera; Md, Malus × domestica; Pa, Pimpinella anisum.
ObEGS1 was the first protein reported to have EGS activity (Koeduka et al., 2006). The multiple alignment shown in Supplemental Figure S3 reveals that the canonical sequence motif for NADPH binding, the residues relevant for the reductive displacement of acetate from the propenyl side chain of coniferyl acetate to produce eugenol, and key invariant residues in previously published EGSs and IGS (Louie et al., 2007) are conserved in most of the enzymes aligned.
Strawberry EGSs Display Different Kinetics
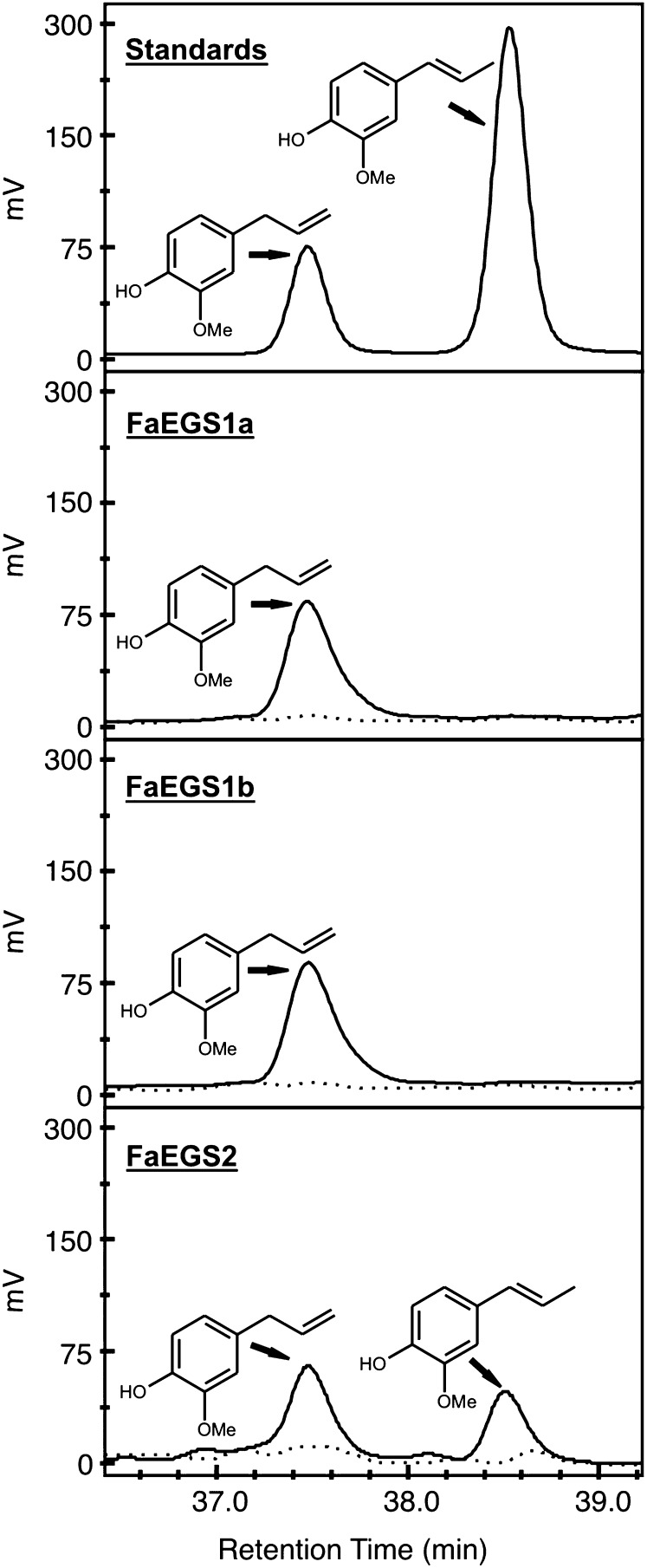
The FaEGS1a, FaEGS1b, and FaEGS2 recombinant proteins were produced in Escherichia coli and purified by affinity chromatography. The enzyme activities were assayed using coniferyl acetate as a substrate and NADPH as a cofactor, as described previously for other EGS enzymes (Koeduka et al., 2006). The optimal temperature and pH for the assays were 30°C and 6.5, respectively. Analysis of the compounds produced demonstrated that FaEGS1a and FaEGS1b catalyze the formation of eugenol. Interestingly, FaEGS2 was able to produce not only eugenol but also isoeugenol from coniferyl acetate, although the production of isoeugenol was lower than that of eugenol (Fig. 3).
Figure 3.
Product analysis by HPLC of the reactions catalyzed by purified FaEGSs. The analysis of eugenol (retention time of 37.5 min) and isoeugenol (retention time of 38.5 min) in a standard sample (25 µg µL−1 of each) and the analysis of products formed in the reactions catalyzed by active (solid lines) and inactive (incubated at 95°C for 5 min; dotted lines) FaEGS1a, FaEGS1b, and FaEGS2 enzymes are shown.
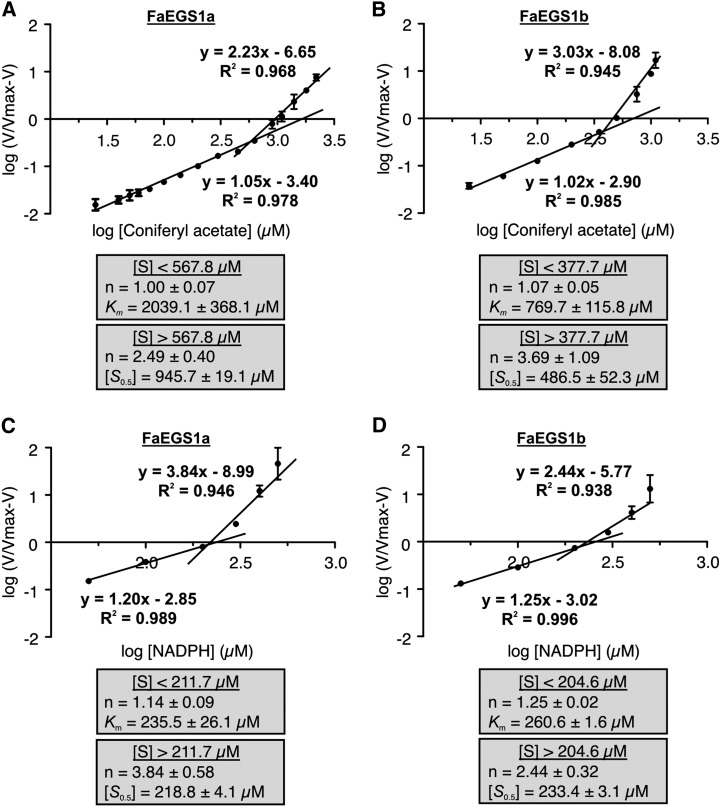
Despite the similarity in the amino acid sequences between FaEGS1a and FaEGS1b, with only six nonidentical amino acids (Supplemental Fig. S3), the two enzymes presented distinct kinetic properties (Fig. 4, A and B). The best fit of the experimental data corresponded to two straight lines in a Hill plot with a change in the slope, indicative of a transition of the kinetics of the enzymes at substrate concentrations of 567.8 µm for FaEGS1a and 377.7 µm for FaEGS1b. At lower substrate concentrations, the enzymes exhibited Michaelis-Menten kinetics with Km values for coniferyl acetate of 2,039.1 and 769.7 µm for FaEGS1a and FaEGS1b, respectively (Fig. 4, A and B). At higher substrate concentrations, both enzymes displayed positive cooperativity but with different kinetic parameters. The Hill coefficients (n) for FaEGS1a and FaEGS1b were 2.49 and 3.69, respectively, and the [S0.5] (substrate concentration for half of the maximum activity) values were 945.7 µm for FaEGS1a and 486.5 µm for FaEGS1b.
Figure 4.
Hill plots of FaEGS1a and FaEGS1b kinetic data measured with respect to coniferyl acetate and NADPH, and apparent kinetic parameters. A and B, Hill plots for coniferyl acetate using 1 mm NADPH. C and D, Hill plots for NADPH using 1 mm coniferyl acetate. Each value is the mean of at least three independent replicates, and error bars indicate sd. The various kinetic parameters are given for each enzyme above and below the substrate concentration [S] in which the kinetic properties change. These values, presented in gray boxes, are means of at least three independent experiments ± sd.
The kinetic properties of FaEGS1a and FaEGS1b were also analyzed relative to NADPH. The Hill plots revealed that, as observed for coniferyl acetate, there was a transition of the kinetics of the enzymes (Fig. 4, C and D). At substrate concentrations lower than 211.7 µm, the enzymes exhibited Michaelis-Menten kinetics with similar values of Km for NADPH: 235.5 and 260.6 µm for FaEGS1a and FaEGS1b, respectively. However, at higher substrate concentrations, both enzymes displayed positive cooperativity: n = 3.84 and n = 2.44 for FaEGS1a and FaEGS1b, respectively; the [S0.5] values were also very similar: 218.8 and 233.41 µm for FaEGS1a and FaEGS1b, respectively (Fig. 4, C and D).
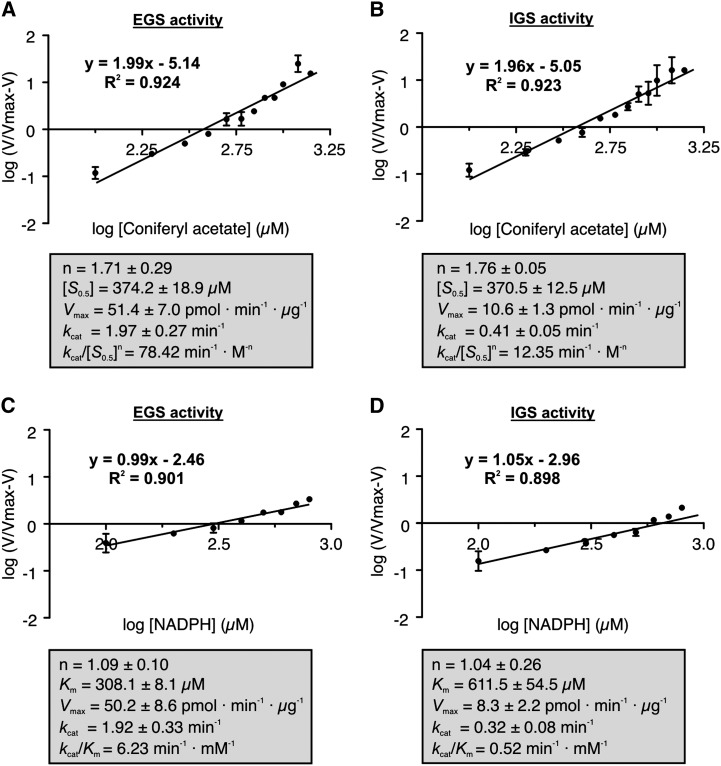
Unlike FaEGS1a and FaEGS1b, FaEGS2 displayed positive cooperativity for the production of both eugenol and isoeugenol in the complete range of coniferyl acetate concentrations studied herein, as illustrated by the corresponding Hill plots (Fig. 5, A and B). The Hill coefficients were approximately 1.7 for the formation of both eugenol and isoeugenol. The [S0.5] values were also very similar: 374.2 and 370.5 µm for EGS and IGS activities, respectively (Fig. 5, A and B). However, the maximum velocity for the formation of eugenol was 5-fold higher than that for isoeugenol formation, resulting in a higher catalytic constant (kcat) and catalytic efficiency (calculated as kcat/[S0.5]n) for the EGS activity. Saturation curves were also constructed for NADPH in the reaction catalyzed by FaEGS2. The corresponding plots indicated that the enzyme followed Michaelis-Menten kinetics for eugenol and isoeugenol formation (Fig. 5, C and D). The Km value for NADPH in the EGS activity was approximately one-half (308.1 µm) of the value in the IGS activity (611.5 µm; Fig. 5, C and D).
Figure 5.
Hill plots of FaEGS2 kinetic data measured with respect to coniferyl acetate and NADPH, and apparent kinetic parameters. A and B, Hill plots for coniferyl acetate using 1 mm NADPH. C and D, Hill plots for NADPH using 1 mm coniferyl acetate. Each value is the mean of at least three independent replicates, and error bars indicate sd. The various kinetic parameters are given for the formation of eugenol (EGS activity) and for the formation of isoeugenol (IGS activity). Vmax, Maximum velocity. These values, presented in gray boxes, are means of at least three independent experiments ± sd.
FaEGS1 and FaEGS2 Have Different Tissue- and Development-Specific Expression Patterns That Correlate with Eugenol Content
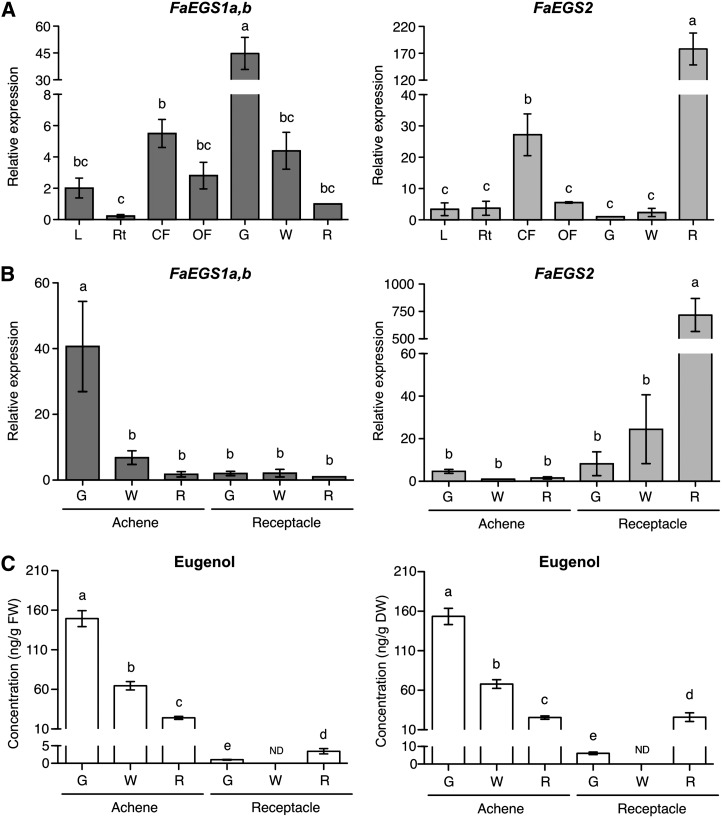
We used quantitative real-time (qRT)-PCR to determine the relative gene expression of FaEGS1 and FaEGS2 in strawberry leaves, roots, closed and open flowers, and along different ripening stages of the fruit (Fig. 6A). Because of the sequence similarity between FaEGS1a and FaEGS1b, we used a set of primers that amplified both cDNAs. The combined transcripts FaEGS1a,b were higher in closed flowers compared with open flowers, leaves, and roots. This pattern was similar for the expression levels of FaEGS2. In fruit, the expression of FaEGS1a,b and FaEGS2 was highest, but the patterns along development were different. Whereas transcripts of FaEGS1a,b were highly abundant in green fruits and significantly lower in white and red fruits, FaEGS2 expression was highest in red fruits and minimal in green and white fruits.
Figure 6.
qRT-PCR analysis of FaEGS1a,b and FaEGS2 expression, and eugenol quantification in different strawberry tissues. A, FaEGS1a,b and FaEGS2 relative transcript levels in vegetative tissues and during fruit development and ripening. L, Leaves; Rt, roots; CF, closed flowers; OF, open flowers; G, green fruit; W, white fruit; R, red fruit. B, Relative transcript levels of FaEGS1a,b and FaEGS2 in green (G), white (W), and red (R) achenes and receptacles. C, Eugenol quantification expressed as nanograms per gram fresh weight (FW; left) and as nanograms per gram dry weight (DW; right) in green (G), white (W), and red (R) achenes and receptacles. Each value is the mean of three independent biological replicates. Error bars indicate se. The different letters above the columns indicate values that differ significantly from each other according to Fisher’s lsd test (P < 0.05). The lsd test was performed separately for achene and receptacle eugenol data presented in C. The statistical analyses were performed with Statgraphics Centurion XVI software. ND, Not detected.
The strawberry fruit is a false fruit that originates from the expansion of the receptacle of the flower base as a pseudocarp, with the one-seeded achenes located on the epidermal layer of the receptacle (Perkins-Veazie, 1995). The achene is the true fruit, and the receptacle, which results from the engrossment of the flower receptacle, constitutes the fleshy part. Therefore, we analyzed the expression of FaEGS1a,b and FaEGS2 separately in achene and receptacle (Fig. 6B). Our study revealed that FaEGS1a,b expression mostly occurs in achenes at the green stage, whereas FaEGS2 expression is almost completely restricted to red receptacles. No expression of FaEGS3 was detected using qRT-PCR either in achenes or in receptacles at the three stages studied.
We examined the quantity of eugenol in achenes and receptacles of strawberry cv Camarosa fruits at the green, white, and red ripening stages. The highest production of eugenol in a fresh weight or in a dry weight basis was found in achenes compared with receptacles (Fig. 6C). Noteworthy, in achenes, the amount of eugenol was more than 6-fold higher at the green stage (149.4 ng g−1 fresh weight, 153.5 ng g−1 dry weight) than at the red stage (24.2 ng g−1 fresh weight, 25.5 ng g−1 dry weight), while in the receptacles, the production of this compound increased from the green stage (1.0 ng g−1 fresh weight, 6.1 ng g−1 dry weight) to the red ripe stage (3.4 ng g−1 fresh weight, 25.9 ng g−1 dry weight).
Eugenol Production by FaEGSs in Transformed Strawberry Fruit
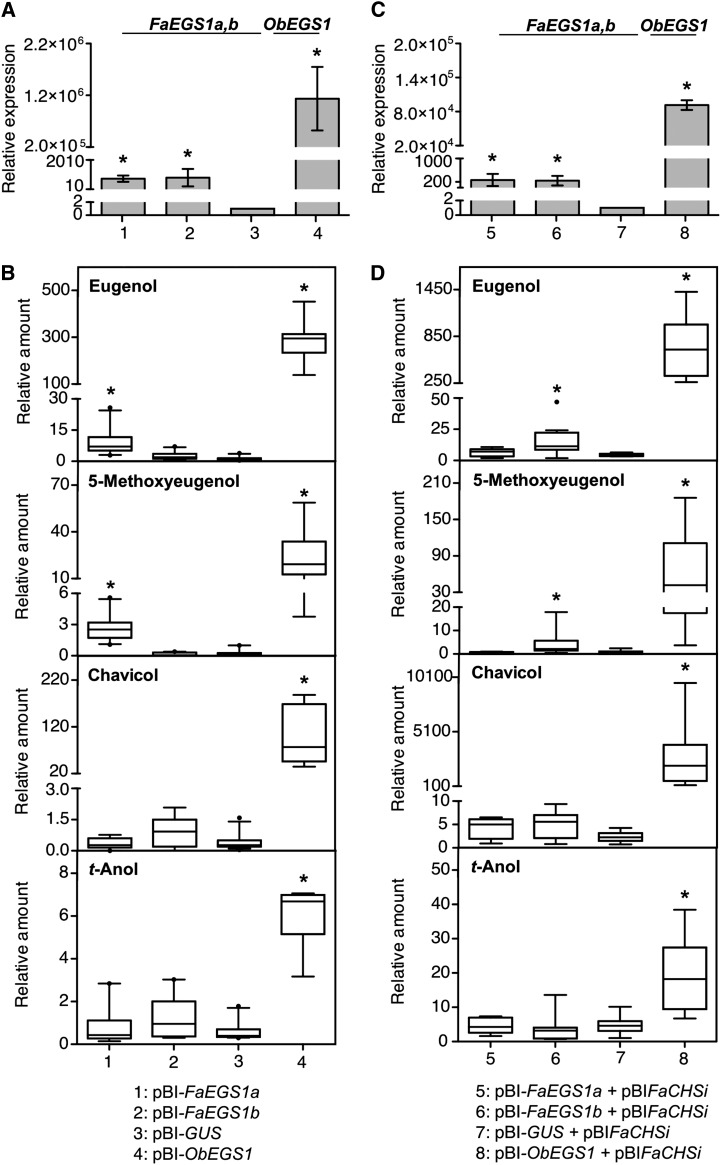
Finally, the in planta function of FaEGSs was investigated first by transient overexpression of FaEGS1a and FaEGS1b under the control of the 2XCaMV 35S promoter in strawberry fruit. Suspensions of Agrobacterium tumefaciens harboring the pBI-FaEGS1a and pBI-FaEGS1b constructs were injected in green strawberry fruits. A pBI-GUS construct was used as a negative control, whereas a pBI-ObEGS1 construct encoding an active EGS (Koeduka et al., 2006) was used as a positive control. The FaEGS1a,b transcripts increased in FaEGS1a- and FaEGS1b-overexpressing fruits relative to the control (Fig. 7A). In the transformed fruits, FaEGS1a overexpression led to an increased production of eugenol and 5-methoxyeugenol (Fig. 7B), likely derived from the acetates of coniferyl and sinapyl alcohols, respectively, the most abundant monolignols in angiosperms (Boerjan et al., 2003). Since the production of 5-methoxyeugenol was also high when transforming with the ObEGS1 from basil (Fig. 7B), its production can be explained as a side activity of the enzyme due to the structural similarities of the substrates. However, FaEGS1b overexpression did not produce a statistically significant accumulation of any of these volatile compounds. We also quantified the amount of chavicol and t-anol (both of them derived from p-coumaryl acetate) produced by the fruits that overexpressed EGSs, and observed that only the overexpression of ObEGS1 led to a significant increase in the accumulation of chavicol (Fig. 7B). The production of eugenol, 5-methoxyeugenol, and chavicol in strawberry fruits overexpressing ObEGS1 was several orders of magnitude higher than that found in fruits overexpressing FaEGS1a and FaEGS1b, most likely a result of the differences in expression observed between these genes (Fig. 7A).
Figure 7.
Effects of the transient overexpression of FaEGSs on gene expression and the production of volatile compounds in strawberry fruits. A and C, qRT-PCR analysis of FaEGS1a,b and ObEGS1 expression in transformed fruits when FaEGS1a, FaEGS1b, GUS, and ObEGS1 were overexpressed (A) and when each gene in A was overexpressed and FaCHS was simultaneously silenced (C). Each column is the mean of the expression of three representative fruits. Error bars indicate se. Asterisks designate statistically significant values (P < 0.05) compared with the values obtained for samples from GUS-expressing fruits (negative control). B and D, Box-plot graphs showing the relative quantification of the volatile compounds eugenol, 5-methoxyeugenol, chavicol, and t-anol in transformed fruits when FaEGS1a, FaEGS1b, GUS, and ObEGS1 were overexpressed (B) and when each gene in B was overexpressed and FaCHS was simultaneously silenced (D). Phenol was used as the internal standard, and the relative amount of each volatile was expressed as the ratio of compound to phenol peak areas in the chromatograms × 1,000. Twelve fruits per construct were individually analyzed by GC-MS. A horizontal line in the boxes indicates the median, and boxes represent the interquartile ranges. The whiskers extend to the 10th and 90th percentiles. Outliers are represented by black dots. The Mann-Whitney (Wilcoxon) W test was used for the nonparametric analysis of the intergroup comparison of data from transformed fruits. The asterisks indicate statistically significant values (P < 0.05) compared with the values of GUS-expressing fruits. The statistical analyses were performed with Statgraphics Centurion XVI software.
In addition to simple transient transformations, cotransformations with a construct silencing the chalcone synthase gene (pBI-FaCHSi) were performed to increase the availability of substrates for the EGS enzymes. Overexpression of the different EGS genes (Fig. 7C) did not produce any visible variation in the color of the fruit (Supplemental Fig. S4A), whereas FaCHS RNA interference-mediated silencing caused a loss of color in the receptacle (Supplemental Fig. S4B), as reported previously (Hoffmann et al., 2006). FaCHS silencing was further confirmed by qRT-PCR (Supplemental Fig. S4C), and our results were of the same order as those reported for the transient silencing of FaCHS (Muñoz et al., 2010). In this case, the overexpression of FaEGS1b caused significant increases in the production of eugenol and 5-methoxyeugenol, whereas FaEGS1a overexpression did not have any significant effect (Fig. 7D). The expression of ObEGS1 together with the silencing of FaCHS increased the production of eugenol and 5-methoxyeugenol more than the expression of FaEGS1b together with the silencing of FaCHS did, most likely owing to the great difference in the magnitude of the expression of the corresponding genes (Fig. 7C). It should be noted that gas chromatography-mass spectrometry (GC-MS) analyses allowed the profiling of different volatile compounds, whose levels did not vary as a consequence of the manipulations of gene expression in the set of experiments in which EGSs were overexpressed (Supplemental Fig. S5A) or in the set of experiments in which EGSs were overexpressed and FaCHS was simultaneously silenced (Supplemental Fig. S5B).
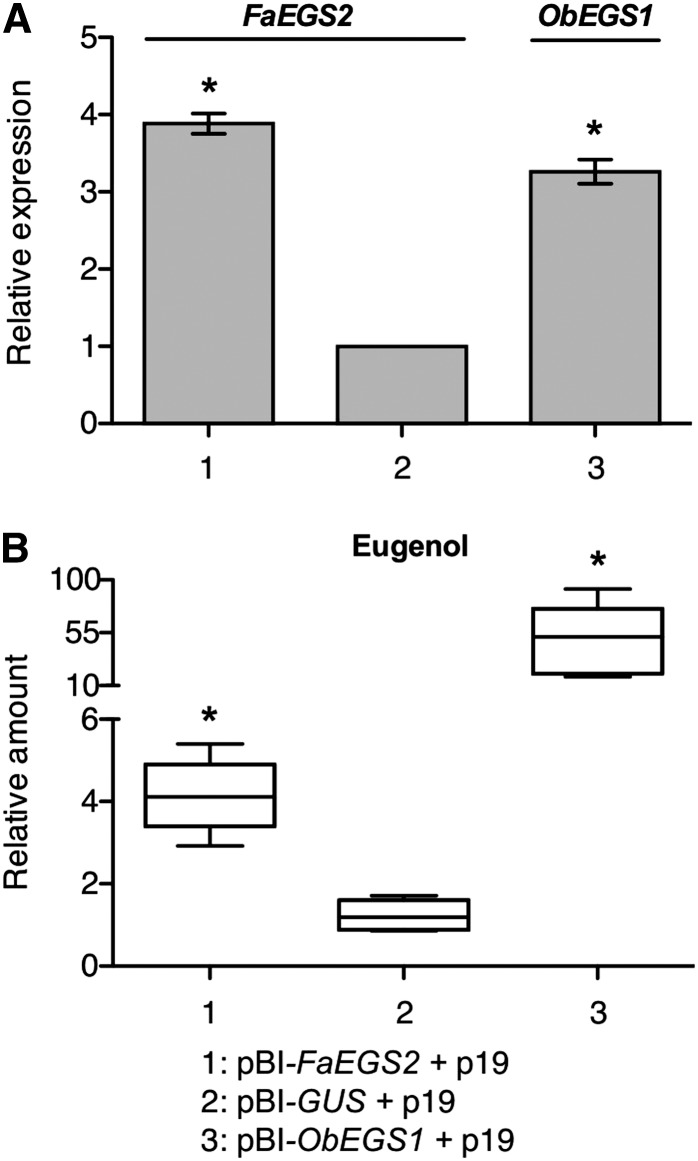
The transient transformation responsible for the overexpression of FaEGS1a and FaEGS1b was used with the aim of increasing the expression of FaEGS2. However, despite many attempts, we were unsuccessful, probably due to a cosuppression effect, given that endogenous FaEGS2 is highly expressed in receptacles (Fig. 6B). Therefore, we coexpressed FaEGS2 jointly with the p19 suppressor of gene silencing (Voinnet et al., 2003), and again we used the transformation with pBI-ObEGS1 as a positive control. This strategy allowed the efficient expression of FaEGS2 (Fig. 8A), which was accompanied by a significant increase in the production of eugenol (Fig. 8B). Taken together, these results confirmed that FaEGS1a, FaEGS1b, and FaEGS2 act as in vivo EGSs in strawberry.
Figure 8.
Results of the overexpression of FaEGS2 in strawberry fruits. A, qRT-PCR analysis of FaEGS2 and ObEGS1 expression in transformed fruits. Each column is the mean of the expression of six representative fruits. Error bars indicate se. The asterisks designate statistically significant values (P < 0.05) compared with the values obtained for samples from GUS-expressing fruits (negative control). B, Box-plot graphs showing the relative quantification of the volatile compound eugenol in transformed fruits. Six fruits per construct were individually analyzed by GC-MS. Fruits were coinfiltrated with constructs to overexpress FaEGS2, GUS, or ObEGS1, together with a construct to overexpress the p19 gene encoding the p19 suppressor of silencing. A horizontal line in the boxes indicates the median, and boxes represent the interquartile ranges. The whiskers extend to the 10th and 90th percentiles. Outliers are represented by black dots. The Mann-Whitney (Wilcoxon) W test was used for the nonparametric analysis of the intergroup comparison of data from transformed fruits. The asterisks indicate statistically significant values (P < 0.05) compared with the values of GUS-expressing fruits. The statistical analyses were performed with Statgraphics Centurion XVI software.
DISCUSSION
Strawberry Has Distinct Synthases for Eugenol Biosynthesis
The analysis of FaEGS1a and FaEGS1b kinetic data indicates that the data fit into two models depending upon the coniferyl acetate concentration: hyperbolic and sigmoidal below and above a certain substrate concentration, respectively. The sigmoidal or cooperative model has often been associated with multimeric enzymes. In fact, several proteins of the PIP family of reductases, including ObEGS1, have been described as forming 2-fold rotationally symmetric homodimers (Louie et al., 2007). The difference between the enzymes encoded by FaEGS1a and FaESG1b is restricted to six residues (Supplemental Fig. S3), which seem to have no direct involvement in the catalytic mechanism described for members of this family (Louie et al., 2007; Koeduka et al., 2008); however, these variations are sufficient to result in minor differences in the kinetic parameters. Globally, the affinity for coniferyl acetate, evaluated by Km and [S0.5] values, is higher for FaEGS1b than for FaEGS1a. On the other hand, the coniferyl acetate affinity parameters for FaEGS1a and FaEGS1b are higher than those reported for CbEGS1 (93 µm), CbEGS2 (310 µm), PhEGS1 (245 µm; Koeduka et al., 2008), and LtCES1 (290 µm; Vassão et al., 2007). While the endogenous concentration of p-coumaryl acetate has already been reported in strawberry fruits (Lunkenbein et al., 2006; Hoffmann et al., 2011), coniferyl acetate was not detected in these studies. It must be noted that there are some difficulties in knowing the cell concentrations of specific metabolites, the kinetic parameters of the enzymes sometimes being an indirect indication of their levels. Thus, the mentioned p-coumaryl acetate concentrations correspond to values in the whole fruit, and it cannot be discarded that its production is restricted to specific cells in this organ. Similarly, the accumulation of coniferyl acetate and eugenol could occur in particular cells, as it has been reported for the production of furaneol in strawberry (Sen et al., 1991). Since the cell concentration of the substrate coniferyl acetate is not known, the meaningfulness of the in vitro values of the affinity parameters of FaEGSs cannot be assessed at present.
FaEGS1a and FaEGS1b display different capacities for producing eugenol in fruits that were infiltrated with the corresponding overexpressing constructs, depending on whether the fruits were coinfiltrated with an RNAi construct of FaCHS. It was expected that CHS silencing would affect the amount of coniferyl acetate available (Lunkenbein et al., 2006), but establishing a direct relationship between the differences in substrate affinity and the in planta activities is not straightforward. It is difficult to infer the physiological relevance of these results, given that the actual concentration of the substrate coniferyl acetate in the cells is unknown. However, it can be concluded that the occurrence of two FaEGS1 enzymes resulting from two possible alleles extends the corresponding range of responsiveness of the enzymes to different substrate concentrations.
The kinetics is different for FaEGS2 relative to FaEGS1a and FaEGS1b. First, the [S0.5] value of FaEGS2 for coniferyl acetate is 370 µm, which is in the range of values obtained for other EGS enzymes, and slightly lower than the values determined for FaEGS1a and FaEGS1b. A major difference between FaEGSs is the capacity of FaEGS2 to produce isoeugenol, although its catalytic efficiency to produce eugenol is over 6-fold higher than that to produce isoeugenol. Differences between these two activities also extend to the Km value for NADPH, which is lower for the eugenol-producing activity compared with the IGS activity. These dissimilarities are probably related to a different positioning of the substrate in the active site.
There are in the literature some examples of monomeric enzymes displaying a sigmoidal kinetics. One of the best characterized is the human glucokinase. Structurally, it is the consequence of a rearrangement induced by substrate binding (Larion and Miller, 2009), being the sharpness of the sigmoidal response related to the number of interactions (Qian, 2012). This structural feature is essential for the in vivo role played by the enzyme in human liver. For FaEGS1a, FaEGS1b, and FaEGS2, beyond its similarity to ObEGS1, whose structure has been determined by x-ray crystallography (Louie et al., 2007), nothing can be concluded about the structural features accounting for their kinetic characteristics. However, the functional consequence of their sigmoidal behavior is directly related to their responsiveness to different substrate concentrations. Since FaEGS1a,b and FaEGS2 are expressed in different tissues, achenes and receptacle, respectively, we expect different endogenous concentrations of coniferyl acetate in these tissues, based on the substrate affinities of the corresponding enzymes and their values of Hill coefficient.
All of the previously characterized native EGSs and IGSs catalyze the formation of only one product from a given substrate, but in vitro mutagenesis of a few residues within the active sites of CbIGS1, CbEGS1, CbEGS2, ObEGS1, PhIGS1, and PhEGS1 resulted in enzymes that produced mixtures of eugenol and isoeugenol (Koeduka et al., 2008). In ObEGS1, residues 85 and 88 (and the corresponding positions in CbEGS1, CbEGS2, CbIGS1, and PhIGS1) have been identified as key amino acids determining product specificity. The amino acids Val-85 and Tyr-88 are conserved in IGSs, whereas Phe-85 and Ile-88 are invariant in EGSs (Koeduka et al., 2008). However, the corresponding amino acids in FaEGSs are not the same as any of those mentioned above (Supplemental Fig. S3). There is also a variant in position 266 of FaEGS2, where a Tyr is found rather than the Ile that is considered to be critical in other EGSs (Louie et al., 2007). In addition, other amino acids in the vicinity of this Tyr are also different in FaEGS2. Overall, all of these amino acid variations must affect the positioning of NADPH in the active site. Thus, FaEGS2 might represent an intermediate form of EGS enzymes with the capacity to produce both eugenol and isoeugenol in vitro, although the capacity of producing isoeugenol has not been demonstrated in vivo. This product versatility is also known in other families of enzymes involved in specialized metabolism, such as the terpene synthase family (Pichersky et al., 2006). The physiological relevance of the ability of FaEGS2 to catalyze the in vitro formation of isoeugenol remains to be established, as isoeugenol is produced only in low amounts in strawberry fruit (Hoffmann et al., 2011), and we have identified in the genome of the wild strawberry F. vesca two putative IGS genes (Fig. 2; Supplemental Fig. S2), which probably have their orthologs in the genome of strawberry.
Expression of FaEGS1 and FaEGS2 in Strawberry Fruit Accounts for Eugenol Production at Specific Tissue/Developmental Stages
The emission by strawberry fruit of both eugenol (Pyysalo et al., 1979; Hoffmann et al., 2011; Zorrilla-Fontanesi et al., 2012) and isoeugenol (Hoffmann et al., 2011) has been reported previously. Our detailed analysis of eugenol content in the two parts of the strawberry fruit revealed a stage-specific profile that was opposite in achenes and receptacle. The major production was found in the achenes at the green stage. Recently, a proteomic study in this organ showed the occurrence of an EGS in green achenes as one of the most abundant proteins (Aragüez et al., 2013). The different patterns of eugenol production reflect the diverse roles played by this phenylpropene during the development of the strawberry fruit. The existence of an EGS gene family in this species must explain the different spatial and temporal production of this volatile. Both expression studies and kinetic analysis of the gene products support a key role for FaEGS1 and FaEGS2 in the production of eugenol by the whole fruit.
The receptacle and the achene are very different in terms of their origin, cell components, identity, and developmental program (Aharoni and O’Connell, 2002; Fait et al., 2008), although the latter is highly connected between these two organs (Csukasi et al., 2011). The achene at the green stage has completed the formation of the embryo and plays a critical role in the growth and viability of the receptacle and, eventually, of the entire fruit (Nitsch, 1950). Some structures present in the mature achene, such as the hard and thick pericarp, have not been developed (Perkins-Veazie, 1995), which makes this organ very sensitive at the green stage. It has been reported that the highest transcript levels of the phenylpropene synthase PaAIS1 are also found in developing fruits of anise (Koeduka et al., 2009). The expression of this gene was coincident with a high content of t-anethole, a methyl derivative of t-anol. The high accumulation of t-anethole, a fungicide, in the reproductive tissues of anise was associated with an adaptation process to increase the protection of the fruit. In strawberry, eugenol treatment at a high level has been reported to protect against postharvest fungal growth (Wang et al., 2007). Although this does not prove a role for eugenol in protection during achene development in planta, this possibility cannot be ignored and deserves to be explored.
Alternatively, the red receptacle corresponds to the mature stage of this part of the fruit, which presents specific cellular and compositional changes (Perkins-Veazie, 1995), including the production of volatiles (Zabetakis and Holden, 1997), which account for the flavor of the commercial fruit. Eugenol is a component of the set of volatiles produced by ripe fruit, but its contribution to the sensory trait has diminished with cultivation (Pyysalo et al., 1979).
CONCLUSION
Two members of the gene family of EGSs have been identified, their function in vivo has been assessed, and the corresponding enzymes have been characterized. These two different EGS genes, which are transcriptionally activated at specific stages of fruit development, account for the production of eugenol in achene and receptacle at different developmental stages. Minor/subtle differences in the sequences of two possible alleles of FaEGS1 affect the kinetic parameters of the encoded enzymes, which must be explained by the fitness of the enzyme activity associated with endogenous concentrations of the substrates. FaEGS2 exhibits product versatility, being able to produce not only eugenol but also isoeugenol in vitro; however, this versatility has no apparent physiological relevance.
MATERIALS AND METHODS
Plant Material
Tissues from the octoploid strawberry (Fragaria × ananassa cv Camarosa) were collected from plants that were grown under field conditions in Huelva, Spain. Achenes and receptacles were carefully separated using a scalpel tip, and samples were obtained at the stages green, white, and red at 12, 21, and 35 days post anthesis as described previously (Csukasi et al., 2011). Strawberry cv Elsanta, used for the transient transformation of fruits, was grown in a chamber at a temperature of 25°C and a 16-h photoperiod under 120 μmol m−2 s−1 irradiance. Strawberry cv Camarosa plants, used for transient transformation of FaEGS2, were grown in greenhouses, under the climatic conditions of Málaga, in the south of Spain.
Eugenol Analysis in Strawberry Fruit Samples by Headspace Solid-Phase Microextraction/GC-MS
Three biological replicates of each type of sample (pools of five to 10 strawberry fruits at different ripening stages) were analyzed. A total of 500 mg of powdered, frozen strawberry fruit (receptacle or achenes) was incubated at 30°C for 5 min in a 7-mL vial in a water bath. Then, 1.25 mL of a saturated NaCl solution was added, together with 7.5 µL of a 5 µg mL−1 solution of isoeugenol as an internal standard. After homogenization, 1 mL of the mixture was transferred to a 10-mL screw-cap headspace vial and analyzed immediately. Volatile compounds were sampled by headspace solid-phase microextraction with a 65-µm polydimethylsiloxane/divinylbenzene fiber (Supelco). Initially, vials were preincubated at 50°C for 10 min; then, volatiles were extracted for 30 min under continuous shaking and heating at 50°C.
GC-MS analyses were performed as described (Orzaez et al., 2009), except that mass spectrometry was performed in simultaneous scan and single ion monitoring (SIM) mode. The monitored ion in SIM for both eugenol and the internal standard was 164. Quantitation was performed by measuring the ratio of the peak areas of eugenol and the internal standard from the SIM data, and these values were interpolated on a calibration curve.
RNA Isolation and Gene Expression Analysis
The total RNA from independent pools of five to 10 strawberry fruits and receptacles at different ripening stages and from roots, leaves, and flowers was extracted according to Manning (1991). The RNA of transiently transformed cv Elsanta fruits was extracted from each fruit individually. To extract RNA from the achenes of five to 10 fruits at different ripening stages, the protocol for genomic DNA extraction described by Medina-Escobar et al. (1997) was followed until the step at which a precipitate of nucleic acids is obtained. At this point, the pellet was washed and dissolved in water, and then a one-third-volume equivalent of 12 m LiCl was added to selectively precipitate the RNA. After incubating overnight at 4°C, the RNA pellet that was obtained after centrifugation at 12,000g for 30 min at 4°C was washed, dissolved in water, and stored at −80°C for later use. Prior to cDNA synthesis, the RNA was treated with amplification-grade DNase I (Sigma-Aldrich) according to the instructions of the manufacturer.
Reverse transcription was carried out starting from 1 µg of total RNA and using the i-Script cDNA synthesis kit (Bio-Rad) according to the instructions of the manufacturer.
Gene expression analyses were performed by qRT-PCR using the fluorescent intercalating dye SYBR Green in a Rotor-Gene Q instrument (Qiagen). Each reaction was performed in triplicate in three biological replicates, and the corresponding cycle threshold values were determined. The absence of primer dimers was confirmed by an examination of melting curves. The cycle threshold values of each qRT-PCR were normalized to the interspacer 26S-18S strawberry RNA housekeeping gene Farib413 (Benítez-Burraco et al., 2003). Relative quantification of target genes was performed using the Pfaffl method (Pfaffl, 2001), and a value of 1 was given to the sample with the lowest expression in each case. The similar efficiencies of the primers used to amplify FaEGS1a,b, FaEGS2, and ObEGS1, together with the use of the same PCR conditions and cDNA dilutions in all the qRT-PCR analyses, allowed us to estimate the relative expression of the different genes.
The forward and reverse primers used for expression analyses are listed in Supplemental Table S2.
Expression and Purification of Heterologous Proteins in Escherichia coli
The pET-FaEGS1a, pET-FaEGS1b, and pET-FaEGS2 overexpression constructs, consisting of the sequences of the FaEGSs in the T7 expression vector pET-28b(+), were used to produce the corresponding proteins in E. coli fused to an N-terminal His6 tag. The primers used to generate the constructs are listed in Supplemental Table S2. After the DNA sequences of the inserts were confirmed, these constructs were transformed into BL21-CodonPlus (DE3)-RIL competent cells (Agilent Technologies) for protein expression. The liquid cultures were induced with 0.5 mm isopropyl-β-thiogalactosidase and grown for 4 h at 18°C. The cells were collected and lysed, and the fusion proteins were purified using a Profinity IMAC Resin charged with Ni2+ (Bio-Rad) following the instructions of the manufacturer. The standard final yields were 4.4 ± 0.4, 5.4 ± 0.6, and 1.9 ± 0.1 µg mL−1 culture in the case of FaEGS1a, FaEGS1b, and FaEGS2 recombinant proteins, respectively.
Enzymatic Assays
A standard assay was performed in a closed vial by incubating 10 µg of purified enzyme solution in a final volume of 200 µL of 0.1 m KH2PO4/K2HPO4 buffer (pH 6.5), 1 mm NADPH, and 1 mm coniferyl acetate. Control assays were performed with heat-inactivated enzymes (95°C for 5 min). After incubation at 30°C for 30 min with shaking, the reactions were stopped by adding 10 µL of 1 n HCl, and the reaction mixture was subjected to HPLC analysis.
Chemoenzymatic Synthesis of Coniferyl Acetate
The synthesis of coniferyl acetate was performed following a procedure reported for the synthesis of p-coumaryl-1-acetate (Lunkenbein et al., 2006) but using coniferyl alcohol (Sigma-Aldrich) as the starting material.
HPLC Analysis
Chromatographic separations were achieved by a Kromasil 100 C18 5-µm, 250- × 3-mm column (Scharlab). The chromatographic assays were performed at room temperature with a mobile phase of methanol and water at a rate of 0.35 mL min−1. The initial separation conditions (20% methanol, 80% water) were maintained for 5 min. Then, the column was eluted with a 45-min linear gradient of methanol-water, such that the methanol percentage increased from 20% to 100%. The UV chromatograms were acquired at 280 nm, and eugenol and isoeugenol in solution were identified by comparing their retention times with those of commercial standard compounds.
Transient Transformation of Strawberry Fruit by Agroinfiltration
The transient overexpression of FaEGS1a, FaEGS1b, GUS, and ObEGS1 in pBI vector, as well as the silencing of FaCHS with the construct pBI-FaCHSi, were carried out in strawberry cv Elsanta fruits as described previously (Hoffmann et al., 2006). Overexpression of FaEGS2 was carried out in strawberry cv Camarosa fruits by the coinfiltration of A. tumefaciens strains harboring pBI-FaEGS2 and the pBin61-p19 construct to overexpress a gene encoding the silencing suppressor p19 protein of Tomato bushy stunt virus under the control of the CaMV 35S promoter (Voinnet et al., 2003).
Overexpression constructs were generated by replacing the GUS gene in the binary vector pBI12135S2×. The primers used are listed in Supplemental Table S2. pBI-ObEGS1 was generated in a similar way from a construct kindly provided by Dr. Eran Pichersky’s group (Koeduka et al., 2006). The pBI-FaCHSi construct was generated as described previously (Hoffmann et al., 2006).
Extraction of Volatile Compounds from Transiently Transformed Strawberry Fruit and GC-MS Analysis
In the case of fruits injected with pBI-FaEGS1a, pBI-FaEGS1b, pBI-GUS, and pBI-ObEGS1, with or without simultaneous silencing of pBI-FaCHSi, a 2-g sample of the tissue powder (stored at −80°C) of each individual fruit was allowed to defrost for 15 min at 25°C. A 10-µg aliquot of phenol was added as an internal standard, and the resulting suspension was homogenized by vortexing. Then, the sample was centrifuged at 10,000g for 25 min at 4°C. After recovering the aqueous phase, 1.5 mL of water was added to the tissue, and the resulting suspension was again homogenized and centrifuged. The aqueous phases were then pooled in a glass vial and extracted twice with 1.5 mL of tert-butyl methyl ether. The resultant extract was concentrated under a nitrogen stream and dissolved in 200 µL of tert-butyl methyl ether. The volatile profile of each fruit was analyzed by GC-MS as reported previously (Lunkenbein et al., 2006).
The volatile compounds of individual fruits injected with pBI-FaEGS2 were analyzed by headspace solid-phase microextraction/GC-MS following the procedure described above.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KF562264 (FaEGS1a), KF562265 (FaEGS1b) and KF562266 (FaEGS2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The proposed reaction mechanism of EGS and IGS involving a quinone-methide intermediate.
Supplemental Figure S2. Analysis of the nucleotide sequences of FaEGSs and putative FvEGSs.
Supplemental Figure S3. Multiple alignment of the amino acid sequences of different EGSs, IGSs, FaEGSs, and FvIGSs.
Supplemental Figure S4. Phenotype of transformed strawberry fruits and FaCHS expression.
Supplemental Figure S5. Analysis of different volatile compounds in fruits that overexpressed FaEGS1a, FaEGS1b, GUS, and ObEGS without and with FaCHS simultaneous silencing.
Supplemental Table S1. Accession numbers of the proteins analyzed in Figure 2.
Supplemental Table S2. Primer sequences used in this study.
Acknowledgments
We thank Dr. Eran Pichersky (Department of Molecular, Cellular and Developmental Biology, University of Michigan) for providing the ObEGS1 clone. We also thank Dr. Rafael Suau (Organic Chemistry Department, University of Málaga) for his assistance in the synthesis of coniferyl acetate.
Glossary
- cDNA
complementary DNA
- qRT
quantitative real-time
- GC-MS
gas chromatography-mass spectrometry
- SIM
single ion monitoring
References
- Aharoni A, O’Connell AP. (2002) Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J Exp Bot 53: 2073–2087 [DOI] [PubMed] [Google Scholar]
- Aragüez I, Cruz-Rus E, Botella MÁ, Medina-Escobar N, Valpuesta V. (2013) Proteomic analysis of strawberry achenes reveals active synthesis and recycling of L-ascorbic acid. J Proteomics 83: 160–179 [DOI] [PubMed] [Google Scholar]
- Aubert C, Pitrat M. (2006) Volatile compounds in the skin and pulp of Queen Anne’s pocket melon. J Agric Food Chem 54: 8177–8182 [DOI] [PubMed] [Google Scholar]
- Benítez-Burraco A, Blanco-Portales R, Redondo-Nevado J, Bellido ML, Moyano E, Caballero JL, Muñoz-Blanco J. (2003) Cloning and characterization of two ripening-related strawberry (Fragaria × ananassa cv. Chandler) pectate lyase genes. J Exp Bot 54: 633–645 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bombarely A, Merchante C, Csukasi F, Cruz-Rus E, Caballero JL, Medina-Escobar N, Blanco-Portales R, Botella MA, Muñoz-Blanco J, Sánchez-Sevilla JF, et al (2010) Generation and analysis of ESTs from strawberry (Fragaria × ananassa) fruits and evaluation of their utility in genetic and molecular studies. BMC Genomics 11: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csukasi F, Osorio S, Gutierrez JR, Kitamura J, Giavalisco P, Nakajima M, Fernie AR, Rathjen JP, Botella MA, Valpuesta V, et al (2011) Gibberellin biosynthesis and signalling during development of the strawberry receptacle. New Phytol 191: 376–390 [DOI] [PubMed] [Google Scholar]
- Dexter R, Qualley A, Kish CM, Ma CJ, Koeduka T, Nagegowda DA, Dudareva N, Pichersky E, Clark D. (2007) Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. Plant J 49: 265–275 [DOI] [PubMed] [Google Scholar]
- Fait A, Hanhineva K, Beleggia R, Dai N, Rogachev I, Nikiforova VJ, Fernie AR, Aharoni A. (2008) Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiol 148: 730–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang DR, Kasahara H, Xia Z-Q, Vander Mijnsbrugge K, Bauw G, Boerjan W, Van Montagu M, Davin LB, Lewis NG. (1999) Evolution of plant defense mechanisms: relationships of phenylcoumaran benzylic ether reductases to pinoresinol-lariciresinol and isoflavone reductases. J Biol Chem 274: 7516–7527 [DOI] [PubMed] [Google Scholar]
- Hoffmann T, Kalinowski G, Schwab W. (2006) RNAi-induced silencing of gene expression in strawberry fruit (Fragaria × ananassa) by agroinfiltration: a rapid assay for gene function analysis. Plant J 48: 818–826 [DOI] [PubMed] [Google Scholar]
- Hoffmann T, Kurtzer R, Skowranek K, Kiessling P, Fridman E, Pichersky E, Schwab W. (2011) Metabolic engineering in strawberry fruit uncovers a dormant biosynthetic pathway. Metab Eng 13: 527–531 [DOI] [PubMed] [Google Scholar]
- Jordán MJ, Tandon K, Shaw PE, Goodner KL. (2001) Aromatic profile of aqueous banana essence and banana fruit by gas chromatography-mass spectrometry (GC-MS) and gas chromatography-olfactometry (GC-O). J Agric Food Chem 49: 4813–4817 [DOI] [PubMed] [Google Scholar]
- Koeduka T, Baiga TJ, Noel JP, Pichersky E. (2009) Biosynthesis of t-anethole in anise: characterization of t-anol/isoeugenol synthase and an O-methyltransferase specific for a C7-C8 propenyl side chain. Plant Physiol 149: 384–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeduka T, Fridman E, Gang DR, Vassão DG, Jackson BL, Kish CM, Orlova I, Spassova SM, Lewis NG, Noel JP, et al. (2006) Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc Natl Acad Sci USA 103: 10128–10133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeduka T, Louie GV, Orlova I, Kish CM, Ibdah M, Wilkerson CG, Bowman ME, Baiga TJ, Noel JP, Dudareva N, et al (2008) The multiple phenylpropene synthases in both Clarkia breweri and Petunia hybrida represent two distinct protein lineages. Plant J 54: 362–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larion M, Miller BG. (2009) 23-Residue C-terminal α-helix governs kinetic cooperativity in monomeric human glucokinase. Biochemistry 48: 6157–6165 [DOI] [PubMed] [Google Scholar]
- Louie GV, Baiga TJ, Bowman ME, Koeduka T, Taylor JH, Spassova SM, Pichersky E, Noel JP. (2007) Structure and reaction mechanism of basil eugenol synthase. PLoS ONE 2: e993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunkenbein S, Coiner H, de Vos CHR, Schaart JG, Boone MJ, Krens FA, Schwab W, Salentijn EMJ. (2006) Molecular characterization of a stable antisense chalcone synthase phenotype in strawberry (Fragaria × ananassa). J Agric Food Chem 54: 2145–2153 [DOI] [PubMed] [Google Scholar]
- Manning K. (1991) Isolation of nucleic acids from plants by differential solvent precipitation. Anal Biochem 195: 45–50 [DOI] [PubMed] [Google Scholar]
- Medina-Escobar N, Cárdenas J, Moyano E, Caballero JL, Muñoz-Blanco J. (1997) Cloning, molecular characterization and expression pattern of a strawberry ripening-specific cDNA with sequence homology to pectate lyase from higher plants. Plant Mol Biol 34: 867–877 [DOI] [PubMed] [Google Scholar]
- Min T, Kasahara H, Bedgar DL, Youn B, Lawrence PK, Gang DR, Halls SC, Park H, Hilsenbeck JL, Davin LB, et al (2003) Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases. J Biol Chem 278: 50714–50723 [DOI] [PubMed] [Google Scholar]
- Muñoz C, Hoffmann T, Escobar NM, Ludemann F, Botella MA, Valpuesta V, Schwab W. (2010) The strawberry fruit Fra a allergen functions in flavonoid biosynthesis. Mol Plant 3: 113–124 [DOI] [PubMed] [Google Scholar]
- Nitsch JP. (1950) Growth and morphogenesis of the strawberry as related to auxin. Am J Bot 37: 211–215 [Google Scholar]
- Ortiz-Serrano P, Gil JV. (2010) Quantitative comparison of free and bound volatiles of two commercial tomato cultivars (Solanum lycopersicum L.) during ripening. J Agric Food Chem 58: 1106–1114 [DOI] [PubMed] [Google Scholar]
- Orzaez D, Medina A, Torre S, Fernández-Moreno JP, Rambla JL, Fernández-Del-Carmen A, Butelli E, Martin C, Granell A. (2009) A visual reporter system for virus-induced gene silencing in tomato fruit based on anthocyanin accumulation. Plant Physiol 150: 1122–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasay C, Mounsey K, Stevenson G, Davis R, Arlian L, Morgan M, Vyszenski-Moher D, Andrews K, McCarthy J. (2010) Acaricidal activity of eugenol based compounds against scabies mites. PLoS ONE 5: e12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins-Veazie P (1995) Growth and ripening of strawberry fruit. In J Janick, ed, Horticultural Reviews, Vol 17. John Wiley & Sons, Oxford, pp 267–297 [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Noel JP, Dudareva N. (2006) Biosynthesis of plant volatiles: nature’s diversity and ingenuity. Science 311: 808–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyysalo T, Honkanen E, Hirvi T. (1979) Volatiles of wild strawberries, Fragaria vesca L., compared to those of cultivated berries, Fragaria × ananassa cv. Senga Sengana. J Agric Food Chem 27: 19–22 [Google Scholar]
- Qian H. (2012) Cooperativity in cellular biochemical processes: noise-enhanced sensitivity, fluctuating enzyme, bistability with nonlinear feedback, and other mechanisms for sigmoidal responses. Annu Rev Biophys 41: 179–204 [DOI] [PubMed] [Google Scholar]
- Sen A, Schieberle P, Grosch W. (1991) Quantitative determination of 2,5-dimethyl-4-hydroxy-3(2H)-furanone and its methyl ether using a stable isotope dilution assay. Lebenson Wiss Technol 24: 364–369 [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassão DG, Kim SJ, Milhollan JK, Eichinger D, Davin LB, Lewis NG. (2007) A pinoresinol-lariciresinol reductase homologue from the creosote bush (Larrea tridentata) catalyzes the efficient in vitro conversion of p-coumaryl/coniferyl alcohol esters into the allylphenols chavicol/eugenol, but not the propenylphenols p-anol/isoeugenol. Arch Biochem Biophys 465: 209–218 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang CY, Wang SY, Yin JJ, Parry J, Yu LL. (2007) Enhancing antioxidant, antiproliferation, and free radical scavenging activities in strawberries with essential oils. J Agric Food Chem 55: 6527–6532 [DOI] [PubMed] [Google Scholar]
- Zabetakis I, Holden MA. (1997) Strawberry flavour: analysis and biosynthesis. J Sci Food Agric 74: 421–434 [Google Scholar]
- Zorrilla-Fontanesi Y, Rambla JL, Cabeza A, Medina JJ, Sánchez-Sevilla JF, Valpuesta V, Botella MA, Granell A, Amaya I. (2012) Genetic analysis of strawberry fruit aroma and identification of O-methyltransferase FaOMT as the locus controlling natural variation in mesifurane content. Plant Physiol 159: 851–870 [DOI] [PMC free article] [PubMed] [Google Scholar]