Abstract
Luminescence reporters have been used successfully in studies of circadian rhythms. Real-time measurements of circadian variations in gene expression were made in living cells, cultured tissues, and whole organisms. Because this technique is relatively easy and continuous noninvasive measurement from tissue cultures allows for a drastic reduction in the number of experimental animals, we believe this method will become a common technique for studying circadian rhythms. Using a multichannel recording apparatus, it may also become a powerful tool for the discovery of new drugs. In the past, measurements were done using hand-made apparatuses or by modifying commercially available equipment. We, along with other investigators, have developed user-friendly equipment for performing circadian rhythms experiments, and these systems are now available commercially. This article describes the use of luminescence reporters in circadian research and provides detailed methods used in these experiments. One of our goals in this article is to reduce experimental variability in different laboratories by proposing standard protocols.
Introduction
Ever since luciferase was introduced in real-time luminescence monitoring of gene expression rhythms in plants and cyanobacteria (Kondo et al., 1993; Millar et al., 1992), luminescence reporter techniques have become a powerful tool used in noninvasive assays of circadian oscillations. This method has faithfully monitored the rhythms of circadian genes in the fly (Brandes et al., 1996), mouse (Asai et al., 2001; Geusz et al., 1997; Wilsbacher et al., 2002; Yoo et al., 2004), rat (Yamazaki et al., 2000), and fungi (Morgan et al., 2003), as well as immortalized cell lines driven from the rat (Izumo et al., 2003; Ueda et al., 2002), zebrafish (Vallone et al., 2004), and human (Maronde and Motzkus, 2003). Using this noninvasive assay, we are able to measure real-time expression of circadian and circadian output genes, as well as the protein dynamic of the circadian genes. Serial measurement of rhythms from individual tissue samples greatly reduces the intersample variability seen using conventional sampling methods and, importantly, reduces the number of tissue samples required. In animal experiments, this has the potential to drastically reduce the number of experimental animals. Circadian rhythms can be measured from whole mice (Collaco and Geusz, 2003) and from cultured tissues (Geusz et al., 1997). By detecting the first peaks in cultured tissues, we can estimate the phase of each tissue in vivo (ex vivo experiment, Stokkan et al., 2001; Yamazaki et al., 2000, 2002). Alternatively, using fiber optics implanted into the animal, rhythms can be measured from freely behaving animals (Yamaguchi et al., 2001). This article describes detailed methods for tissue culture and recording apparatuses used in circadian rhythms studies in mammals.
Animals and Cells
Transgenic animals with a DNA construct in which the promoter of the gene is fused to the luciferease reporter gene were made for monitoring the circadian oscillation of the transcription. c-fos::luc mice, CMV::luc mice, Per1::luc rats, and Per1::luc mice have been examined in circadian rhythm studies (Asai et al., 2001; Geusz et al., 1997; Herzog et al., 2004; Wilsbacher et al., 2002; Yamazaki et al., 2000, referencing only the original publication for each animal). In Per1::luc mice, the phase of Per1 transcription rhythm in the suprachiasmatic nucleus (SCN) in vivo matched the rhythm of luciferase mRNA in vivo, as well as the luminescence rhythm from cultured SCN (Wilsbacher et al., 2002). This indicates that the luminescence reporter can be used in real-time monitoring of Per1 transcription and that the first peak in cultured SCN reflects the peak of in vivo Per1 mRNA rhythm.
The circadian rhythm of the PER2 protein can also be monitored using a luminescence reporter (Yoo et al., 2004). Using homologous recombination in embryonic stem cells, we knocked in the reporter (luciferase) into the C terminus of the Per2 gene to create a PER2::LUCIFERASE fusion protein. Using this knockin mouse line, we were able to record the PER2 protein oscillations from cultured tissues. Luminescence rhythms from cultured SCN matched the PER2 protein rhythm in the SCN in vivo, but not the Per2 mRNA rhythm (there is a time lag between the mRNA and protein rhythms). Because homozygous knockin mice (both copies of Per2 are replaced with the Per2::luc fusion gene) exhibited completely normal activity rhythms and behavioral phase shifts in response to a light pulse, we concluded that the fusion of luciferase to the PER2 protein did not disrupt the normal clock function of the PER2 protein (Yoo et al. 2004). It is known that the loss of functional PER2 protein altered these circadian parameters (Bae et al., 2001; Zheng et al., 1999).
Circadian rhythms of transcription can also be monitored from immortalized cell lines. Both stably and transiently transfected cell lines have been used successfully (Izumo et al., 2003; Maronde and Motzkus, 2003; Ueda et al., 2002). To measure circadian oscillations in cell cultures, a number of stimuli such as forskolin (10μM, 30 min), horse serum (50%, 120 min), or other stimuli must be used to induce circadian oscillations (Balsalobre et al., 2000; Tsuchiya et al., 2003).
Tissue Culture
Using sterile technique, tissue explants can be made from adult mice or rats, aged rats (2 years old), or neonates (Asai et al., 2001; Yamaguchi et al., 2003; Yamazaki et al., 2002). Exposing animals to light at night (light pulse) or terminating light during the day (dark pulse) induces both transient and permanent changes in their circadian organization. To avoid this phase shift, we usually perform tissue preparation at just before (within 1 h) lights off. Preparing the samples immediately after lights on is also acceptable. If the animal has been kept in constant darkness, sampling should be done in the dark, using an infrared viewer without exposing the animal to visible light. Because the retina is known as the only circadian photoreceptive tissue in mammals, we anesthetize and enucleate in the dark and perform subsequent procedures in the light. The animals should be anesthetized with CO2 [or anesthesia suggested by Institutional Animal Care and Use Committee (IACUC) in each institution] and euthanized by decapitation (cervical dislocation without anesthesia is also accepted by IACUC at some institutions). Due to both animal welfare and scientific reasons, stress during euthanasia should be kept to a minimum (it is known that stress can alter the phase of circadian rhythms). Brain, eyes, pineal, pituitary, liver, lung, and kidney, as well as any other tissues to be used in the experiment, should be removed quickly and be kept in chilled Hanks’ buffered salt solution (HBSS, Table I). For a short period of time, they can be also kept in warm HBSS to minimize temperature-induced phase shifts.
TABLE I.
Contents for HBSS (Final Volume of 1 Liter)a
|
All contents should be dissolved in autoclaved Milli-Q water; the total volume should be adjusted to 1 liter and kept at 4°.
Coronal sections of the brain (200–400 μm) should be made shortly after removal using a Vibroslicer (horizontal or sagittal slices can be used as well; a tissue chopper is not appropriate for acute recordings because it disrupts rhythms). The suprachiasmatic nucleus (master circadian pacemaker and phase organizer), retrochiasmatic area, arcuate nucleus, or any other brain areas can be dissected and cultured separately on Millicell culture plate inserts (PICMORG50, Millipore, Bedford, MA). The dissected brain area should be smaller than 15 mm2, as the culture will not survive for a long period of time using this static organotypic method. Long-lasting high-amplitude oscillations can be measured by photon-counting devices from the paired SCN, which contains minimal volumes of the non-SCN brain area. Luminescence imaging with image analysis can also be used to measure the SCN rhythmicity from a hypothalamic slice preparation, which contains the SCN and surrounding non-SCN area. Pineal, pituitary, and retina should be cultured on Millicell inserts. Pineals need to be cut halfway through, flattened, and placed on the culture inserts. Pituitaries taken from rats should be hand sliced and reduced to a small piece (about 1–4mm2; whole pituitary can be used in mouse experiments). Small pieces of retina (about 1 mm2) should be placed on Millicell inserts to obtain better rhythms.
Other peripheral tissues can be cultured without the use of culture inserts. However, when using a carousel unit, these tissues should be placed on a polypropylene mesh sheet (Spectra/Mesh, Medical Industries, Inc., Los Angeles, CA), as movement of the tissue will produce a huge noise on the baseline. Hand-sliced liver, lung, kidney, or other peripheral tissues should be dissected into small pieces (1–9mm2). Whole corneas dissected from the eyes can also be cultured.
Each tissue should be cultured in a 35-mm petri dish with culture medium (see Table II). The volume of the medium is important when using Millicell culture inserts; we obtain the best results using either 1.2 ml of culture medium in Falcon dishes or 1.0 ml of medium in Corning dishes. Peripheral tissue can be cultured with 1.0–3.0 ml of culture medium. Culture dishes need to be airtight to prevent evaporation of the culture medium. Autoclaved high-vacuum grease (Dow Corning; #14-635-5D, Fisher) should be pasted around the edge of the culture dish using a 3-ml disposable syringe (Becton Dickinson and Co., Franklin Lakes, NJ) and air sealed with a 40-mm microscope glass cover (40 Circle #1, Fisher or VWR).
TABLE II.
Contents for the Recording Medium (Final Volume of 1 Liter)a
|
All contents (except Nos. 5 and 6) should be dissolved in autoclaved Milli-Q water, and total volume should be adjusted to 1 liter. pH should be stable at around 7 in a few days and osmolality should be around 300. Medium should be kept light protected at 4°. Luciferin should be added, and necessary amounts of the medium should be warmed up to 37°.
We used this medium in all experiments published in 2000–2004. However, DMEM powder has been discontinued and is only available through custom orders. The DMEM powder (#13000-021 Gibco) can be replaced with DMEM power (#D-2902, with L-glutamine and 1000 mg glucose, without phenol red and sodium bicarbonate, Sigma) and 3.5 g of D-glucose powder (G7021, Sigma).
Recording Medium
In our experience, high-glucose, high-glutamine Dulbeccos Modified Eagles Medium works for most of the tissues. Because most of the equipment we have used (except LM-2400, Hamamatsu, see later) requires the use of airtight static cultures, the concentration of sodium bicarbonate needs to be adjusted to the optimal concentration (350 mg/L), which can be buffered with the CO2 concentration of the air (0.03%). We also add 10 mM of HEPES and antibiotics. B27 supplement or fetal bovine serum (5%) can be used for most cultures. SCN cultures can be maintained without a B27 supplement or fetal bovine serum. Because phenol red reduces light signal penetration, the culture media should not contain phenol red. The medium must be sterilized by filtering. Luciferin (final concentration 0.1 mM, beetle luciferin, potassium salt, Promega Co., Madison, WI) should be added just before the luciferase activity assay. The medium should be warmed at 37° before being used.
Recording Apparatuses
Instruction for Custom-Built (Hand-Made) Apparatus
Choosing the Optimal Sensor for Photon Counting
Although many types of photosensitive devices are available, photomultiplier tubes (PMTs) remain the preferred light detectors for many applications. This vacuum tube photodetector has a potential advantage for photon counting. Because luminescence from cultured SCN is extremely weak, we must choose highly sensitive and low-noise (low dark count, nonspecific noise) PMTs. We have been using two different types of photon-counting units (side-on and head-on), and both have enough sensitivity to detect signals from SCN explants. Relatively inexpensive PMTs may be used for peripheral tissue and cell cultures because the luminescence signal can be intensified by increasing the number of cells or the size of cultured tissue.
The H6240 photon-counting head (Hamamatsu, Bridgewater, NJ) consists of a side-on PMT, voltage divider, amplifier, discriminator, and high-voltage power supply, and it works by supplying 5 V without any adjustment. TTL pulses with corresponding photons (at 560 nm, about 8% of the photons reaching the PMT will convert to TTL pulses) can be recorded by personal computer via a TTL counting board (eight-channel counter board, PCI 6602 with SCB-68 connector box; National Instruments, Austin, TX). For SCN explants, we asked the manufacturer to change the standard PMT to R7518P, which is hand selected with dark counts of less than 10 counts per second (cps) at room temperature. Although we have not yet tested the R4220P (selected <10 cps), it has a similar (although slightly less sensitive) performance and can be used in the H6240 photon-counting head.
The HC135-01 (Hamamatsu) is a self-contained head-on photon-counting module with a microprocessor and requires only 5 V power. Counts can be stored on a personal computer using the RS-232C interface. For SCN explants, modifications by the manufacturer (PMT replaced with R3550 < 10 cps, reduce rescale factor to 2) should be made.
In both the H6240 and the HC135-01, there is a linear relationship between the incident number of photons and the count rate of up to 10,000,000 cps. Therefore, no data correction is required in most of luminescence measurements.
Setting up PMTs
To ensure stable recordings, it is better to place the PMT about 3–10 mm above the culture sample. Recoding from the bottom of the culture sample is possible; however, heat insulation between the PMT and the sample will be required because a slightly increased temperature from the bottom will produce condensation on top of the culture dish and osmolality changes in the medium will disturb the culture sample. To prevent condensation, the cover of the culture dish should be a little warmer than the culture medium. Because the PMT produces heat, placing it on top of the culture dish will create optimal conditions. Both the PMT and the culture sample need to be placed in absolute darkness at 36–37°. Shielding the inside of the incubator with black cardboard or building a light-tight box inside the incubator is necessary (Fig. 1). Because each PMT produces a little heat, the inside of the light-tight box will become warmer than the inside of the incubator. Therefore, when building a light-tight box inside the incubator, the incubator temperature should be set a little below the optimal temperature. A plywood box with black matte paint will be sufficient for a permanent setup. When the power of the PMT is on, exposing the PMT to visible light will immediately damage the PMT. Therefore, the power must be tuned off before loading the samples. Because the HC-135 contains a microprocessor and memory, turning off the power of the HC135 will terminate the recording. The PMT can be inactivated using a command from the personal computer, which will allow for continuous recording. Turning off the power of the H6240 will not terminate the TTL counting because the TTL pulse counting is operated by an independent circuit located within the personal computer.
Figure 1.
Examples of custom-built PMT setup for luminescence recordings. (A) Example of the light-tight box for PMT housing. Four HC135 photon-counting modules are located inside the light-tight box. The box is kept in an environmental chamber in which the temperature is set at 35.5°. A small fan with baffles is used for circulating the air inside the light-tight box and temperature inside of the box stays at 36.5°. (B) Example of the use of a light-tight incubator. Eight HC135 photon-counting modules are located inside the incubator. Black cardboard is used for the shielding of light, and black plastic sheet is placed between the inside glass door and the metal outside door to prevent light leaks.
Flow-Through System
For a continuous supply of nutrition and substrate (luciferin) or for pharmacological stimulation, a flow-through system has potential advantages. Because the tubes with the medium can act as fiber-optic conductors, both intake and drain tubes, as well as the pumps, need to be kept in the light-tight boxes. Because the slightest movement of the water surface produces a noise, the culture chamber needs to be an air-free closed system and oxygen supply into the medium is required.
Commercially Available Circadian Luminescence Recording Apparatuses
LumiCycle
The LumiCycle (Actimetrics Inc., Evanston, IL) is a 32channel carousel unit designed by our group to be used in circadian experiments. Four manufacture-modified H6240 units (see earlier) count photons from 32 cultures. This unit has no thermo-controller and needs to be placed in an incubator. Generally, the incubator temperature should be set 1° below the optimal temperature. However, because temperature uniformity of each incubator is different, temperature inside the Lumi-Cycle should be confirmed by a temperature data logger (HOBO H8 Pro, Onset, Pocasset, MA). One of the unique features of this unit is that it is designed for multiusers. Loading new samples, taking out old samples, and/or pharmacological stimulations can be done without disrupting the recordings on other channels. One personal computer can handle up to two units. Using data analysis software, circadian parameters (including phase, period, amplitude, and damping rate) can be obtained after the baseline subtraction. To perform pharmacological stimulation at a specific circadian time, the phase of the next cycle can be estimated by the software. Both low and baseline subtracted data can be exported to ClockLab software (Actimetrics Inc.) for detailed analysis. For dual reporting with two different wavelengths of luminescence reporters, optical filters can be installed on the front of two PMTs and the luminescence of 16 samples can be measured by those two PMTs.
LM-2400
The weak light detection unit (LM-2400, Hamamatsu) is a 24-channel carousel unit designed by Dr. Tei’s group for use in circadian rhythm experiments. Two head-on PMTs (low dark counts selected R1924; similar to R3550) measure the luminescence from 24 cultures. This unit can be used in a regular CO2 incubator. Therefore, CO2-buffered culture medium can be used in the open, in a water-saturated atmosphere. All 24 channels of recordings need to be started and terminated at the same time. Although we have not yet tested this unit, Dr. Tei’s group has been using it for Per 1<luc measurement from cultured SCN. Temperature inside this unit is less than 1° higher than the environmental temperature.
LM-300
The low-light detection unit (LM-300, C8801–01, Hamamatsu) is a one-channel unit, which can be used for SCN culture samples. Because this unit is not a carousel, it is suitable for experiments requiring higher time resolution. One personal computer can handle up to five units. The incubator temperature should be set about 1–2° lower than optimal culture temperature.
Kronos
The Kronos (AB-2500, Atto Co., Tokyo, Japan) is an eight-channel carousel unit. Because we have yet not tested this apparatus, we describe the characteristics based on data in the manufacturer’s brochure. This unit contains a temperature controller and can be used without an incubator. However, temperature accuracy of ±0.5° (per brochure) remains a concern because luciferase activity changes due to small changes in temperature. An automated controller of the optical filters can be used for dual reporters coding different light wavelengths. One personal computer can handle up to five units.
POLARstar
The POLARstar Optima (BMG Labtechnologies Inc, Durham, NC) is a plate reader that can be used for circadian rhythms studies. The TopCount (a plate reader, Packard Instrument Co., Meriden, CT) has been used to measure circadian luminescence rhythms of fly and zebrafish at room temperature (Stanewsky et al., 1997; Vallone et al., 2004). However, because of condensation issues (it is very difficult to set the temperature inside the machine to be exactly the same as the temperature outside the machine and because the plates stay outside the machine most of the time except during luminescence reading and small temperature changes produce condensation on top of the culture sample; see earlier discussion), the TopCount is a difficult unit to use in mammalian tissue culture experiments at 36 °. The POLARstar has two heaters; one is located on the top and the other at the bottom of the culture plate. Because the top heater is set a little higher than the bottom heater and the culture plate stays between the heaters at all times, no condensation is observed (we noticed a little condensation on the #1 well, home position of the plate). The POLARstar uses H6240 with fiber optics, and the signal can be recorded from either the top or the bottom of the culture sample. To increase the sensitivity (especially in SCN culture recordings) we asked the manufacturer to direct the positioning of the PMT on top of the culture plate without using fiber optics. We have successfully measured luminescence rhythms from rat-1 cells cultured in 24-well plates using this modified apparatus (Izumo et al., 2003).
Luminescence Imaging
Luminescence Imaging allows us to obtain spatial information that we are not able to obtain using the photon-counting methods described earlier. Although luminescence from luciferase reporter is extremely weak, several CCD cameras can detect the signals. The Per1<luc oscillations from single SCN neurons have been measured (Yamaguchi et al., 2003).
We briefly describe the luminescence imaging methods in this chapter. Principles and detailed methods for imaging are described in Welsh et al. (2005).
Cameras
An intensified CCD camera (ICCD or VIM, C2741-35, Hamamatsu) with a photon-counting mode has been used in living cell imaging expressing luciferase (Brandes et al., 1996; Geusz et al., 1997; Millar et al., 1992). Because a microarray of narrow cylinder-shaped PMTs (photo intensifier) converts each photo to electrons, this camera has high sensitivity to light, but has fewer pixels compared with cooled CCDs.
A back-thinned, back-illuminated CCD camera can be used with cooling devices. Liquid nitrogen is often used for cooling (Yamaguchi et al., 2003). Electrical cooling devices, such as Peltier or Cryotiger, are also used. An electron bombardment (special vacuum chamber with photocathode) CCD camera is also available with a cooling device (Water-cooled EB-CCD camera, C7190-10, Hamamatsu). Single-cell imaging from SCN explants can be captured by this camera using a microscope with high NA, long working-distance objectives (H. Tei et al., personal communication).
Optics
The most efficient way to collect a signal from a culture sample is by direct coupling of 35-mm camera lenses to the CCD. We used two coupled 50-mm f1.2 Nikkor lenses (HLN50-12 Nikon) face to face using a connecting ring attached directly to the ICCD camera using a Nikon F/c adaptor. Although the magnification of this setup is 1 (unilateral SCN covers approximately 375 pixels), we were able to obtain an image from the SCN using a 25-min exposure setting (Fig. 2). High NA, long working-distance microscope objectives can also be used for higher-magnification images using an extension barrel attached directly to the camera (Geusz, 2001). A microscope can also possibly be used; however, a longer integration time is required.
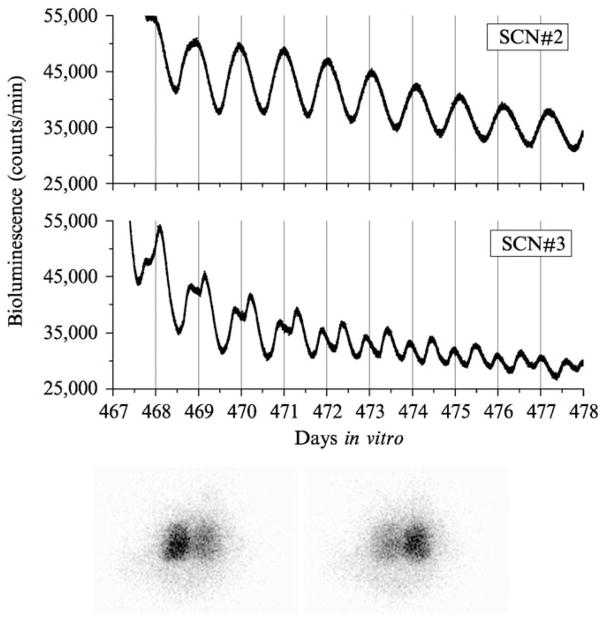
Figure 2.
Circadian rhythms of Per1<luc activity from long-term cultured rat SCN. Four SCN explants were made and maintained in vitro for a long period of time. Two examples of the luminescence rhythms are shown (top). Although SCN#2 showed robust unimordal rhythms all the time, the peak of the rhythms in SCN#3 started breaking down into two components at 468 days in culture and both peaks were phase locked at 180° of the phase (bimodal). At the end of the recording (478 days), SCN#3 explants were transferred to the luminescence imaging setup and images were obtained using an ICCD camera (VIM, photon counting mode with 25-min exposure) at 30-min intervals. We were able to confirm the paired SCN structure in 478-day-old SCN explants. On the first image (bottom left), the left side of the SCN showed a significantly stronger signal than the one on the right side, indicating that the peak, which occurred on the vertical lines (top) came from the left side of the SCN. The image 12 h later (bottom right) shows that the right side of the SCN is brighter than the left side.
Recording Stages
Although heating the camera may increase dark currents, putting the whole setup inside the incubator (or environmental chamber) is a simple way to control the temperature of the culture sample.
A Sykes-Moore type of chamber can also be used to control the temperature with good optical access (Geusz, 2001).
Data Analysis
Phase Map
By measuring the peak time between 12 and 36 h in culture, we can estimate the in vivo phase of each tissue. We usually use an adjacent averaging method with 2-h running means (moving average) to smooth data. The peak is calculated as the highest point of smoothed data. By plotting peaks of each tissue, we are able to draw the “phase map” and we have been successful using this method to observe the effects of aging and in vivo treatments (such as shifting the light and dark cycle or restricting feeding) on circadian organization (Stokkan et al., 2001; Yamazaki et al., 2000, 2002).
Circadian Parameters, Period, Amplitude, Waveform, Damping Rate
Several circadian analysis programs can be used to analyze luminescence rhythms: Chrono (courtesy of T. Roenneberg, University of Munich, Munich, Germany), LumiCycle (Actimetrics Inc.), and ClockLab (Actimetrics). To analyze the circadian parameters, we first need to remove baseline changes because there are often drastic changes in the baseline in the first few days in culture, and these changes are different in each tissue sample. The baseline drift can be obtained by either adjacent averaging method with 24 or 48 h (Chrono) or by fitting a polynomial curve (Lumi-Cycle). This can be used for trend correction by subtracting it from data. Because most circadian analysis programs cannot handle negative values, adding a minimum value to detrended data is necessary. The period can be obtained from regression analysis of various circadian markers (peak, trough, half-maximum, half-minimum) or other periodgram analyses. Other circadian parameters, such as amplitude, bandwidth, and damping rate, can also be calculated by those programs.
Conclusion
We expect that luminescence reporting with an organotypic culture will become a powerful system for circadian studies. SCN explants have been maintained for up to 681 days in vitro. The organization in long-term cultured SCN explants was well maintained and the oscillation of Per1::luc activity continued (Fig. 2). The initial phase in culture can be used for estimation of the in vivo phase. Tissues taken from adult animals or even from aged animal can be cultured. Therefore, tissue cultures shortly after in vivo treatments can be used to measure the effect of in vivo treatments on circadian organization. Immortalized cells (and primary cell cultures) with a transfected reporter construct can be used for drug screening and molecular analysis. Previously, transfection assays were done only to monitor acute activation. This can now be extended to measure circadian parameters, such as changes in period, phase, and amplitude of circadian oscillations.
Acknowledgments
We thank Susan J. McMahon for reading of the manuscript.
References
- Asai M, Yamaguchi S, Isejima H, Jonouchi M, Moriya T, Shibata S, Kobayashi M, Okamura H. Visualization of mPer1 transcription in vitro: NMDA induces a rapid phase shift of mPer1 gene in cultured SCN. Curr Biol. 2001;11:1524–1527. doi: 10.1016/s0960-9822(01)00445-6. [DOI] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- Brandes C, Plautz JD, Stanewsky R, Jamison CF, Straume M, Wood KV, Kay SA, Hall JC. Novel features of drosophila period Transcription revealed by real-time luciferase reporting. Neuron. 1996;16:687–692. doi: 10.1016/s0896-6273(00)80088-4. [DOI] [PubMed] [Google Scholar]
- Collaco AM, Geusz ME. Monitoring immediate-early gene expression through firefly luciferase imaging of HRS/J hairless mice. BMC Physiol. 2003;19:8. doi: 10.1186/1472-6793-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geusz ME. Methods in Cellar Imaging. Oxford University Press; Oxford: 2001. Bioluminescence imaging of gene expression in living cells and tissues; pp. 395–408. [Google Scholar]
- Geusz ME, Fletcher C, Block GD, Straume M, Copeland NG, Jenkins NA, Kay SA, Day RN. Long-term monitoring of circadian rhythms in c-fos gene expression from suprachiasmatic nucleus cultures. Curr Biol. 1997;7:758–766. doi: 10.1016/s0960-9822(06)00334-4. [DOI] [PubMed] [Google Scholar]
- Izumo M, Johnson CH, Yamazaki S. Circadian gene expression in mammalian fibroblasts revealed by real-time luminescence reporting: Temperature compensation and damping. Proc Natl Acad Sci USA. 2003;100:16089–16094. doi: 10.1073/pnas.2536313100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog ED, Aton SJ, Numano R, Sakaki Y, Tei H. Temporal precision in the mammalian circadian system: A reliable clock from less reliable neurons. J Biol Rhythms. 2004;19:35–46. doi: 10.1177/0748730403260776. [DOI] [PubMed] [Google Scholar]
- Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH. Circadian rhythms in prokaryotes: Luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90:5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maronde E, Motzkus D. Oscillation of human period 1 (hPER1) reporter gene activity in human neuroblastoma cells in vivo. Chronobiol Int. 2003;20:671–681. doi: 10.1081/cbi-120022413. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua NH, Kay SA. A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell. 1992;4:1075–1087. doi: 10.1105/tpc.4.9.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan LW, Greene AV, Bell-Pedersen D. Circadian and light-induced expression of luciferase in Neurospora crassa. Fung Genet Biol. 2003;38:327–332. doi: 10.1016/s1087-1845(02)00562-5. [DOI] [PubMed] [Google Scholar]
- Stanewsky R, Jamison CF, Plautz JD, Kay SA, Hall JC. Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila. EMBO J. 1997;16:5006–5018. doi: 10.1093/emboj/16.16.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Akashi M, Nishida E. Temperature compensation and temperature resetting of circadian rhythms in mammalian cultured fibroblasts. Genes Cells. 2003;8:713–720. doi: 10.1046/j.1365-2443.2003.00669.x. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Vallone D, Gondi SB, Whitmore D, Foulkes NS. E-box function in a period gene repressed by light. Proc Natl Acad Sci USA. 2004;101:4106–4111. doi: 10.1073/pnas.0305436101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Imaizumi T, Kay SA. Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods Enzymol. 2005;393 (11):2005. doi: 10.1016/S0076-6879(05)93011-5. (this volume) [DOI] [PubMed] [Google Scholar]
- Wilsbacher LD, Yamazaki S, Herzog ED, Song EJ, Radcliffe LA, Abe M, Block GD, Spitznagel E, Menaker M, Takahashi JS. Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice in vivo. Proc Natl Acad Sci USA. 2002;99:489–494. doi: 10.1073/pnas.012248599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kobayashi M, Mitsui S, Ishida Y, van der Horst GT, Suzuki M, Shibata S, Okamura H. View of a mouse clock gene ticking. Nature. 2001;409:684. doi: 10.1038/35055628. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Effects of aging on central and peripheral mammalian clocks. Proc Natl Acad Sci USA. 2002;99:1081–1086. doi: 10.1073/pnas.152318499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2:: LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101(15):5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]