Abstract
Interferon beta (IFNβ) reduces disease burden in relapsing-remitting multiple sclerosis (MS) patients. In this study, IFNβ-1b-treated MS patient gene expression profiles and biological knowledgebases were integrated to study IFNβ's pleiotropic mechanisms of action. Genes involved in immune regulation, mitochondrial fatty acid metabolism and antioxidant activity were discovered. Plausible mediators of neuronal preservation included NRF2, downregulation of OLA1, an antioxidant suppressor, and the antioxidant gene ND6, implicated in optic neuropathy and MS-like lesions. Network analysis highlighted IKBKE, which likely has a role in both viral response and energy metabolism. A comparative analysis of therapy-naive MS- and IFNβ-associated gene expression suggests an IFNβ insufficiency in MS. We observed more gene expression changes in long-term treatment than during acute dosing. These distinct short- and long-term effects were driven by different transcription factors. Multi-gene biomarker signatures of IFNβ treatment effects were developed and subsequently confirmed in independent IFNβ-1b-treated MS studies, but not in glatiramer acetate-treated patients.
Keywords: interferon β-1b, biomarker signatures, multiple sclerosis, oxidative stress, IKK epsilon, NRF2
Introduction
Multiple sclerosis (MS) is an autoimmune-mediated progressive neurodegenerative disease that results in permanent disability and a significant decrease in quality of life.1, 2 Current standard of care MS drugs provide similar clinical benefit but vary considerably in side effects, dosing, route of administration and mechanism of action.3 There is an increasing need to provide guidance in treatment options to MS patients and their physicians. Treatment monitoring using biomarkers could provide assertive management of disease progression.4, 5, 6, 7
Interferon beta (IFNβ) is a highly pleiotropic member of a family of cytokines that regulate innate and adaptive immune responses.8, 9 IFNβ suppresses activation of T and B cells, prevents translocation of immune cells from the periphery into the central nervous system, inhibits astrocyte and microglial cell differentiation, and is potentially neuroprotective.10, 11, 12, 13 The mechanism of action of IFNβ in MS remains uncertain but elucidating effects underlying treatment could provide guidance in treatment choices.
Previously, we have characterized14 and confirmed15 differential gene expression in short-term and long-term IFNβ therapy.16, 17 Gene expression microarrays were employed here to discover biological information relevant to the mechanism of action of IFNβ-1b. Analysis built on an approach previously applied to study IFNβ-1b effects in patients14 and was applied to a broader set of genes measured in mononuclear cells obtained from MS patients and healthy individuals using the Affymetrix HuEx1.0ST (‘all exon') platform. As multi-gene effects are expected to be more stable across microarray platforms than single-gene effects,18 multi-gene IFN-biomarker signatures were characterized and subsequently confirmed in independently sampled, previously published patient data.
Gene expression microarray analysis presents a challenge to make sense of high-dimensional data comprised of measurements on tens of thousands of genes. We approached this challenge by bringing together genes into pre-defined biological contexts. Three biological themes were integrated into this study to infer protein effects upstream, downstream and interacting with the measured genes. First, for transcription factor binding, sets of genes that were annotated as bound by a specific transcription factor implicated events upstream of gene expression and facilitated pre-expression inference. Second, gene sets defined by canonical pathways and functions provided post-expression inference for biological effects downstream of drug-mediated gene expression changes. Finally, gene sets encoding the interaction neighbors of a given protein used gene expression to implicate proteins potentially influenced by a changing molecular environment. This protein network analysis supports peri-expression inference, or guilt-by-association. In this study, specific connections among these realms of knowledge and MS scientific literature were highlighted and permitted development of multi-gene IFNβ-bioactivity signatures that were evaluated across larger groups of independently sampled patients.
Materials and methods
Study population
Fifty-two human subjects, including healthy individuals and clinically defined MS patients diagnosed according to McDonald criteria,19 were recruited to the study (Table 1). The study was approved by the Internal Review Board at the University of Chicago, and all patients gave written informed consent.
Table 1. Patient characteristics.
| N | Gender | Mean age±s.d. (years) | Mean EDSS±s.d. | |
|---|---|---|---|---|
| Control | 9 | 4 F, 5 M | 40.1±13.9 | NA (0) |
| Stable RRMS | 11 | 7 F, 4 M | 38.2±13.8 | 2.3±1.48 |
| Active RRMS | 10 | 6 F, 4 M | 43.4±11.5 | 4.72±1.3 |
| PPMS | 7 | 1 F, 6 M | 49.9±13.6 | 5.43±1.79 |
| SPMS | 8 | 5 F, 3 M | 46.5±7.1 | 5.91±1.56 |
| IFNβ treated | 7 | 6 F, 1 M | 42.6±14.3 | 4.4±1.0 |
Abbreviations: EDSS, expanded disability status scale; F, female; IFNβ, interferon beta; M, male; MS, multiple sclerosis; NA, not applicable; PPMS, primary-progressive MS; RRMS, relapsing-remitting MS; SPMS, secondary-progressive MS.
Seven patients were treated with IFNβ-1b, and their expanded disability status scale (EDSS)20 scores had a mean of 4.4 and ranged from 2.5 to 5.5. To obviate effects of disease activity on IFN responses, treated clinically ‘stable' patients were free of exacerbations for at least 10 months before and 4 months after blood collection, and none were in the progressive phase of MS. No patients had received corticosteroids or immunosuppressants for 1 year before or during the study. Six of the seven patients had received high-dose, high-frequency Betaseron IFNβ-1b therapy for >3 years. Three patients had received prior treatment with other IFNβ therapies (that is, IFNβ-1a). Patients had received IFNβ for an average of 6.9 (0.5–10.5) years and had discontinued therapy for at least 64 h before beginning this study. For this kinetic study, Betaseron IFNβ-1b (8 million international units (IU), specific activity 2.3 × 107 IU mg–1) was self-administered subcutaneously under a physician's supervision. Blood was drawn by the physician just before administration of drug (t=0) and at precisely controlled times at 4, 18 and 42 h post-administration.
Forty-five treatment-naive subjects were studied. Ten relapsing-remitting MS (RRMS) patients had blood drawn during a clinical exacerbation (active RRMS), and 11 RRMS patients were free of exacerbations 6 months before and after sample draw and showed no EDSS progression for 1 year (stable RRMS). Seven treatment-naive patients had primary-progressive MS, and eight had secondary-progressive MS. Nine healthy individuals were included. Table 1 summarizes patient information.
RNA isolation and gene chip analysis
Heparinized blood (35 ml) was collected by venipuncture and peripheral blood mononuclear cells were immediately isolated using Ficoll–Paque Plus density gradients (Amersham Biosciences, NJ, USA). RNA was isolated using an RNeasy Midi Kit (Qiagen, Santa Clara, CA, USA). Quality and quantity of the RNA were determined using a Bioanalyzer 2100 (Agilent Instruments, Foster City, CA, USA). Samples having a 28S/18S ratio below 1.5 were excluded from this study. RNA was hybridized to GeneChip HuEx1.0ST arrays (Affymetrix, Santa Clara, CA, USA) containing approximately 1.4 million probe sets recognizing over 1 million exon clusters.
Comparative gene expression analysis using microarrays in a two other data sets
Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) record GSE16214 provided data from the Comprehensive Longitudinal Investigation of MS at the Brigham and Women's Hospital (CLIMB study), which included untreated (n=82), IFNβ-treated (n=94), and glatiramer–acetate-treated (n=64) (Copaxone) patients. Timing of dosing relative to phlebotomy was not captured in the public CLIMB data. GEO record GSE24427 by Goertsches provided independent data from IFNβ-1b-treated patients.21 We inferred that patients in the Goertsches study were sampled 48 h after administration.
Probe set (3′ arrays) and transcript (HuEx1.0ST arrays) expression summarization was performed using the robust multichip analysis (RMA)22 implementation in Affymetrix Power Tools version 1.12.0. Probe set and transcript annotations used Affymetrix's NetAffx Release 30 (November 2009).
Knowledgebases
Gene sets by transcription factor binding were obtained from MSigDB (http://www.broadinstitute.org/gsea/msigdb/index.jsp), specifically the c3 TFT gene sets, and by searching the Biomolecular Object Network Databank (http://bond.unleashedinformatics.com/) for DNA–protein interactions. Pathway Commons23 provided canonical pathways and a network of interacting proteins. Genes in these annotations were identified by Entrez Gene identifiers.
Data handling
Data handling was performed using Perl and PostgreSQL. Statistical analysis was performed in R.24
Short- and long-term differential gene expression
The coincident extreme ranks in numerical observations method14 used a gene set analysis approach to investigate IFNβ-1b short-term effects (hours post-administration), long-term effects (comparing samples from patients treated for years to those from therapy-naive MS patients), and MS disease gene expression (therapy-naive MS compared with healthy individuals). To facilitate a large number of gene set tests, a nested coincident extreme ranks in numerical observation testing routine replaced the Monte Carlo approach in Yamaguchi et al.14 Statistical testing including false discovery rate analysis details are provided as Supplementary Methods.
An integrated, cross-microarray platform database enabled flexible querying of microarray results to select genes regulated by IFNβ-1b and genes annotated with specific functions or pathways. Selection of 3′ array probe sets, with the intent to capture the effects measured on the HuEx1.0ST array, were completed before running tests on the published Goertsches21 data (see Supplementary Methods).
Signatures were selected based on statistical evidence derived from short- and long-term IFN-β-induced gene expression, and grouped according to treatment-affected biological functions and MS-relevant biology described in published literature. Supplementary Information provides in-depth descriptions of signature definition criteria, as well as the lists of the selected genes and probe sets (Supplementary Tables 1–10).
Signature testing was performed using Global Test,25 a multiple regression method that used only the gene expression results within a set of transcript measurements, and thus explicitly ignored expression levels estimated from the entire microarray. Global Test does not specifically model multiple transcript measurements per gene, so only one transcript or probe set measurement per gene was permitted in any signature.
Coincident extreme ranks in numerical observation was used to identify the most important differential expression effects of IFNβ-1b in the research sample profiles. Global Test was used to evaluate whether signatures were significantly regulated in particular studies.
Results
IFNβ-1b treatment: differential expression and timescales
Differential gene expression in MS patient peripheral blood mononuclear cells at pre-dose levels was compared with 4, 18 and 43 h post-IFNβ-1b (‘short-term' comparisons). Pharmacokinetics of differential gene expression after a single dose of IFNβ-1b using HuEx1.0ST arrays were similar to those previously observed.15 Classic acute IFN response genes were among the most differentially expressed (OAS1/2/3, MX1/MxA, CXCL10/IP-10 and others).
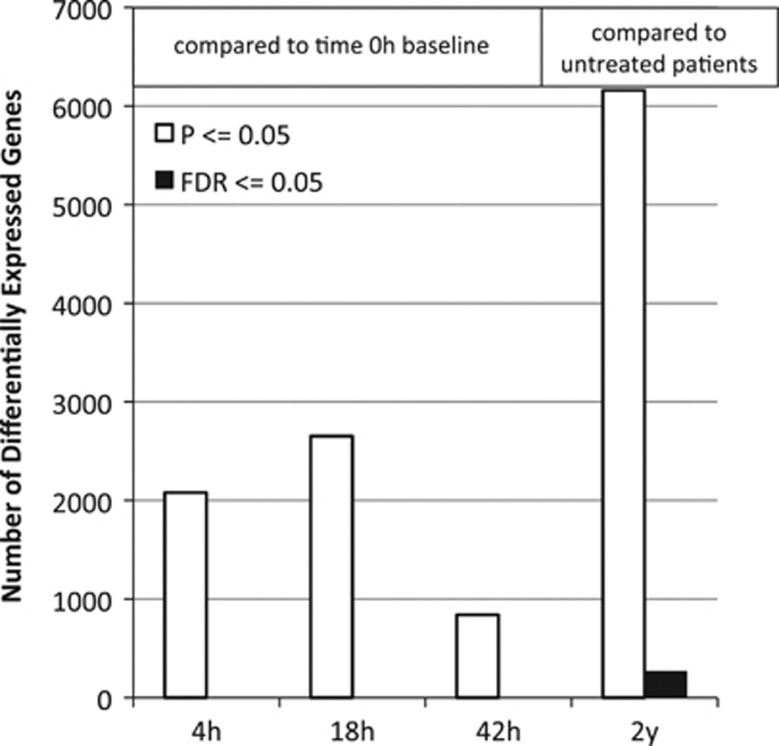
Short-term changes were contrasted with long-term treatment changes. ‘Long-term' changes were defined by the difference between gene expression in therapy-naive patients and patients after an average of 6.9 years of IFNβ-1b therapy. The long-term comparison revealed a larger number of differentially expressed genes than did short-term comparisons, whether differential expression was measured by a nominal P-value cutoff or by false discovery rate (FDR) analysis (Figure 1). Although the range of expression ratios was similar for short- and long-term effects, more significant up and downregulation was observed in the long term. Moreover, for most transcripts, long-term treatment reversed the effects of short-term dosing, with larger changes in magnitude in the long term (Supplementary Figure 1).
Figure 1.
Differentially expressed genes at the P⩽0.05 cutoff are roughly threefold more abundant in the long-term comparison than in the short-term comparisons. No probe sets exhibit a false discovery rate (FDR) of 0.05 or less in the short-term comparisons. In all, 264 genes have at least one probe set that exhibits a t-test FDR of 0.05 in the long-term comparison (black bar). In all, 715 probe sets map to these 264 genes. The FDR analysis tells us that, even considering the large number of tests (312 023) for transcript differential expression, there is an excess of probe sets yielding small P-values, such that we expect only 5% to be false discoveries. Thus, there is confidence that the vast majority of the 264 genes are regulated by long-term interferon beta (IFNβ)-1b treatment. h, hours; y, years.
Pathways and network analysis of long-term IFNβ-1b effects in patients
Three gene set analyses were computed to infer protein effects upstream, downstream and interacting with the measured long-term gene expression changes. The inferred upstream effects of transcription factors clearly showed a contrast between short-term and long-term transcriptional changes (Figure 2). The multi-gene analysis approach revealed distinct short-term and long-term gene expression patterns. The independently sampled CLIMB study data, which comprises patient samples drawn at various times after beginning IFNβ therapy, shows a mixture of the patterns.
Figure 2.
Gene sets characterized by transcription factor binding (rows) show excessive differential expression in sample comparisons (columns) as indicated by false discovery rate (FDR) coincident extreme ranks in numerical observation (CERNO) values. The most significant differentially expressed gene sets (dark orange) are distinct between short-term and long-term comparisons. Independent data from the Comprehensive Longitudinal Investigation of MS at the Brigham and Women's Hospital (CLIMB) study (Affymetrix 3′ array HG-U133 Plus 2.0) also show evidence of both short- and long-term treatment effects, which corroborates the multi-gene analysis of the HuEx1.0ST data.
Downstream effects revealed a large number of canonical pathways influenced by long-term IFNβ-1b treatment (224 pathways at FDR<0.05 of 953 tested). To relate biological pathways to single-gene changes, 28 of the 264 long-term differentially expressed genes were identified as members of the top-100 differentially expressed pathways. A cluster analysis of these 28 genes was used to group the observed pathway effects (Figure 3). Three major effects of IFNβ-1b long-term treatment appeared within the top-100 differentially expressed pathways. These effects included gene transcription, apoptosis and metabolism. Metabolism pathways included mitochondrial beta-oxidation of fatty acids, depicted as the intersection of a set of annotated pathways and a distinct pattern of gene membership. Canonical pathways spotlighted mitochondrial function and energy metabolism, both of which have an important role in maintaining neuronal integrity26 as well as immune cell function.
Figure 3.
The 100 most significantly changed functional pathways, as detected with coincident extreme ranks in numerical observation (CERNO), are shown. In all, 28 of the 264 long-term interferon beta (IFNβ)-1b-regulated genes map to these 100 pathways. Membership of the genes in the pathways is indicated by a red square. The membership matrix was ordered using hierarchical clustering, revealing a pattern that includes a cluster of functions and genes related to beta-oxidation of fatty acids.
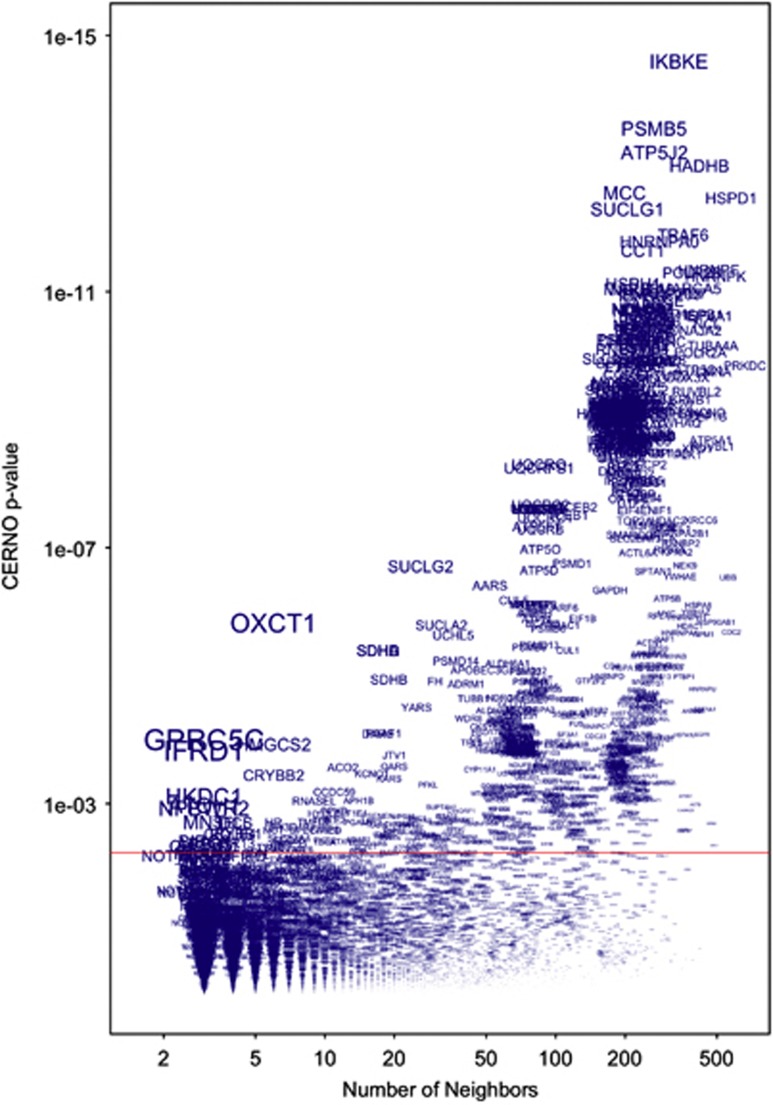
Peri-expression analysis of protein interaction neighborhoods revealed additional regulatory proteins, not necessarily ones in named pathways (Figure 4). Disrupted levels of gene transcripts were significantly associated with neighbors of particular proteins. As larger signature sets provided more power to detect smaller effects on test ranks, we employed a plotting technique to determine P-values associated with a given protein neighbor that stood out when viewed as a function of set size. We explored three of the most differentially expressed neighborhoods of different sizes.
Figure 4.
Specific protein neighborhoods, such as those surrounding IKBKE and GPRC5C, exhibit excessive change, as implied by the long-term interferon beta (IFNβ)-1b treatment comparison. Each name represents a gene set of the proteins that interact with the named protein. The results of the multi-gene test for excessive differential expression are represented by the coincident extreme ranks in numerical observation (CERNO) P-value (y axis) plotted against the size of the gene sets (x axis). The sizes of the text names on the plot are scaled to emphasize more significant test results adjusted for the number of genes in the multi-gene test. The red line indicates the false discovery rate (FDR) 0.05 cutoff for the P-values adjusted for the 7433 protein neighborhood tests.
The most significant long-term differentially expressed protein neighborhood, IKBKE (343 genes, FDR=2e−11), highlighted the IκB kinase (IKK)-related kinase IKKɛ. Another notable interaction neighborhood was GPRC5C (Figure 4, toward lower left, three loci, FDR=0.002). All three neighbors of GPRC5C encode genes regulated by IFNβ-1b, and individually exhibited transcripts differentially expressed at P<0.001. One, OLA1, yielded an FDR of 0.02 and thus was also identified individually by differential expression analysis. Another neighborhood, around OXCT1, was regulated by IFNβ-1b treatment (Figure 4, six genes, FDR=5e−5). SCOT, encoded by the OXCT1 gene, is a homodimeric mitochondrial matrix enzyme that catalyzes the reversible transfer of coenzyme A from succinyl-CoA to acetoacetate and has a central role in extrahepatic ketone body catabolism.
Long-term IFNβ-1b treatment changes compared with changes in therapy-naive MS
Long-term-regulated genes were used to explore the relationship between effects of treatment versus intrinsic MS disease activity in therapy-naive patients. We plotted the expression ratios comparing long-term-treated patients with therapy-naive patients against ratios comparing healthy controls with therapy-naive patients. Significant long-term IFNβ-1b-induced gene expression changes tended to reverse disease-modified gene expression, in contrast to short-term IFNβ-1b effects that enhanced this gene expression (Figure 5).
Figure 5.
Long-term, but not short-term, interferon beta (IFNβ) effects counter-regulate disease-related changes. (a) Short-term changes: genes exhibiting the most significant changes (t-test P⩽0.002) observed 4-h after injection tend to be either unchanged in (stable) disease or change in the same direction as characteristic of multiple sclerosis (MS) patients, which is especially true of those probe sets more significantly changed in disease (P⩽0.05, red points show positive slope). (b) Long-term changes: expression ratios in the long-term-treated patients versus therapy-naive patients compared with the ratios found in therapy-naive patients relative to healthy controls. The long-term-regulated gene expression for these differentially expressed probe sets (false discovery rate (FDR)⩽0.05 based on t-test) tends to reverse the expression changes that characterize disease, especially when focusing on the probe sets exhibiting more significant changes in disease (P⩽0.05, red points show negative slope).
Selecting genes for multi-gene signatures
Our observation of distinct long-term treatment effects suggested that long-term-specific differential gene expression signatures could be identified. As signatures were intended for cross-platform use (to evaluate independent studies), criteria included significance of change observed in the HuEx 1.0 short-term data and, in some cases, a lower bound on expression change ratio. Further details are described in Materials and methods section and Supplementary Information (Supplementary Tables 1–10).
Two long-term IFNβ-1b signatures designated long-term (no short-term regulation in the same direction as long-term regulation) and long-term 2 (none of the transcripts being regulated in the short term) were defined. Two additional signatures were defined to target the mitochondrial (MT-OX) and IKBKE (IKE) effects observed in the long-term comparison. A fifth signature (disease reversal) was defined as genes that change in an opposite direction in long-term treatment as compared with therapy-naive MS. Therapy-naive MS gene expression changes were determined by comparing stable therapy-naive RRMS patients and healthy individuals.
IFNβ-regulated genes involved in blood–brain barrier, important in MS etiology, were selected as an additional signature. NFE2L2 (NRF2), a key regulator of antioxidant response,27, 28 provides a mechanism of neuroprotection.29 Observed changes in NRF2-related gene expression suggested IFNβ effects could be mediated by NRF2, and a signature was defined. Finally, three short-term signatures designed to capture IFNβ biomarkers common to 4- and 18-h comparisons (short-term: 4 h+18 h), and differential gene expression effects specific to 4 h (short-term: 4 h) or 18 h (short-term: 18 h) post-IFNβ-1b treatment.
IFNβ-1b gene expression signatures were validated in an independent study and distinct from GA
As the 10 signatures were developed using the HuEx1.0ST data set and adapted to the HG-U133 Plus 2.0 microarray platform using (in part) observations in the CLIMB study, results from testing the signatures in these same data were largely confirmatory (Supplementary Table 1). In all, 8 of the 10 signatures were significant in the CLIMB IFNβ data at P⩽0.05, with signatures NRF2 and disease reversal as the exceptions. The Goertsches data set21 provided an entirely independent set of IFNβ-1b-treated patient data in which to test the 10 signatures (Table 2). All 10 signatures were significant at P⩽0.05 in at least one of the time-point comparisons.
Table 2. Independent confirmation of IFNβ-1b gene expression signatures.
|
Goertsches |
CLIMB | ||||
|---|---|---|---|---|---|
| 2 d vs 0 m | 1 m vs 0 m | 12 m vs 0 m | 24 m vs 0 m | GA vs untreated | |
| ST-4 h+18 h | 4.33E−06 | 2.51E−15 | 1.17E−05 | 4.88E−08 | 5.10E−01 |
| ST-4 h | 2.46E−03 | 2.52E−12 | 7.00E−05 | 7.92E−06 | 1.27E−01 |
| ST-18 h | 8.50E−02 | 3.18E−09 | 3.18E−02 | 8.80E−07 | 3.37E−01 |
| LT | 1.16E−03 | 2.57E−10 | 2.43E−05 | 9.39E−03 | 1.08E−01 |
| LT2 | 2.26E−02 | 3.44E−05 | 3.88E−04 | 5.72E−02 | 9.96E−02 |
| MT-OX | 5.10E−03 | 3.54E−04 | 1.56E−02 | 6.21E−01 | 5.02E−02 |
| NRF2 | 1.47E−01 | 8.94E−03 | 3.51E−01 | 5.90E−01 | 2.89E−01 |
| BBB | 1.41E−06 | 4.03E−11 | 4.91E−06 | 2.59E−05 | 8.68E−01 |
| IKE | 4.17E−02 | 1.67E−05 | 1.12E−01 | 4.86E−02 | 3.16E−03 |
| DR | 2.60E−02 | 6.96E−07 | 2.78E−02 | 5.53E−02 | 7.47E−02 |
Abbreviations: BBB, blood–brain barrier; CLIMB, Comprehensive Longitudinal Investigation of MS at the Brigham and Women's Hospital; DR, disease reversal; GA, glatiramer acetate; IFNβ, interferon beta; IKE, nuclear factor kappa-B kinase subunit epsilon related; LT, long-term; m, months; MT-OX, mitochondrial; NRF2, nuclear factor erythroid 2-related factor; ST, short-term. Values are globaltest P-values for multi-gene signatures. Values in bold are individually significant at P⩽0.05.
To investigate the specificity of the 10 signatures for IFNβ treatment, significance in the GA-treated arm of the CLIMB study was tested (Table 2). Only the IKE signature reached significance after GA (P=0.0032).
Signatures evaluated in treatment-naive MS subtypes (Supplementary Table 2) revealed that the disease reversal signature reached significance in all four. In addition, the blood–brain barrier signature was significantly changed in both active RRMS and primary-progressive MS. The short-term 18 h, long-term and mitochondrial fatty acid-beta-oxidation signatures all reached significance in primary-progressive MS (P⩽0.05), drawing attention to a possible range of distinguishing features of this MS subtype.
Discussion
IFNβ-bioactivity markers have been proposed,6, 30, 31, 32, 33 yet validation in independent studies and links to treatment outcomes that are relevant to clinical endpoints such as disability, exacerbation rate, cognition loss, brain atrophy and EDSS remain limited. A small number of studies have suggested that certain IFNβ-bioactivity markers, such as transient, neutralizing antibody effects, may predict magnetic resonance imaging outcomes.34 However, treatment response markers associated with MS-relevant regulation of biological functions are lacking.1, 2, 11, 12, 35 Treatment outcome biomarkers from differential gene expression studies in IFNβ-treated MS patients have been a challenge to confirm.6 Mechanistically themed gene expression patterns that are confirmed across studies should better represent treatment-mediated changes. To identify such biomarkers, we integrated biological knowledgebases with the effects of IFNβ-1b treatment in a well-controlled differential gene expression study14, 15 and defined 10 multi-gene signatures to measure specific treatment effects. Statistical power compared with single-gene models is increased using this approach, which allowed for rigorous confirmatory hypothesis testing in independent IFNβ-treated MS patient data (Table 2).
The incorporation of biological themes relevant in MS therapy into these confirmed signatures, in combination with other measures (such as relapse rate, number of Gd-enhancing lesions, presence of neutralizing antibodies), can lead to mechanistically specific biomarkers able to facilitate treatment decisions. In addition, multi-gene RNA signatures measured before treatment provide an advantage over genome-wide association study biomarkers because the latter yield odds ratios often too low for predicative engineering.36, 37 An obvious advantage to RNA signatures measured during treatment is that these characterize actual treatment-mediated changes in patients.
The finding that long-term IFNβ-1b treatment reversed particular MS-related gene expression abnormalities (Figure 5), together with the observation that short-term treatment effects are similar to some MS-related changes, raises the possibility that these MS patients exhibited an IFNβ deficiency38 that was corrected over time by repeated IFNβ-1b dosing. Long-term IFNβ-1b treatment effects were clearly distinct from short-term effects. Short-term effects influenced immune regulation and antiviral responses as expected.35 Long-term treatment instead influenced metabolic functions, mitochondrial energetics and specific anti-oxidant mechanisms, which can preserve neuronal homeostasis.39 Fatigue and cognitive function are associated with decreased brain metabolism in MS patients.40, 41 This study presents multi-gene evidence for IFNβ-1b regulation of mitochondrial function, in particular metabolic processes that control the balance of fatty acids and their derivative biomolecules. Mitochondrial dysfunction and neuroprotection in MS is extensively reviewed in Mao and Reddy39 and Guzmán and Blázquez.42 IFNβ-1b effects on the central nervous system could be mediated by carnitine regulation, as we observed dysregulation of the specific function ‘import of palmitoyl-CoA into the mitochondrial matrix' (Figure 3). If MS brain exhibits defects in mitochondrial function, IFNβ-1b effects on extrahepatic ketone body catabolism/formation may influence alternative energy sources for neurons.
MS relapses, magnetic resonance imaging activity, cognition and even survival33, 43 are influenced by IFNβ treatment. However, no convincing association is observed between relapse reduction and long-term disease progression,44 which suggests there exist different mechanisms for neurodegenerative disease progression. Susceptibility of neurons to degeneration through mechanisms other than acute inflammation warrants consideration. Many energy metabolism genes are influenced by IFNβ and are likely to be important in MS pathology.39 As in Parkinson's disease and amyotrophic lateral sclerosis (ALS), energy metabolism and cytoprotective genes are important in chronic degenerative MS, and likely mediate beneficial effects of treatment.
This study's results suggest NADH dehydrogenase 6 (ND6), GPRC5C, OLA1 and NRF2 could potentially mediate long-term antioxidant effects of treatment. IFNβ-1b-treated patients exhibited significantly higher expression of the mitochondrially encoded ND6 gene than treatment-naive patients. Wild-type mt-ND6 maintains levels of reactive oxygen species,45 potentially reducing oxidative damage. Mutated mt-ND6 is causative of Leber hereditary optic neuropathy,46 a disease with MS-like white matter lesions.47 This suggests a mechanism by which IFNβ-1b treatment-modified mitochondria-mediated oxidative-stress responses could influence oligodendroglia in MS. GPRC5C was itself somewhat increased by IFNβ-1b treatment (23% increase in long-term comparison, P=0.015) as well as defining a neighborhood significantly regulated by treatment. This G protein-coupled receptor family protein has been suggested to mediate retinoic acid effects on G protein signal transduction cascade,48 but is otherwise little studied. OLA1 is an NRF2-independent, transcription-independent regulator of oxidative stress.49 OLA1, identified in the long-term treatment comparison, was a contributing neighbor of GPRC5C and was significantly decreased by treatment. The observation was confirmed in the CLIMB study data (P=0.002). This particular oxidative-stress-reducing effect of IFNβ-1b has not been previously identified. In addition, our observed treatment effects related to NRF2, a major mediator of oxidative control provides a neuroprotective mechanism that requires further investigation.50
IKKɛ (encoded by IKBKE) is a link between the protein network analysis, in which IKKɛ was the hub of the most significantly regulated network neighborhood, and the canonical pathway analysis, in which mitochondrial energy metabolism was highlighted. IKKɛ regulates energy balance.51 A role for IKKɛ in long-term IFNβ treatment is consistent with the anti-inflammatory synergy between IKKɛ and IFNβ in vivo.52 IFNβ induces rapid phosphorylation and activation of IKKɛ.53 IKKɛ is critical to IFNβ's role in response to viral infection53 as it integrates important signals from pattern recognition receptors that detect viral nucleic acids,54 particularly notable here in light of the association of viruses and exacerbations in MS.
All 10 RNA signatures developed in this study were significantly regulated in the Goertsches study data,21 indicating that both long-term and short-term effects were captured in the Goertsches patient profiles and that signatures of IFNβ-1b-controlled biological functions were confirmed. The temporal relationship between IFN-induced differential gene expression and physiological response can now be defined across different studies. The 10 signatures described provide a family of IFN-bioactivity markers that target distinct molecular mechanisms of action that can be studied further, especially in regard to patient response and clinical or magnetic resonance imaging outcomes.
Treatment of MS patients with GA reduces disease burden and relapses.55 However, as with IFNβ treatment, a cohort of patients appears to be non-responsive to treatment. With the sole exception of the IKE signature that characterized IKKɛ-related expression changes, signatures derived from IFNβ treatment effects were not changed by GA treatment, confirming that the signatures developed in this study measure IFNβ-1b-specific treatment effects. Monitoring the activity of these signatures should now serve as a foundation to predict effective combinational drug use and provide guidance in selecting treatment options. In addition, the IKBKE signature may point to an important common treatment mechanism and warrants further investigation.
Large multi-gene expression patterns reflect biological effects observable across different gene expression platforms and different MS patient treatment studies. More focused multi-gene signatures permitted confirmation of diverse IFNβ-1b effects in independent study data. Analysis revealed distinct long-term and short-term effects of IFNβ-1b treatment that actually contrasted in direction of change. Protein interaction network analysis strongly underscored a key role for IKKɛ in long-term IFNβ-1b treatment effects. Long-term IFNβ-1b treatment affected numerous pathways, including oxidative control and mitochondrial mechanisms important in maintaining normal brain homeostasis.
Acknowledgments
We thank Daniel Perez and Peter Bringmann for their support of the Systems Biology Program at Berlex Biosciences that generated the data analyzed in this study, Susanne Schwenke and Wilmar Igl of Bayer Schering Pharma AG for discussions on implementing Global Test, and D Allan Butterfield at the University of Kentucky for enlightening discussions on oxidative stress and NRF2 regulation.
The efforts of HS and KDY were compensated through a contract funded by Bayer HealthCare Pharmaceuticals, EC and VK were employed by Bayer HealthCare Pharmaceuticals, and ATR has consulted for Bayer, Biogen, Novartis, Serono and Teva Pharmaceuticals.
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Supplementary Material
References
- Hafler DA, Slavik JM, Anderson DE, O'Connor KC, De Jager P, Baecher-Allan C. Multiple sclerosis. Immunol Rev. 2005;204:208–231. doi: 10.1111/j.0105-2896.2005.00240.x. [DOI] [PubMed] [Google Scholar]
- Weiner HL. The challenge of multiple sclerosis: how do we cure a chronic heterogeneous disease. Ann of Neurol. 2009;65:239–248. doi: 10.1002/ana.21640. [DOI] [PubMed] [Google Scholar]
- Markowitz CE. The current landscape and unmet needs in multiple sclerosis. Am J Managed Care. 2010;16 (Suppl 8:S211–S218. [PubMed] [Google Scholar]
- Wagner JA. Innovations driving biomarker evaluation and use. Biomarkers Med. 2010;4:779–781. doi: 10.2217/bmm.10.114. [DOI] [PubMed] [Google Scholar]
- Micheel C.Evaluation of Biomarkers and Surrogate Endpoints in Chronic Disease(Micheel C, Ball J (eds)), National Acad Press, 1 September2010 [PubMed]
- Croze E. Differential gene expression and translational approaches to identify biomarkers of interferon beta activity in multiple sclerosis. J IFN Cyto Res. 2010;30:743–749. doi: 10.1089/jir.2010.0022. [DOI] [PubMed] [Google Scholar]
- Vandenbroeck K, Urcelay E, Comabella M. IFN-beta pharmacogenomics in multiple sclerosis. Pharmacogenomics. 2010;11:1137–1148. doi: 10.2217/pgs.10.108. [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Ann Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Mogensen KE, Lewerenz M, Reboul J, Lutfalla G, Uzé G. The type I interferon receptor: structure, function, and evolution of a family business. J IFN Cyto Res. 1999;19:1069–1098. doi: 10.1089/107999099313019. [DOI] [PubMed] [Google Scholar]
- Gok B, Okutan O, Beskonakli E, Palaoglu S, Erdamar H, Sargon MF. Effect of immunomodulation with human interferon-beta on early functional recovery from experimental spinal cord injury. Spine. 2007;32:873–880. doi: 10.1097/01.brs.0000259841.40358.8f. [DOI] [PubMed] [Google Scholar]
- Dhib-Jalbut S, Marks S. Interferon-beta mechanisms of action in multiple sclerosis. Neurology. 2010;74 (Suppl 1:S17–S24. doi: 10.1212/WNL.0b013e3181c97d99. [DOI] [PubMed] [Google Scholar]
- Agrawal SM, Yong VW. Immunopathogenesis of multiple sclerosis. Int Rev Neurobiol. 2007;79:99–126. doi: 10.1016/S0074-7742(07)79005-0. [DOI] [PubMed] [Google Scholar]
- Filippi M, Rocca MA, Camesasca F, Cook S, O'Connor P, Arnason BGW, et al. Interferon β-1b and glatiramer acetate effects on permanent black hole evolution. Neurology. 2011;76:1222–1228. doi: 10.1212/WNL.0b013e3182143577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi KD, Ruderman DL, Croze E, Wagner TC, Velichko S, Reder AT, et al. IFN-beta-regulated genes show abnormal expression in therapy-naïve relapsing-remitting MS mononuclear cells: gene expression analysis employing all reported protein-protein interactions. J Neuroimmunol. 2008;195:116–120. doi: 10.1016/j.jneuroim.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Reder AT, Velichko S, Yamaguchi KD, Hamamcioglu K, Ku K, Beekman J, et al. IFN-beta1b induces transient and variable gene expression in relapsing-remitting multiple sclerosis patients independent of neutralizing antibodies or changes in IFN receptor RNA expression. J IFN Cyto Res. 2008;28:317–331. doi: 10.1089/jir.2007.0131. [DOI] [PubMed] [Google Scholar]
- Croze E, Knappertz V, Yamaguchi KD, Reder A, Salamon H.Integration of molecular mechanism studies and biological knowledge-bases reveals the presence of a possible neuronal preservation process linked to mitochondrial dysfunction and oxidative stress in IFNB-1b-treated MS patients Multiple Sclerosis 201016(Suppl 5(abstract P565). [Google Scholar]
- Croze E, Knappertz V, Yamaguchi KD, Reder AT, Salamon H.Long-term interferon beta treatment in MS patients elevates brain-specific carnitine palmitoyltransferase, a key regulator of mitochondrial fatty acid oxidation Multiple Sclerosis 201016(Suppl 5(abstract P884). [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50:121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- Kurtzke JF. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS) Neurology. 1983;33:1444–1452. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- Goertsches RH, Hecker M, Koczan D, Serrano-Fernandez P, Moeller S, Thiesen HJ, et al. Long-term genome-wide blood RNA expression profiles yield novel molecular response candidates for IFN-beta-1b treatment in relapsing remitting MS. Pharmacogenomics. 2010;11:147–161. doi: 10.2217/pgs.09.152. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:685–690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . A Language and Environment for Statistical Computing: R Foundation for Statistical Computing. Vienna, Austria; 2010. [Google Scholar]
- Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- Gonsette RE. Neurodegeneration in multiple sclerosis: the role of oxidative stress and excitotoxicity. J Neurol Sci. 2008;274:48–53. doi: 10.1016/j.jns.2008.06.029. [DOI] [PubMed] [Google Scholar]
- Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinogenesis. 2009;48:91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linker RA, Lee DH, Ryan S, van Dam AM, Conrad R, Bista P, et al. Fumeric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf antioxidant pathway. Brain. 2011;134:678–692. doi: 10.1093/brain/awq386. [DOI] [PubMed] [Google Scholar]
- Johnson JA, Johnson DA, Kraft AD, Calkins MJ, Jakel RJ, Vargas MR, et al. The Nrf2-ARE pathway: an indicator and modulator of oxidative stress in neurodegeneration. Ann NY Acad Sci. 2008;1147:61–69. doi: 10.1196/annals.1427.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comabella M, Lünemann JD, Río J, Sánchez A, López C, Julià E, et al. A type I interferon signature in monocytes is associated with poor response to interferon-beta in multiple sclerosis. Brain. 2009;132:3353–3365. doi: 10.1093/brain/awp228. [DOI] [PubMed] [Google Scholar]
- van Baarsen LGM, Vosslamber S, Tijssen M, Baggen JMC, van der Voort LF, Killestein J, et al. Pharmacogenomics of interferon-beta therapy in multiple sclerosis: baseline IFN signature determines pharmacological differences between patients. PloS One. 2008;3:e1927. doi: 10.1371/journal.pone.0001927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosslamber S, van Baarsen LGM, Verweij CL. Pharmacogenomics of IFN-beta in multiple sclerosis: towards a personalized medicine approach. Pharmacogenomics. 2009;10:97–108. doi: 10.2217/14622416.10.1.97. [DOI] [PubMed] [Google Scholar]
- Reder AT. MxA: a biomarker for predicting multiple sclerosis disease activity. Neurology. 2010;75:1222–1223. doi: 10.1212/WNL.0b013e3181f6466f. [DOI] [PubMed] [Google Scholar]
- Goodin DS, Hartung HP, O'Connor P, Filippi M, Arnason B, Comi G, et al. Neutralizing antibodies to interferon beta-1b multiple sclerosis: a clinico-radiographic paradox in the BEYOND trial. Multiple Sclerosis. 2011;18:181–195. doi: 10.1177/1352458511418629. [DOI] [PubMed] [Google Scholar]
- Arnason BG, Dayal A, Qu ZX, Jensen MA, Genç K, Reder AT. Mechanisms of action of interferon-beta in multiple sclerosis. Sem Immunopath. 1996;18:125–148. doi: 10.1007/BF00792613. [DOI] [PubMed] [Google Scholar]
- De Jager PL, Chibnik LB, Cui J, Reischl J, Lehr S, Simon KC, et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8:1111–1119. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Conssortium (IMSGC) IL12A, MPHOSPH9/CDK2AP1 and RGS1 are novel multiple sclerosis susceptibility loci. Genes Immunity. 2010;11:397–405. doi: 10.1038/gene.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Petraglia AL, Chen M, Byskosh PV, Boos MD, Reder AT. Low expression of interferon-stimulated genes in active multiple sclerosis is linked to subnormal phosphorylation of STAT1. J Neuroimmunol. 2002;129:205–215. doi: 10.1016/s0165-5728(02)00182-0. [DOI] [PubMed] [Google Scholar]
- Mao P, Reddy PH. Is multiple sclerosis a mitochondrial disease. Biochim Biophys Acta. 2010;1802:66–79. doi: 10.1016/j.bbadis.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelcke U, Kappos L, Lechner-Scott J, Brunnschweiler H, Huber S, Ammann W, et al. Reduced glucose metabolism in the frontal cortex and basal ganglia of multiple sclerosis patients with fatigue: a 18F-fluorodeoxyglucose positron emission tomography study. Neurology. 1997;48:1566–1571. doi: 10.1212/wnl.48.6.1566. [DOI] [PubMed] [Google Scholar]
- Sørensen PS, Jønsson A, Mathiesen HK, Blinkenberg M, Andresen J, Hanson LG, et al. The relationship between MRI and PET changes and cognitive disturbances in MS. J Neurol Sci. 2006;245:99–102. doi: 10.1016/j.jns.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Guzmán M, Blázquez C. Ketone body synthesis in the brain: possible neuroprotective effects. Prostaglandins, Leukot Essent Fatty Acids. 2004;70:287–292. doi: 10.1016/j.plefa.2003.05.001. [DOI] [PubMed] [Google Scholar]
- Reder AT, Ebers GC, Traboulsee A, Li D, Langdon D, Goodin DS, et al. Cross-sectional study assessing long-term safety of interferon-beta-1b for relapsing-remitting MS. Neurology. 2010;74:1877–1885. doi: 10.1212/WNL.0b013e3181e240d0. [DOI] [PubMed] [Google Scholar]
- Scalfari A, Neuhaus A, Degenhardt A, Rice GP, Muraro PA, Daumer M, et al. The natural history of multiple sclerosis: a geographically based study 10: relapses and long-term disability. Brain. 2010;133:1914–1929. doi: 10.1093/brain/awq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo R, Garcia-Arumi E, Llige D, Marti R, Solano A, Montoya J, et al. Free radicals-mediated damage in transmitochondrial cells harboring the T14487C mutation in the ND6 gene of mtDNA. FEBS Lett. 2005;579:6909–6913. doi: 10.1016/j.febslet.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Fauser S, Leo-Kottler B, Besch D, Luberichs J. Confirmation of the 14568 mutation in the mitochondrial ND6 gene as causative in Leber's hereditary optic neuropathy. Ophthalmic Genet. 2002;23:191–197. doi: 10.1076/opge.23.3.191.7881. [DOI] [PubMed] [Google Scholar]
- Kovács GG, Höftberger R, Majtényi K, Horváth R, Barsi P, Komoly S, et al. Neuropathology of white matter disease in Leber's hereditary optic neuropathy. Brain. 2005;128:35–41. doi: 10.1093/brain/awh310. [DOI] [PubMed] [Google Scholar]
- Robbins MJ, Michalovich D, Hill J, Calver AR, Medhurst AD, Gloger I, et al. Molecular cloning and characterization of two novel retinoic acid-inducible orphan G-protein-coupled receptors (GPRC5B and GPRC5C) Genomics. 2000;67:8–18. doi: 10.1006/geno.2000.6226. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rubio V, Lieberman MW, Shi ZZ. OLA1, an Obg-like ATPase, suppresses antioxidant response via nontranscriptional mechanisms. Proc Natl Acad Sci USA. 2009;106:15356–15361. doi: 10.1073/pnas.0907213106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellrichmann G, Petrasch-Parwez E, Lee DH, Reick C, Arning L, Saft C, et al. Efficacy of fumaric acid esters in the R6/2 and YAC128 models of Huntington's disease. PloS One. 2011;6:e16172. doi: 10.1371/journal.pone.0016172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SH, Bazuine M, Lumeng CN, Geletka LM, Mowers J, White NM, et al. The protein kinase IKKepsilon regulates energy balance in obese mice. Cell. 2009;138:961–975. doi: 10.1016/j.cell.2009.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corr M, Boyle DL, Ronacher L, Flores N, Firestein GS. Synergistic benefit in inflammatory arthritis by targeting I kappaB kinase epsilon and interferon beta. Ann Rheum Dis. 2009;68:257–263. doi: 10.1136/ard.2008.095356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenoever BR, Ng SL, Chua MA, McWhirter SM, García-Sastre A, Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- Hiscott J. Another detour on the Toll road to the interferon antiviral response. Nature. 2004;11:1028–1030. doi: 10.1038/nsmb1104-1028. [DOI] [PubMed] [Google Scholar]
- Carter NJ, Keating GM. Glatiramer acetate: a review of its use in relapsing-remitting multiple sclerosis and in delaying the onset of clinically definite multiple sclerosis. Drugs. 2010;70:1545–1577. doi: 10.2165/11204560-000000000-00000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.