Abstract
Background
High serum uric acid levels are associated with gout, atherosclerosis and cardiovascular disease. Three genes (SLC2A9, ABCG2, and SLC17A3) were reported to be involved in the regulation of uric acid levels.
Research
Design and Methods: SNPs rs2231142 (ABCG2) and rs1165205 (SLC17A3) were genotyped in three cohorts (n = 4492) and combined with previously genotyped SNPs within SLC2A9 (rs6855911, rs7442295, rs6449213, rs12510549).
Results
Each copy of the minor allele decreased uric acid levels by 0.30–0.38 mg/dL for SLC2A9 (p values: 10−20–10−36) and increased levels by 0.34 mg/dL for ABCG2 (p = 1.1×10−16). SLC17A3 influenced uric acid levels only modestly. Together the SNPs showed graded associations with uric acid levels of 0.111 mg/dL per risk allele (p = 3.8×10−42). In addition, we observed a sex-specific interaction of age with the association of SLC2A9 SNPs with uric acid levels, where increasing age strengthened the association of SNPs in women and decreased the association in men.
Conclusions
Genetic variants within SLC2A9, ABCG2 and SLC17A3 show highly significant associations with uric acid levels, and for SNPs within SLC2A9 this association is strongly modified by age and sex.
Keywords: Epidemiology, Genetics, Uric acid, Copy number variation, Sex-specific effect, Genetic risk score
1. Introduction
Elevated serum uric acid (UA) levels are associated with gout, hypertension, diabetes mellitus, metabolic syndrome, and cardiovascular diseases (CVD) [1]. It is a long-lasting discussion whether elevated uric acid levels are a cause or a consequence for the association with diseases. In the meanwhile several prospective studies have demonstrated an association between high UA levels and especially atherosclerosis complications such as CVD, stroke, and peripheral arterial disease [2–4]. According to the rules of Mendelian randomization, association studies of genetic variants that are associated with a life-long increase in UA levels and their association with atherosclerosis outcomes became a strong tool to support causality of an intermediate phenotype such as UA levels and atherosclerosis outcome. It is therefore of importance to identify those variants with certainty which are associated with increased UA levels.
Recent genome-wide association studies identified SLC2A9, ABCG2 and SLC17A3 to be most strongly associated with UA concentrations [5–7]. Our study aimed to replicate the highest-scoring SNPs within ABCG2 and SLC17A3 in three epidemiological studies of different design including even a group of severe obese individuals and focused on sex-specific effects, differences in explained variance and possible interactions of copy number variations (CNV) within these regions. We combined these data with previously genotyped SLC2A9 SNPs [8] to provide an overall picture on the most important genes influencing UA levels.
2. Materials and methods
The Bruneck Study (n = 800) is a prospective population-based sex- and age-stratified random sample. SAPHIR is an observational study conducted in a healthy working population (n = 1732). The Utah Obesity Case-Control Study composes 1106 severely obese subjects (mean BMI 46.1±7.6 kg/m2) and 854 controls. All participants were of West-Eurasian origin and signed an informed consent. Baseline characteristics are summarized in Table 1.
Table 1.
Baseline characteristics of the analyzed populations.
| Bruneck (n = 800) | SAPHIR (n = 1732) | Utah controls (n = 854) | Utah obesity (n = 1106) | |
|---|---|---|---|---|
| Age, years | 62.7 ± 11.1 | 51.8 ± 6.1 | 52.6 ± 8.6 | 43.9 ± 11.4 |
| Males (%) | 49.8 | 63.0 | 48.0 | 18.1 |
| BMI | 25.6 ± 3.8 | 26.8 ± 4.1 | 27.6 ± 4.9 | 46.1 ± 7.6 |
| Uric acid [mg/dL] | 4.7 ± 1.3 | 5.9 ± 1.4 | 5.5 ± 1.5 | 6.3 ± 1.5 |
| Total cholesterol [mg/dL] | 230.0 ± 42.6 | 228.8 ± 40.1 | 187.1 ± 34.0 | 186.8 ± 36.2 |
| LDL cholesterol [mg/dL] | 145.5 ± 37.9 | 145.6 ± 36.6 | 105.1 ± 27.7 | 108.2 ± 27.6 * |
| HDL cholesterol [mg/dL] | 58.7 ± 16.2 | 59.6 ± 15.7 | 50.1 ± 15.0 | 45.9 ± 11.0 * |
| Triglycerides [mg/dL] | 131.7 ± 71.9 | 125.6 ± 87.7 | 156.0 ± 105.0 | 186.0 ± 106.0 |
| Systolic blood pressure [mmHg] | 148 ± 21 | 134 ± 13 | 121 ± 17 | 127 ± 18 * |
| Diastolic blood pressure [mmHg] | 87 ±9 | 82 ±8 | 73 ± 10 | 72 ± 11 ** |
| Glucose [mg/dL] | 102 ± 24 | 94 ± 18 | 91 ± 18 | 104 ± 33 * |
| Diabetes mellitus, n (%) | 76(9.5) | 54 (3.2) | 55(6.6) | 225(21.7) * |
| Use of gout medication, n (%) | 13(1.6) | n.a. | 7(0.8) | 11(1.1) ** |
| Use of lipid lowering drugs, n (%) | 25(3.1) | 77 (4.5) | 64(7.7) | 122(11.9) * |
Values are provided as mean and standard deviation if not indicated otherwise.
p < 0.05
p < 0.001 for comparison between severely obese subjects and controls from Utah.
Blood samples were collected after overnight fasting. UA levels were measured using enzymatic–colorimetric methods. Top-scoring SNPs rs2231142 (ABCG2) and rs1165205 (SLC17A3) from recent GWAS [5, 6] were selected and genotyped using TaqMan® SNP Genotyping Assays on a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). The SLC2A9 SNPs (rs12510549, rs6449213, rs6855911, rs7442295) were also geno-typed before [5, 8] using Taqman assays. The fractions of samples that were genotyped twice for quality assurance were 9.5% for Utah Obesity Case-Control Study, 14.5% for SAPHIR Study and 11.5% for Bruneck Study. The discordance rate in each population was 0.
General linear regression adjusted for age, sex and BMIwas used for inferring the influence of the SNPs on UA levels assuming an additive genetic model. Data analyses combining subjects from all populations were conducted using linear mixed effects models with random intercepts.
To describe sex differences in the genetic effects and in the amount of variance explained by each SNP, UA levels were standardized according to sex and study population. A linear mixed model including a SNP*sex interaction term was applied. The percentage of explained variance per SNP was obtained as the difference of R2 of a full linear model (regressing BMI, age and each SNP) to the R2 of a reduced linear model ignoring the respective SNP. Differences of sex-specific explained variances were tested with a z-test. To infer the interaction of the SNPs with sex and age, a SNP*age*sex-interaction term was introduced in a general linear model adjusted for BMI on standardized UA levels.
A genetic risk score was generated by counting the number of alleles associated with high UA concentrations (rs1165205 A, rs2231142 A, rs12510549 T, rs6449213 T, rs6855911 A, and rs7442295 A; range 0–12). Statistical analyses were performed with R.
The potential functional effects of rs2231142 and rs1165205 were evaluated using several bioinformatic tools. These included information on the occurrence of CNVs, ESPERR regulatory potential and tissue-specific expression patterns [17].
For extended methodological details see Online Appendix.
3. Results
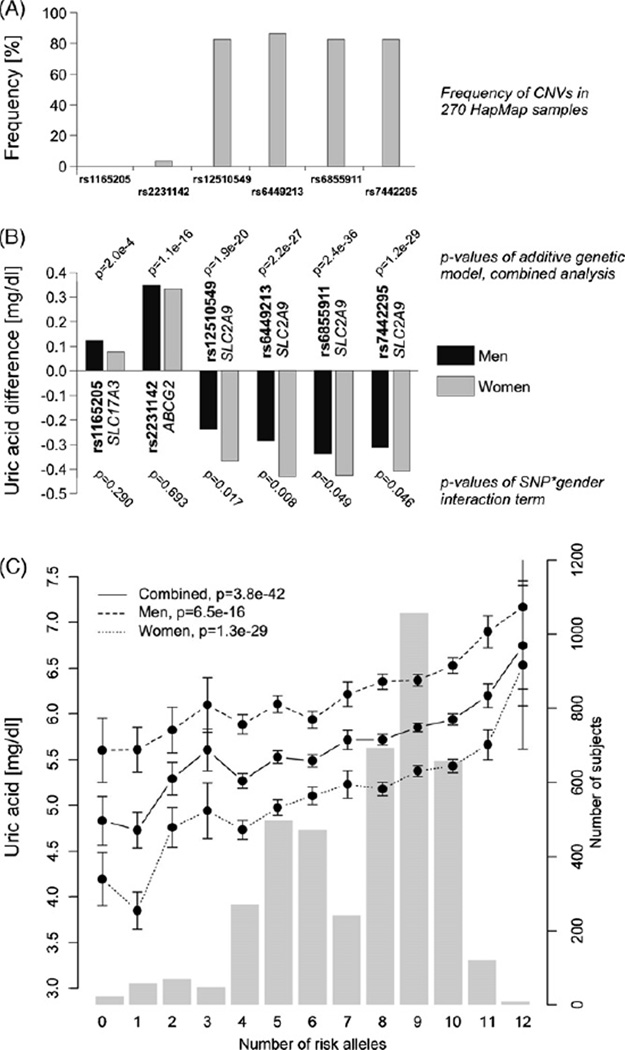
rs1165205 A (SLC17A3) was least pronounced but nevertheless significantly associated with UA levels (p = 0.0002). The minor allele of rs2231142 (ABCG2) was markedly associated with higher UA levels (p = 1.1×10−16). The four SNPs within SLC2A9 were most strongly associated with lower UA levels (pmin = 2.4×10−36). Detailed results are provided in Figure 1B and Suppl.-Tables S1–S3. It was of interest that the observed associations for the investigated SNPs with UA levels were not only seen in the general population but also in severely obese individuals, where the association was even stronger for the SNPs in SLC2A9 when compared to the control group (Suppl.-Tables S3).
Fig. 1.
(A) The frequency of copy number variations (CNVs) in 270 HapMap samples in the region of the analyzed SNPs. (B) Sex-specific effects of the analyzed SNPs on uric acid concentrations. The bars show the difference in uric acid levels per minor allele compared to the wild-type stratified for men and women. The p values above the graphs are derived from the linear mixed effect model of the combined analysis of the additive model using standardized uric acid levels (adjusted for age and BMI). The p values below the graph are for the SNP*sex interaction terms. (C) Sex-specific effects of an additive genetic risk score of the number of risk alleles within ABCG2, SLC2A9 and SLC17A3 that are associated with increased uric acid levels. The dots indicate mean values (±standard errors) of uric acid levels as indicated on the left y-axis; the histogram represents the distribution of number of subjects in each group of risk alleles as indicated in the right y-axis. The risk alleles for this score were allele A for rs1165205, rs2231142, rs6855911, and rs7442295 as well as allele T for rs12510549 and rs6449213. The range of the score was from 0 to 12.
3.1. Sex- and age-specific associations
Together the six SNPs explained ~5% of the observed variance with a difference of 2.2% between men and women (3.9% versus 6.1%). Strong differences in the explained variance were especially observed for SLC2A9 SNPs: in women the explained variance was 2–3 times higher than inmen(pmin = 0.0049) (Suppl.-Table S2). This resulted in significant SNP*sex interactions in all four SLC2A9 SNPs (Fig. 1B).
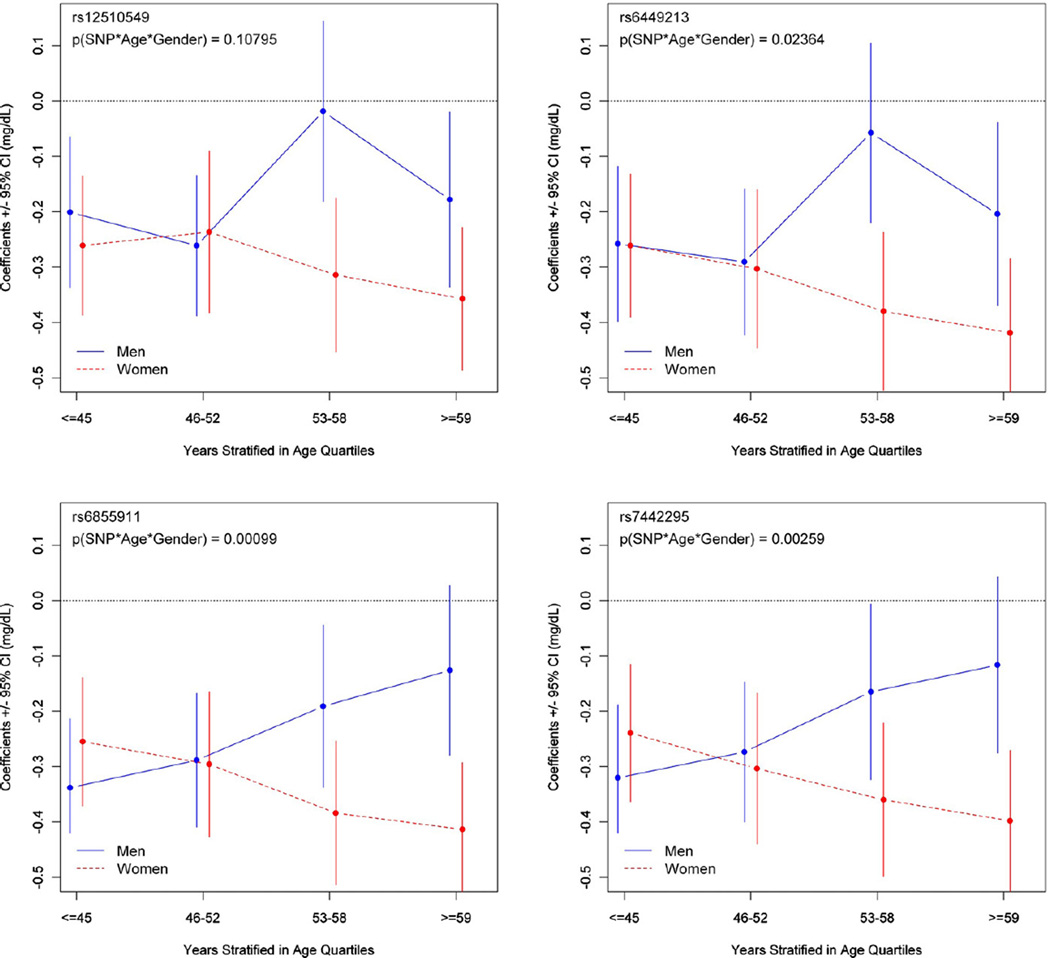
When further stratifying the analysis by age in quartiles, it became evident that the SNP effects within SLC2A9 on UA levels and the UA variance explained by these SNPs were not only modified by gender, but additionally by age (Supp.-Table S4): in women, the effects of the SLC2A9 SNPs increased nearly linearly with age (Fig. 2). For example, for rs6855911, the effect in women 59 years or older was 1.6-fold higher than in women 45 years or younger (ß =−0.413 versus ß =−0.255). On the other hand, increasing age diminished the effects of the SNPs in men. Therefore, the interaction term SNP*age*sex was highly significant for rs6855911 and rs7442295, and significant for rs6449213 (Fig. 2). No effect modification by age was observed for the two SNPs within SLC17A3 and ABCG2 (Supp.-Table S4).
Fig. 2.
Interaction of age and gender on the genetic association of SLC2A9 SNPs with uric acid levels. The x-axis corresponds to the age in quartiles; the y-axis represents the ß-estimates +/−95% confidence intervals (CI) of the additive effect of the minor allele of each SNP. The p value of the SNP*age*sex interaction term is indicated on the upper left corner of each graph.
3.2. Genetic risk score
Mean UA concentrations increased linearly with the number of UA-increasing risk alleles (Fig. 1C). Each additional copy of a risk allele was associated with an elevation of UA levels by 0.111 mg/dL (p = 3.8×10−42).
3.3. Bioinformatic analysis
The ABCG2 SNP rs2231142 (Q141K) was recently shown to be a loss-of-function mutation that causes hyperuricemia and gout [9]. Interestingly, rs2231142 is located in a region that was described several times to harbor a copy number-variable region (Fig. 1A and Suppl.-Figure S2).
SLC17A3 encodes a sodium phosphate transporter (NPT4). In humans, the gene is highly expressed in the kidney, with a 30-fold median expression compared to all other tissues (Suppl.-Fig. S3). The SLC17A3 SNP rs1165205 is intronic, does not lie in a region with an elevated ESPERR score and is not correlated with other HapMap SNPs. No CNV were described for SLC17A3.
SLC2A9, a facilitative urate transporter, shows a higher expression in CD33+ myeloid cells and CD14 monocytes. Only rs7442295 is located in a region with high regulatory potential (Suppl.-Fig. S4). The SNPs rs12510549, rs6855911, rs6449213 and rs7442295 are located on two CNVs with rs12510549 only lying on their very edges; rs6449213 additionally lies on the boundary of another CNV (Suppl.-Fig. S5).
4. Discussion
Despite considerable therapeutic advances over the past 50 years, CVD is the leading cause of death worldwide. This is mainly a result of the increasing prevalence of atherosclerosis, owing to the aging population, and the widespread under-recognition and under-treatment of individuals with risk factors for atherosclerosis [10]. Besides classical risk factors, hyperuricemia is considered an independent risk factor for atherosclerosis [2–4]. Apart from lifestyle factors [11], genetic variation influences UA concentrations through regulation of UA synthesis, excretion, or reabsorption [9].
The variance explained by the investigated SNPs (~5%) is low given the high heritability of UA concentrations of 40–63% [12, 13]. What causes the high portion of UA heritability? Rare variants could have considerable effects in singular individuals. Another, yet unexplored possibility is the influence of CNVs, which are involved in the genetic predisposition to common diseases or quantitative phenotypes [14]. GIANT found that the SNP most strongly associated with BMI was linked to a 45-kb deletion polymorphism [15]. Interestingly, the SNPs that were most strongly associated with UA levels were located in CNV-rich regions. SLC17A3, which is not linked to a CNV, was weakly associated with UA levels.
For UA levels, sex-specific genetic effects were described to be strong [5–8]. In this study, we could further corroborate that the genetic influence of SLC2A9 on UA levels is more pronounced in women than in men. In addition, we observed a sex-specific interaction of age with the association of SLC2A9 SNPs with UA levels, where increasing age strengthened the association of SNPs in women and decreased the association in men. This is especially interesting as serum uric acid levels among women with menopause were reported to be higher than in premenopausal women [16]. Although we do not know the menopausal status of the women in our study population, it can be anticipated that in the third and fourth age quartiles (age 53 yrs and higher), the proportion of post-menopausal women steadily increases towards 100%. This observed effect modification by the menopausal status might be an explanation for the changing association effects with age. However, the possible underlying mechanism in men is unclear.
We could not replicate the sex-specific effect of the SNP within ABCG2 [6, 7]. This might be explained by a too low statistical power in our study or by the applied normalization of UA levels, which has not been done in other studies.
Recent genetic studies have demonstrated that the size of the expected risk associated with a disease end-phenotype such as CVD is rather small with odds ratios between 1.10 and 1.40. Therefore the use of a single risk allele for individual risk prediction is strongly limited. For this reason, the search for additive risk allele panels for risk predictions continues and it is expected that the identification of more and more risk alleles combined in an additive genetic risk score will lead to a markedly better precision of risk prediction, which will be useful in clinical practice in the future. In our study an additive genetic risk score based on the number of risk alleles for higher UA levels was strongly and linearly associated with UA concentrations. The power of our study would have been too small to use this risk score for cardiovascular risk prediction. However, this panel of SNPs should be tested in large, well-powered studies with atherosclerosis outcomes. In case they are predictive, the investigated risk alleles (1) would support causality of the UA-CVD association and (2) might be worthwhile to include them in future genetic CVD risk prediction scores. As atherosclerosis is a process that occurs slowly and ‘silently’ over decades, the knowledge of the risk alleles could help to identify individuals at risk of developing atherosclerosis long before the onset of clinical features. The genetic risk score is a stable predictor, whereas UA levels are influenced by physiological variability over time.
It might be considered a limitation of our study that we did not genotype CNVs that were reported to occur especially in SLC2A9. This can be explained by the current lack of stable and valid high-throughput CNV genotyping application that allows an allele-specific CNV genotyping.
5. Conclusions
The observed associations between genetic variants in ABCG2, SLC2A9 and SLC17A3 with UA levels were strong and could be influenced by the occurrence of CNVs described in these genetic regions. The associations that were modified by sex and age highlight essential pathways in UA regulation.
Supplementary Material
Acknowledgments
Financial support
This project was supported by the Medizinische Forschungsförderung Innsbruck (Grant 2007-402) and the Österreichische Nationalbank (Grant 13059) to A. Brandstätter and by a grant from the Austrian GEN-AU-Programme “GOLD” to F. Kronenberg.
Footnotes
Conflicts of interest
None to declare.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.atherosclerosis.2009.12.013.
References
- 1.Richette P, Bardin T. Gout Lancet. in press. [Google Scholar]
- 2.Chen JH, Chuang SY, Chen HJ, Yeh WT, Pan WH. Serum uric acid level as an independent risk factor for all-cause, cardiovascular, and ischemic stroke mortality: a Chinese cohort study. Arthritis Rheum. 2009;61:225–232. doi: 10.1002/art.24164. [DOI] [PubMed] [Google Scholar]
- 3.Fang J, Alderman MH. Serumuric acid and cardiovascular mortality theNHANES I epidemiologic follow-up study, 1971–1992. National health and nutrition examination survey. JAMA. 2000;283:2404–2410. doi: 10.1001/jama.283.18.2404. [DOI] [PubMed] [Google Scholar]
- 4.Hozawa A, Folsom AR, Ibrahim H, et al. Serum uric acid and risk of ischemic stroke: the ARIC Study. Atherosclerosis. 2006;187:401–407. doi: 10.1016/j.atherosclerosis.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 5.Döring A, Gieger C, Mehta D, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 6.Dehghan A, Köttgen A, Yang Q, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolz M, Johnson T, Sanna S, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandstätter A, Kiechl S, Kollerits B, et al. Sex-specific association of the putative fructose transporter SLC2A9 variants with uric acid levels is modified by BMI. Diabetes Care. 2008;31:1662–1667. doi: 10.2337/dc08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riches PL, Wright AF, Ralston SH. Recent insights into the pathogenesis of hyperuricaemia and gout. Hum Mol Genet. 2009;18:R177–R184. doi: 10.1093/hmg/ddp369. [DOI] [PubMed] [Google Scholar]
- 10.Sanz J, Fayad ZA. Imaging of atherosclerotic cardiovascular disease. Nature. 2008;451:953–957. doi: 10.1038/nature06803. [DOI] [PubMed] [Google Scholar]
- 11.Choi HK, Curhan G. Soft drinks, fructose consumption, and the risk of gout in men: prospective cohort study. BMJ. 2008;336:309–312. doi: 10.1136/bmj.39449.819271.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voruganti VS, Nath SD, Cole SA, et al. Genetics of variation in serum uric acid and cardiovascular risk factors in Mexican Americans. J Clin Endocrinol Metab. 2009;94:632–638. doi: 10.1210/jc.2008-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Q, Guo CY, Cupples LA, et al. Genome-wide search for genes affecting serum uric acid levels: the Framingham heart study. Metabolism. 2005;54:1435–1441. doi: 10.1016/j.metabol.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Brandstätter A, Lingenhel A, Zwiauer K, Strobl W, Kronenberg F. Decrease of Lp(a) during weight reduction in obese children is modified by the apo(a) kringle-IV copy number variation. Int J Obes (Lond) 2009;33:1136–1142. doi: 10.1038/ijo.2009.144. [DOI] [PubMed] [Google Scholar]
- 15.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hak AE, Choi HK. Menopause, postmenopausal hormone use and serum uric acid levels in US women–the third national health and nutrition examination survey. Arthritis Res Ther. 2008;10:R116. doi: 10.1186/ar2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coassin S, Brandstätter A, Kronenberg F. Lost in the space of bioinformatic tools: Aconstantly updated survival guide for genetic epidemiology. The GenEpi Toolbox. Atherosclerosis. doi: 10.1016/j.atherosclerosis.2009.10.026. (in press) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.