Abstract
Hydrogenases are the most active molecular catalysts for hydrogen production and uptake on earth1,2 and are thus extensively studied with respect to their technological exploitation as noble metal substitutes in (photo)electrolysers and fuel cells3-5. In [FeFe]-hydrogenases catalysis takes place at a unique diiron center (the [2Fe] subsite) featuring a bridging dithiolate ligand, as well as three CO and two CN− ligands (Figure 1)6,7. Through a complex and as yet poorly understood multienzymatic biosynthetic process, this [2Fe] subsite is first assembled onto a maturation enzyme, HydF. From there, it is delivered to the apo-hydrogenase for activation8. Synthetic chemistry has allowed the preparation of remarkably close mimics of that subsite1 but failed to reproduce the natural enzymatic activities so far. Here we show that three such synthetic mimics (with different bridging dithiolate ligands) can be loaded onto HydF and then transferred to apo-HydA1, one of the hydrogenases of Chlamydomonas reinhardtii. Remarkably, full activation of HydA1 was achieved exclusively using the HydF hybrid protein containing the mimic with an azadithiolate bridge, confirming the presence of this ligand in the active site of [FeFe]-hydrogenases9,10. This is the first example of controlled metalloenzyme activation using the combination of a specific protein scaffold and active site synthetic analogues. This simple methodology provides both new mechanistic and structural insight into hydrogenase maturation and a unique tool for producing recombinant wild-type and variant [FeFe]-hydrogenases, with no requirement for the complete maturation machinery. It opens the possibility to engineer the [FeFe]-hydrogenase active site through synthetic chemistry.
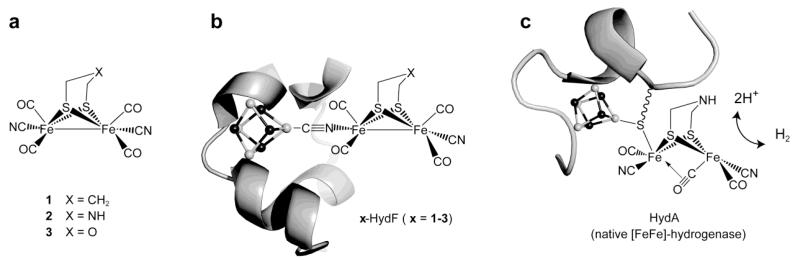
Complexes 111-13, 214, and 315 (Figure 1) represent the closest synthetic mimics of the [2Fe] subsite in HydA1. They all share the same primary coordination sphere with four CO, two CN− and a bridging dithiolato ligand. They do however differ in the nature of the central bridgehead atom of the dithiolate: carbon in 1, nitrogen in 2 and oxygen in 3. The nature of this atom in the enzyme [2Fe] subsite has been a matter of controversy7,9,10,16. Anaerobic reaction of HydF from Thermotoga maritima (expressed in E. coli), containing a [4Fe-4S] cluster17 and named “HydF” in the following, with a 10-fold molar excess of complex 1, 2 or 3, led to new hybrid species x-HydF (x = 1, 2 or 3 respectively), that could be isolated in pure form and characterized. In all cases, iron quantification indeed showed an increase from 3.9 ± 0.4 to 5.6 ± 0.4 iron atoms per protein and the UV-visible spectrum of these hybrids displayed features consistent with a ~1:1 ratio of the synthetic complexes and the HydF protein (Figure S1a-c).
Figure 1. Structures of the diiron clusters discussed in the study.
Left: the synthetic mimics 111-13, 214, and 315; Middle: proposed structure for the x-HydF (x = 1-3) hybrid proteins; Right: the H-cluster (active site) of [FeFe]-hydrogenase. The protein ribbon and the [4Fe-4S] clusters (shown as balls and sticks with Fe shown as white spheres) are shown only schematically.
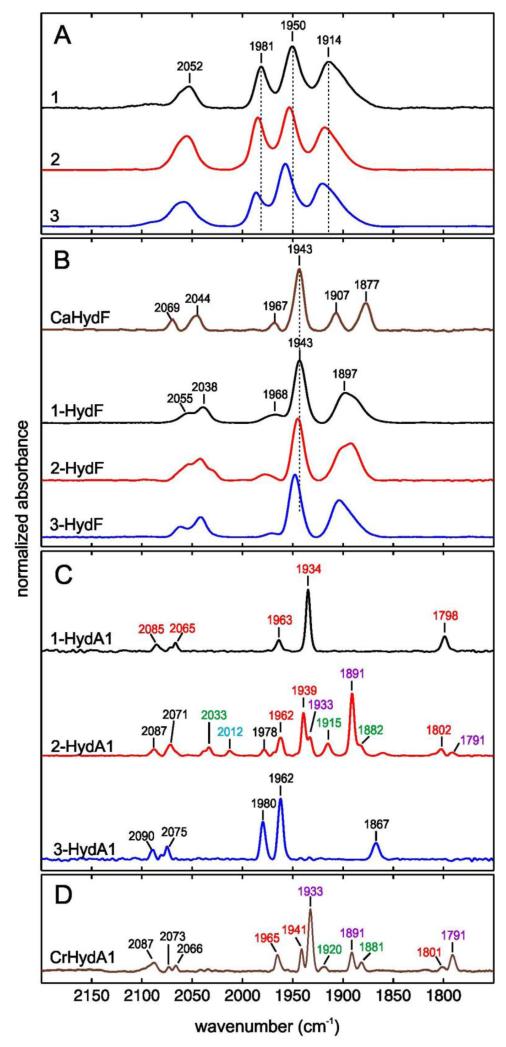
FTIR spectroscopy is a convenient method for characterizing metalloproteins such as hydrogenases containing CO and CN− ligands18. Thus, further evidence for the incorporation of synthetic complexes in HydF was obtained from their FTIR spectra which contained CN− stretching bands between 2000 and 2100 cm−1 and four partly overlapping CO-stretching bands in the 1800-2000 cm−1 range (Figure 2B and Table S1). The high-energy bands underwent a 40 cm−1 shift upon 13C-labeling of the CN− ligands (Figure S2). Interestingly, the width of the FTIR bands is still identical to those of the unbound complexes (Figure 2A) but their positions show strong similarities with those of CaHydF (Figure 2B and Table S1), a HydF preparation isolated from a strain of Clostridium acetobutylicum expressing the complete maturase machinery (including HydE and HydG)19. CaHydF contains, in addition to a [4Fe-4S] cluster, a still-undefined [2Fe] center and is capable of activating the apo form of HydA119. While the width of the FTIR bands of the hybrids would suggest a ligand conformational freedom similar to the unbound complexes, the position of the FTIR bands is a clear indication that the synthetic complexes closely mimic the natural [2Fe] subsite in HydF.
Figure 2. Normalized Fourier-transform infrared (FTIR) spectra recorded in liquid solution at 15°C.
Panel A: complexes 1-3; Panel B: CaHydF (from ref 19) and x-HydF (x = 1-3) hybrid species; Panel C: HydA1 after treatment of apo-HydA1with 1-HydF (1-HydA1), 2-HydF (2-HydA1) and 3-HydF (3-HydA1). Peak positions in the spectrum of 2-HydA1 are color coded to indicate the contributions from Hox (red), Hred (violet), Hsred (green) and Hox-CO (blue) (Figure S6 for a complete dataset). Panel D: HydA1 from C. rheinhardtii expressed in C. acetobutylicum (CrHydA1) (ref 18). Color code as in panel C. Samples of complexes 1-3 and x-HydF (x = 1-3) were prepared in HEPES buffer (20 mM, 100 mM KCl) pH 7.5. Samples of x-HydA1 have been prepared in 10 mM Tris-HCl buffer pH 8 containing sodium dithionite (2mM).
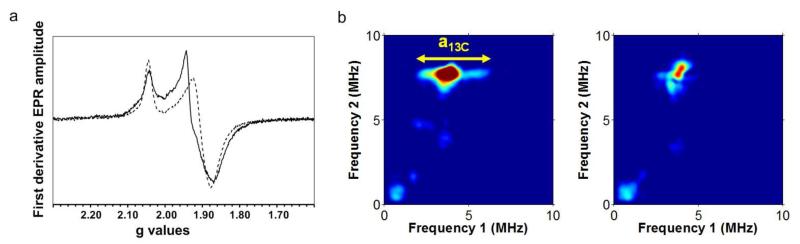
The arrangement in which the synthetic complexes are bound to HydF and its [4Fe-4S] cluster is not evident from the FTIR spectra. In particular FTIR spectroscopy does not allow to definitively distinguish between terminal and bridging cyanide ligands (see below and supplementary discussion)20. EPR and HYSCORE spectroscopies are more powerful in this respect and demonstrate a close interaction between the cluster and the synthetic complex as revealed for the case of 1-HydF. First, the EPR spectrum of the S=1/2 [4Fe-4S]1+ cluster in dithionite-reduced 1-HydF was markedly different from that of the reduced cluster in unloaded HydF, with the high-field feature at g = 1.90 in HydF shifted to g = 1.93 in 1-HydF (Figure 3A). A comparable shift was observed in the case of hybrids 2-HydF and 3-HydF (Figure S3). The absence of additional signals indicated that, in all cases, the synthetic complex remained in the EPR-silent FeIFeI state (both iron centers are in a low spin S=1/2 configuration but are antiferromagnetically coupled leading to a diamagnetic S=0 ground state). Second, pulsed EPR spectroscopy unambiguously demonstrated that the [4Fe-4S] cluster and the [2Fe] subsite analogue shared a CN− ligand in 1-HydF. For this purpose we used a nuclear coherence–transfer experiment (CF-NF)21, correlating the combination frequencies (CF) with the nuclear frequencies (NF), which is more sensitive than HYSCORE spectroscopy for disordered systems and best adapted for the observation of 13C signals in the presence of weakly coupled14N atoms. Interestingly this is the first time that a metalloprotein is characterized by such an experiment. The CF-NF spectrum of 1-HydF (Figure 3B right) displayed peaks from distant 13C carbon atoms present in natural abundance. When 1 was prepared with 13C-labelled CN−, the spectrum of 1-HydF displayed a new feature reflecting coupling of the unpaired electron in the [4Fe-4S] cluster with the 13C nucleus, characterized by a hyperfine coupling constant of 4±0.2 MHz (Figure 3B left). As shown in Figure S4, the HYSCORE spectrum of reduced 1-HydF displayed an additional feature consistent with the presence of a nitrogen atom weakly coupled to the [4Fe-4S] cluster in 1-HydF. The hyperfine coupling constant (aN ≤ 1 MHz) is significantly smaller than those (aN in the range of 4-7 MHz) generally obtained when a N atom is directly coordinated to an Fe-S cluster17,22,23. These coupling constants are consistent with a CN− ligand bridging one iron of the [4Fe-4S] cluster and one iron of 1, as nicely established by Density Functional Theory (DFT) calculations (a detailed description of the DFT calculations is provided as supplementary discussion and Tables S2-S5 in the Supplementary Information). More precisely, computed hyperfine coupling constants indicate that the cyanide C atom is bound to one iron atom belonging to a mixed-valence (Fe2.5+) iron of the [4Fe-4S] cluster while the N atom is bound to the diiron complex, implying cyanide linkage isomerism upon formation of 1-HydF, as precedented in the synthesis of Prussian blue analogues24 and other molecular metal clusters.20,25 Furthermore, DFT calculated values of CO and CN frequencies (2010 and 2060 cm−1) of a 1-HydF model, containing a CN ligand bridging the [4Fe-4S] cluster and complex 1, are well in the range of the experimental values (2038 and 2055 cm−1) (see Supplementary discussion and Table S6).
Figure 3. Continuous wave and pulsed EPR spectra on 1-HydF.
a: X-band EPR spectra recorded at 10K for dithionite-reduced 1-HydF (black line) and HydF (dashed line) in 50 mM Tris-HCl buffer, 150 mM NaCl, 5 mM DT, pH 8. Microwave power = 100 μW, mod. amp. = 1 mT, mwfreq. = 9.39 GHz. The shoulder observed at g = 1.90 in the 1-HydF spectrum, corresponding to a few percent of the total signal intensity, is assigned to a small fraction of HydF lacking 1. b: X-band 2D pulsed ESEEM spectroscopy (CF-NF) of 1-HydF labelled with 13CN– (left) and unlabelled 1-HydF (right). The horizontal ridge seen at (ν2 = 2·ν13C = 7.7 MHz) is attributed to a hyperfine interaction between a 13C nucleus and the paramagnetic [4Fe-4S] cluster. Its extension yields the magnitude of the coupling (Δν1 = 4.1 MHz = a13C). This feature is absent from the unlabelled 1-HydF spectrum.
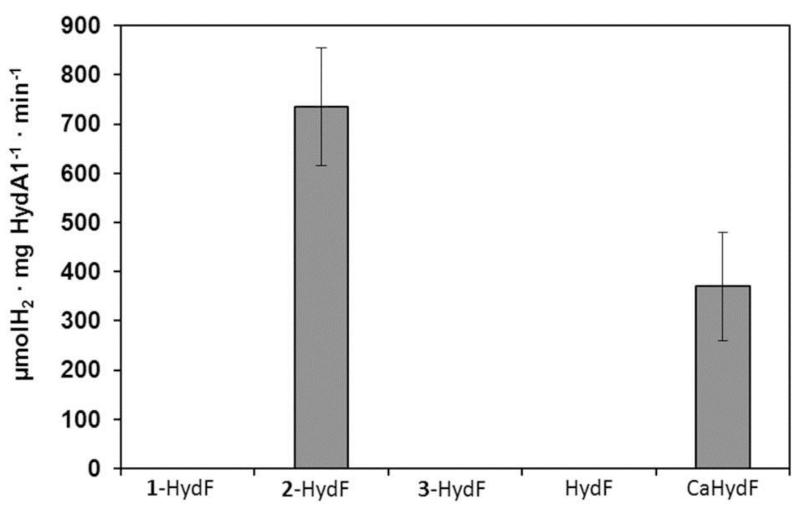
The hybrid x-HydF proteins were finally studied for their potential to activate apo-HydA1 from Chlamydomonas reinhardtii containing a single [4Fe-4S] cluster and no [2Fe] subsite26. A pure preparation of apo-HydA1 was incubated anaerobically with 10 equivalents of the hybrid protein, the optimal excess ratio (Figure S5), in phosphate buffer pH 6.8 at 37°C for 30 min. and hydrogen evolution was monitored under standard conditions (see the Methods section)26. No H2 evolution could be detected using HydF, 1-HydF or 3-HydF (Figure 4). In contrast, vigorous H2 evolution was observed using 2-HydF, corresponding to a specific activity of 700-800 μmol H2 min−1 mg HydA1−1, comparable to the activity values reported for wild type HydA127, thus indicating complete maturation/activation of HydA1 by 2-HydF28 (Figures 4 and S5). Furthermore, activation by 2-HydF was more efficient than by CaHydF (specific activity = 350-400), assayed under the same conditions (Figure 4). Indeed, CaHydF provided full activation only when present in larger excess.19 2-HydF by itself did not show any hydrogenase activity. Finally, apo-HydA1 was treated with a four-fold excess of x-HydF (x= 1, 2 or 3), under reducing conditions, then separated from HydF by affinity chromatography and analyzed by FTIR spectroscopy. In all cases, the presence of characteristic narrow Fe-CO and Fe-CN bands demonstrated that the synthetic complex has been transferred from HydF to HydA1 (Figure 2C and S6). The FTIR spectrum of HydA1 after treatment with 2-HydF shows a strong correspondence to that of fully active wild type HydA1 (Figure 2D)18. Specifically, both species exist as a mixture of the Hox, Hred and Hsred redox states that all participate to the catalytic cycle18. Furthermore, after flushing with CO, a complete conversion to Hox-CO occurred (Figure S7). These data exquisitely demonstrate that 2 is efficiently transferred from HydF to apo-HydA1 in which it acquires the structure of the natural active [2Fe] subsite. This implies isomerisation of one CN− ligand, replacement of one CO by a cysteinate ligand of the proximal [4Fe-4S] cluster in HydA1 and conformational rearrangement to adopt the inverted square pyramid structure required for opening a substrate binding site on the distal iron atom of the [2Fe] subsite (Figure 1).29 Interestingly, also 1-HydA1 and 3-HydA1 show “H-cluster like” FTIR signatures. In fact, the FTIR of 1-HydA1 has strong similarities with the Hox-state (Figure 2C and S6) whereas the FTIR of 3-HydA1 does not resemble any known H-cluster redox state, but seems to indicate a pure redox state and even shows a band assigned to a bridging CO.
Figure 4. Specific hydrogenase activity of reconstituted HydA1.
Activity (μmol H2 ·min−1 ·mg HydA1−1 with standard deviation) of HydA1 was measured in the presence of methyl viologen (10 mM) and sodium dithionite (100 mM) after in vitro maturation of apo-HydA1 for 30 minutes at 37°C with 10 equivalents of x-HydF (x = 1-3), HydF or CaHydF. The value for the latter was obtained after 60 min reaction and was taken from reference 23.
Besides unequivocally demonstrating that nitrogen is the bridgehead atom in the dithiolate ligand of the H-cluster, these results shed light on a number of important questions regarding hydrogenase maturation. They strongly support the hypothesis that HydF transiently binds a diiron precursor of the active [2Fe] subsite of HydA1 and suggest stabilization through interactions with the [4Fe-4S] cluster. The structure of this natural precursor is likely to be very close to that of 2. Further investigation of HydA1 maturation by the hybrid system, combining site-directed mutagenesis experiments and synthetic manipulation of the [2Fe] subsite (for example isotopic labelling as shown here with 13CN), will likely provide additional insight into the transfer mechanism and the structure of both HydF and HydA1 binding sites. These data also demonstrate the unique properties of the HydA1 protein binding pocket in converting the otherwise inactive complex 2 into an active catalyst. More importantly, this novel artificial hybrid maturase system provides a unique, simple and straightforward biotechnological tool for producing active recombinant hydrogenases, with no requirement for coexpression with the still incompletely characterized complex biosynthetic machinery. Since this procedure has been shown to work perfectly with proteins (HydF from T. maritima and HydA1 from C. reinhardtii) from two completely different organisms, it is very likely that [FeFe]-hydrogenases from other microorganisms, overexpressed in their apo form in E. coli, which lacks the maturation machinery, can be simply activated through reaction with 2-HydF. This reaction can thus be used for exploring a large variety of [FeFe] hydrogenases, e.g. from different species or derived from directed mutagenesis, with the aim of finding the most active and stable enzymes for their exploitation in biotechnological processes of H2 production30 as well as in bioelectrodes in (photo)electrolyzers or fuel-cells3-5
Methods
All chemicals were purchased from Sigma-Aldrich and used as received unless otherwise stated. NMR was recorded on a Bruker AC300 using the residual solvent peak as internal standard. Complex (Et4N)2[Fe2(adt)(CO)4(CN)2]) (2, adt2– = 2-azapropanedithiolate)14 was synthesized following literature procedure, whilst (Et4N)2[Fe2(pdt)(CO)4(CN)2] (1, pdt2– = propanedithiolate)11, (Et4N)2[Fe2(pdt)(CO)4(13CN)2] 31 2 and (Et4N)2[Fe2(odt)(CO)4(CN)2] (3, odt2– = 2-oxopropanedithiolate)15 were prepared by modified literature procedures (see the Supplementary Information) and their thin film solution FTIR spectra were recorded on a Perkin Elmer Spectrum-100 spectrometer. TmHydF (referred to as HydF throughout the text) was over-expressed, purified and its [4Fe-4S] cluster reconstituted as previously described17. Apo-CrHydA1 (referred to as apo-HydA1 throughout the text) was over-expressed in E. coli BL21 DE3 ΔiscR using growth conditions and a pET plasmid as previously published for the production of active HydA1 in E. coli 32.
Protein purity was assessed by gel electrophoresis by loading samples on Any kD™ Mini-Protean® TGX precast gels (Biorad) alongside Precision Plus Protein™ standards (Biorad). Migration was achieved on a Mini-Protean apparatus (Biorad) at 200 V for 30 min. Protein concentrations were determined with the Biorad Protein Assay, using bovine serum albumin as a standard as well as by optical absorption measurements. Aerobic UV visible absorption spectra were recorded on a Cary 1Bio spectrophotometer (Varian) and anaerobic measurements were made with a fiber-optic fitted UvikonXL spectrophotometer (BioTek Instruments). Iron and sulfur quantification were performed following the methods of Fish33 and Beinert34, respectively. The specific hydrogenase activity was determined as described previously35.
Spectroscopic characterization
FTIR spectra of protein samples were recorded on Bruker IFS 66 v/s FTIR spectrometer equipped with a Bruker MCT (mercury cadmium telluride) detector. The spectrometer was controlled by the Bruker Opus software. All measurements were performed at 15°C with a resolution of 2 cm−1. The spectra were accumulated in the double-sided, forward-backward mode with 1000 scans. Data were processed using home written routines in the MATLAB® programing environment. FTIR-samples of complexes 1-3 and x-HydF (x = 1-3) were prepared in HEPES buffer (20mM, 100 mM KCl) pH 7,5. FTIR-samples of x-HydA1 have been prepared in 10 mM TrisHCl buffer pH 8.0 containing sodium dithionite (10 mM). For the FTIR measurement of maturated HydA1, apo-HydA1 was washed twice with 10 mM Tris-HCl pH8.0, 2 mM NaDT (a buffer referred to in the following as TPW2) by concentration and dilution to remove any trace of desthiobiotin originating from the prior purification of apo-HydA1 by strep-tag affinity chromatography. 100 μl TPW2 containing 100 μM apo-HydA1 and 4-fold molar excess of the hybrid protein (1-HydF, 2-HydF or 3-HydF, respectively) was incubated for 60 minutes at 37°C. Afterwards, 500 μl of TPW2 was added, the solution was loaded on a 750 μl Strep-Tactin Superflow (IBA, Göttingen, Germany) column and the HydA1 protein obtained after treatment of apo-HydA1with 1-HydF (1-HydA1), 2-HydF (2-HydA1) or 3-HydF (3-HydA1) was separated again by affinity chromatography using 10 mM Tris-HCl pH 8.0, 2 mM NaDT, 200 mM NaCl as washing buffer and TPW2, 2.5 mM desthiobiotin for elution. The isolated protein was concentrated using Amicon Ultra centrifugal filters 10K (Millipore) and stored as described previously36. For the FTIR spectra shown in Figure S7, the preparation was done as described above with 2-HydF but without the final purification step.
X-band EPR spectra were recorded on a Bruker ESP 300D spectrometer equipped with an Oxford Instruments ESR 900 flow cryostat. Protein samples were anaerobically reduced with 10 molar equivalents of sodium dithionite before freezing. Hyperfine sublevel correlation (HYSCORE) spectra and their Combination Frequency (CF) – Nuclear Frequency (NF) variants were recorded on a Bruker Elexsys E-580 X band (frequency = 9.71 GHz) pulsed spectrometer with a Bruker ER4118X dielectric resonator and a continuous flow He cryostat (Oxford Instrument CF935) controlled by an Oxford Instrument temperature controller ITC 503. Experiments (128×128 dataset) were performed at 8 K using the standard four-pulse sequence (π/2-τ-π/2-t1-π-t2-π/2-echo) with a nominal pulse width of 16 ns for π/2 pulses, a τ value of 132 ns and a pulse repetition rate of 1 kHz. Unwanted echoes were removed by four-step phase cycling. The background decay in both dimensions was subtracted using a linear fit followed by apodization with a Hamming window and zero-filling to 2048 points in each dimension. The 2D Fourier Transform magnitude spectrum was then calculated. The static magnetic field was set at 3600 G (g⊥). In the HYSCORE experiment t1 and t2 were incremented in 20 ns steps from an initial value (tini = 200 ns) according to the following formula: t1 = tini + d1 and t2 = tini + d2. In the CF-NF experiment, t1 and t2 were incremented in 20 ns steps according to the following formula: t1 = tini +d1 and t2 = tini +d1+d2. The value of tini was chosen as long as 1000 ns to remove as much as possible the broad features arising from 14N quadrupole coupling.
DFT calculations
DFT calculations were performed using the ADF2012 quantum chemistry code (See the supplementary discussion in the Supplementary Information). Hyperfine coupling constants have been computed using the parameter-free PBE0 exchange-correlation potential with triple-zeta basis sets (+ two polarization functions) and unfrozen cores.
Supplementary Material
Acknowledgments
GB gratefully acknowledges Bengt Lundqvist minnesfond, FORMAS (contract number 213-2010-563) and the Swedish royal academy of sciences. This work was supported by the French National Research Agency (ANR) through Grant 07-BLAN-0298-01 and Labex program (ARCANE, 11-LABX-003). V. A. acknowledges support from the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013/ERC Grant Agreement n.306398). T.H. gratefully acknowledges support from the Deutsche Forschungsgemeinschaft (HA 255/2-1), the BMBF (Bio-H2), and the Volkswagen foundation (LigH2t). AA, ER, and WL gratefully acknowledge the Max Planck Society for financial support and Dr. Alexey Silakov for providing the FTIR processing software
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interest.
“Reprints and permissions information is available at www.nature.com/reprints”.
Published paper: www.nature.com/nature/journal/v499/n7456/full/nature12239.html
References
- 1.Tard C, Pickett CJ. Structural and Functional Analogues of the Active Sites of the [Fe]-, [NiFe]-, and [FeFe]-Hydrogenases. Chemical Reviews. 2009;109:2245–2274. doi: 10.1021/cr800542q. [DOI] [PubMed] [Google Scholar]
- 2.Cracknell JA, Vincent KA, Armstrong FA. Enzymes as working or inspirational electrocatalysts for fuel cells and electrolysis. Chemical Reviews. 2008;108:2439–2461. doi: 10.1021/cr0680639. [DOI] [PubMed] [Google Scholar]
- 3.Hambourger M, et al. [FeFe]-Hydrogenase-Catalyzed H2 production in a photoelectrochemical biofuel cell. Journal of the American Chemical Society. 2008;130:2015–2022. doi: 10.1021/ja077691k. [DOI] [PubMed] [Google Scholar]
- 4.Krishnan S, Armstrong FA. Order-of-magnitude enhancement of an enzymatic hydrogen-air fuel cell based on pyrenyl carbon nanostructures. Chemical Science. 2012;3:1015–1023. [Google Scholar]
- 5.Ciaccafava A, et al. An innovative powerful and mediatorless H2/O2 biofuel cell based on an outstanding bioanode. Electrochemistry Communications. 2012;23:25–28. [Google Scholar]
- 6.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (Cpl) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 7.Nicolet Y, Piras C, Legrand P, Hatchikian CE, Fontecilla-Camps JC. Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Structure. 1999;7:13–23. doi: 10.1016/s0969-2126(99)80005-7. [DOI] [PubMed] [Google Scholar]
- 8.Mulder DW, et al. Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(Delta EFG) Nature. 2010;465:248–U143. doi: 10.1038/nature08993. [DOI] [PubMed] [Google Scholar]
- 9.Nicolet Y, et al. Crystallographic and FTIR spectroscopic evidence of changes in Fe coordination upon reduction of the active site of the Fe-only hydrogenase from Desulfovibrio desulfuricans. Journal of the American Chemical Society. 2001;123:1596–1601. doi: 10.1021/ja0020963. [DOI] [PubMed] [Google Scholar]
- 10.Silakov A, Wenk B, Reijerse E, Lubitz W. 14N HYSCORE investigation of the H-cluster of [FeFe] hydrogenase: evidence for a nitrogen in the dithiol bridge. Physical Chemistry Chemical Physics. 2009;11:6592–6599. doi: 10.1039/b905841a. [DOI] [PubMed] [Google Scholar]
- 11.Le Cloirec A, et al. A di-iron dithiolate possessing structural elements of the carbonyl/cyanide sub-site of the H-centre of Fe-only hydrogenase. Chemical Communications. 1999:2285–2286. [Google Scholar]
- 12.Lyon EJ, Georgakaki IP, Reibenspies JH, Darensbourg MY. Carbon monoxide and cyanide ligands in a classical organometallic complex model for Fe-only hydrogenase. Angewandte Chemie-International Edition. 1999;38:3178–3180. [PubMed] [Google Scholar]
- 13.Schmidt M, Contakes SM, Rauchfuss TB. First generation analogues of the binuclear site in the Fe-only hydrogenases: [Fe2–2(μ-SR)2(CO)4(CN)2 ]2−. Journal of the American Chemical Society. 1999;121:9736–9737. [Google Scholar]
- 14.Li HX, Rauchfuss TB. Iron carbonyl sulfides, formaldehyde, and amines condense to give the proposed azadithiolate cofactor of the Fe-only hydrogenases. Journal of the American Chemical Society. 2002;124:726–727. doi: 10.1021/ja016964n. [DOI] [PubMed] [Google Scholar]
- 15.Song LC, Yang ZY, Bian HZ, Hu QM. Novel single and double diiron oxadithiolates as models for the active site of [Fe]-Only hydrogenases. Organometallics. 2004;23:3082–3084. [Google Scholar]
- 16.Pandey AS, Harris TV, Giles LJ, Peters JW, Szilagyi RK. Dithiomethylether as a Ligand in the Hydrogenase H-Cluster. Journal of the American Chemical Society. 2008;130:4533–4540. doi: 10.1021/ja711187e. [DOI] [PubMed] [Google Scholar]
- 17.Brazzolotto X, et al. The [Fe-Fe]-hydrogenase maturation protein HydF from Thermotoga maritima is a GTPase with an iron-sulfur cluster. Journal of Biological Chemistry. 2006;281:769–774. doi: 10.1074/jbc.M510310200. [DOI] [PubMed] [Google Scholar]
- 18.Adamska A, et al. Identification and Characterization of the “Super-Reduced” State of the H-Cluster in [FeFe] Hydrogenase: A New Building Block for the Catalytic Cycle? Angewandte Chemie International Edition. 2012;51:11458–11462. doi: 10.1002/anie.201204800. [DOI] [PubMed] [Google Scholar]
- 19.Czech I, Silakov A, Lubitz W, Happe T. The [FeFe]-hydrogenase maturase HydF from Clostridium acetobutylicum contains a CO and CN− ligated iron cofactor. FEBS Letters. 2010;584:638–642. doi: 10.1016/j.febslet.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 20.Geiss A, Vahrenkamp H. M(μ-CN)Fe(μ-CN)M’ chains with phthalocyanine iron centers: Preparation, structures, and isomerization. Inorganic Chemistry. 2000;39:4029–4036. doi: 10.1021/ic0001412. [DOI] [PubMed] [Google Scholar]
- 21.Hubrich M, Jeschke G, Schweiger A. The generalized hyperfine sublevel coherence transfer experiment in one and two dimensions. Journal of Chemical Physics. 1996;104:2172–2184. [Google Scholar]
- 22.Gambarelli S, Luttringer F, Padovani D, Mulliez E, Fontecave M. Activation of the anaerobic ribonucleotide reductase by S-adenosylmethionine. ChemBioChem. 2005;6:1960–1962. doi: 10.1002/cbic.200500182. [DOI] [PubMed] [Google Scholar]
- 23.Chen DW, Walsby C, Hoffman BM, Frey PA. Coordination and mechanism of reversible cleavage of S-adenosylmethionine by the [4Fe-4S] center in lysine 2,3-aminomutase. Journal of the American Chemical Society. 2003;125:11788–11789. doi: 10.1021/ja036120z. [DOI] [PubMed] [Google Scholar]
- 24.Coronado E, et al. Pressure-Tuning of Magnetism and Linkage Isomerism in Iron(II) Hexacyanochromate. Journal of the American Chemical Society. 2005;127:4580–4581. doi: 10.1021/ja043166z. [DOI] [PubMed] [Google Scholar]
- 25.Shatruk M, et al. Properties of prussian blue materials manifested in molecular complexes: Observation of cyanide linkage isomerism and spin-crossover behavior in pentanuclear cyanide clusters. Journal of the American Chemical Society. 2007;129:6104–6116. doi: 10.1021/ja066273x. [DOI] [PubMed] [Google Scholar]
- 26.Happe T, Naber JD. Isolation, Characterization and N-Terminal Amino-Acid-Sequence of Hydrogenase from the Green-Alga Chlamydomonas-Reinhardtii. European Journal of Biochemistry. 1993;214:475–481. doi: 10.1111/j.1432-1033.1993.tb17944.x. [DOI] [PubMed] [Google Scholar]
- 27.Kamp C, et al. Isolation and first EPR characterization of the [FeFe]-hydrogenases from green algae. Biochimica Et Biophysica Acta-Bioenergetics. 2008;1777:410–416. doi: 10.1016/j.bbabio.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Sybirna K, et al. Shewanella oneidensis: a new and efficient system for expression and maturation of heterologous [Fe-Fe] hydrogenase from Chlamydomonas reinhardtii. Bmc Biotechnology. 2008;8 doi: 10.1186/1472-6750-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darensbourg MY, Lyon EJ, Zhao X, Georgakaki IP. The organometallic active site of [Fe]hydrogenase: Models and entatic states. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3683–3688. doi: 10.1073/pnas.0536955100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mertens R, Liese A. Biotechnological applications of hydrogenases. Current Opinion in Biotechnology. 2004;15:343–348. doi: 10.1016/j.copbio.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Fiedler AT, Brunold TC. Combined spectroscopic/computational study of binuclear Fe(I)-Fe(I) complexes: Implications for the fully-reduced active-site cluster of Fe-only hydrogenases. Inorganic Chemistry. 2005;44:1794–1809. doi: 10.1021/ic048739n. [DOI] [PubMed] [Google Scholar]
- 32.Kuchenreuther JM, et al. High-Yield Expression of Heterologous [FeFe] Hydrogenases in Escherichia coli. Plos One. 2010;5:e15491. doi: 10.1371/journal.pone.0015491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fish WW. Rapid Colorimetric Micromethod for the Quantitation of Complexed Iron in Biological Samples. Methods in Enzymology. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 34.Beinert H. Semi-Micro Methods for Analysis of Labile Sulfide and of Labile Sulfide Plus Sulfane Sulfur in Unusually Stable Iron Sulfur Proteins. Analytical Biochemistry. 1983;131:373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- 35.Hemschemeier A, Melis A, Happe T. Analytical approaches to photobiological hydrogen production in unicellular green algae. Photosynthesis Research. 2009;102:523–540. doi: 10.1007/s11120-009-9415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stripp ST, et al. How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17331–17336. doi: 10.1073/pnas.0905343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.