Abstract
Intramembrane metalloproteases are nearly ubiquitous in living organisms and they function in diverse processes ranging from cholesterol homeostasis and the unfolded protein response in humans to sporulation, stress responses, and virulence of bacteria. Understanding how these enzymes function in membranes is a challenge of fundamental interest with potential applications if modulators can be devised. Progress is described toward a mechanistic understanding, based primarily on molecular genetic and biochemical studies of human S2P and bacterial SpoIVFB and RseP, and on the structure of the membrane domain of an archaeal enzyme. Conserved features of the enzymes appear to include transmembrane helices and loops around the active site zinc ion, which may be near the membrane surface. Extramembrane domains such as PDZ (PSD-95, DLG, ZO-1) or CBS (cystathionines-β-synthase) domains govern substrate access to the active site, but several different mechanisms of access and cleavage site selection can be envisioned, which might differ depending on the substrate and the enzyme. More work is needed to distinguish between these mechanisms, both for enzymes that have been relatively well-studied, and for enzymes lacking PDZ and CBS domains, which have not been studied.
Keywords: intramembrane metalloprotease, S2P, SpoIVFB, RseP, PDZ domain, CBS domain
1. Introduction
The cloning of human Site-2 protease (S2P) in 1997 marked the beginning of our understanding of intramembrane proteases (IPs) [1]. Mutational analysis supported the idea that S2P is a metalloprotease that cleaves sterol-regulatory element-binding proteins (SREBPs) within a transmembrane segment (TMS), releasing the NH2-terminal domain of the SREBP from a membrane so it can enter the nucleus and activate genes encoding the low density lipoprotein (LDL) receptor and enzymes for cholesterol and fatty acid biosynthesis. Sequence analyses suggested that S2P is a polytopic membrane protein that is conserved in animals and archaea. Additional sequence comparisons identified similar proteins in the plant Arabidopsis thaliana and in diverse bacteria [2]. Shortly thereafter, mutational studies provided evidence that bacterial proteins SpoIVFB of Bacillus subtilis and YaeL (subsequently renamed RseP) of Escherichia coli are intramembrane metalloproteases (IMMPs) that cleave Pro-σK and RseA, respectively [3–7]. Pro-σK is membrane-associated and inactive as a σ factor, and cleavage by SpoIVFB releases σK into the mother cell where it directs RNA polymerase (RNAP) to transcribe genes involved in spore formation. RseA is an anti-σ factor with one TMS and an NH2-terminal cytoplasmic domain that sequesters σE until a proteolytic cascade that includes RseP causes RseA to be degraded, allowing σE to direct transcription of genes involved in the response to cell envelope stress.
After the early work on S2P, SpoIVFB, and RseP, mounting genomic sequence data underscored the near-ubiquitous conservation of putative IMMPs in living organisms [8, 9]. Phylogenetic analysis defined four subfamilies. S2P and RseP are members of the largest subfamily, which contain one or more PDZ domains, and SpoIVFB is a member of the large CBS domain-containing subfamily. The PDZ and CBS domains will be described in section 2. The other two subfamilies lack PDZ or CBS domains and have different numbers of TMSs, but none of these putative enzymes have been characterized. The evolution of IPs is discussed by Grishin and Kinch [10] in this issue.
In addition to the roles mentioned above for S2P, SpoIVFB, and RseP, subsequent molecular genetic studies revealed diverse functions of IMMPs. Bacterial IMMPs are implicated in production of mating signals [11], polar morphogenesis [12], cell division [13], pathogenesis (reviewed in [14, 15]), and clearance of signal peptides from the cell membrane [16]. These functions are described by Glickman {Glickman, 2012 #1605} in this issue. Human S2P has at least two other substrates besides SREBPs, the transcription factors ATF6 and CREBH, which mediate the unfolded protein and acute phase responses, respectively [17, 18]. These and other roles of IMMPs in animals and disease are discussed by Rawson {Rawson, 2012 #1606} in this issue. It seems likely that many other functions of IMMPs remain to be discovered, since the few studied so far have diverse functions and some bacterial genomes code for more than 10 of these enzymes.
Here, we focus on mechanistic insights from molecular genetic and biochemical studies primarily of S2P, SpoIVFB, and RseP, since these three IMMPs have been studied most extensively. We describe their membrane topology, extramembrane domains, regulation of activity, and substrate sequence specificity, emphasizing progress since prior reviews [19–21] and questions that remain, even in light of the crystal structure of part of an archaeal IMMP [22].
2. Membrane topology and extramembrane domains
Figure 1 depicts the biological membranes in which S2P, SpoIVFB, and RseP function as well as their predicted membrane topology and extramembrane domains.
Figure 1.
Cellular location, membrane topology, and extramembrane domains of S2P, SpoIVFB, and RseP. A) Human S2P is located in the membranes of the Golgi apparatus and has 8 predicted TMSs of which TMSs 4–6 make up the conserved core (orange). The positions of a Ser-rich loop, the HEXXH motif, the lumenal PDZ domain with its Cys-rich insert, and D467 in TMS 6 are shown. When substrate SREBPs are transported from the endoplasmic reticulum to the Golgi apparatus, S1P cleaves a lumenal loop, allowing S2P to cleave TMS 1 and release the N-terminal basic helix-loop-helix (bHLH) leucine-zipper (zip) domain into the cytosol. The bHLH-zip domain enters the nucleus and activates transcription. The regulatory domain (Reg.) interacts with SCAP (not shown). B) SpoIVFB is located in the outermost membrane surrounding the forespore during B. subtilis sporulation. Features analogous to those noted above in S2P are indicated. The CBS domain is in the mother cell, as is most of Pro-σK, whose pro-sequence is depicted to interact peripherally and loop into the membrane, although this is speculative. SpoIVFB cleaves Pro-σK, releasing σK into the mother cell to direct transcription. C) RseP is located in the E. coli inner membrane with its tandem PDZ domains in the periplasm. DegS cleaves the periplasmic C-terminal domain of RseA, then RseP cleaves the TMS, releasing the RseA N-terminal domain in complex with σE into the cytosol, where ClpXP degrades the rest of RseA, releasing σE to direct transcription.
2.1 S2P
Based on studies in Chinese hamster ovary cells, S2P functions in the Golgi apparatus, cleaving SREBPs within a TMS or near the membrane surface after an initial cleavage by Site-1 protease (S1P) (reviewed in [23]). The membrane topology of S2P shown in Figure 1A is based partly on protease protection experiments that suggest the two ends of the protein are exposed to the cytosol and on glycosylation studies consistent with three substantial lumenal loops [24]. The first lumenal loop is hydrophilic but otherwise unremarkable whereas the second loop consists of 76% Ser residues and the third loop, which is by far the longest at 188 residues, contains a PDZ domain with a Cys-rich insert [8] and a site that appears to be glycosylated although this is not necessary for activity of the enzyme [24]. Functions of the Ser-rich loop and the PDZ domain with its Cys-rich insert are unknown. The number of TMSs is uncertain but we favor the 8-TMS model proposed by Ha [19] in which TMSs 4, 5, and 6 form a conserved core predicted by sequence comparisons [2, 8]. A 3-TMS core was observed in the archaeal IMMP crystal structure [22]. TMS 4 is predicted to contain the HEXXH metalloprotease motif in which the two His residues would coordinate the metal ion and Glu would activate a water molecule for peptide bond hydrolysis. Predicted TMS 6 contains Asp467, which would provide a third metal ligand, and substitution of this residue with Asn abolished S2P activity [24]. Less certain is whether residues 71–107 form TMSs 2 and 3, and residues 492–518 form TMSs 7 and 8, but this seems plausible since both regions are hydrophobic and may be long enough to span the membrane twice (since the membrane may be slightly compressed around the enzyme, see section 5). Residues 186–214 between predicted TMSs 4 and 5 are quite hydrophobic and are depicted in Figure 1A to form a loop in the membrane, based on analogy with the archaeal IMMP crystal structure [22].
2.2 SpoIVFB
SpoIVFB is localized to the outermost membrane surrounding the forespore during B. subtilis endospore formation [25–28], where it cleaves membrane-associated Pro-σK [29], releasing active σK into the mother cell compartment (Fig. 1B). The predicted membrane topology of SpoIVFB, based on hydropathy analysis and the distribution of charged residues in hydrophilic segments [30], was supported by the results of topological analysis with fusion proteins to alkaline phosphatase and β-galactosidase in E. coli [31]. Further support for the 6-TMS model shown in Figure 1B came from the archaeal IMMP crystal structure [22]. Predicted TMSs 2, 3, and 4 comprise the conserved core, with the HEXXH motif in TMS 2, and Asp137 in TMS 4 providing the third metal ligand, as supported by mutational studies [3, 4]. Residues 57–83 between predicted TMSs 2 and 3 contain a hydrophobic stretch (residues 61–70) that might loop into the membrane based on the archaeal IMMP crystal structure [22]. Predicted TMS 6 is followed by a 92-residue hydrophilic region that sequence comparisons identified as a CBS domain [8]. As noted in section 1, the CBS domain is the defining feature of a large subfamily of IMMPs. The CBS domain of SpoIVFB is exposed to the mother cell cytosol, where it is proposed to bind ATP and regulate enzyme activity, coupling cleavage of Pro-σK to mother cell energy status [32].
2.3 RseP
Cell fractionation experiments have shown that RseP is a cytoplasmic membrane protein of E. coli [6]. Similar to human S2P cleavage of its substrates after initial cleavage by S1P, RseP catalyzes the second cleavage of RseA, a membrane-associated anti-σE protein, within or near its TMS, following the first cleavage in its periplasmic region by DegS (Fig. 1C). This latter protein is a trimeric serine protease of the cytoplasmic membrane [5, 7, 33, 34]. The amino acid sequence of RseP contains four hydrophobic stretches that would act as TMSs [6]. While topology prediction based on amino acid sequence information yields ambiguous results [35], experimental analysis using PhoA fusions has indicated that RseP spans the membrane four times with periplasmically-exposed N-and C-termini [7]. This was confirmed by another fusion assay using GFP as a reporter [35]. The predicted TMS 1 and 3 sequences contain the HEXXH motif and a possible third metal ligand, Asp402, respectively (Fig. 1C). Functionally important roles of these residues were demonstrated by mutational analyses [6, 7]. TMS 1, 2 and 3 form the conserved core and the expected membrane orientation of the conserved core region supports the topology suggested by the experimental data. The predicted first cytoplasmic region between TMS 1 and 2 contains a hydrophobic stretch (residues 60–73) that could form a membrane-embedded loop, similar to the corresponding regions of human S2P and SpoIVFB (Fig. 1). Early studies suggested that the central periplasmic domain of RseP (between TMS 2 and 3) contains a single PDZ domain [6, 36, 37]. The PDZ domain is a ubiquitous protein module generally involved in protein-protein interactions [38]. Later, more detailed sequence analyses predicted that RseP contains two tandemly arranged PDZ domains (PDZ-N/PDZ1 and PDZ-C/PDZ2) with circularly-permutated primary sequences [8]. This was confirmed by crystal structure analyses of the separately expressed PDZ domains [39, 40]. In contrast, S2P probably possesses only one PDZ domain (Fig. 1). Genetic and biochemical studies suggest that the PDZ domains of RseP are involved in regulation of its proteolytic functions (see section 3.3) [36, 37, 39].
3. Regulation of activity
3.1 S2P
Very little is known about the regulation of S2P, except that cleavage of SREBPs, ATF6, and CREBH by S2P requires prior cleavage by S1P [17, 18, 41]. The initial cleavage by S1P in the lumenal loop of SREBPs causes separation of its two TMSs [42] and this was proposed to allow partial unfolding of the TMS that is cleaved by S2P [43]. The cleavage by S1P is regulated at the level of transport of the substrate from endoplasmic reticulum membranes to the Golgi apparatus, where S1P and S2P are located (reviewed in [23]) (Fig. 1A). Insig proteins retain SREBP-cleavage-activating protein (SCAP) in complex with SREBPs in the endoplasmic reticulum until SCAP senses a drop in sterol concentration, releasing SCAP·SREBP complexes for vesicular transport to the Golgi apparatus.
3.2 SpoIVFB
SpoIVFB is inhibited by BofA and SpoIVFA until a signal from the forespore relieves the inhibition [44] (Fig. 2). The primary signal from the forespore is SpoIVB, a serine protease made under σG RNA polymerase control and secreted into the space between the two membranes surrounding the forespore [45, 46]. SpoIVB cleaves SpoIVFA and this is crucial to relieve inhibition of SpoIVFB [47–49]. A second serine protease, CtpB, made in both the mother cell and the forespore, and secreted from both into the space between the membranes surrounding the forespore, can cleave both SpoIVFA and BofA but appears to be a fine-tuning mechanism since absence of CtpB only delays SpoIVFB cleavage of Pro-σK slightly [47, 49, 50]. During sporulation of B. subtilis, loss of SpoIVFA and BofA in pulse-chase immunoprecipitation experiments correlated with cleavage of Pro-σK to σK by immunoblot [49]. How BofA is removed from the complex remains an open question since the BofA level still decreases, albeit more slowly, in a sporulating ctpB mutant. Removal of BofA is important since BofA alone is sufficient to substantially inhibit SpoIVFB cleavage of Pro-σK upon coexpression of these proteins in E. coli [51]. SpoIVFA alone failed to inhibit SpoIVFB, but in combination with BofA, SpoIVFB was completely inhibited in E. coli coexpression experiments [49]. Likewise, SpoIVFA alone was insufficient to inhibit SpoIVFB in sporulating B. subtilis, but SpoIVFA facilitates assembly of SpoIVFB with its inhibitor BofA [26] and localizes the complex to foci in the outermost membrane surrounding the forespore [28, 52] (Fig. 3).
Figure 2.
Regulation of SpoIVFB activity. During B. subtilis sporulation, the forespore is surrounded by two membranes upon the completion of engulfment (top part). The bottom parts depict a series of proteolytic cleavages. First, σG RNAP in the forespore causes expression of serine proteases SpoIVB and CtpB (also expressed under σE RNAP control in the mother cell), which are translocated into the intermembrane space, where they cleave the C-terminal domain of SpoIVFA to initiate its degradation (dashes). SpoIVFA is in complex with BofA and SpoIVFB in the outermost membrane surrounding the forespore, after these proteins are expressed in the mother cell under σE RNAP control. In the second step, CtpB and one or more other proteases (not shown) cleave BofA to initiate its degradation (dashes). BofA is the primary inhibitor of SpoIVFB. Finally, SpoIVFB cleaves Pro-σK, releasing σK into the mother cell.
Figure 3.
A model for ATP transport and accumulation during B. subtilis sporulation, and localization of the SpoIVFB complex with channel and engulfment complexes. A complex of SpoIIM, SpoIIP, and SpoIID proteins (red) interacts with the cell wall and causes the mother cell membrane to engulf the forespore (top left). During engulfment, channels (green) composed of SpoIIQ expressed in the forespore and SpoIIIA proteins expressed in the mother cell are formed. The channels span the intermembrane space and have been proposed to allow small molecules like ATP to move from the mother cell into the forespore. Upon completion of engulfment, the channels undergo reorganization and some components are degraded (top right). We propose that the ATP concentration rises in the mother cell and this is sensed by the CBS domain of SpoIVFB. SpoIVFA facilitates assembly of SpoIVFB with its inhibitor BofA and localizes the complex (magenta) to foci that during engulfment include the channel and engulfment complexes (bottom part), although whether SpoIVFA interacts directly with a protein(s) in the other complexes or interacts indirectly is unknown (dashed arrows).
The significance of localizing SpoIVFB in complex with its inhibitor BofA and the assembly factor SpoIVFA to foci is not clear, but it could relate to the ATP dependence of SpoIVFB [32]. The foci contain at least five other proteins, including SpoIID, SpoIIM, and SpoIIP, which are normally required for engulfment [53, 54]. These proteins appear to form a complex that interacts with the cell wall and pulls the mother cell membrane around the forespore [55] (Fig. 3). The other two proteins, SpoIIQ and SpoIIIAH, have extracellular domains that interact and zipper the mother cell membrane around the forespore during engulfment [56, 57]. The interaction of SpoIIQ with SpoIIIAH forms channels that connect the mother cell and forespore cytoplasms [58, 59]. The channels have been proposed to allow the mother cell to nurture the forespore by providing small molecules for biosynthetic activity [60]. In addition to SpoIIIAH, several other proteins encoded in the spoIIIA operon resemble components of secretion systems [58, 59], and SpoIIIAA is similar to secretion ATPases and its ATPase motifs are important for sporulation [61]. The ATPase activity of SpoIIIAA, and perhaps secretion of ATP from the mother cell, through the channels, into the forespore, might result in a relatively low ATP concentration in the vicinity of channels, ensuring that channel-associated SpoIVFB remains inactive in case it escapes BofA inhibition (Fig. 3). Upon completion of engulfment, the channels undergo reorganization and some components are degraded [59, 62], perhaps allowing the ATP concentration to rise in the mother cell, at least in the vicinity of the outermost membrane surrounding the forespore. Binding of ATP to the CBS domain of SpoIVFB would activate the enzyme [32]. In this way, activation of SpoIVFB would be coupled not only to channel-dependent activation of σG in the forespore, leading to secretion of SpoIVB and CtpB proteases that target SpoIVFA and BofA (Fig. 2), but also to completion of engulfment and destruction of the channels, resulting in a rise in ATP in the mother cell sensed by the CBS domain of SpoIVFB (Fig. 3). Since SpoIVFB cleaves Pro-σK and since ensuing transcription by σK RNAP primarily leads to production of spore coat proteins that assemble on the forespore surface, a mechanism to ensure completion of engulfment and destruction of channels seems desirable.
How might ATP activate SpoIVFB? Biochemical studies with the CBS domain of SpoIVFB suggest it forms dimers, as expected [63, 64], but that ATP binds better to a monomer and that ATP changes the interaction between the CBS domain and Pro-σK [32]. These findings led to the proposal that ATP binding to the CBS domain influences oligomerization of SpoIVFB, which was estimated to be tetrameric after detergent solubilization from membranes, and/or changes the conformation of SpoIVFB so that Pro-σK gains access to its active site. Binding of adenosine-containing ligands such as ATP, ADP, AMP, and/or S-adenosylmethionine to CBS domains in metabolic enzymes, kinases, and channels regulates their activity by inducing a conformational change, based on structural and biochemical studies (reviewed in [65]), allowing these proteins to function as sensors of cellular energy status [66]. By analogy, it was proposed that the CBS domain of SpoIVFB regulates its activity in response to the ATP concentration in the mother cell [32]. Whether the ATP concentration rises in the mother cell upon completion of engulfment and destruction of the channels, as proposed above (Fig. 3), remains to be determined, as does the ATP concentration dependence of SpoIVFB, which might indicate whether it serves as a switch that is sensitive to a small increase in ATP concentration or as a rheostat that regulates production of σK and therefore expression of its regulon over a wider range of ATP concentrations. Possible effects of ADP or AMP in combination with ATP also remain to be tested.
3.3 RseP
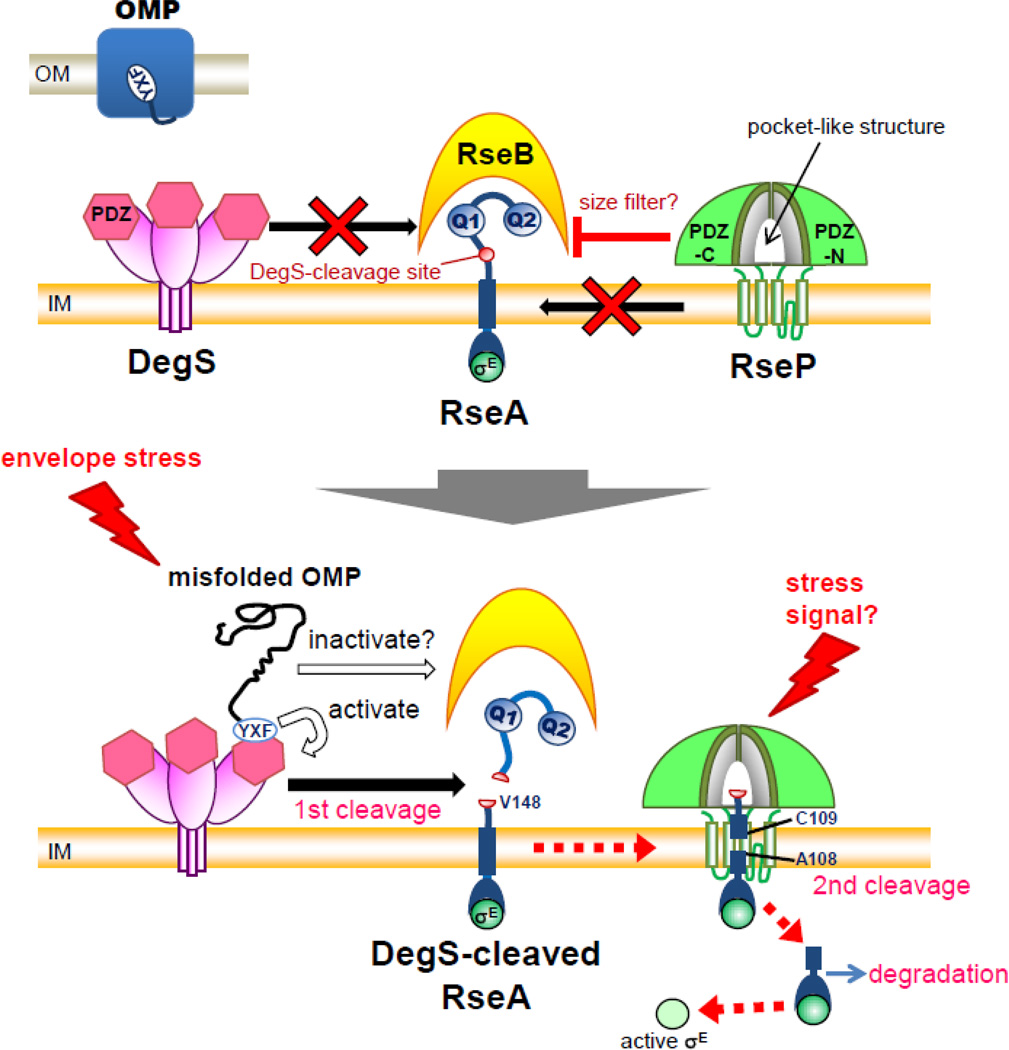
RseP is part of a proteolytic cascade that responds to extracytoplasmic stress. In E. coli, several pathways of the extracytoplasmic stress response serve to maintain homeostasis of cell surface components. Among these, the σE pathway is thought to play a major role in quality control of outer membrane proteins [67]. The most well-characterized stress cue that triggers activation of the σE pathway is misfolded outer membrane proteins (OMPs) [68, 69]. Many OMPs share a conserved motif, YXF, at their C-terminus that is normally buried within a folded β-barrel structure [70] (Fig. 4). The YXF motif is exposed by unfolding of OMPs and is directly recognized by the periplasmic PDZ domain of DegS. This stimulates DegS to cleave a peptide bond between Val148 and Ser149 in the periplasmic region of RseA [70]. This first (site-1) cleavage by DegS triggers a second cleavage (site-2 cleavage), mediated by RseP, between Ala108 and Cys109, which releases the cytoplasmic domain of RseA in a complex with σE from the membrane [5, 7, 71, 72]. Finally, the RseA cytoplasmic domain is degraded by cytoplasmic ATP-dependent proteases such as ClpXP, allowing σE to regulate transcription of stress-responsive genes [72, 73]. Full induction of the σE response requires not only OMP-dependent activation of DegS but also OMP-dependent inactivation of RseB, a periplasmic protein that binds to the periplasmic domain of RseA [74, 75] (Fig. 4). RseB binding to RseA inhibits DegS cleavage in vitro [76], probably by masking the DegS cleavage site in RseA [76, 77]. Potential β-strand-forming sequences in OMPs are suggested to act directly or indirectly to inactivate RseB and/or release it from RseA [75]. Such dual signal recognition involving DegS and RseB would ensure a specific response of the σE pathway to abnormalities in OMP folding. Although accumulation of modified or intermediate forms of lipopolysaccharide (LPS) [78] and abnormal cytoplasmic membrane proteins [79] induce the σE stress response, the underlying mechanisms remain unknown. Normally, the rseP and degS genes are essential for cell growth, but are dispensable upon down-regulation or loss of two major outer membrane proteins, OmpA and OmpC [80], since this probably reduces the requirement for σE.
Figure 4.
A model for RseP function in transmembrane signaling of the σE stress response. Upper part) Under resting conditions, DegS remains inactive as to RseA cleavage. Cleavage of intact RseA by RseP is prevented by several negative regulators including RseB, the Gln-rich sequences (Q1 and Q2) in RseA, and the PDZ domains (PDZ-N and PDZ-C) of RseP. The RseP PDZ domains could act as a size-exclusion filter to block the access of RseB-bound intact RseA into the active site of RseP. Lower part) Extracytoplasmic stresses cause accumulation of misfolded OMPs, which trigger DegS-catalyzed cleavage of the RseA periplasmic region through activation of DegS and inactivation of RseB. This first cleavage releases negative regulation and allows intramembrane cleavage of DegS-processed RseA by RseP. σE is finally activated in the cytoplasm and promotes transcription of stress-responsive genes. Some stress signal(s) might be directly recognized by the RseP PDZ domains to induce cleavage of intact RseA, resulting in σE activation.
The proteolytic functions of RseP are modulated or affected by several factors, including RseB, the Gln-rich regions in the RseA periplasmic domain, and the PDZ domains of RseP. Usually, site-2 cleavage of RseA by RseP strictly depends on prior site-1 cleavage by DegS [5, 7, 36]. However, elimination of RseB allows significant cleavage of intact RseA by RseP in the absence of DegS and suppresses lethality of the degS disruption mutant. This suggests that RseB acts to inhibit DegS-independent RseA cleavage by RseP [81]. This inhibition depends on the PDZ domains of RseP, although it is not known whether RseB directly interacts with the RseP PDZ domains. Further, systematic deletion analyses of the RseA periplasmic domain suggested that two Gln-rich regions, Q1 (residue 162–169) and Q2 (residues 190–200), prevent DegS-independent cleavage of RseA by RseP [36] (Fig. 4). However, these Gln-rich regions are not conserved among bacterial RseA homologues [82]. The crystal structure of the complex between RseB and the periplasmic domain of RseA, as well as biochemical analyses, revealed that RseB binds to neighboring regions of Q1 and Q2 [76, 77]. However, it is unclear whether RseB and the Gln-rich regions act independently or have a functional relationship.
Although the PDZ domain was initially suggested as essential for RseP function, later studies showed that mutation of conserved residues or deletion of the entire PDZ-N and/or PDZ-C domains do not inactivate RseP [36, 37, 39, 83]. Partial deletion of the PDZ domains deregulates RseP and enables it to cleave intact RseA without prior site-1 cleavage by DegS [36, 37]. Most of the mutations identified during screening for deregulated RseP mutants mapped within the PDZ domains, thus demonstrating the crucial and specific roles of the PDZ domains in the regulation of RseA cleavage [39]. In Salmonella, a bacterium closely related to E. coli, acid stress causes DegS-independent cleavage of intact RseA by RseP and the resulting activation of σE [84]. This acid-induced RseA cleavage depends on intact RseP PDZ domains. Collectively, these observations raise an intriguing possibility that, aside from the conventional OMP-dependent DegS-RseP protease cascade, there exists a novel signaling pathway(s) in which a stress signal(s) is directly recognized via the RseP-PDZ domains. Interestingly, most of the strong deregulating mutations affected conserved PDZ-N domain residues that are suggested to be critical for ligand binding [39]. The PDZ domain generally recognizes 3 to 5 C-terminal residues of a peptide ligand [38]. However, the putative ligand-binding grooves of the RseP PDZ domains are too narrow to accommodate ordinary ligands. Furthermore, the ligand-binding groove of PDZ-N is partially covered by a small helix [39, 40]. Whether ligand binding to PDZ-N is involved in modulation of RseP function remains to be determined.
An S2P homolog from Methanocaldococcus jannaschii, the only S2P family protein whose structure has been elucidated [22], possesses no PDZ domain. Although structural data are available for both of the PDZ-N and -C domains of E. coli RseP, the structures have been solved as individual domains [39, 40]. Thus, the exact disposition of the periplasmic PDZ domains relative to the membrane domain is not known. Recently, the structure of the PDZ-N/PDZ-C tandem of aaRseP (an RseP homolog from hyperthermophile Aquifex aeolicus VF5) was determined (Y. Hizukuri, S. Tabata, K. Tamura-Kawakami, T. Oda, M. Sato, J. Takagi, Y. Akiyama, and T. Nogi, unpublished results). The structure suggests a possible role of the PDZ domains in substrate discrimination. In the aaRseP PDZ tandem, the putative ligand-binding grooves face each other in an anti-parallel orientation to form a single pocket-like structure. In this configuration, PDZ domains may be positioned over the transmembrane domain such that the putative ligand-binding grooves point toward the transmembrane domain (Fig. 4). In this manner, the PDZ tandem would prevent access of substrates with a bulky periplasmic domain to the proteolytic active site. Consistent with this notion, complete removal of the entire PDZ domains (both PDZ-N and PDZ-C) renders E. coli RseP capable of cleaving intact RseA [83] (Y. Hizukuri, S. Tabata, K. Tamura-Kawakami, T. Oda, M. Sato, J. Takagi, Y. Akiyama, and T. Nogi, unpublished results). These findings raise the possibility that PDZ domains serve as a size-exclusion filter to discriminate between intact and DegS-processed RseA proteins. According to this model, DegS cleavage would facilitate RseP-catalyzed intramembrane proteolysis of RseA by reducing the size of the periplasmic domain of RseA, and by releasing the negative regulation mediated by RseB and the Gln-rich sequences. The periplasmic domain of RseA may be largely disordered when RseB is not bound to it [70, 77]. The possible cleavage of intact RseA by RseP in the absence of RseB [81] could also be explained by the size-exclusion model if RseA with a disordered, flexible periplasmic domain could pass through the PDZ filter. Mutations in PDZ-N that enable DegS-independent RseA cleavage render RseP susceptible to trypsin digestion around the PDZ-N and PDZ-C linker region [39]. This suggests that a conformational change is induced in the PDZ domains, which modulates the size-exclusion function of the PDZ filter. This in turn permits intact RseA to access the active site. Although this model can account for many previous observations, further studies are required in order to validate it.
A genetic study has suggested that DegS is also able to inhibit RseP-mediated cleavage of intact RseA [81], although little is known regarding the inhibitory mechanism, and there is no evidence of a direct interaction between DegS and RseP.
4. Substrate sequence specificity
4.1 S2P
Early mutational studies of SREBPs identified two sequence features important for cleavage by S2P, but neither of the features appears to be part of a recognition sequence that dictates the specific peptide bond to be cleaved. The sequence DRSR immediately precedes TMS 1 of human SREBP-1a and SREBP-2 (Fig. 5A shows SREBP-2). Changing DRSR to AS severely reduced cleavage within TMS 1 by S2P [85]. Changing the Asp residue to Ala in SREBP-1a (i.e., D484A) abolished cleavage, but the corresponding change in SREBP-2 (D478A) did not affect cleavage. Several additional changes to the DRSR sequence of SREBP-2 were reported in a study that also mapped the S2P cleavage site in SREBP to the peptide bond that joins L484 to C485 [86] (summarized in Fig. 5A). Changing DRSR to AAAA or DAAA abolished cleavage, but interestingly changes to DASR or DRSA did not alter the amount or position of cleavage, suggesting that neither R479 nor R481 is part of a recognition sequence that determines the cleavage site. It was subsequently mentioned that SR of the DRSR sequence is sufficient for cleavage, but the data was not shown [17]. Taken together, it appears that S480 and/or one of R479 or R481 of SREBP-2 is important for cleavage by S2P, but probably not as part of a recognition sequence dictating the cleavage site. Indeed, substitutions of Phe or Ala for residues near the cleavage site in SREBP-2 did not impair cleavage [86] (summarized in Fig. 5A), so S2P might not recognize a specific sequence near the cleavage site. The cleavage sites in two other substrates of S2P, ATF6 and CREBH [17, 18], have not been mapped, so an alignment based on cleavage sites is not possible; however, human ATF6 has the sequence PKRR preceding the target TMS and human CREBH has the sequence QTGT in the corresponding position, so R or T residues in these sequences could fulfill the role played by S and/or R residues in SREBPs. It is worth noting that B. subtilis YluC (subsequently renamed RasP) is a PDZ domain-containing IMMP that cleaves the anti-σ factor RsiW [87] and the cell division protein FtsL [13], which share the sequence KKRAS preceding their target TMSs, and substitutions in this sequence appear to impair FtsL cleavage, so RasP and S2P might both recognize one or more residues preceding the target TMS.
Figure 5.
Residues in SREBP-2 important for cleavage by S2P and a model for partial α-helix unwinding of SREBPs. A) Effect of substitutions in TMS 1 of SREBP-2 on cleavage by S2P. Residues 478–502 include four residues (blue) preceding TMS 1 (black). The cleavage site is indicated by an arrow. Single-residue substitutions having no effect (green) or reducing cleavage (orange) are shown immediately above and below the sequence, respectively. Multi-residue substitutions having no effect (green) or abolishing cleavage (red) are separated from the sequence by lines that indicate which residues were substituted. B) Model for partial α-helix unwinding of SREBPs. Left) S1P cleaves the lumenal loop of an SREBP (see Fig. 1A for domain abbreviations). Right) Separation of the two TMSs is proposed to allow the N-terminal part of TMS 1 to unwind, exposing the cleavage site to the cytosolic face of the membrane for cleavage by S2P.
The second sequence feature shown to be important for cleavage by S2P is the sequence NP within TMS 1 of SREBP-2 [43]. The substitutions N495F and P496L independently reduced cleavage, and together the double substitution abolished cleavage (summarized in Fig. 5A). Interestingly, moving the NP sequence five residues N-terminally did not change the position of cleavage, suggesting the sequence does not interact directly with S2P [43]. Rather, it was proposed that the NP sequence allows the N-terminal part of TMS 1 to unwind, after separation of the two TMSs of SREBP-2 following S1P cleavage of the lumenal loop (Fig. 5B). Substrate bending or unwinding is believed to be necessary for all types of IPs to hydrolyze a peptide bond normally inaccessible to nucleophilic attack when present in an α-helix [88]. The S2P substrate ATF6 has the sequence NYGP within its target TMS and the double substitution, N391F and P394L, abolished cleavage [17]. CREBH has a Pro residue within its target TMS that aligns with the important Pro residues of SREBP-2 and ATF6 [18], but the importance of the Pro residue and other nearby helix-destabilizing residues for CREBH cleavage by S2P remains to be tested.
In summary, one or two Arg or polar (Ser or Thr) residues preceding the target TMS, and two helix-destabilizing residues within the TMS, appear to be important for cleavage by S2P, but neither of these features appears to be part of a recognition sequence that dictates the site to be cleaved, and to the extent it has been examined, S2P might not recognize a specific sequence near the cleavage site.
4.2 SpoIVFB
Little has been reported about sequence features of Pro-σK required for cleavage by SpoIVFB. The N-terminal residue of mature σK is Y21 of Pro-σK, implying that SpoIVFB cleaves the peptide bond that joins S20 to Y21 [89]. Confirmation of the cleavage site includes N-terminal sequencing of a cleavage product from E. coli engineered to coexpress Pro-σK and SpoIVFB [51] and from an in vitro reaction with purified substrate and enzyme [32]. Subcellular fractionation experiments showed that the majority of Pro-σK is membrane-associated in sporulating B. subtilis, whereas the majority of σK is associated with core RNAP [29]. Removal of the pro-sequence appeared to release σK from the outermost membrane surrounding the forespore, allowing σK RNAP holoenzyme to form, and the results of immunofluorescence microscopy also supported this model. High salt (0.5 M NaCl or 0.6 M KCl) released much of the Pro-σK from membranes, suggesting a peripheral association. SpoIVFB was not required for association of Pro-σK with membranes. Residues 1–27 of Pro-σK, when fused to the green fluorescent protein (GFP), targeted the fusion protein to membranes in a B. subtilis spoIVF null mutant that had been starved to initiate sporulation [90]. However, the fusion protein was not cleaved in wild-type cells undergoing sporulation. A series of C-terminal GFP fusions to Pro-σK revealed that the N-terminal 117 residues of Pro-σK, but not the N-terminal 109 residues, are sufficient for cleavage to occur in sporulating wild-type B. subtilis. Replacing GFP with a His6 tag resulted in poor accumulation of the 117-residue version, although it was still cleaved, whereas the 109-residue version of Pro-σK C-terminally tagged with His6 failed to accumulate in sporulating B. subtilis. Coexpression of the His6-tagged versions of Pro-σK with SpoIVFB in E. coli yielded similar results; the 109-residue version barely accumulated and cleavage was not detected, whereas the 117-residue version accumulated but was cleaved poorly, in this case because most of the fusion protein accumulated in inclusion bodies. In contrast, a 126-residue version of Pro-σK C-terminally tagged with His6 accumulates and is cleaved very well in both B. subtilis and E. coli [51, 90]. This version, Pro-σK(1–126)-H6, includes all of the predicted sigma factor region 2.4, whereas the shorter versions lack part of this predicted α-helical region and might partially or completely misfold. It seems likely that proper folding of region 2 is required for Pro-σK to interact with SpoIVFB, but the interacting surfaces remain to be defined.
The ability of Pro-σK(1–126)-H6 to accumulate and be cleaved upon coexpression with SpoIVFB in E. coli has been utilized to investigate features of the pro-sequence that might be important for cleavage. Addition or deletion of five residues near the N terminus allowed accurate cleavage of the peptide bond joining S20 to Y21 [90]. Hence, SpoIVFB does not measure the distance from the N terminus to the cleavage site. An S20G substitution enhanced cleavage of Pro-σK(1–126)-H6 by SpoIVFB in E. coli and in vitro, perhaps because a residue with a small side chain at the position preceding the cleavage site improves access to the target peptide bond [32]. Other residues near the cleavage site influence the abundance and accuracy of cleavage by SpoIVFB (R. Zhou and L. K., unpublished results), suggesting that SpoIVFB differs from S2P in the mechanism of cleavage site selection. The predominantly hydrophobic pro-sequence contains two residues with charged side chains (K13 and E14) that might prevent it from inserting into a membrane like a typical TMS. As noted above, high salt releases much of the Pro-σK from membranes, suggesting a peripheral association [29]. However, this association does not require SpoIVFB and may not require any other protein, since Pro-σK(1–126)-H6 S20G associated readily with preformed liposomes made from E. coli lipids [32]. Perhaps K13 and/or E14 interact with the membrane surface, and/or hydrophobic parts of the pro-sequence loop into the membrane (Fig. 1B). The charge reversal substitution E14K in Pro-σK(1–126)-H6 did not affect cleavage upon coexpression with SpoIVFB in E. coli, but a K13E substitution prevented cleavage [90]. The same K13E substitution in full-length Pro-σK-H6 resulted in poor accumulation and undetectable cleavage in sporulating B. subtilis, although the variant protein appeared to be membrane associated. Whether the K13E substitution alters interaction with the membrane, with another part of Pro-σK (e.g., region 2.4, which also affects accumulation of the protein), or with SpoIVFB is unknown.
Much remains to be learned about how SpoIVFB interacts with Pro-σK The CBS domain of SpoIVFB interacts with Pro-σK [32] but the details of this interaction and the effects of ATP remain to be elucidated. Nevertheless, it seems clear already that Pro-σK interacts differently with membranes than other IMMP substrates that have been studied and SpoIVFB selects its cleavage site differently than PDZ-domain containing IMMPs. The PDZ-domain containing IMMPs that have been studied most, S2P (see section 4.1) and RseP (see section 4.3), interact with substrates in which a typical TMS likely must unwind partially in order to be cleaved, and the mechanism of cleavage site selection, to the extent it has been examined, does not appear to rely on recognition of certain residues near the cleavage site.
4.3 RseP
RseP efficiently cleaves HA-MBP-RseA140, an RseA-derived model substrate in which the cytoplasmic domain of RseA has been replaced by an unrelated sequence, HA-MBP, and the periplasmic domain is truncated to mimic the DegS-processed form [71]. This suggests that the cytoplasmic domain of any particular substrate does not play a critical role in cleavage by RseP. To date, RseA is the only established physiological substrate of E. coli RseP. However, the first and fifth TMSs (LacY TMS 1 and LacY TMS 5, respectively) of LacY (lactose permease) were found to be cleaved by RseP when the TMS of HA-MBP-RseA140 was individually replaced by these sequences [71]. A recent study has revealed that RseP can cleave a variety of signal peptides of secretory and membrane proteins in vivo, at least in the context of an N-terminally HA-MBP-attached construct [16]. The native signal peptide of periplasmic protein LivK, which is generated upon in vitro translocation of a precursor protein into inverted inner membrane vesicles, was susceptible to RseP-dependent degradation. Degradation of a signal peptide by RseP required prior processing of the precursor protein by a signal peptidase, Lep, which removes the large C-terminal mature domain. This fits with the PDZ size-exclusion filter model described in section 3.3. These observations suggest that RseP is involved in clearance of remnant signal peptides from the cytoplasmic membrane. RasP, another IMMP of B. subtilis, can also cleave signal peptides of B. subtilis secretory proteins [16]. IMMPs might generally play a role in signal peptide degradation in both gram-negative and gram-positive bacteria that do not have signal peptide peptidase homologs.
Amino acid sequences of the target TMSs of RseP substrates (RseA TMS 1, LacY TMS 1 and 5, and signal peptides) are highly variable and apparently do not share any specific motif for recognition by RseP and determination of the cleavage site, although the exact cleavage site has been definitively determined only for RseA (between A108 and C109) and LacY TMS 1 (mainly between F16 and F17, but with a small amount of cleavage between F15 and F16; the numbering is according to the original LacY protein) [71]. While RseP seems to have low sequence specificity for target TMSs, it has been shown that helix-destabilizing residues, such as Pro, Asn, Gly and Ser, in a target TMS can facilitate RseP cleavage [71]. As described in section 4.1, a pair of helix-destabilizing residues within the target TMS also promote cleavage by S2P. These residues were proposed to induce partial unwinding of the target TMS after S1P proteolysis, thereby exposing the otherwise membrane-embedded cleavage site to the cytosolic side of the membrane at which S2P cleavage occurs [43] (Fig. 5B). However, helix-destabilizing residues in a target TMS promote RseP cleavage even under detergent-solubilized conditions, and RseP can cleave intact RseA in several situations (see section 3.3). Thus, the helix-destabilizing residues of RseP substrates might play a different role from those in S2P substrates. Pull-down experiments have shown that helix-destabilizing residues in target TMSs stabilize substrate binding to RseP [91]. Similar experiments revealed that several residues (highly conserved Asn394 and Pro397 and less conserved Asn389 and Pro399) in TMS 3 of RseP are important for substrate binding. Specific combinations of Cys substitutions in RseP TMS 3 and in the target TMS of RseA, engender disulfide bond formation, suggesting that TMS 3 directly binds a substrate. In addition to facilitating nucleophilic attack of a sessile peptide bond, unwinding of a substrate’s transmembrane helix might be required for it to interact with RseP TMS 3. Thus, several properties of substrates, including the presence of helix-destabilizing residues within the target TMS and the size of the periplasmic domain may determine their cleavability by RseP in a combinational manner.
The C-terminal residue exposed by DegS cleavage has been proposed to be another feature of RseA recognized by RseP [40]. OMP signal-dependent sequential cleavage of RseA by DegS and RseP has been reconstituted in vitro using purified proteins under detergent-solubilized conditions [39, 40]. Biochemical analysis using an in vitro reconstitution system showed that mutational alterations of V148 of RseA to several charged or dissimilar amino acids compromised site-2 cleavage [40]. In addition, structural work demonstrated that the C-terminal residue of PDZ-C or its derivative is accommodated by the shallow putative ligand-binding groove of a neighboring PDZ-C molecule, and perturbation of this interaction by a mutation (I304A) at the putative ligand binding site abolishes RseP-dependent cleavage of RseA. On the basis of these results, a model was proposed suggesting that binding of PDZ-C to the newly exposed C-terminal residue (Val148) of RseA, generated by DegS-cleavage, is essential for cleavage of RseA by RseP. Although this model plausibly explains why site-2 cleavage by RseP requires the preceding site-1 cleavage, it is not consistent with the results of recent in vivo experiments in which substitutions in the putative ligand-binding regions of the RseP PDZ domains, or deletion of the PDZ domains, had little effect on RseA cleavage [83]. Moreover, alterations of the C-terminal residue of the DegS-processed form of RseA had a minor impact on in vivo substrate cleavage. Furthermore, strains having a mutant form of the chromosomal rseP gene lacking either of the two PDZ domains grew normally and exhibited almost normal RseA cleavage and σE-activation in response to an extracytoplasmic stress. These results strongly suggest that binding of the RseA C-terminus to RseP PDZ domains is not essential for RseP-catalyzed site-2 cleavage. The previously observed strong dependence of RseP cleavage on the RseA C-terminal residue [40] is reproducible but is evident only under detergent-solubilized conditions [83], highlighting the importance of investigating IMMPs and their substrates in the membrane-integrated state in vivo to discern their physiological functions.
5. Structural and biochemical insights
The crystal structure of part of an archaeal IMMP was an important advance toward a deeper understanding of these fascinating enzymes [22]. As noted in section 2, the structure confirmed the 3-TMS conserved core predicted by sequence comparisons [2, 8] and supported the 6-TMS model for the membrane-embedded domain of SpoIVFB (Fig. 1B) that had been proposed on the basis of hydropathy and topological analyses [30, 31]. The M. jannaschii enzyme used in the structure determination was C-terminally truncated to remove a tandem pair of CBS domains, leaving the 6-TMS domain referred to as mjS2P, which cleaved an artificial protein substrate and contained one zinc atom per monomer [22]. As expected, the catalytic zinc atom was coordinated by two His residues of the HEXXH motif in TMS 2 and by one Asp residue in TMS 4 (Fig. 6). Unexpectedly, TMS 4 was interrupted by a 6-residue loop that is preceded by a conserved Asn residue, contains a conserved Pro residue, and is followed by the conserved Asp residue that coordinates zinc. The side chain of the conserved Asn residue was located above the active site (as oriented in Fig. 6) and was suggested to play a critical role in catalysis such as contributing to substrate binding and/or formation of the oxyanion hole [22]. Substitutions at the corresponding position in SpoIVFB (N129A) or RseP (N394C) reduced but did not abolish substrate cleavage [3, 91]. As noted in section 4.3, several residues in TMS 3 of RseP, which corresponds to TMS 4 in the mjS2P structure (Fig. 6) or in SpoIVFB (Fig. 1B), have been implicated in substrate binding [91]. The N394C substitution in RseP interfered with substrate binding, as did several substitutions for N389 predicted to be in the TMS preceding the loop and substitutions for two Pro residues (P397C and P399C) predicted to be in the loop. RseP variants with a single Cys at P397 or P399, but not at N389 or N394, could form disulfide crosslinks with substrate RseA variants with a single Cys at P5-P1 (i.e., the five residues preceding the cleavage site N-terminally). These results indicate that P397 or P399 predicted to be in the loop of RseP (based on the mjS2P structure) are in close proximity to the substrate near its cleavage site, although there appears to be flexibility in the interaction. The results also indicate that N389 and N394 of RseP participate indirectly in substrate binding (e.g., by destabilizing α-helical structure in the enzyme). Helix-destabilizing residues in the substrate TMS were also shown to be important for substrate binding by the enzyme [91], and had previously been shown to be important for cleavage [71]. Precisely how the enzyme positions the substrate so that predominantly one peptide bond is cleaved remains unclear. To address this question, the structure of an enzyme·substrate complex would likely provide great insight. Nevertheless, the mjS2P structure both confirmed expectations from previous studies and revealed unexpected features like the TMS interrupted by a loop with conserved residues near the active site. Future studies of other IMMPs can take advantage of predictions based on the mjS2P structure [22].
Figure 6.
Structure of the membrane domain of an archaeal IMMP. The 6-TMS domain of the M. jannaschii enzyme, referred to as mjS2P, is shown in the open conformation with each α-helix a different color and other parts blue (PDB ID: 3B4R). The three residues that coordinate a zinc atom, which together with E55 activates a water molecule to catalyze hydrolysis of a substrate peptide bond, are shown. Part of the loop connecting TMSs 2 and 3 is predicted to enter the membrane and a 6-residue loop interrupts TMS 4. These loops are the basis for predicting analogous loops in S2P, SpoIVFB, and RseP as depicted in Figure 1.
Because the mjS2P structure was solved for a protein fragment that had been solubilized from membranes, the position of the active site with respect to the membrane surface is unknown. The catalytic zinc atom was predicted to be located ~14 Å from the cytosolic face of the membrane surface [22]. A channel leading from the cytosolic side was proposed to allow water access to the active site. However, Ha [19] has noted residues with charged side chains in portions of TMSs 2–6 that are oriented toward the cytosol and has argued that mjS2P, like the E. coli rhomboid GlpG, compresses the membrane. Molecular dynamics simulations suggest a modest thinning (~4 Å) of the lipid bilayer around GlpG [92, 93]. We note that S2P, SpoIVFB, and RseP have residues with charged side chains at positions corresponding to those noted by Ha in mjS2P (Fig. 7). Hence, the active site of IMMPs might not be buried quite as deeply in the membrane as predicted by Feng et al. [22]. RseP variants with a single Cys were used in experiments that measured Cys accessibility to membrane-impermeable thiol-alkylating reagents, and residues near the active site were inaccessible to probes in the 500–700 Da range, indicating that the active site is not completely exposed at the membrane surface [94]. Residues near the active site became partially accessible in the presence of protein denaturant, suggesting that the active site is sequestered in a proteinaceous structure that is partially exposed to the aqueous environment. How do substrates gain access to the active sites of IMMPs? The mjS2P structure suggested a lateral gating mechanism because the protein crystallized as a dimer with one subunit in a relatively closed conformation and the other in a more open conformation with TMSs 1 and 6 from 10 to 12 Å farther apart [22]. The open conformation was envisioned to allow a portion of the substrate in an extended conformation to enter laterally and gain access to the active site of the enzyme (Fig. 8A). However, the dimer and the two conformations could be crystal-packing artifacts. It seems likely that removal of tandem CBS domains from the M. jannaschii enzyme to produce the mjS2P fragment, together with crystallization, produced an artifactual dimer in which the two molecules are antiparallel. Whether the two conformations reflect a substrate gating mechanism of the M. jannaschii enzyme remains to be tested. In any case, RseP and many other IMMPs do not have the TMSs corresponding to TMSs 1 and 6 of mjS2P, so a different mechanism of controlling access to the active site would be needed for these IMMPs. Other mechanisms of substrate access to the active site can be envisioned. Interaction of the substrate with the enzyme and thinning of the membrane around the enzyme might allow entry of a portion of the substrate in an extended conformation from the solvent-exposed juxtamembrane region into a solvent-exposed cavity containing the active site of the enzyme (Fig. 8B). This model is similar to one proposed for substrate access to the E. coli rhomboid protease GlpG [95–97] and work on other rhomboids also supports the model [98]. However, in the case of GlpG, these and other studies indicate the importance of loop-5 (also called the L5 cap) and TMS 5 in gating substrate access to the active site (reviewed in [19, 20, 99]). A recent biochemical study of rhomboids reconstituted into proteoliposomes or expressed in cells revealed the importance of the membrane in gate dynamics as well as in unwinding of the substrate [100]. Crystal structures of GlpG·inhibitor complexes suggest that the L5 cap likely moves when substrate interacts with the enzyme [101–103]. This has been proposed to allow a portion of the substrate in an extended conformation to enter the active site via a lateral opening [102]. A similar model can be envisioned for IMMPs (Fig. 8C), especially for substrates with a cleavage site in a very hydrophobic region, since extensive solvent exposure might be too unfavorable energetically.
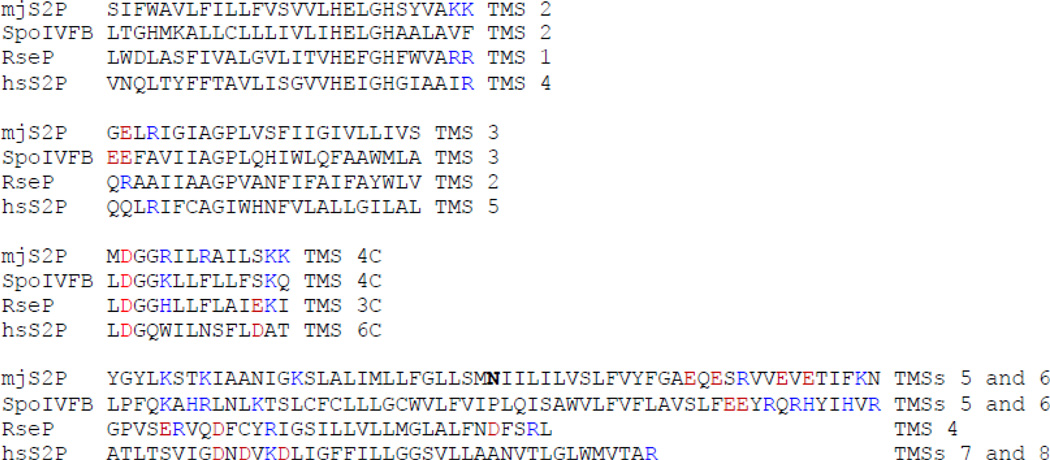
Figure 7.
Residues with charged side chains near the cytosolic ends of predicted TMSs in IMMPs. The indicated TMSs of mjS2P (numbered as in Fig. 6) are aligned with corresponding predicted TMSs of B. subtilis SpoIVFB, E. coli RseP, and human S2P (hsS2P). Residues with positively (blue) or negatively (red) charged side chains that are near the cytosolic ends of predicted TMSs are shown in color. The Asn residue at the turn separating mjS2P TMSs 5 and 6 is shown in bold.
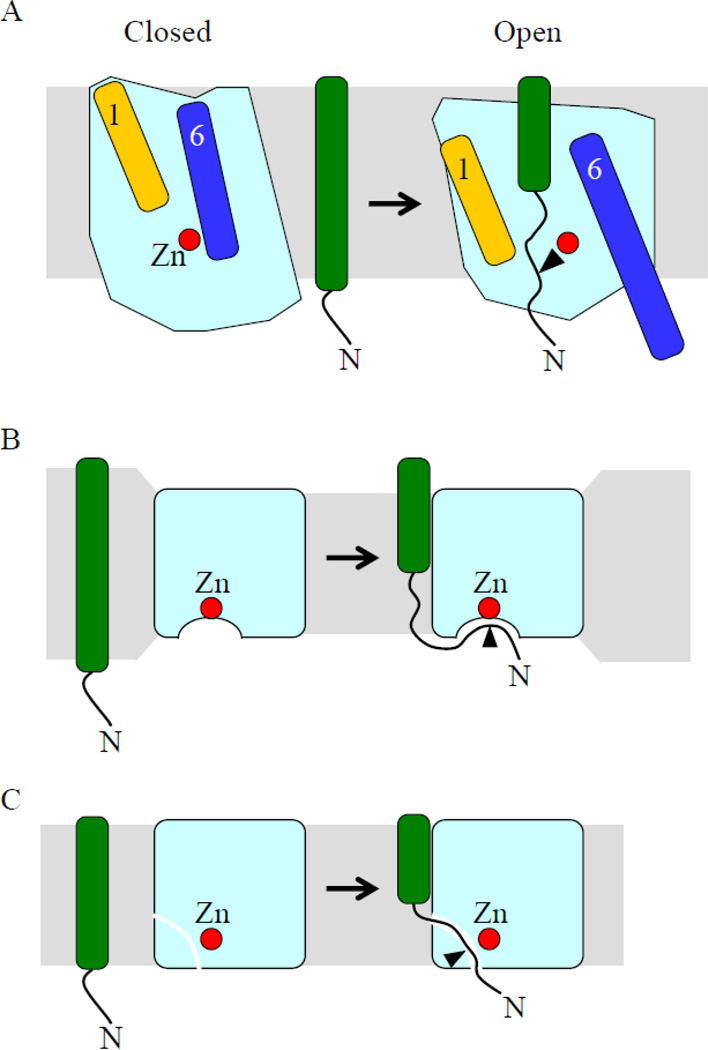
Figure 8.
Models for substrate access to the active sites of IMMPs. A) Based on closed and open conformations observed in crystals of mjS2P, TMSs 1 and 6 have been proposed to function as lateral gates that in the open conformation allow substrate access to the active site (red zinc atom). The substrate presumably must be unwound in the vicinity of the cleavage site in order for cleavage to occur (arrowhead). B) The membrane is proposed to be compressed around the enzyme so that when substrate interacts with the enzyme, a portion of the target TMS unwinds, allowing it to access the enzyme active site from the solvent-exposed juxtamembrane region. C) When substrate interacts with the enzyme, a portion of the target TMS unwinds, allowing it to access the active site via a lateral opening in the enzyme.
In the case of RseA access to the active site of RseP, the PDZ domains of RseP might normally act as a size-exclusion filter that monitors site-1 cleavage or other signals (e.g., acid-induced stress) and helix-destabilizing residues in the target TMS of RseA might allow an extended conformation to form and interact with TMS 3 of RseP, positioning the target peptide bond near the active site for cleavage via a mechanism like that depicted in Figure 8B or 8C. SpoIVFB might use a mechanism like that depicted in Figure 8B, especially since the pro-sequence of its substrate, Pro-σK, does not appear to insert into a membrane like a typical TMS, so lateral access to the active site might not be necessary even though SpoIVFB has TMSs 1 and 6 that could function as lateral gates. Rather, ATP binding to the CBS domain might change the conformation and/or oligomeric state of SpoIVFB so that the pro-sequence gains access to its active site from the solvent-exposed juxtamembrane region (Fig. 8B). Whether residues near the target peptide bond in Pro-σK interact with residues predicted to be in the loop interrupting TMS 4 of SpoIVFB (analogous to RseA interacting with TMS 3 of RseP) and whether helix-destabilizing residues in Pro-σK (e.g., N24, N25, P28, and P30) play a role (as in RseA) remains to be seen. The importance of helix-destabilizing residues within target TMSs of S2P substrates has been established but the functions of its predicted loop interrupting TMS 6, its PDZ domain, and potential lateral gates (presumably TMSs 3 and 8) (Fig. 1A) remain to be explored with insights from the mjS2P structure and from studies of RseP and SpoIVFB.
6. Conclusions and future directions
Over a decade of work on IMMPs has provided considerable mechanistic insight. IMMPs have a conserved core of three TMSs and one of these is interrupted by a loop that plays a role in substrate binding based on studies of RseP [22, 91] (Figs. 1 and 6). Presumably, the loop helps position the substrate for cleavage, but precisely how it does so and whether this function is broadly conserved in IMMPs remains to be tested. The mjS2P structure revealed a second loop that enters the membrane and is located near the active site [22] (Fig. 6). This loop appears to be conserved (Fig. 1) but its function is unknown. In addition to the conserved 3-TMS core, IMMPs have a variable number of additional TMSs and they have extramembrane domains such as PDZ and CBS domains that govern substrate access to the active site (Fig. 1). Recent work suggests the PDZ domains of RseP normally act as a size-exclusion filter that ensures substrate has undergone site-1 cleavage (Y. Hizukuri, S. Tabata, K. Tamura-Kawakami, T. Oda, M. Sato, J. Takagi, Y. Akiyama, and T. Nogi, unpublished results) (Fig. 4), yet this function of the PDZ domains can be circumvented in multiple ways. More work is needed to understand both how under certain circumstances RseP with intact PDZ domains manages to cleave RseA that has not undergone site-1 cleavage and how RseP lacking PDZ domains is prevented from cleaving membrane proteins that are not usually substrates. Likewise, more work is needed to understand how the CBS domain of SpoIVFB regulates its activity in response to ATP [32]. In addition, many putative IMMPs lack CBS or PDZ domains, but none have yet been studied. What are the substrates of these enzymes and how are they brought to the active site for cleavage?
While RseP can cleave RseA that has not undergone site-1 cleavage under certain circumstances, IMMPs typically function in pathways with other proteases. In response to misfolded OMPs, DegS cleaves RseA at site 1, RseP then cleaves RseA, releasing its N-terminal domain in complex with σE into the cytosol, and finally the N-terminal domain of RseA is degraded by ATP-dependent Clp proteases to release σE (Fig. 4). Design principles of this proteolytic cascade have begun to be revealed by detailed studies [73]. The other known substrates of RseP are remnant signal peptides left in the membrane after cleavage by signal peptidase [16]. RasP is a homolog of RseP that also cleaves signal peptides [16], but RasP has a few twists in its proteolytic cascades involving other substrates. The anti-σ factor RsiW appears to be trimmed by one or more extracellular proteases after site-1 cleavage by PrsW [104]. The need for trimming before RasP can cleave at site 2 might reflect size exclusion by the PDZ domain of RasP. On the other hand, some data suggests that RasP cleaves the cell division protein FtsL without a site-1 cleavage [13]; however, intermediates that reflect site-1 cleavage and/or trimming, could be unstable in vivo, so a definitive answer will require in vitro biochemical reconstitution. So far, this has been achieved only for RseP [71] and SpoIVFB [32]. Although SpoIVFB does not require prior cleavage of its substrate, SpoIVFB nevertheless functions in a proteolytic cascade, since the serine proteases SpoIVB and CtpB relieve inhibition of SpoIVFB by SpoIVFA and BofA (Fig. 2). Association of these proteins with channel proteins and engulfment proteins (Fig. 3) likely couples cleavage of Pro-σK by SpoIVFB to physiological and morphological changes during development in ways that remain to be understood. Human S2P cleaves its known substrates after initial cleavage by S1P (Figs. 1A and 5B). In the case of SREBPs, the sterol concentration regulates transport to the Golgi apparatus where S1P and S2P are located, but it would be surprising if the cleavages were not subject to additional regulation that remains to be elucidated, as do the regulatory mechanisms for other substrates of S1P and S2P.
Understanding how IMMPs interact with their substrates and select their cleavage sites could guide efforts to modulate cleavage by IMMPs and thus control vital signaling pathways. Early studies revealed the importance of helix-destabilizing residues in the target TMS of substrates for S2P [17, 43] and RseP [71]. For SpoIVFB, the target peptide bond in Pro-σK does not appear to be within a typical TMS, and the importance of nearby helix-destabilizing residues remains to be seen. Early studies of S2P also revealed the importance of one or two Arg or polar (Ser or Thr) residues preceding the target TMS [85, 86]. This feature is observed in RseA (i.e., R95), but its importance for cleavage by RseP has not been tested. Other RseP substrates, signal peptides, typically have one or more residues with a positively-charged side chain preceding the target TMS. The RasP substrates FtsL and RsiW share the sequence KKRAS preceding the target TMS, which has been shown to be important in the case of FtsL [13] and remains to be tested for RsiW. The SpoIVFB substrate Pro-σK has a polar Thr residue at position 2, but residues 2–6 can be deleted and cleavage still occurs [90]. SpoIVFB instead appears to recognize residues near the cleavage site (R. Zhou and L. K., unpublished data), suggesting that it differs from S2P and RseP in its mechanism of cleavage site selection. The CBS domain of SpoIVFB interacts with Pro-σK [32] and the details of this interaction are important to understand, because it might provide clues about how Pro-σK gains access to the active site of SpoIVFB, which might be different than the mechanisms of substrate access to the active sites of PDZ domain-containing IMMPs like S2P and RseP (Fig. 8). Solving the structures of IMMP·substrate complexes would likely provide additional clues about mechanisms of substrate access and cleavage site selection, as well as aiding in rational design of IMMP modulators for therapeutic purposes.
Highlights.
Progress toward understanding intramembrane metalloprotease mechanisms is described
The enzymes have a conserved core of three transmembrane helices
Two loops near the active site are also conserved and one of these binds substrate
Extramembrane domains govern substrate access to the active site
Mechanisms of substrate access and cleavage site selection remain to be elucidated
Acknowledgements
Research in the authors’ laboratories is supported by National Institutes of Health Grant GM43585 (to L.K.), by Michigan State University AgBioResearch, and by research grants from the Institute for Fermentation, Osaka (to. Y.A.), JSPS KAKENHI Grant Number 24370054 (to Y.A.) and MEXT KAKENHI Grant Number 19058007 (to Y.A.). We thank T. Nogi for helpful comments and Y. Hizukuri for assistance in preparing a figure.
Abbreviations
- S2P
Site-2 protease
- PDZ
(PSD-95, DLG, ZO-1)
- CBS
(cystathionines-β-synthase)
- IP(s)
intramembrane protease(s)
- SREBP(s)
sterol-regulatory element-binding protein(s)
- TMS(s)
transmembrane segment(s)
- IMMP(s)
intramembrane metalloprotease(s)
- RNAP
RNA polymerase
- S1P
Site-1 protease
- SCAP
SREBP-cleavage-activating protein
- OMP(s)
outer membrane protein(s)
- GFP
green fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rawson R, Zelenski N, Nijhawan D, Ye J, Sakai J, Hasan M, Chang T, Brown M, Goldstein J. Complementation cloning of SP2, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 2.Lewis A, Thomas P. A novel clan of zinc metallopeptidases with possible intramembrane cleavage properties. Protein Sci. 1999;8:439–442. doi: 10.1110/ps.8.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudner D, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu Y-TN, Kroos L. Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. J. Bacteriol. 2000;182:3305–3309. doi: 10.1128/jb.182.11.3305-3309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the σE-dependent extracytoplasmic stress response. Genes Dev. 2002;16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanehara K, Akiyama Y, Ito K. Characterization of the yaeL gene product and its S2P-protease motifs in Escherichia coli . Gene, 2001;281:71–79. doi: 10.1016/s0378-1119(01)00823-x. [DOI] [PubMed] [Google Scholar]
- 7.Kanehara K, Ito K, Akiyama Y. YaeL (EcfE) activates the σE pathway of stress response through a site-2 cleavage of anti-σERseA. Genes Dev. 2002;16:2147–2155. doi: 10.1101/gad.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinch LN, Ginalski K, Grishin NV. Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade. Protein Sci. 2006;15:84–93. doi: 10.1110/ps.051766506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen K, Gu L, Xiang X, Lynch M, Zhou R. Identification and characterization of five intramembrane metalloproteases in Anabaena variabilis . J. Bacteriol. 2012;194:6105–6115. doi: 10.1128/JB.01366-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grishin NV, Kinch LN. Bioinformatic insights into intramembrane protease evolutio. BBA Biomemb. 2012 this issue. [Google Scholar]
- 11.An FY, Sulavik MC, Clewell DB. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 1999;181:5915–5921. doi: 10.1128/jb.181.19.5915-5921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol. Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- 13.Bramkamp M, Weston L, Daniel RA, Errington J. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis . Mol. Microbiol. 2006;62:580–591. doi: 10.1111/j.1365-2958.2006.05402.x. [DOI] [PubMed] [Google Scholar]
- 14.Makinoshima H, Glickman MS. Site-2 proteases in prokaryotes: regulated intramembrane proteolysis expands to microbial pathogenesis. Microbes Infect. 2006;8:1882–1888. doi: 10.1016/j.micinf.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 15.Urban S. Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nat. Rev. Microbiol. 2009;7:411–423. doi: 10.1038/nrmicro2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito A, Hizukuri Y, Matsuo E, Chiba S, Mori H, Nishimura O, Ito K, Akiyama Y. Post-liberation cleavage of signal peptides is catalyzed by the site-2 protease (S2P) in bacteria. Proc. Natl. Acad. Sci. USA. 2011;108:13740–13745. doi: 10.1073/pnas.1108376108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 18.Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 19.Ha Y. Structure and mechanism of intramembrane protease, Semin. Cell Dev. Biol. 2009;20:240–250. doi: 10.1016/j.semcdb.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urban S, Shi Y. Core principles of intramembrane proteolysis: comparison of rhomboid and site-2 family proteases. Curr. Opin. Struct. Biol. 2008;18:432–441. doi: 10.1016/j.sbi.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolfe MS. Intramembrane proteolysis. Chem. Rev. 2009;109:1599–1612. doi: 10.1021/cr8004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, Shi Y. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 23.Rawson RB. The SREBP pathway--insights from Insigs and insects. Nat. Rev. Mol. Cell Biol. 2003;4:631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- 24.Zelenski N, Rawson R, Brown M, Goldstein J. Membrane topology of S2P, a protein required for intramembranous cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 1999;274:21973–21980. doi: 10.1074/jbc.274.31.21973. [DOI] [PubMed] [Google Scholar]
- 25.Resnekov O, Alper S, Losick R. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis . Genes Cells. 1996;1:529–542. doi: 10.1046/j.1365-2443.1996.d01-262.x. [DOI] [PubMed] [Google Scholar]
- 26.Rudner DZ, Losick R. A sporulation membrane protein tethers the pro-σK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 2002;16:1007–1018. doi: 10.1101/gad.977702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudner DZ, Pan Q, Losick RM. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci U S A. 2002;99:8701–8706. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang X, Rubio A, Chiba S, Pogliano K. Engulfment-regulated proteolysis of SpoIIQ: evidence that dual checkpoints control sigma activity. Mol. Microbiol. 2005;58:102–115. doi: 10.1111/j.1365-2958.2005.04811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B, Hofmeister A, Kroos L. The pro-sequence of pro-σK promotes membrane association and inhibits RNA polymerase core binding. J. Bacteriol. 1998;180:2434–2441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cutting S, Roels S, Losick R. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis . J. Mol. Biol. 1991;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- 31.Green D, Cutting S. Membrane topology of the Bacillus subtilis Pro-σK processing complex. J. Bacteriol. 2000;182:278–285. doi: 10.1128/jb.182.2.278-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou R, Cusumano C, Sui D, Garavito RM, Kroos L. Intramembrane proteolytic cleavage of a membrane-tethered transcription factor by a metalloprotease depends on ATP. Proc. Natl. Acad. Sci. USA. 2009;106:16174–16179. doi: 10.1073/pnas.0901455106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ades SE, Connolly LE, Alba BM, Gross CA. The Escherichia coli σE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T. Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease. Cell. 2004;117:483–494. doi: 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
- 35.Drew D, Sjostrand D, Nilsson J, Urbig T, Chin CN, de Gier JW, von Heijne G. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. USA. 2002;99:2690–2695. doi: 10.1073/pnas.052018199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehara K, Ito K, Akiyama Y. YaeL proteolysis of RseA is controlled by the PDZ domain of YaeL and a Gln-rich region of RseA. EMBO J. 2003;22:6389–6398. doi: 10.1093/emboj/cdg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bohn C, Collier J, Bouloc P. Dispensable PDZ domain of Escherichia coli YaeL essential protease. Mol. Microbiol. 2004;52:427–435. doi: 10.1111/j.1365-2958.2004.03985.x. [DOI] [PubMed] [Google Scholar]
- 38.Jelen F, Oleksy A, Smietana K, Otlewski J. PDZ domains - common players in the cell signaling. Acta Biochim. Pol. 2003;50:985–1017. [PubMed] [Google Scholar]
- 39.Inaba K, Suzuki M, Maegawa K, Akiyama S, Ito K, Akiyama Y. A pair of circularly permutated PDZ domains control RseP, the S2P family intramembrane protease of Escherichia coli . J. Biol. Chem. 2008;283:35042–35052. doi: 10.1074/jbc.M806603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Wang B, Feng L, Kang H, Qi Y, Wang J, Shi Y. Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage. Proc. Natl. Acad. Sci. USA. 2009;106:14837–14842. doi: 10.1073/pnas.0903289106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL. Sterol-regulated release of SREBP-2 from cell membrane requires two sequential cleavages, one within a transmembrane domain. Cell. 1996;85:1037–1048. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 42.Sakai J, Nohturfft A, Cheng D, Ho YK, Brown MS, Goldstein JL. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J. Biol. Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- 43.Ye J, Dave UP, Grishin NV, Goldstein JL, Brown MS. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc. Natl. Acad. Sci. USA. 2000;97:5123–5128. doi: 10.1073/pnas.97.10.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother-cell gene expression during development in Bacillus subtilis . Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- 45.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-σK processing in Bacillus subtilis . Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 46.Wakeley PR, Dorazi R, Hoa NT, Bowyer JR, Cutting SM. Proteolysis of SpoIVB is a critical determinant in signalling of Pro-σK processing in Bacillus subtilis . Mol. Microbiol. 2000;36:1336–1348. doi: 10.1046/j.1365-2958.2000.01946.x. [DOI] [PubMed] [Google Scholar]
- 47.Campo N, Rudner DZ. A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Mol. Cell. 2006;23:25–35. doi: 10.1016/j.molcel.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 48.Dong TC, Cutting SM. SpoIVB-mediated cleavage of SpoIVFA could provide the intercellular signal to activate processing of Pro-σK in Bacillus subtilis . Mol. Microbiol. 2003;49:1425–1434. doi: 10.1046/j.1365-2958.2003.03651.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhou R, Kroos L. Serine proteases from two cell types target different components of a complex that governs regulated intramembrane proteolysis of pro-σK during Bacillus subtilis development. Mol. Microbiol. 2005;58:835–846. doi: 10.1111/j.1365-2958.2005.04870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campo N, Rudner DZ. SpoIVB and CtpB are both forespore signals in the activation of the sporulation transcription factor sigmaK in Bacillus subtilis . J. Bacteriol. 2007;189:6021–6027. doi: 10.1128/JB.00399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou R, Kroos L. BofA protein inhibits intramembrane proteolysis of pro-σK in an intercompartmental signaling pathway during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. USA. 2004;101:6385–6390. doi: 10.1073/pnas.0307709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol. Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- 53.Abanes-De Mello A, Sun YL, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aung S, Shum J, Abanes-De Mello A, Broder DH, Fredlund-Gutierrez J, Chiba S, Pogliano K. Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation. Mol. Microbiol. 2007;65:1534–1546. doi: 10.1111/j.1365-2958.2007.05887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morlot C, Uehara T, Marquis KA, Bernhardt TG, Rudner DZ. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis . Genes Dev. 2010;24:411–422. doi: 10.1101/gad.1878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blaylock B, Jiang X, Rubio A, Moran CP, Jr, Pogliano K. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 2004;18:2916–2928. doi: 10.1101/gad.1252704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broder DH, Pogliano K. Forespore engulfment mediated by a ratchet-like mechanism. Cell. 2006;126:917–928. doi: 10.1016/j.cell.2006.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Camp AH, Losick R. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol. Microbiol. 2008;69:402–417. doi: 10.1111/j.1365-2958.2008.06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meisner J, Wang X, Serrano M, Henriques AO, Moran CP., Jr A channel connecting the mother cell and forespore during bacterial endospore formation. Proc. Natl. Acad. Sci. USA. 2008;105:15100–15105. doi: 10.1073/pnas.0806301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Camp AH, Losick R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis . Genes Dev. 2009;23:1014–1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, Jr, Rudner DZ. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis . PLoS Genet. 2009;5:e1000566. doi: 10.1371/journal.pgen.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiba S, Coleman K, Pogliano K. Impact of membrane fusion and proteolysis on SpoIIQ dynamics and interaction with SpoIIIAH. J. Biol. Chem. 2007;282:2576–2586. doi: 10.1074/jbc.M606056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- 64.Ignoul S, Eggermont J. CBS domains: structure, function, and pathology in human proteins. Am. J. Physiol. Cell Physiol. 2005;289:C1369–C1378. doi: 10.1152/ajpcell.00282.2005. [DOI] [PubMed] [Google Scholar]
- 65.Baykov AA, Tuominen HK, Lahti R. The CBS domain: a protein module with an emerging prominent role in regulation. ACS Chem. Biol. 2011;6:1156–1163. doi: 10.1021/cb200231c. [DOI] [PubMed] [Google Scholar]
- 66.Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ades SE. Regulation by destruction: design of the σE envelope stress response. Curr. Opin. Microbiol. 2008;11:535–540. doi: 10.1016/j.mib.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 68.Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. The activity of σE, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 69.Sohn J, Sauer RT. OMP peptides modulate the activity of DegS protease by differential binding to active and inactive conformations. Mol. Cell. 2009;33:64–74. doi: 10.1016/j.molcel.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 71.Akiyama Y, Kanehara K, Ito K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 2004;23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 73.Chaba R, Grigorova IL, Flynn JM, Baker TA, Gross CA. Design principles of the proteolytic cascade governing the σE-mediatedand envelope stress response in Escherichia coli: keys to graded, buffered, and rapid signal transduction. Genes Dev. 2007;21:124–136. doi: 10.1101/gad.1496707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chaba R, Alba BM, Guo MS, Sohn J, Ahuja N, Sauer RT, Gross CA. Signal integration by DegS and RseB governs the σE-mediated envelope stress response in Escherichia coli . Proc. Natl. Acad. Sci. USA. 2011;108:2106–2111. doi: 10.1073/pnas.1019277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kulp A, Kuehn MJ. Recognition of beta-strand motifs by RseB is required for σE activity in Escherichia coli . J. Bacteriol. 2011;193:6179–6186. doi: 10.1128/JB.05657-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cezairliyan BO, Sauer RT. Inhibition of regulated proteolysis by RseB. Proc. Natl. Acad. Sci. USA. 2007;104:3771–3776. doi: 10.1073/pnas.0611567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim DY, Kwon E, Choi J, Hwang HY, Kim KK. Structural basis for the negative regulation of bacterial stress response by RseB. Protein Sci. 2010;19:1258–1263. doi: 10.1002/pro.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tam C, Missiakas D. Changes in lipopolysaccharide structure induce the σE-dependent response of Escherichia coli . Mol Microbiol. 2005;55:1403–1412. doi: 10.1111/j.1365-2958.2005.04497.x. [DOI] [PubMed] [Google Scholar]
- 79.Shimohata N, Nagamori S, Akiyama Y, Kaback HR, Ito K. SecY alterations that impair membrane protein folding and generate a membrane stress. J. Cell Biol. 2007;176:307–317. doi: 10.1083/jcb.200611121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Douchin V, Bohn C, Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli . J. Biol. Chem. 2006;281:12253–12259. doi: 10.1074/jbc.M600819200. [DOI] [PubMed] [Google Scholar]
- 81.Grigorova IL, Chaba R, Zhong HJ, Alba BM, Rhodius V, Herman C, Gross CA. Fine-tuning of the Escherichia coli σE envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev. 2004;18:2686–2697. doi: 10.1101/gad.1238604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim DY, Jin KS, Kwon E, Ree M, Kim KK. Crystal structure of RseB and a model of its binding mode to RseA. Proc. Natl. Acad. Sci. USA. 2007;104:8779–8784. doi: 10.1073/pnas.0703117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hizukuri Y, Akiyama Y. PDZ domains of RseP are not essential for sequential cleavage of RseA or stress-induced σE activation in vivo . Mol. Microbiol. 2012;86:1232–1245. doi: 10.1111/mmi.12053. [DOI] [PubMed] [Google Scholar]
- 84.Muller C, Bang IS, Velayudhan J, Karlinsey J, Papenfort K, Vogel J, Fang FC. Acid stress activation of the σE stress response in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2009;71:1228–1238. doi: 10.1111/j.1365-2958.2009.06597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hua X, Sakai J, Brown MS, Goldstein JL. Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J. Biol. Chem. 1996;271:10379–10384. doi: 10.1074/jbc.271.17.10379. [DOI] [PubMed] [Google Scholar]
- 86.Duncan E, Dave U, Sakai J, Goldstein J, Brown M. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J. Biol. Chem. 1998;273:17801–17809. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]
- 87.Schobel S, Zellmeier S, Schumann W, Wiegert T. The Bacillus subtilis σW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 2004;52:1091–1105. doi: 10.1111/j.1365-2958.2004.04031.x. [DOI] [PubMed] [Google Scholar]
- 88.Wolfe MS, Kopan R. Intramembrane proteolysis: theme and variations. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- 89.Kroos L, Kunkel B, Losick R. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science. 1989;243:526–529. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- 90.Prince H, Zhou R, Kroos L. Substrate requirements for regulated intramembrane proteolysis of Bacillus subtilis pro-σK . J. Bacteriol. 2005;187:961–971. doi: 10.1128/JB.187.3.961-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koide K, Ito K, Akiyama Y. Substrate recognition and binding by RseP, an Escherichia coli intramembrane protease. J. Biol. Chem. 2008;283:9562–9570. doi: 10.1074/jbc.M709984200. [DOI] [PubMed] [Google Scholar]
- 92.Bondar AN, del Val C, White SH. Rhomboid protease dynamics and lipid interactions. Structure. 2009;17:395–405. doi: 10.1016/j.str.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou Y, Moin SM, Urban S, Zhang Y. An internal water-retention site in the rhomboid intramembrane protease GlpG ensures catalytic efficiency. Structure. 2012;20:1255–1263. doi: 10.1016/j.str.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koide K, Maegawa S, Ito K, Akiyama Y. Environment of the active site region of RseP, an Escherichia coli regulated intramembrane proteolysis protease, assessed by site-directed cysteine alkylation. J. Biol. Chem. 2007;282:4553–4560. doi: 10.1074/jbc.M607339200. [DOI] [PubMed] [Google Scholar]
- 95.Akiyama Y, Maegawa S. Sequence features of substrates required for cleavage by GlpG, an Escherichia coli rhomboid protease. Mol. Microbiol. 2007;64:1028–1037. doi: 10.1111/j.1365-2958.2007.05715.x. [DOI] [PubMed] [Google Scholar]
- 96.Maegawa S, Koide K, Ito K, Akiyama Y. The intramembrane active site of GlpG, an E. coli rhomboid protease, is accessible to water and hydrolyses an extramembrane peptide bond of substrates. Mol. Microbiol. 2007;64:435–447. doi: 10.1111/j.1365-2958.2007.05679.x. [DOI] [PubMed] [Google Scholar]
- 97.Wang Y, Maegawa S, Akiyama Y, Ha Y. The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG. J. Mol. Biol. 2007;374:1104–1113. doi: 10.1016/j.jmb.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Strisovsky K, Sharpe HJ, Freeman M. Sequence-specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol. Cell. 2009;36:1048–1059. doi: 10.1016/j.molcel.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lemberg MK, Freeman M. Cutting proteins within lipid bilayers: rhomboid structure and mechanism. Mol. Cell. 2007;28:930–940. doi: 10.1016/j.molcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 100.Moin SM, Urban S. Membrane immersion allows rhomboid proteases to achieve specificity by reading transmembrane segment dynamics. elife. 2012;1:e00173. doi: 10.7554/eLife.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vinothkumar KR, Strisovsky K, Andreeva A, Christova Y, Verhelst S, Freeman M. The structural basis for catalysis and substrate specificity of a rhomboid protease. Embo J. 2010;29:3797–3809. doi: 10.1038/emboj.2010.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xue Y, Chowdhury S, Liu X, Akiyama Y, Ellman J, Ha Y. Conformational change in rhomboid protease GlpG induced by inhibitor binding to its S' subsites. Biochemistry. 2012;51:3723–3731. doi: 10.1021/bi300368b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xue Y, Ha Y. Catalytic mechanism of rhomboid protease GlpG probed by 3,4-dichloroisocoumarin and diisopropyl fluorophosphonate. J. Biol. Chem. 2012;287:3099–3107. doi: 10.1074/jbc.M111.310482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Heinrich J, Hein K, Wiegert T. Two proteolytic modules are involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW. Mol. Microbiol. 2009;74:1412–1426. doi: 10.1111/j.1365-2958.2009.06940.x. [DOI] [PubMed] [Google Scholar]