SUMMARY
Sinorhizobium meliloti is a soil bacterium that invades the root nodules it induces on Medicago sativa, whereupon it undergoes an alteration of its cell cycle and differentiates into nitrogen-fixing, elongated and polyploid bacteroid with higher membrane permeability. In Caulobacter crescentus, a related alphaproteobacterium, the principal cell cycle regulator, CtrA, is inhibited by the phosphorylated response regulator DivK. The phosphorylation of DivK depends on the histidine kinase DivJ, while PleC is the principal phosphatase for DivK. Despite the importance of the DivJ in C. crescentus, the mechanistic role of this kinase has never been elucidated in other Alphaproteobacteria.
We show here that the histidine kinases DivJ together with CbrA and PleC participate in a complex phosphorylation system of the essential response regulator DivK in S. meliloti. In particular, DivJ and CbrA are involved in DivK phosphorylation and in turn CtrA inactivation, thereby controlling correct cell cycle progression and the integrity of the cell envelope. In contrast, the essential PleC presumably acts as a phosphatase of DivK. Interestingly, we found that a DivJ mutant is able to elicit nodules and enter plant cells, but fails to establish an effective symbiosis suggesting that proper envelope and/or low CtrA levels are required for symbiosis.
INTRODUCTION
Caulobacter crescentus and Sinorhizobium meliloti belong to the class of Alphaproteobacteria, which includes plant endosymbionts (e.g., Rhizobium, Sinorhizobium, Mesorhizobium and Azorhizobium), animal pathogens (e.g., Brucella, Rickettsia) and plant pathogens (e.g., Agrobacterium). Sinorhizobium meliloti, one of the most intensively studied of these organisms, is able to elicit the formation of nodules on the roots of plants of the genera Medicago, Melilotus and Trigonella (Horvath et al., 1986). S. meliloti induces nodule formation, invades plant cells in the interior of the nodule and then undergoes a cellular differentiation process in order to become a nitrogen-fixing bacteroid. In this differentiation, the cells become elongated and polyploid as a result of endo-reduplication of the genome, which suggests that a cell cycle change may be inherent to the differentiation process (Mergaert et al., 2006; Kobayashi et al., 2009; Van de Velde et al., 2010; Wang et al., 2010).
The cell cycle machinery responsible for DNA replication, cell division, and morphogenesis of polar structures is the engine of every organism and has been extensively studied in C. crescentus (reviewed by Curtis and Brun, 2010). Many factors are known to regulate cell cycle progression, most of which are members of the family of two-component signal transduction proteins, which is comprised of histidine kinases and their response regulator substrates. Among these, the essential response regulator CtrA is the master regulator and its activity varies as a function of the cell cycle (Quon et al., 1996; Laub et al., 2002).
In C. crescentus, CtrA regulates gene expression of key players in the cell cycle and other processes, and it also blocks DNA replication by binding the origin of replication and thus making it inaccessible to the replication initiation factors. The regulon directly controlled by CtrA comprises genes involved in cell division (ftsZ, ftsA, ftsQ and ftsW), proteolysis (clpP), DNA methylation (ccrM), flagellar biogenesis (e.g. flgBC, fliE and fliLM), stalk biogenesis (tacA), pili biogenesis (pilA), and chemotaxis (Skerker and Shapiro, 2000; Wortinger et al., 2000; S E Jones et al., 2001; Laub et al., 2002; Biondi, Jeffrey M Skerker, et al., 2006; Collier et al., 2007). The essential role of CtrA has also been demonstrated in other Alphaproteobacteria, such as Brucella (Bellefontaine et al., 2002) and S. meliloti (Barnett et al., 2001), while in several other species, cells can survive without CtrA. In these cases, this protein only controls dispensable functions, such as motility and chemotaxis (e.g. in Rhodospirillum and Magnetospirillum) (Bird and MacKrell, 2011; Greene et al., 2012).
In C. crescentus, CtrA activity peaks at the predivisional stage (Domian et al., 1997), thanks to a combination of transcriptional, proteolytic and phosphorylation control. CtrA is activated through phosphorylation in a cell-cycle dependent fashion; this is accomplished by an essential phosphorelay, comprised of the hybrid histidine kinase CckA and the histidine phosphotransferase ChpT (Biondi, Reisinger, et al., 2006). ChpT can also shuttle the phosphate from CckA to CpdR, a second response regulator that, together with RcdA, is involved in CtrA proteolysis mediated by the ClpP-ClpX protease (Jenal and Fuchs, 1998; Hung and Shapiro, 2002; Ryan et al., 2002; Ryan et al., 2004; McGrath et al., 2006; Iniesta et al., 2006). The phosphorylated response regulator DivK promotes cell cycle progression because it acts at the top of the phosphorelay, interrupting the phosphate flow towards CtrA and thus promoting DNA replication (Hecht et al., 1995; Wu et al., 1998).
Two histidine kinases, DivJ and PleC, are known to interact with DivK. DivJ plays a role in controlling the length and location of the stalk and the cell division plane (Ohta et al., 1992), while a null Caulobacter pleC mutant produces almost symmetric cells at division and shows abnormal polar development (Burton et al., 1997). Phosphorylated DivK also acts as an allosteric activator for DivJ and PleC, triggering PleD-dependent production of cyclic-di-GMP, which ultimately modulates CtrA proteolysis in the stalked compartment (Paul et al., 2008; Abel et al., 2011). In Caulobacter, DivJ and PleC are the principal kinase and phosphatase of DivK, respectively (Wheeler and Shapiro, 1999). It should be noted that, although DivK has an essential role and its activation by phosphorylation is crucial, the non-essentiality of DivJ and PleC in C. crescentus is still inexplicable.
In other Alphaproteobacteria, histidine kinases similar to DivJ/PleC have been described, such as CbrA and PleC in S. meliloti and PdhS in B. abortus (Gibson et al., 2006; Hallez et al., 2007; Gibson et al., 2007; Mignolet et al., 2010; Fields et al., 2012; Sadowski et al., 2013). Although two-hybrid experiments have shown that PdhS binds DivK in Brucella, no direct biochemical demonstration have been provided yet for the other species. Recently, CbrA has been connected to the positive control of DivK phosphorylation in S. meliloti (Sadowski et al., 2013), as it is positively responsible for the control of DivK localization, which in turn depends on its phosphorylation state. The investigation of the cell cycle’s genetic architecture in Alphaproteobacteria has been recently explored using bioinformatics, revealing the conservation of the regulatory network of CtrA and DivK in Caulobacterales and the Rhizobiales (Brilli et al., 2010), although no direct experimental evidence has been provided.
Here we studied the S. meliloti phosphorylation system, consisting of several putative kinases, that controls the essential cell cycle factor DivK. We integrated both in vivo and in vitro approaches to dissect its architecture and understand its function. Our results indicate that the kinases involved in phosphorylation/dephosphorylation of DivK are essential in S. meliloti, a major difference with respect to Caulobacter despite the similarities concerning their cell cycle networks. In addition to the defects in the cell cycle caused by loss of DivJ, we show that the absence of DivJ strongly affects the ability of Sinorhizobium meliloti to function as an efficient symbiont of M. sativa, suggesting a link between cell cycle regulators and symbiosis.
RESULTS AND DISCUSSION
DivJ in S. meliloti is involved in cell cycle regulation
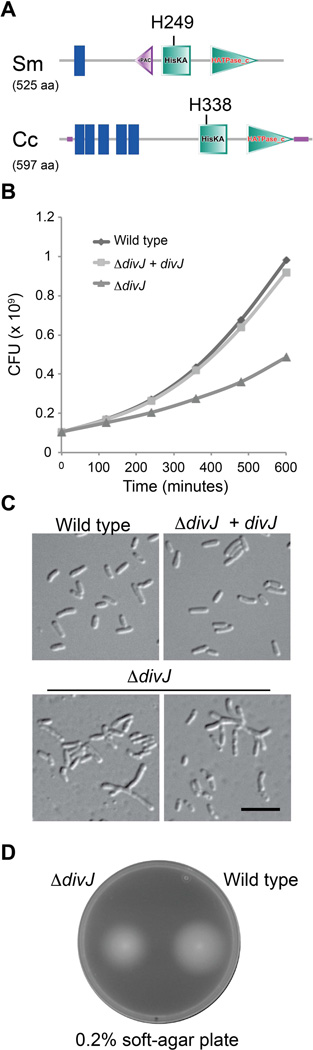
In S. meliloti, the putative DivJ ortholog is a histidine kinase that is anchored to the membrane and has a sensor region that is divergent from that of the C. crescentus DivJ. Instead of having several membrane spanning domains, the sensor region of S. meliloti DivJ only contains one (Fig. 1A). In order to study its function, we constructed a S. meliloti strain carrying the deletion of the gene SMc00059, encoding DivJ (Hallez et al., 2004; Brilli et al., 2010). The ΔdivJ (BM253) mutant was viable, but it showed a severe reduction of its doubling time (Fig. 1B). We confirmed the deletion by PCR and excluded the possibility that the phenotypes were caused by polar mutations by using the phage ΦM12 (Finan et al., 1984) to transduce the deletion cassette from BM253 into a strain carrying a plasmid-borne divJ+ and showing that the divJ+ plasmid is indeed able to fully complement all the mutant phenotypes (Fig. 1C). Most of the cells of ΔdivJ were abnormally shaped (long, branched or short morphologies > 60 %, sampling of 100 cells) and in particular we observed a branched phenotype in 10% of the cells (Fig. 1C), which usually suggests cell division and polarity defects. As in C. crescentus, S. meliloti ΔdivJ cells were still motile (assayed by soft agar plates and directly observed by light microscopy, Fig. 1D). The slightly smaller halo of the divJ mutant in the soft agar could be due to the slower growth of the mutant and/or the branched phenotype of cells, which usually retards the motility. Confirming the functional annotation, the putative divJ of S. meliloti was able to complement deletion of divJ in C. crescentus. In particular, the C. crescentus ΔdivJ growth defect was rescued by expressing S. meliloti divJ, which resulted in a change from a doubling time in rich medium of 140 ± 10 min. to 102 ± 8 min (derived from three independent growth curves in each strain), the same as the wild type doubling time of ca. 100 ± 5 min. Moreover the overall morphology of this complemented strain (Fig. 2A) closely resembled that of the wild type cells (cell length corresponding to 90% ± 10% of wild type cells), as compared to ΔdivJ cell (180% ± 20% of wild type, analyzing 100 cells) and stalk length (120% ± 15% of wild type), as compared to ΔdivJ (240% ± 20% of normal stalks, analyzing 100 stalked cells).
Figure 1. S. meliloti ΔdivJ is viable but shows a cell cycle phenotype.
A. Schematics of domain organization of DivJs in C. crescentus and S. meliloti. Blue bars are the predicted transmembrane regions, the pink triangle is a predicted PAC domain, green squares are the HisKA domains that include the phosphorylated histidine residue, purple horizontal lines are intrinsically disordered regions and finally the HATPase_c domains are the green triangles (analysis performed using SMART database) (Letunic et al., 2102); B. Colony Forming Units (CFU) of wild type, ΔdivJ (BM253), ΔdivJ + divJ (BM224). Doubling time (30°C, 180rpm) of BM224 is 200 ± 15 min (similar to wild type cells, 190 ± 13 min), while BM253 doubling time is 284 ± 21 min (standard errors). In figure S7 the same curve is represented in logarithmic scale; C. Cell morphology of the S. meliloti wild type, divJ mutant and ΔdivJ + divJ. Black bar corresponds to 4 µm; D. Soft agar swarmer assay (wild type is 5.6 ± 0.2 cm, while ΔdivJ is 5.4 ± 0.3 cm after 5 days, standard errors).
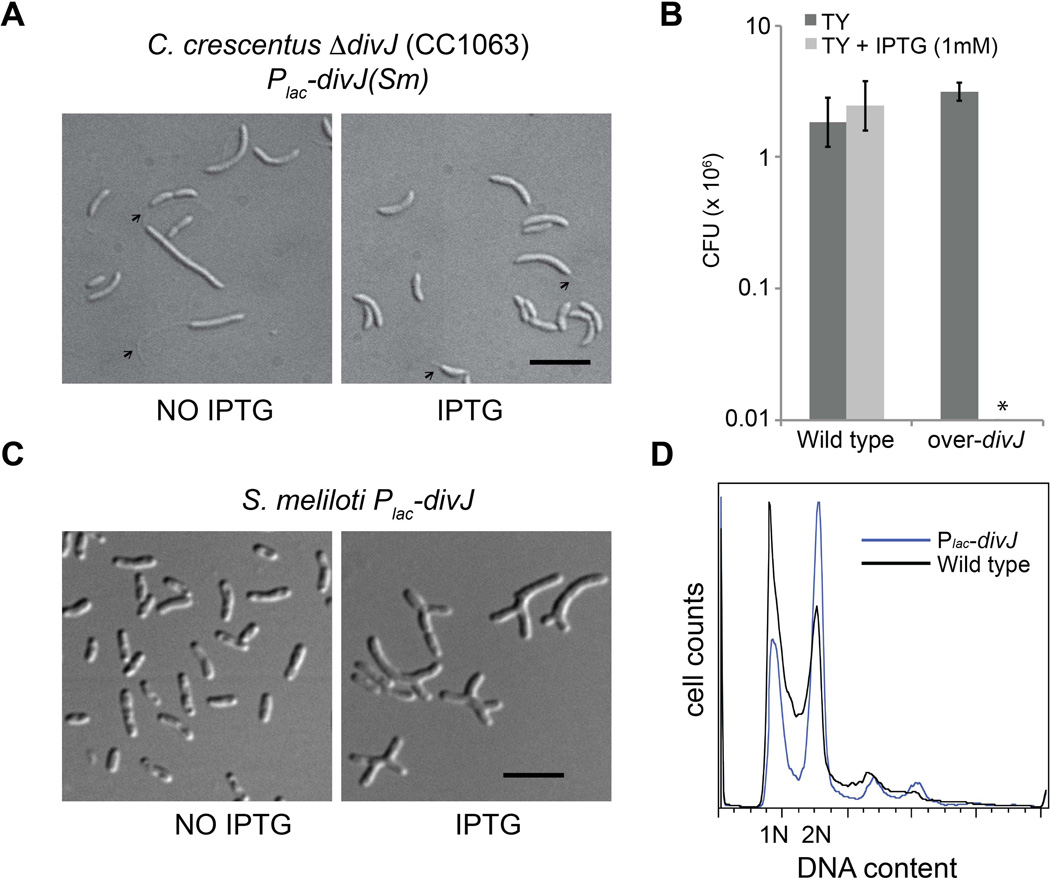
Figure 2. Complementation of C. crescentus ΔdivJ by the S. melilotigene.
A. Morphology of C. crescentus ΔdivJ (Skerker et al., 2005) (BM331) and ΔdivJ complemented (BM333) by an IPTG-inducible copy of S. meliloti divJ (100 µM IPTG). Black bar corresponds to 4 µm. Small black arrows indicate stalks. The presence of S. meliloti divJ was indeed able to partially rescue the growth defect and the abnormal morphology of C. crescentus ΔdivJ as shown by normal cell morphologies (see text for details). Also the C. crescentus ΔdivJ containing the empty vector (BM331) in IPTG conditions did not show any complementation (Data not shown); B. CFUs of over-divJ (BM317) in comparison with wild type cells containing the empty over-expression vector; Cells of cultures grown for 4 hours with or without IPTG, were plated at different dilutions (minimum detectable CFU/ml is 104 cells) without IPTG in order to measure the viability (CFU). Clearly the overexpression of divJ (IPTG) shows a CFU/ml < 104; C. Morphology of over-divJ. D. FACS analysis of over-divJ in comparison with wild type cells.
Next we tested the effect of divJ overexpression in S. meliloti by constructing a strain in which divJ was under the control of an IPTG inducible Plac promoter (pSRK derivatives) (Khan et al., 2008) (BM317). Overexpression of divJ caused a severe growth defect as implied by the absence of colony forming units in TY medium with IPTG (Fig. 2B). The strain overexpressing DivJ also showed an elongated cell morphology suggesting a negative effect on cell division (Fig. 2C). Finally, we checked alterations of the DNA content by using flow cytometry analysis (Fig. 2D). This investigation revealed that, after 4 h of overexpression of divJ, cells with two genome copies accumulated in comparison with wild type, suggesting a block of cell division at the G2 stage. Overexpression of divJ(H249A) (EB775), which is mutated in the conserved histidine putatively required for phosphorylation showed no over-expression phenotypes, suggesting that the histidine in position 249 is necessary for DivJ activity (data not shown). divJ(H249A) was also unable to complement the deletion phenotype of the divJ mutant (data not shown). As we were unable to obtain a good preparation of DivJ antibodies, we cannot exclude the formal possibility that that instability of the DivJ(H249A) mutant protein may be responsible for the absence of an overexpression phenotype, but we consider this to be very unlikely.
The observation that PleC, the phosphatase that dephosphorylates DivK in C. crescentus, is essential in S. meliloti (Fields et al., 2012) suggests that severity of divJ overexpression may be because higher levels of DivK phosphorylation are not well-tolerated in S. meliloti. We speculated then that deletion of pleC should be similar to overexpression of divJ, and that the over-expression of divJ is lethal in S. meliloti due to the high levels of DivK-P. This explanation requires that DivJ would be able to transfer phosphate groups to DivK.
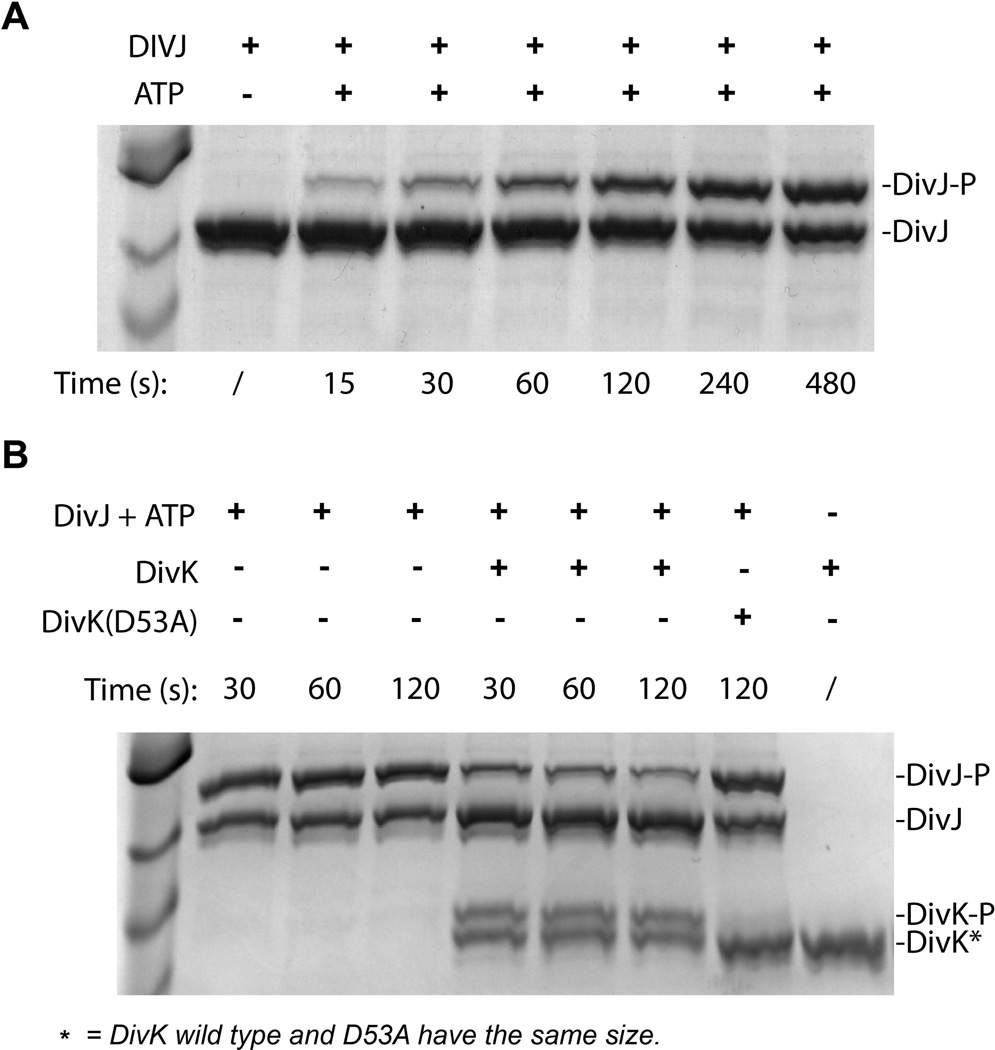
In order to confirm that DivJ is in fact a histidine kinase (HK) able to phosphorylate DivK, we purified its HK domain, as predicted by SMART database and used it for phosphorylation biochemical assays. After incubating the DivJ HK domain with ATP, we were able to separate the phosphorylated form of DivJ-HK by Phos™-Tag SDS-PAGE electrophoresis. The phosphorylated form of DivJ-HK accumulated over time indicating auto-kinase activity (Fig. 3A). This auto-phosphorylation is dependent on the presence of the histidine residue H249 since mutation of this residue abolished the autokinase activity (Fig. S1). In order to test the ability of DivJ-P to transfer phosphate to DivK, we removed ATP after DivJ-P had accumulated and added purified DivK or DivK(D53A), incubating at different time points. Results in Fig. 3B showed that DivJ can phosphorylate DivK and that the predicted aspartate receiver residue is required for phosphotransfer. These in vitro phosphotransfer experiments indicate that purified DivJ-HK is able to autophosphorylate and transfer the phosphate to DivK, and that both conserved sites of phosphorylation (H249 and D53 respectively) are required.
Figure 3. DivJ is a kinase that phosphorylates DivK in vitro.
A. Purified DivJ histidine kinase domain is able to auto-phosphorylate using ATP as the phosphate source. DivJ in presence of ATP gives two distinct bands in a SDS-PAGE Phos-tag™ gel (Coomassie blue). In particular the amount of phosphorylated band (upper band) increases over time; B. The mutant D53A is not able to receive the phosphate from DivJ.
DivJ represses CtrA phosphorylation and activity in S. meliloti
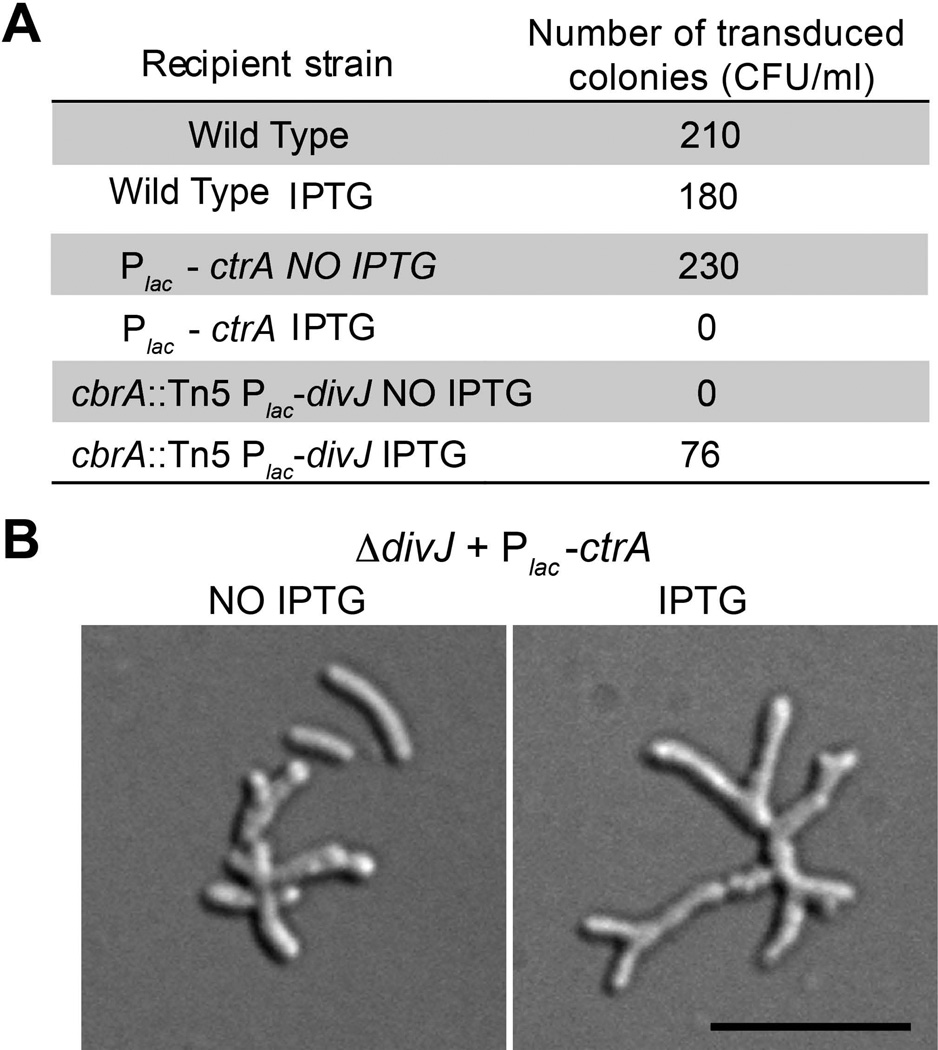
In C. crescentus, DivK inhibits CtrA via DivL/CckA (Biondi, Reisinger, et al., 2006; Tsokos et al., 2011) and triggers c-di-GMP production via PleD (Paul et al., 2008; Abel et al., 2011). If DivJ/DivK function to inhibit CtrA phosphorylation in S. meliloti, deletion of divJ should lead to a more severe/lethal phenotype when CtrA levels are increased. Using an M12 phage lysate of strain BM253 (ΔdivJ carrying the resistance cassette for tetracycline), we attempted to transduce the divJ deletion, into a strain with ctrA under an inducible promoter (BM240), creating the strain BM264. Transductants were recovered only without IPTG in the selective medium (Fig. 4A) indicating that a strain carrying a deletion of divJ does not tolerate high levels of CtrA. We further analyzed the strain BM264 grown first without IPTG and then switched to a medium supplemented with IPTG; the strain developed a highly branched and elongated phenotype confirming that overproduction of CtrA results in severe cell cycle defect(s) (Fig. 4B).
Figure 4. High CtrA levels are lethal in combination with ΔdivJ.
A. Transduction table, in which over-expression of ctrA (IPTG) in combination with the ΔdivJ is lethal; B. Morphology of S. meliloti strain BM264 (ΔdivJ + over-ctrA) with and without induction by 1 mM IPTG. The black bar is 3 µm.
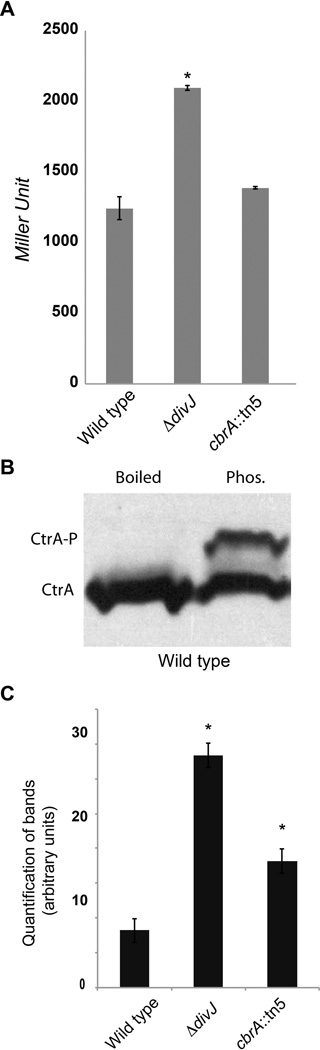
To investigate this further, we measured the expression levels of the pilA promoter in a ΔdivJ strain in comparison with wild type cells. In C. crescentus, pilA expression is directly controlled by CtrA (Skerker and Shapiro, 2000). We measured the expression of pilA by fusing a pilA promoter to the lacZ gene and measuring β-galactosidase activity. Our results (Fig. 5A) demonstrate higher levels in the divJ mutant (EB638), suggesting that CtrA is also more active, consistent with the model of DivJ and DivK inhibiting CtrA activity.
Figure 5. DivJ and CbrA are inhibiting CtrA phosphorylation and activity in vivo.
A. β-galactosidase activity of a CtrA-controlled promoter in wild type (EB594), ΔdivJ (EB638) and cbrA∷ Tn5 (EB593) genetic backgrounds. Experiments were performed in biological triplicates, errors are calculated as standard deviations. Asterisk corresponds to significant statistical difference with wild type conditions (Student’s test); B. Phos-tag™ SDS-PAGE gel shows phosphorylation of CtrA in vivo in wild type. A lane with SDS-lysed material (Phos.) and another with boiled sample are shown. The phosphorylated band disappears after boiling; C. Quantification of phosphorylation levels of CtrA in vivo in wild type, ΔdivJ and cbrA∷ Tn5 genetic backgrounds. The average of three experiments using samples at the same OD600 is showed, errors are calculated as standard deviations. The amount of CtrA-P was normalized for the number of cells. Asterisk corresponds to significant statistical difference with wild type conditions (Student’s test).
We further tested this model by measuring phosphorylation levels of CtrA in different genetic backgrounds. In order to quantify CtrA-P levels in vivo, we used the Phos-Tag system in combination with immunoblots with anti-CtrA antibodies (Fig. 5B). To the best of our knowledge, is the first time in the S. meliloti field that in vivo measurements of phosphorylation of a protein have been successfully performed. Cell lysates are loaded on SDS-Page electrophoresis gels and, in contrast to measurements using radioactivity, no specific culture medium is required, as Phos-Tag detects unlabeled wild type proteins. We measured levels of CtrA-P (Figure 5C) in three biological replicates of wild type, ΔdivJ and cbrA∷ Tn5 cells (this latter case is discussed in the following sections). Consistent with the increased activity of the CtrA-controlled promoter of pilA, levels of phosphorylated CtrA were significantly increased in the ΔdivJ strain compared to wild type.
In summary, the results discussed in this section show that DivJ, which is able to phosphorylate DivK in vitro, is also required for down-regulation of CtrA phosphorylation and subsequently its activity as transcriptional activator. However, this raises the question of whether DivJ is the only histidine kinase controlling DivK phosphorylation in S. meliloti.
In silico analysis of histidine kinases predicted to interact with DivK in S. meliloti
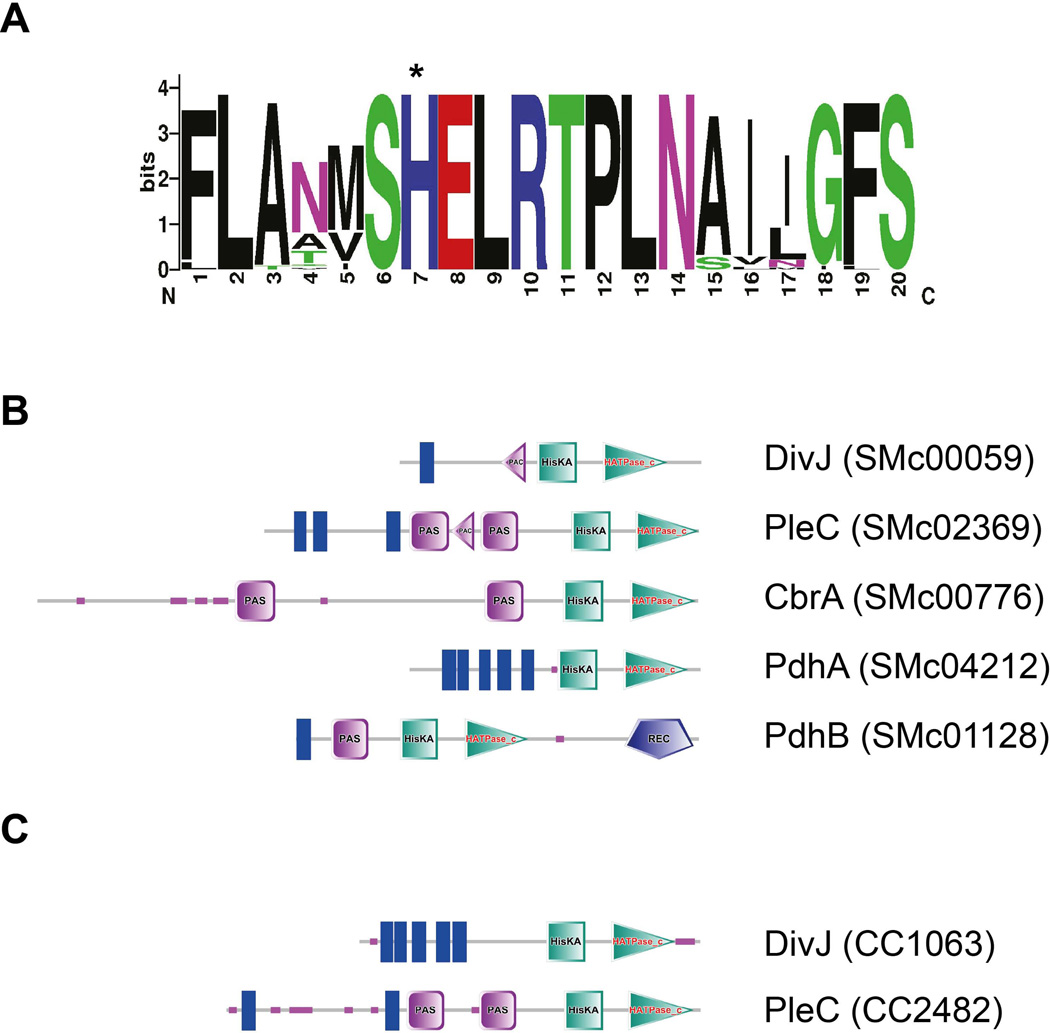
We employed an in silico strategy to identify other genes in S. meliloti (extended also to genomes of other Alphaproteobacteria) that encode for proteins that belong to the family of histidine kinases that controls DivK phosphorylation and dephosphorylation. This family was named pleC/divJ homolog sensor family (PdhS) as previously suggested (Hallez et al., 2004). In order to predict the kinases interacting with a response regulator, we took advantage of a previous analysis that defined the regions of the histidine kinase that make contact with the response regulator and that are responsible for the specificity of this interaction (Skerker et al., 2008). This approach was integrated with the hypothesis that all Alphaproteobacterial DivK and PleC proteins are able to interact with DivK (Brilli et al., 2010). The fragment of the HK responsible for the specific interaction with the response regulator DivK comprises helix 1 and helix 2 of the two-helix bundle that surrounds the histidine residue (Ohta and Newton, 2003; Skerker et al., 2008). Helix 1 of the C. crescentus DivJ corresponds to residues 332 to 351 and helix 2 corresponds to 369 to 395. Results of the alignment of DivJs and PleCs are shown in figure S2. From this alignment, in which we used both helices, we derived a probability model describing the variability at each position of the most conserved helix (helix 1) in DivJ and PleC proteins from organisms possessing DivK (Fig. 6A). We scanned for HKs in Alphaproteobacteria genomes using a probability matrix that allowed us to assign a score to each of them, while a threshold chosen to include known DivK partners allowed identifying additional putative DivK interactors. Notably, S. meliloti CbrA (Gibson et al., 2007) and the B. abortus PdhS (Hallez et al., 2007), which have been hypothesized to interact with DivK, were in fact detected with this bioinformatic analysis. The list of accession numbers of PdhS family members is shown in table S1. S. meliloti showed five PdhS kinases including CbrA (Gibson et al., 2006), DivJ and PleC (Fields et al., 2012) and two other histidine kinases putatively belonging to the PdhS family that we named PdhSA and PdhSB (Fig. 6B), SMc04212 and SMc01128 respectively.
Figure 6. In silico analysis of histidine kinases of the PdhS family interacting with DivK.
A. PdhS (PleC DivJ Homolog) family specificity consensus based on the alignment in figure S2. Asterisk corresponds to the phosphorylated histidine; B. PdhS-family members in S. meliloti; domains were predicted by SMART (Letunic et al., 2012); C. Domains organization of DivJ and PleC in C. crescentus predicted by SMART (Letunic et al., 2012). Blue bars are the predicted transmembrane regions, the pink squares are predicted PAS domains, the pink triangles are predicted PAC domains, green squares are the HisKA domains that include the phosphorylated histidine residue, purple horizontal lines are intrinsically disordered regions and finally the HATPase_c domains are the green triangles (analysis performed using SMART database) (Letunic et al., 2012).
DivJ and CbrA are in vivo kinases of DivK while PleC acts as a phosphatase
Our prediction identified 5 putative histidine kinases able to interact with DivK, but are those proteins really involved in control of DivK phosphorylation? Previous studies showed that CbrA controls DivK localization by controlling the phosphorylation of DivK (Sadowski et al., 2013). Several other altered phenotypes of the cbrA null mutant were reported, such as abnormal EPS production and nodulation defects in alfalfa plants (Gibson et al., 2006; Gibson et al., 2007). PleC is essential in S. meliloti, influencing the septum localization and interactions with PodJ (Fields et al., 2012), but no evidence of DivK control by PleC has ever been provided. Mutants of PdhSA and PdhSB, previously generated by mini Tn5 mutagenesis (Pobigaylo et al., 2006), were viable and did not show any abnormal growth or cell cycle phenotype (data not shown). Hence these two latter factors were not analyzed further and we focused on the putative interactions of DivJ, PleC and CbrA with DivK in vivo.
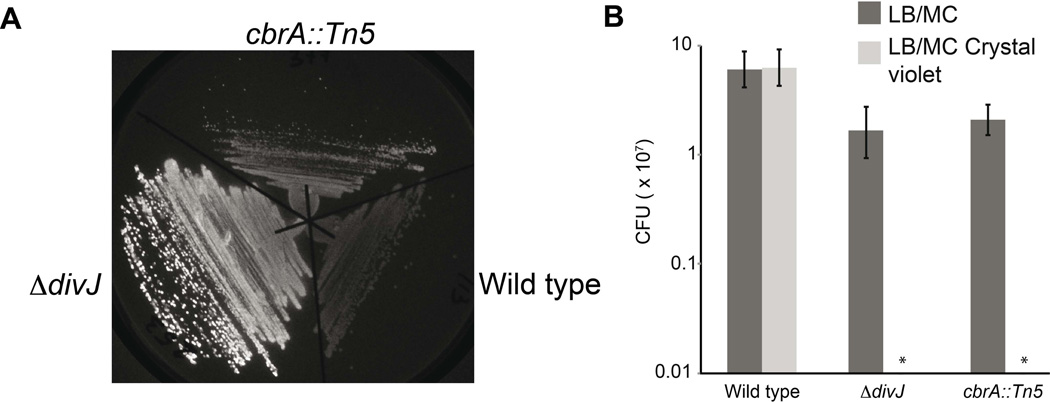
First we compared the phenotypes of the cbrA and the divJ mutants. We tested the ability to bind calcofluor (Gibson et al., 2006), revealing that like the cbrA∷ Tn5 strain (KEG2016), the ΔdivJ strain is brighter than wild type; in fact, the divJ deletion is much brighter than the cbrA∷ Tn5 strain (Fig. 7A). Since calcofluor is an indicator of alterations in envelope composition we tested the integrity of the cell envelope /resistance to osmotic stresses of ΔdivJ by assaying the sensitivity of ΔdivJ to the hydrophobic dye crystal violet in comparison with the cbrA∷ Tn5 and wild type cells. Both mutant strains were unable to form single colonies in LB supplemented with the crystal violet, while wild type cells could survive, suggesting an alteration of the cell envelope composition (Fig. 7B). This is interesting because permeability of membranes and resistance to oxidative stress are important factors during the infection of legume hosts (Sharypova et al., 2003; Campbell et al., 2003).
Figure 7. Envelope-related phenotypes of ΔdivJ and cbrA∷Tn5.
A. Calcofluor staining of deletion of divJ in comparison with wild type and cbrA∷ Tn5reveals that both cbrA and divJ mutants are brighter than wild type cells; B. Viable counts of the deletion of divJ in comparison with wild type and cbrA∷ Tn5 with Crystal violet dye revealed that both divJ and cbrA mutants are sensitive to crystal violet; Cells were plated at different dilutions in LB/MC or LB/MC plus crystal violet in order to measure the viability (CFU) (minimum detectable CFU/ml is 105 cells). Clearly ΔdivJ and cbrA∷ Tn5 in LB/MC crystal violet show a CFU/ml < 105.
In order to gain more information about the functions controlled by DivJ, transcriptome profile analysis of the divJ mutant was performed and compared with the transcriptome profile of cbrA∷ Tn5 (Gibson et al., 2007). We first determined genes differentially expressed in the divJ mutant compared to the wild type (Table 1). The analysis revealed genes that had altered expression in the divJ mutant compared with wild type cells; log2 ratios of the mutant vs. wild type are shown. A total of 16 genes were lower in the ΔdivJ cells, including several flagellar genes (fliE, flgG, flaA and flab), as well as chemotactic genes (mcpU, mcpZ and cheR) and genes encoding putative manganese transporters (sitB and sitC). Also four genes encoding conserved hypothetical proteins, a putative transcription factor gene of the family of merR, and gcvT, possibly involved in catabolism of glycine were down regulated. Eighteen genes were more highly expressed in the ΔdivJ cells, ten of which code for hypothetical proteins. Among the genes with an assigned function, feuP and five FeuP-controlled genes (Smb20838, SMc00198, Smc01557, SMc01586 and ndvA), and the exoN2 and pilA genes, the latter encoding a pilin subunit, were upregulated. FeuP has previously been shown to control several genes such as SMc00198, SMc03900 (ndvA), SMc01586, SMc01557 that are required for cyclic glucan export and symbiosis (Griffitts et al., 2008).
Table 1.
Transcriptome profile of the S. meliloti divJ deletion with log ratios in comparison with wild type (see Experimental Procedures)
| Gene code (Rm1021) |
Annotation | Log2-Ratio | CbrA array* | CtrA bs$ |
|---|---|---|---|---|
| Genes with expression reduced in ΔdivJ | ||||
| SMa0281 | Putative regulator, MerR family | −0.61 | ||
| SMc00888 | Response regulator | −0.99 | y (−) | y |
| SMc00765 | mcpZ | −0.65 | y (−) | |
| SMc00975 | mcpU | −0.52 | y (−) | |
| SMc03009 | cheR | −0.63 | y (−) | |
| SMc02047 | gcvT | −0.51 | ||
| SMc02507 | sitC | −0.54 | ||
| SMc02508 | sitB | −0.64 | ||
| SMc03029 | fliE | −0.58 | y (−) | |
| SMc03030 | flgG | −0.72 | y (−) | |
| SMc03037 | flaA | −0.51 | y (−) | y |
| SMc03038 | flaB | −0.78 | y (−) | |
| SMc02104 | Conserved hypothetical protein | −0.57 | ||
| SMc00360 | Conserved hypothetical protein | −0.62 | y (−) | y |
| SMc03013 | Conserved hypothetical protein | −0.66 | y (−) | |
| SMc03057 | Conserved hypothetical protein | −0.51 | y (−) | |
| Genes with expression increased in ΔdivJ | ||||
| SMb20838 | putative secreted calcium-binding protein |
0.57 | y (+) | |
| SMc00949 | Conserved hypothetical protein | 0.76 | y (+) | |
| SMc01557 | Hypothetical signal peptide protein | 0.74 | y (+) | |
| SMa1043 | Hypothetical protein | 1.45 | ||
| SMb21069 | Hypothetical protein | 2.16 | ||
| SMb21440 | Hypothetical protein | 1.16 | y (+) | |
| SMc00198 | Hypothetical protein | 0.59 | y (+) | |
| SMc01586 | Hypothetical protein | 0.75 | y (+) | |
| SMc03999 | Hypothetical protein | 0.65 | ||
| SMc02051 | Conserved hypothetical protein | 0.56 | y | |
| SMc02052 | Conserved hypothetical protein | 0.56 | ||
| SMc02266 | Conserved hypothetical protein | 0.83 | y (+) | |
| SMc02900 | Conserved hypothetical protein | 0.96 | ||
| SMc03100 | Conserved hypothetical protein | 0.66 | ||
| SMc00458 | feuP | 1.48 | ||
| SMc03900 | ndvA | 0.54 | y (+) | |
| SMc04023 | exoN2 | 0.55 | ||
| SMc04114 | pilA1 | 0.54 | y | |
y(−) means that the same gene was downregulated in the cbrA mutant arrays (Gibson et al., 2007); y(+) means that the gene was upregulated also in the cbrA arrays.
= bs., binding site, the prediction is based on Brilli et al., 2010.
A selection of genes suggested to be differentially expressed by the transcriptome data were tested in wild type and ΔdivJ by fusing the lacZ reporter gene to the promoter regions of these genes. We measured β-galactosidase activity in these strains and the results confirmed our transcriptome data (Fig. S3).
Since we had discovered that CtrA activity is higher in a ΔdivJ strain, (Fig. 5), it was interesting to find that several genes upregulated in this mutant are preceded by a putative CtrA binding site (Brilli et al., 2010). This included the pilA promoter whose expression levels are higher in the ΔdivJ strain. This observation is consistent with the discovery that CtrA is upregulated in the ΔdivJ mutant, although the presence of the consensus CtrA site does not establish a regulatory role.
The limited number of genes discovered by the transcriptomic analysis could be explained by the hypothesis that the effects of DivJ may be limited to a short portion of the cell cycle, in which case transcriptional differences in the window of DivJ activity will be blurred in a mixed population.
Many of the genes (19 genes out of 34) putatively controlled by DivJ were also found to be influenced by CbrA (Gibson et al., 2007), indicating a common pathway between the two histidine kinases possibly involving DivK. This observation is also consistent with the observation that both DivJ and CbrA appear to be involved in CtrA activity repression (Figure 5). Additionally, cbrA∷ Tn5 showed significantly higher levels of CtrA-P (Fig. 5C) suggesting that both DivJ and CbrA participate in similar functions.
Since in C. crescentus DivK is essential, we investigated whether DivK was also essential in S. meliloti. Using a two-step recombination strategy, we first constructed an S. meliloti strain in which divK coding sequence was replaced by tetracycline resistance cassette, complemented by the divK locus including the promoter. We then selected for excision of the integrative plasmid by plating on sucrose medium. We were able to select sucrose resistant colonies only when the complementing plasmid was present, suggesting the essentiality of DivK (data not shown). To gain additional support for the conclusion that DivK is essential in S. meliloti, we also attempted to transduce the divK deletion into several genetic backgrounds as reported in Table S2. Again, we were only successful in introducing the divK deletion when an extra copy of divK was present. These data confirm that is divK is essential in S. meliloti. We confirmed in vivo the importance of the putative phosphorylated site of DivK, the aspartate in position 53 (D53), by overexpressing divK and divK(D53A). Overexpression of divK, but not overexpression of divK(D53A) caused cell cycle defects in S. meliloti (Fig. S4, Fig. S4B). DivK overexpression produced cells with abnormal morphologies, resembling the morphological phenotype produced by DivJ overexpression (Fig. S4A).
Next we tested the hypothesis that DivJ and CbrA were synergic, by attempting to combine the deletion of divJ with the cbrA∷ Tn5 mutant, using a phage lysate produced by infection of BM253. Although cbrA∷ Tn5 is sensitive to phage infection (data not shown), allowing transduction, we could not recover any colonies when we attempted to transduce the divJ deletion allele, while the same transduction with wild type as recipient yielded hundreds of colonies (Fig. 4A). This result suggests that the combination of the divJ and cbrA mutations is lethal in S. meliloti. However, we were able to create a double conditional mutant ΔdivJ and cbrA∷ Tn5 that was able to survive by expressing divJ from an inducible promoter (Plac) in the presence of IPTG, while the transduction without IPTG did not yield any colonies, thereby confirming the lethality of the divJ cbrA double mutation (EB602).
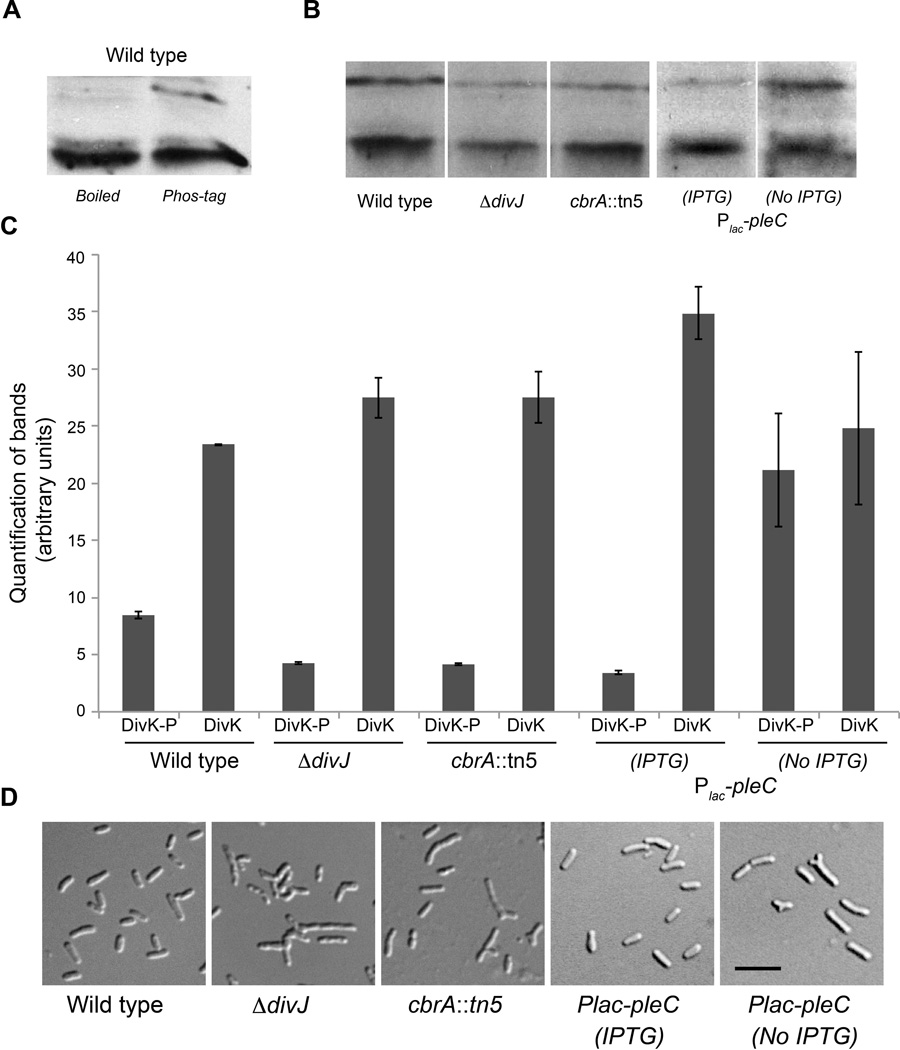
To determine the in vivo activity of each predicted DivK kinase/phosphatase we measured the DivK phosphorylation levels in different backgrounds (ΔdivJ, cbrA∷ Tn5, ΔpleC+Plac-pleC), as described for CtrA, this time using anti-DivK antibodies raised in rabbit. The anti-DivK antibodies were able to detect two bands in Phos-Tag SDS-Page gels, one of which corresponded to the phosphorylated form that disappeared by boiling the sample, which destroys the labile phosphate bond (Fig. 8A) (Barbieri and Stock, 2008).
Figure 8. DivJ and CbrA are required for DivK phosphorylation, while PleC acts as a phosphatase.
A. SDS-PAGE Phos-tag™ gel detects phosphorylation of DivK in vivo in wild type; boiling step (“boiled”), which breaks the phosphate bond, specifically affected the upper band; B. SDS-PAGE Phos-tag™ gel shows phosphorylation of DivK in vivo in wild type, ΔdivJ cbrA∷ Tn5 and pleC depletion (After 7 h) genetic backgrounds; C. Quantification of phosphorylation levels of DivK in vivo in wild type, ΔdivJ cbrA∷ Tn5 and pleC depletion genetic backgrounds. The average of three experiments is shown, errors are calculated as standard deviations; D. Morphologies of cells of ΔdivJ cbrA∷ Tn5 and pleC depletion genetic backgrounds. This latter condition is shown with 1mM IPTG and after washes, without IPTG for 7 hours. The black bar corresponds to 3 µm.
Phosphorylation of DivK in vivo (Fig. 8B), together with previous results, demonstrated that DivJ is a kinase of DivK and also that CbrA is involved in this DivK phosphorylation, as the level of DivK-P dropped by about half in both strains (Fig. 8C). This result is consistent with analyses showed in the previous sections for DivJ and similar to the conclusion recently published for CbrA (Sadowski et al., 2013). It also suggests that, as the combination of DivJ and CbrA mutations is lethal, phosphorylation of the essential factor DivK is also essential in S. meliloti cells.
We also measured in vivo phosphorylation of DivK in the pleC depletion strain (EB601). Using 100 uM IPTG, the pleC depletion strain showed a mild overexpression of PleC, but had DivK phosphorylation levels similar to ΔdivJ. After 6 hours of pleC depletion DivK-P levels were 2 fold higher than wild type, demonstrating that in S. meliloti PleC plays an opposite role of DivJ and is involved in maintaining low levels of DivK-P, as observed in C. crescentus.
Next we tested whether it was possible to rescue the lethal phenotype of ΔpleC by transducing this deletion in cbrA∷ Tn5 or ΔdivJ backgrounds (Table S3). We found that it was only possible to transduce the pleC deletion into a strain carrying the cbrA mutation (EB630). The observation that only CbrA mutation (not DivJ) is able to rescue the lethality of pleC deletion is puzzling but it could be explained by introducing other regulatory levels of these kinases besides the simple contribution to the chemical equilibrium of DivK/DivK-P. For example DivJ, CbrA and PleC could be expressed at different times and/or present in different subcellular locations during the cell cycle. This regulation in time and space could suggest that DivJ is never together PleC while the soluble kinase CbrA could be co-localized with PleC therefore influencing DivK/DivK-P levels at the same time/space as PleC. If this were the case, one possible model is that CbrA functions coordinately with PleC in the same time/space, but that DivJ does not.
In order to test the enzymatic capability of PleC and CbrA to phosphorylate DivK in vitro, as we did for DivJ, we attempted to purify both HK domains. We cloned the HK domains of both PleC and CbrA and expressed them in E. coli cells. PleC was soluble and purified well, as shown in Fig. S5. In contrast, several preparations of CbrA were all insoluble; therefore no in vitro experiments were performed with CbrA. In contrast to the DivJ HK, the preparation of PleC HK did not show any autokinase activity both with either ATP and GTP, suggesting that for PleC the sensor part of the protein and/or specific signals are required to activate this kinase in vitro. Alternatively, the kinase domain may not be well folded.
Nevertheless, taken together the in vivo results, the genetic experiments, the high degree of homology of PleC-CbrA-DivJ, and the recent results with CbrA (Sadowski et al., 2013) strongly support a direct role of CbrA and PleC in controlling DivK-P levels.
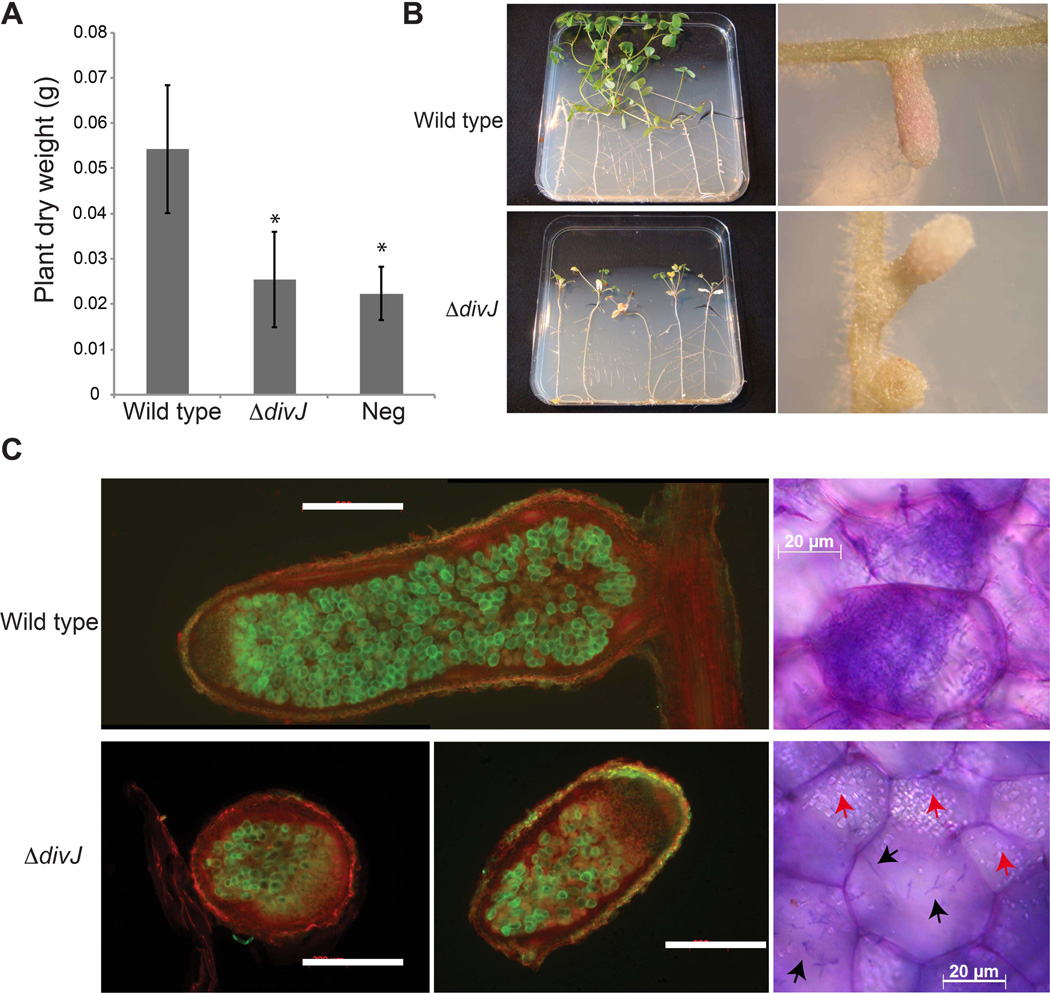
DivJ activity is required for efficient symbiosis
The alteration of the cell cycle that occurs during bacteroid differentiation in the symbiotic process suggests a possible role for cell cycle regulators, a conjecture supported by previous experiments on the cell cycle regulators CbrA (Gibson et al., 2006) and CpdR (Kobayashi et al., 2009). We therefore tested the ability of the divJ mutant (BM253) to nodulate and fix nitrogen in M. sativa (Fig. 9). Plants inoculated with the ΔdivJ mutant had similar appearance and dry weight to non-infected plants, suggesting that nitrogen fixation was impaired (Fig. 9A). The ΔdivJ mutant was able to induce nodule formation but, compared to the nodules elicited by the wild type strain, these were more abundant, smaller, white, and abnormal in shape (Fig. 9B). Therefore we tested if cells lacking DivJ were able to invade the nodule cells. We infected alfalfa plants using GFP-tagged wild type and ΔdivJ strains (Fig. 9C). Both nodules of wild type and ΔdivJ showed GFP signal inside the internal part of the nodule tissue, suggesting infection by bacteria. Sections of nodules containing wild type or ΔdivJ cells were also stained with the bacteria-specific Toluidine blue and observed under the microscope (Fig. 9D) in order to understand if mutants were able to enter the plant cells and their ability to proliferate inside. It was evident that ΔdivJ bacteria were able to infect plant cells inside nodule, however starch accumulation was present, which is usually a sign of inefficient symbiosis. Normally, starch accumulates in root cells before the infection and then when symbiosis is established the granules are quickly metabolized (Hirsch et al., 1983). As expected, the ΔdivJ strain complemented with wild type divJ (BM224) gave a normal symbiotic phenotype (data not shown).
Figure 9. Symbiotic efficiency of ΔdivJ.
A. Histogram with the dry weight of alfalfa plants infected by S. meliloti wild type and ΔdivJ (neg = un-inoculated control). Errors are calculated as standard deviations; B. Pictures of five plants and details on nodules; C. Nodules from an infection of alfalfa plants using GFP-tagged strains (green), wild type is strain Rm1021G and ΔdivJ is BM253G (Table S4); on the left (white bars correspond to 500 µm) and Toluidine blue staining on the right. Black arrows indicate bacteria inside plants cells, red arrows indicate starch granules.
This result suggests that ΔdivJ is not able to infect efficiently alfalfa plants possibly due to high CtrA levels in ΔdivJ cells; high CtrA levels may be responsible for the severe symbiotic defects, impairing the ability of the cell to grow, differentiate, or survive in plant cells. As showed before, CtrA protein and phosphorylation levels are both high in ΔdivJ. Similar results involving a strain with putative high levels of CtrA, were also documented for the null mutant of CpdR, a response regulator required for proper CtrA proteolysis in C. crescentus (Kobayashi et al., 2009).
In order to test this hypothesis that CtrA levels or activity should be low in bacteria infecting plant cells, we isolated bacteroids from mature nitrogen fixing nodules and measured CtrA protein levels by immunoblot (Figure S6). The same number of bacteroids and wild type cells was loaded in the SDS-Page gel. Our results clearly showed that CtrA in bacteroids, although more protein content was loaded, was absent. This observation may explain why the deletion mutant of divJ, the cbrA∷ Tn5 mutant, and also the null mutant of cpdR, which have all high CtrA levels, are compromised in the establishment of an efficient symbiosis.
Conclusions
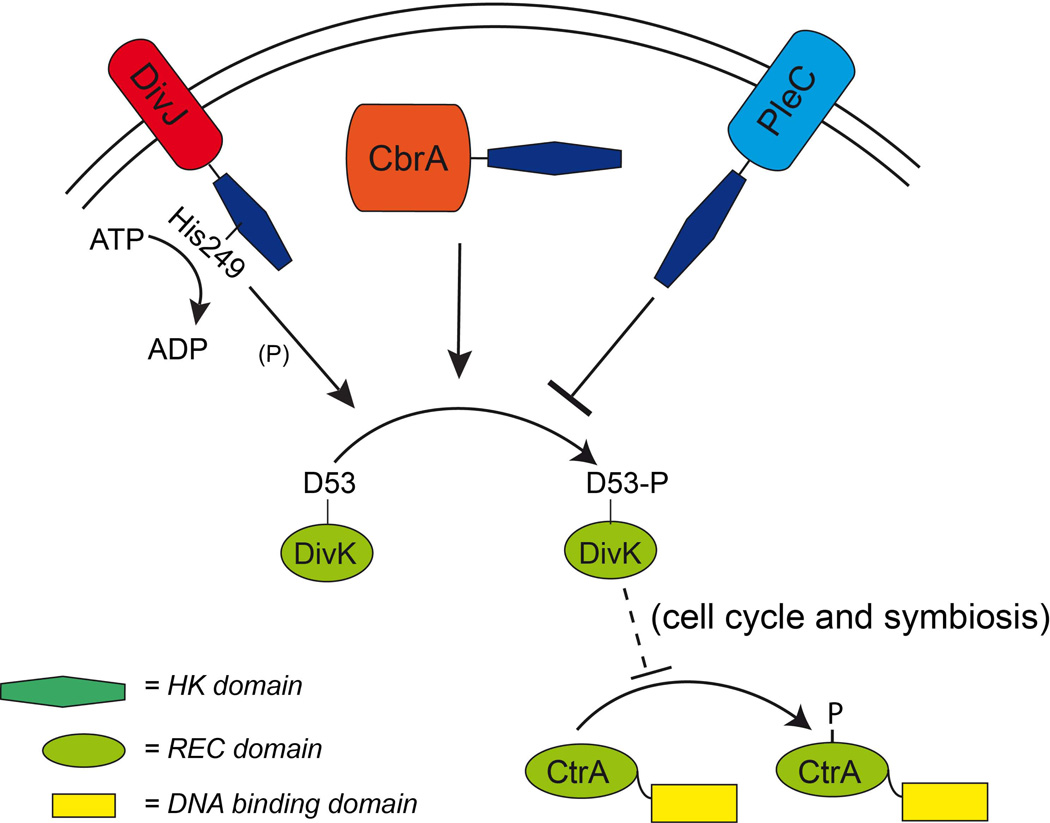
Our biochemical and genetic investigation of the histidine kinase DivJ, and its relationship with CbrA and PleC, sheds light on the DivK cell cycle regulatory module in S. meliloti and unveils an association between cell cycle regulation and symbiosis. We propose here a model (Fig. 10) in S. meliloti where DivJ is a kinase of the essential response regulator DivK. CbrA is also involved in DivK phosphorylation, while PleC, as in C. crescentus, may act as phosphatase. Although the putative cell cycle regulated activity (regulation in time) of CbrA, DivJ and PleC and their subcellular localization (regulation in space) have not been completely investigated, we can hypothesize that these kinases create a complex sensory module that is able to coordinate the phosphorylation levels of the essential factor DivK in time and space.
Figure 10. Functional scheme of DivK control system of CtrA.
DivK is phosphorylated presumably on the aspartate 53 by the membrane histidine kinase DivJ (using the conserved residue H249 and ATP) and likely by the soluble histidine kinase CbrA. The absence of both kinases from S. meliloti is a lethal condition, abolishing DivK phosphorylation. Also the deletion of the membrane histidine kinase PleC is lethal (Fields et al., 2012); results presented here show that PleC is involved in dephosphorylation of DivK. Finally we showed here that DivJ is negatively acting on CtrA and apparently CtrA inactivation is required for an efficient symbiosis, presumably through degradation of the protein. In fact, mature bacteroids do not show detectable CtrA levels, suggesting that one of the symbiotic problems of ΔdivJ may be the high level of activity of CtrA.
As in C. crescentus the phosphorylated DivK acts negatively on CtrA, which in turn plays a positive role for the biogenesis of polar structures and cell division. Unlike in C. crescentus, both DivK phosphorylation and dephosphorylation are essential in S. meliloti. This essentiality is indicated by the lethality of both the divJ-cbrA double mutant and the pleC deletion mutant and our in vivo phosphorylation data. The comparison between S. meliloti and C. crescentus suggests that the genetic architecture that controls cell cycle regulation in Alphaproteobacteria, although similar in all Alphaproteobacteria, also exhibits certain differences, possibly due to different levels of redundancy of feedbacks and regulatory connections. This has been recently observed in Agrobacterium tumefaciens, in which cell cycle regulation shows specific characteristics despite being generally similar to the other Alphaproteobacteria (Kim et al., 2013).
An interesting feature of the cell cycle defects discovered in S. meliloti is the high degree of branching that has been observed when levels of phosphorylated DivK are low. This feature, which is absent in C. crescentus cell cycle mutants, may be related to the polar asymmetric growth of peptidoglycan observed in Rhizobiales (Brown et al., 2012).
The analysis presented here sheds light on the role of DivJ as kinase of DivK, ultimately inhibiting, as in C. crescentus, the CtrA activity as cell cycle transcription factor. However, we cannot exclude that some of the phenotypes we observed in the S. meliloti ΔdivJ may be explained by CtrA abnormally acting on the origin of replication, as described in C. crescentus, or by a reduction of the phosphorylation levels of PleD, which is involved in key steps of cell development in C. crescentus.
Our investigation of the role of the DivK module during symbiosis revealed that bacteroids are deficient of CtrA and strains with putative high CtrA levels, as the ΔdivJ in this study, ΔcbrA (Sadowski et al., 2013) or the CpdR mutant (Kobayashi et al., 2009) are directly impaired in establishing an efficient symbiosis. As mentioned in the introduction, CpdR is a response regulator that is required for CtrA proteolysis in C. crescentus. The S. meliloti cpdR mutant showed the ability to penetrate into the nodule and infect plant cells, but it failed to differentiate in bacteroids. Previous studies indicated that bacteroids have an interrupted cell cycle, associated with the multiplication of the chromosome number, a block of cell division, inducing enlargement of cell bodies, and the consequent loss of the ability to multiply (Mergaert et al., 2006).
The ability of the ΔdivJ mutant to infect alfalfa plant cells and enter the cytoplasm of nodule cells suggest that DivJ is not required in early steps of the infection process outside the roots or inside the infection thread. The symbiotic efficiency, however, is impaired since plants infected by the ΔdivJ mutant are similar in size to non-inoculated ones and the histology of the nodule tissue revealed many starch granules, typical of inefficient nitrogen fixation. Also the low number of bacteria inside plant cells in comparison with plants infected by wild type suggests problems in the inside the infection thread or endocytosis or multiplication inside the plant cell cytoplasm or problems in the differentiation process. Those problems may be related to the growth defects of the divJ mutant observed in the free-living state. Also this symbiotic defect of the divJ mutant could be associated with the phenotypes correlated to envelope integrity we observed in this work, such as increased envelope material detected by calcofluor staining or increased sensitivity to oxidative stresses (Fig. 7). However, combined with previous studies, our results suggest that mutations in the cell cycle factors that play a negative role on CtrA (CpdR, DivJ, CbrA) result in a symbiotic defect. Since, DivJ and CbrA, are involved in the inhibition of CtrA, it appears that a high level of CtrA may interfere negatively with the symbiotic process leading to the speculation that bacteroid differentiation requires the down-regulation of CtrA. Direct support for this hypothesis is provided by our discovery that that mature bacteroids have no CtrA. Perhaps plants are able to block cell cycle of infectious rhizobia by affecting the master regulator CtrA. Proper regulation of CtrA may be required to respond to plant inhibitory activity. Alternatively, one could speculate that strains with lower CtrA activities may show a higher symbiotic activity that could be exploited to increase symbiosis efficiency.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids, cloning and growth conditions
The bacterial strains and plasmids used in this study are described in Table S4. Escherichia coli strains were grown in liquid or solid Luria-Bertani (LB) broth (Sigma Aldrich) (Sambrook et al., 1989) at 37°C supplemented with appropriate antibiotics: kanamycin (50 µg/ml in broth and agar), tetracycline (10µg/ml in broth and agar). S. meliloti strains were grown in broth or agar TY (Beringer, 1974) supplemented when necessary with kanamycin (200 µg/ml in broth and agar), streptomycin (500 µg/ml in broth and agar), tetracycline (1 µg/ml in liquid broth, 2 µg/ml in agar), nalidixic acid (10 µg/ml in broth and agar) as necessary. For negative selection 10% sucrose was added to agar plates. For calcofluor analyses, LB agar was buffered with 10 mM MES (morpholine-ethane-sulfonic acid), pH 7.5, and calcofluor white MR2 Tinopal UNPA-GX (Sigma Aldrich) was added at a final concentration of 0.02%.
For conjugation experiments, 1×109 S. meliloti and 0.5×109 E. coli S17-1 cells (Simon et al., 1983) were used and incubated 24h at 30° C. For creating the deletion of divJ, divK and pleC, two fragments of about 1000-bp long amplifying the upstream (P1-P2) and downstream (P3-P4) regions respectively of the target genes were amplified by PCR using specific oligonucleotides (Table S5). For divJ the deletion cassette was constructed as previously described (Skerker et al., 2005). For pleC and divK, instead, restriction enzymes sites for directional forced cloning with the tetracycline resistance cassette were used. All plasmids were then sequenced for verification. The first six and last 12 codons of each gene deleted were left intact to protect against disruption of possible regulatory signals for adjacent genes. Two-step recombination of deletion cassettes was conducted as previously described using integrative plasmid pNPTS138 (Skerker et al., 2005). Deletion of genes was verified by PCR using primers pSmc00059_P1tris, pSmc00059_P4tris, pSmc02369_Pext_fw, pSmc02369_Pext_rv and pSmc02369_Pint_fw, pSmc01371_P1ext and pSmc01371_P4ext‥
For transduction, phage and bacteria (in LB containing 2.5 mM CaCl2 and 2.5 mM MgSO4) were mixed to give a multiplicity of infection 1/2 (phage/cell). The mixture was incubated at 30°C for 30 min.
For construction of the complementation plasmid, divJ and pleC and their putative promoter regions were amplified by PCR using the Rm1021 genomic DNA as template and primers named P1 and P4, for divK P1 and P6 primers were used (Table S5). Fragments were gel purified and cloned into the low copy vector pMR10 (Roberts et al., 1996). Plasmids obtained were introduced in S. meliloti strains by electroporation (Ferri et al., 2010).
The divJ gene for overexpression in vivo was amplified from genomic DNA of S. meliloti Rm1021 by PCR using pSmc00059_P0 and pSmc00059_P6, digested by restriction (NdeI and XhoI) and ligated in pSRKKm (previously restricted with the same enzymes), generating pSRKKmdivJ, which was transferred to Rm1021 by electroporation. Similarly pSRKGm-divJ, pSRKKmpleC, pSRKKmctrA and pSRKKmdivJ(D249A), pSRKKmdivK and pSRKKmdivk (D53A) were constructed.
The divJ H249A mutant was constructed with the primers pSmc00059_H249A_sense and pSmc00059_H249A_antisense (Table S5) on the plasmid pSRKKmdivJ using PfuTurbo DNA polymerase (Stratagene) as previously described (Biondi, Reisinger, et al., 2006).
For β-galactosidase assay, plasmids were constructed by directional forced cloning of pRKlac290 (Alley et al., 1991) digested with BamHI and XbaI with fragments (600bp) of the Smc0360 and Smc0949 promoter regions amplified with the primers pSmc0360_prom_XbaI, pSmc0360_prom_BamHI, pSmc0949_prom_XbaI and pSmc0949_prom_BamHI. β-galactosidase assay was performed as previously described (Fioravanti et al., 2013).
For the efficiency-of-plating (EOP) assays showed in figure 7, cultures were grown to exponential phase (OD600, ≈0.5) in LB/MC medium and then diluted to an OD600 of 0.1 of LB. Each sample was serially diluted up to 10−6 in LB, and spread onto LB agar containing either crystal violet (Sigma) or IPTG (1mM). After 4 to 5 days of growth at 30°C, the number of CFU was determined, with the exception of the ?divJ and cbrA∷ Tn5 mutant, which required an additional 48 h of growth at 30°C for colonies to appear. The average and standard deviation for each strain were derived from two independent cultures.
FACS analysis
Cells were cultured into LB/MC and grown to OD600 ca. 0.1–0.2 with the appropriate antibiotic. Samples were taken and fixed in 70% ethanol overnight. Fixed cells were centrifuged at and resuspended in 1mL of 50mM sodium citrate buffer plus 100 mg/mL RNaseA and then incubated for two hours at 50° C. After the RNaseA treatment 1uL of a 1:6 dilution of Sytox Green dye (Invitrogen) was added to each sample. Each sample was then read using a FACScan flow cytometer and results were plotted using Flojo software.
Transcriptome analysis: microarray-based gene expression profiling
In this study, we applied the Sm14kOLI microarray carrying 50mer to 70 mer oligonucleotide probes directed against coding and intergenic regions of the S. meliloti Rm1021 genome (Galibert et al., 2001). Each of the 6208 coding regions predicted by Galibert et al. (2001) were represented by a single oligonucleotide whereas both strands of the intergenic regions were covered by 8080 oligonucleotides. Intergenic oligonucleotides mapped at distances of ∼50 to 150 nucleotides to the intergenic regions. The microarray layout and oligonucleotide sequences are available at ArrayExpress accession no. A-MEXP-1760.
Production and processing of microarrays were done as described in (Brune et al., 2006). Four biological replicates of control strain 1021 or experiment strain BM253 were grown in 100 ml TY supplemented with nalidixic acid medium to an OD600 of 0.6. RNA isolation, cDNA synthesis, labeling, hybridization, image acquisition and data analysis were done as described in (Serrania et al., 2008). To identify significant up- or down-regulated genes, EMMA 2.2 microarray data analysis software (Dondrup et al., 2003) was used for LOWESS normalization and t statistics. Genes were classified as differentially expressed if p ≤ 0.05 and M ≥ 0.5 or ≤ −0.5. The M value represents the log2 ratio between both channels. Microarray data were submitted to ArrayExpress (Accession number).
In vitro and in vivo phosphorylation
We use Phos-tag™ Acrylamide (Nard Chemicals, LTD, Japan) in order to separate and visualize in SDS-Page gels the phosphorylated form (on histidine and aspartate residues) of CtrA, DivJ and DivK as previously described (Barbieri and Stock, 2008). Bands corresponding to the phosphorylated forms of CtrA and DivK were empirically recognized by a simple boiling step that affects specifically the stability of phosphate. Due to this instability all samples were lysed and directly loaded on gels unless specifically indicated.
For biochemical assays (Fig. 3), S. meliloti DivJ (just the kinase domain), DivJ H249A (the kinase domain), PleC (the kinase domain), DivK and Divk D53A were PCR amplified using specific primers (Table S5) expressed in E. coli BL21 and purified as previously described (Fioravanti et al., 2012). The divK D53A mutant was prepared from a plasmid containing wild-type divK performing a site-directed mutagenesis with the primers Smc01371_D53A_sense and Smc01371_D53A_antisense (Table S5) using PfuTurbo DNA polymerase (Stratagene) as previously described (Biondi, Reisinger, et al., 2006). Several clones were sequence verified to confirm the presence of the mutation.
Phos-tag™ Acrylamide SDS-PAGE gels (29:1 acrylamide:N,N”-methylene-bis-acrylamide) were prepared with 50 µM Phos-tag™ acrylamide and 100 µM MnCl2 for in vitro phosphorylation assays (figure 3) or 25 µM Phos-tag™ acrylamide and 50 µM MnCl2 for in vivo phosphorylation analysis (Figures 5 and 8). All gels were run at 4°C under constant voltage (100 V).
In vitro phosphorylation assays were performed using HK 10 µM, ATP 1 µM, MgCl2 5 mM in HKEDG buffer (10 mM HEPES-KOH pH 8.0, 50 mM KCl, 10% glycerol, 0.1 mM EDTA, 2 mM DTT). Incubation was performed RT and removal of ATP was done by filtration 4 times with HKEDG/Mg buffer using Amicon Ultra 0.5 10 KDa (Millipore).
For in vivo analysis, strains were grown to mid-log phase, and then 2ml of the cells were pelleted and stored at −80°C. Pellets were resuspended using a lysis buffer with 10 mM Tris-Cl, pH 7.5 and 4% SDS and incubated at RT for 5 min, then the loading dye was added. Samples were stored on ice for a short time (<10 min) prior to loading onto Phos-tag™ acrylamide gels. Gels were fixed for 10 min in transfer buffer (50 mM Tris-Cl, 40 mM glycine, 15% (v/v) ethanol,) with 1 mM EDTA to remove Mn2+ from the gel. Gels were washed 3 times in transfer buffer without EDTA to remove the chelated metal. Immunoblots were performed using Western Blot Signal Enhancer (Thermo Pierce) with rabbit anti-CtrA (1:5000) or anti-DivK (1:2500) primary antibodies. Chemiluminescent detection was performed using Super Signal West Pico Chemiluminescent Substrate (Thermo-Pierce). Bands intensities were analyzed using ImageJ (Schneider et al., 2012).
Nodulation assays and GFP Strains construction
To observe infected cells using eGFP-expressing bacteroids, a mutated constitutive S. meliloti sinR promoter region was amplified by PCR from the pSRmig (McIntosh et al., 2009) derivative pSRmigPsinR171mutcc (Matthew McIntosh) using primer pairs psinRmut_fwd and psinRmut_rev. The fragment was inserted into KpnI and XbaI sites of the integrating pG18mob derivative pGEE upstream of EGFP, resulting in vector pGECE. The construct was transferred by E. coli S17-1-mediated conjugation to S. meliloti Rm1021 or BM253 and integrated into the chromosome by homologous recombination.
Medicago sativa seeds (cv. Eugenia seeds, Samen-Frese, Osnabrück) were surface sterilized and germinated as described (Müller et al., 1988). 48h-old seedlings were transferred to square petri plates containing buffered nodulation medium (BNM) agar (Ehrhardt et al., 1992). The seedlings were inoculated with 200ul bacteria culture, which was grown to logarithmic phase in TY medium supplemented with nalidixic acid and washed in BNM medium. Plant growth and nodule development were screened over the duration of four weeks. After 28 days, plant height, plant dry weight and number of nodules per plant were measured. Images of plant plates and nodules were acquired, and microscopy images of nodule thin cuts were taken.
Bacteroids were extracted from alfalfa mature nodules as previously described (Finan et al., 1983).
Microscopy
S. meliloti cells were grown to mid-log phase, fixed in 70% ethanol, washed, and concentrated with saline solution (0.85% NaCl). Samples were deposited on microscope slides coated with 0.1% poly-L-lysine. Differential interference contrast and fluorescence imaging of nodules was done on a Zeiss Observer Z1 inverted microscope using Zeiss Axiovision software. Exponential phase bacteria were immobilized on 1% agarose slides and imaged using an alpha Plan-Apochromat 100x/1.46 OilDIC objective and Zeiss AxiocamMR3 camera. Nodule thin sections (100 µm) were stained with 16 µM FM4-64 membrane stain, and imaged using an EC Plan-Neofluar 5x/0.16 Ph1 objective and AxiocamMR3 camera, or using a Plan-Apochromat 40x/0.95 DICII objective and AxiocamHRc color camera. Images were processed with ImageJ (Schneider et al., 2012).
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge support from the Ente Cassa di Risparmio di Firenze (Italy), Accademia dei Lincei (Italy) and Fondazione Buzzati-Traverso (Italy). EGB lab was also supported by ANR (ANR_11_SVJ3_003_01, CASTACC), the Region Nord-Pas-de-Calais (France) and the CPER-CIA. AB lab is supported by the LOEWE program of the State of Hesse (Germany). G.C.W. and N.J.D. were supported by NIH grant GM31010. G.C.W. is an American Cancer Society Professor.
REFERENCES
- Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, et al. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell. 2011;43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley MR, Gomes SL, Alexander W, Shapiro L. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics. 1991;129:333–341. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CM, Stock AM. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal Biochem. 2008;376:73–82. doi: 10.1016/j.ab.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett MJ, Hung DY, Reisenauer A, Shapiro L, Long SR. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J Bacteriol. 2001;183:3204–3210. doi: 10.1128/JB.183.10.3204-3210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefontaine A-F, Pierreux CE, Mertens P, Vandenhaute J, Letesson J-J, Bolle XDe. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol Microbiol. 2002;43:945–960. doi: 10.1046/j.1365-2958.2002.02777.x. [DOI] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker Jeffrey M, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- Biondi EG, Skerker Jeffrey M, Arif M, Prasol MS, Perchuk BS, Laub MT. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol Microbiol. 2006;59:386–401. doi: 10.1111/j.1365-2958.2005.04970.x. [DOI] [PubMed] [Google Scholar]
- Bird TH, MacKrell A. A CtrA homolog affects swarming motility and encystment in Rhodospirillum centenum. Arch Microbiol. 2011;193:451–459. doi: 10.1007/s00203-011-0676-y. [DOI] [PubMed] [Google Scholar]
- Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol. 2010;4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJB, Pedro MAde, Kysela DT, Henst CVan der, Kim J, Bolle XDe, et al. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA. 2012;109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune I, Becker Anke, Paarmann D, Albersmeier A, Kalinowski J, Pühler Alfred, Tauch A. Under the influence of the active deodorant ingredient 4-hydroxy-3-methoxybenzyl alcohol, the skin bacterium Corynebacterium jeikeium moderately responds with differential gene expression. J Biotechnol. 2006;127:21–33. doi: 10.1016/j.jbiotec.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Burton GJ, Hecht GB, Newton A. Roles of the histidine protein kinase pleC in Caulobacter crescentus motility and chemotaxis. J Bacteriol. 1997;179:5849–5853. doi: 10.1128/jb.179.18.5849-5853.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GRO, Sharypova LA, Scheidle H, Jones KM, Niehaus K, Becker Anke, Walker Graham C. Striking complexity of lipopolysaccharide defects in a collection of Sinorhizobium meliloti mutants. J Bacteriol. 2003;185:3853–3862. doi: 10.1128/JB.185.13.3853-3862.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J, McAdams HH, Shapiro Lucy. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci USA. 2007;104:17111–17116. doi: 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis PD, Brun Yves V. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Dondrup M, Goesmann A, Bartels D, Kalinowski J, Krause L, Linke B, et al. EMMA: a platform for consistent storage and efficient analysis of microarray data. J Biotechnol. 2003;106:135–146. doi: 10.1016/j.jbiotec.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Atkinson EM, Long SR. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science. 1992;256:998–1000. doi: 10.1126/science.10744524. [DOI] [PubMed] [Google Scholar]
- Ferri L, Gori A, Biondi EG, Mengoni A, Bazzicalupo M. Plasmid electroporation of Sinorhizobium strains: The role of the restriction gene hsdR in type strain Rm1021. Plasmid. 2010;63:128–135. doi: 10.1016/j.plasmid.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Fields AT, Navarrete CS, Zare AZ, Huang Z, Mostafavi M, Lewis JC, et al. The conserved polarity factor podJ1 impacts multiple cell envelope-associated functions in Sinorhizobium meliloti. Mol Microbiol. 2012;84:892–920. doi: 10.1111/j.1365-2958.2012.08064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan TM, Hartweig E, LeMieux K, Bergman K, Walker GC, Signer ER. General transduction in Rhizobium meliloti. J Bacteriol. 1984;159:120–124. doi: 10.1128/jb.159.1.120-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan TM, Wood JM, Jordan DC. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J Bacteriol. 1983;154:1403–1413. doi: 10.1128/jb.154.3.1403-1413.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioravanti A, Fumeaux C, Mohapatra SS, Bompard C, Brilli M, Frandi A, et al. DNA Binding of the Cell Cycle Transcriptional Regulator GcrA Depends on N6-Adenosine Methylation in Caulobacter crescentus and Other Alphaproteobacteria. PLoS Genet. 2013;9:e1003541. doi: 10.1371/journal.pgen.1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- Gibson KE, Barnett Melanie J, Toman CJ, Long Sharon R, Walker Graham C. The symbiosis regulator CbrA modulates a complex regulatory network affecting the flagellar apparatus and cell envelope proteins. J Bacteriol. 2007;189:3591–3602. doi: 10.1128/JB.01834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson KE, Campbell GR, Lloret J, Walker Graham C. CbrA is a stationary-phase regulator of cell surface physiology and legume symbiosis in Sinorhizobium meliloti. J Bacteriol. 2006;188:4508–4521. doi: 10.1128/JB.01923-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene SE, Brilli M, Biondi EG, Komeili A. Analysis of the CtrA pathway in Magnetospirillum reveals an ancestral role in motility in alphaproteobacteria. J Bacteriol. 2012;194:2973–2986. doi: 10.1128/JB.00170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffitts JS, Carlyon RE, Erickson JH, Moulton JL, Barnett Melanie J, Toman CJ, Long Sharon R. A Sinorhizobium meliloti osmosensory two-component system required for cyclic glucan export and symbiosis. Mol Microbiol. 2008;69:479–490. doi: 10.1111/j.1365-2958.2008.06304.x. [DOI] [PubMed] [Google Scholar]
- Hallez R, Bellefontaine A-F, Letesson J-J, Bolle XDe. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 2004;12:361–365. doi: 10.1016/j.tim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Hallez R, Mignolet J, Mullem VVan, Wery M, Vandenhaute J, Letesson J-J, et al. The asymmetric distribution of the essential histidine kinase PdhS indicates a differentiation event in Brucella abortus. EMBO J. 2007;26:1444–1455. doi: 10.1038/sj.emboj.7601577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht GB, Lane T, Ohta N, Sommer JM, Newton A. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch AM, Bang M, Ausubel FM. Ultrastructural analysis of ineffective alfalfa nodules formed by nif∷ Tn5 mutants of Rhizobium meliloti. J Bacteriol. 1983;155:367–380. doi: 10.1128/jb.155.1.367-380.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath B, Kondorosi E, John M, Schmidt J, Török I, Györgypal Z, et al. Organization, structure and symbiotic function of Rhizobium meliloti nodulation genes determining host specificity for alfalfa. Cell. 1986;46:335–343. doi: 10.1016/0092-8674(86)90654-9. [DOI] [PubMed] [Google Scholar]
- Hung Dean Y, Shapiro Lucy. A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression. Proc Natl Acad Sci USA. 2002;99:13160–13165. doi: 10.1073/pnas.202495099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, McGrath PT, Reisenauer Ann, McAdams HH, Shapiro Lucy. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci USA. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Ferguson NL, Alley MR. New members of the ctrA regulon: the major chemotaxis operon in Caulobacter is CtrA dependent. Microbiology (Reading, Engl) 2001;147:949–958. doi: 10.1099/00221287-147-4-949. [DOI] [PubMed] [Google Scholar]
- Khan SR, Gaines J, Roop RM, Farrand SK., 2nd Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol. 2008;74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Heindl JE, Fuqua C. Coordination of Division and Development Influences Complex Multicellular Behavior in Agrobacterium tumefaciens. PLoS ONE. 2013;8:e56682. doi: 10.1371/journal.pone.0056682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Nisco NJDe, Chien P, Simmons LA, Walker Graham C. Sinorhizobium meliloti CpdR1 is critical for co-ordinating cell cycle progression and the symbiotic chronic infection. Mol Microbiol. 2009;73:586–600. doi: 10.1111/j.1365-2958.2009.06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Chen SL, Shapiro Lucy, McAdams HH. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci USA. 2002;99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Iniesta AA, Ryan KR, Shapiro Lucy, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- McIntosh M, Meyer S, Becker Anke. Novel Sinorhizobium meliloti quorum sensing positive and negative regulatory feedback mechanisms respond to phosphate availability. Mol Microbiol. 2009;74:1238–1256. doi: 10.1111/j.1365-2958.2009.06930.x. [DOI] [PubMed] [Google Scholar]
- Mergaert P, Uchiumi T, Alunni Benoît, Evanno G, Cheron A, Catrice O, et al. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc Natl Acad Sci USA. 2006;103:5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignolet J, Henst CVan der, Nicolas C, Deghelt M, Dotreppe D, Letesson J-J, Bolle XDe. PdhS, an old-pole-localized histidine kinase, recruits the fumarase FumC in Brucella abortus. J Bacteriol. 2010;192:3235–3239. doi: 10.1128/JB.00066-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Hynes M, Kapp D, Niehaus K, Pühler Alfred. Two classes of Rhizobium meliloti infection mutants differ in exopolysaccharide production and in coinoculation properties with nodulation mutants. Mol Gen Genet. 1988;211:17–26. [Google Scholar]
- Ohta N, Lane T, Ninfa EG, Sommer JM, Newton A. A histidine protein kinase homologue required for regulation of bacterial cell division and differentiation. Proc Natl Acad Sci USA. 1992;89:10297–10301. doi: 10.1073/pnas.89.21.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Noriko, Newton Austin. The core dimerization domains of histidine kinases contain recognition specificity for the cognate response regulator. J Bacteriol. 2003;185:4424–4431. doi: 10.1128/JB.185.15.4424-4431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Jaeger T, Abel S, Wiederkehr I, Folcher M, Biondi EG, et al. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell. 2008;133:452–461. doi: 10.1016/j.cell.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobigaylo N, Wetter D, Szymczak S, Schiller U, Kurtz S, Meyer F, et al. Construction of a large signature-tagged mini-Tn5 transposon library and its application to mutagenesis of Sinorhizobium meliloti. Appl Environ Microbiol. 2006;72:4329–4337. doi: 10.1128/AEM.03072-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- Roberts RC, Toochinda C, Avedissian M, Baldini RL, Gomes SL, Shapiro L. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J Bacteriol. 1996;178:1829–1841. doi: 10.1128/jb.178.7.1829-1841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KR, Huntwork S, Shapiro Lucy. Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc Natl Acad Sci USA. 2004;101:7415–7420. doi: 10.1073/pnas.0402153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KR, Judd EM, Shapiro Lucy. The CtrA response regulator essential for Caulobacter crescentus cell-cycle progression requires a bipartite degradation signal for temporally controlled proteolysis. J Mol Biol. 2002;324:443–455. doi: 10.1016/s0022-2836(02)01042-2. [DOI] [PubMed] [Google Scholar]
- Sadowski C, Wilson D, Schallies K, Walker G, Gibson KE. The Sinorhizobium meliloti sensor histidine kinase CbrA contributes to free-living cell cycle regulation. Microbiology (Reading, Engl) 2013 doi: 10.1099/mic.0.067504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrania J, Vorhölter F-J, Niehaus K, Pühler Alfred, Becker Anke. Identification of Xanthomonas campestris pv. campestris galactose utilization genes from transcriptome data. J Biotechnol. 2008;135:309–317. doi: 10.1016/j.jbiotec.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Sharypova LA, Niehaus K, Scheidle H, Holst O, Becker Anke. Sinorhizobium meliloti acpXL mutant lacks the C28 hydroxylated fatty acid moiety of lipid A and does not express a slow migrating form of lipopolysaccharide. J Biol Chem. 2003;278:12946–12954. doi: 10.1074/jbc.M209389200. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat Biotech. 1983;1:784–791. [Google Scholar]
- Skerker JM, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker Jeffrey M, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker Jeffrey M, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokos CG, Perchuk BS, Laub MT. A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev Cell. 2011;20:329–341. doi: 10.1016/j.devcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velde WVan de, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, Kevei Z, et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science. 2010;327:1122–1126. doi: 10.1126/science.1184057. [DOI] [PubMed] [Google Scholar]
- Wang D, Griffitts J, Starker C, Fedorova E, Limpens E, Ivanov S, et al. A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science. 2010;327:1126–1129. doi: 10.1126/science.1184096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- Wortinger M, Sackett MJ, Brun YV. CtrA mediates a DNA replication checkpoint that prevents cell division in Caulobacter crescentus. EMBO J. 2000;19:4503–4512. doi: 10.1093/emboj/19.17.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Ohta N, Newton A. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc Natl Acad Sci USA. 1998;95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.