Abstract
Foxp3+ T-regulatory (Treg) cells maintain immune homeostasis and limit autoimmunity, but can also curtail host immune responses to various types of tumors1,2. Foxp3+ Tregs are therefore considered promising targets to enhance anti-tumor immunity, and efforts are underway to develop approaches for their therapeutic modulation. However, while studies showing that Foxp3+ Treg depletion experimentally can enhance anti-tumor responses provide proof-of-principle, they lack clear translational potential and have various shortcomings. Histone/protein acetyltransferases (HATs) promote chromatin accessibility, gene transcription and the function of multiple transcription factors and non-histone proteins3,4. We now report that conditional deletion or pharmacologic inhibition of one HAT, p300 (Ep300, KAT3B), in Foxp3+ Tregs, increased TCR-induced apoptosis in Tregs, impaired Treg suppressive function and peripheral Treg induction, and limited tumor growth in immunocompetent, but not in immunodeficient, hosts. Our data thereby demonstrate that p300 is important for Foxp3+ Treg function and homeostasis in vivo and in vitro, and identify novel mechanisms by which appropriate small molecule inhibitors can diminish Treg function without overtly impairing T-effector (Teff) cell responses or inducing autoimmunity. Collectively, these data suggest a new approach for cancer immunotherapy.
Foxp3+ Treg cells are pivotal to limiting host autoimmunity, but their increased numbers within blood, tumors or lymphoid tissues of patients with cancers are often associated with poor prognosis1,2. Experimental strategies to deplete Tregs and promote anti-tumor immunity have transient efficacy, are non-specific, and often provoke autoimmunity5–7. Epigenetic approaches are being developed to pharmacologically modulate Treg functions, especially for autoimmunity8. Thus, deacetylase inhibitors increase acetylation of the key Treg transcription factor, Foxp3, and enhance Foxp3+ Treg suppressive function in vivo and in vitro9–12, but there are no data concerning HAT inhibitors and Tregs. Of the three main HAT families (GNAT, p300/CBP, MYST13), p300 can acetylate and stabilize Foxp3 expression in transfected cells3,4. The current studies assessed whether p300 targeting could affect Treg homeostasis or function, and thereby promote antitumor immunity.
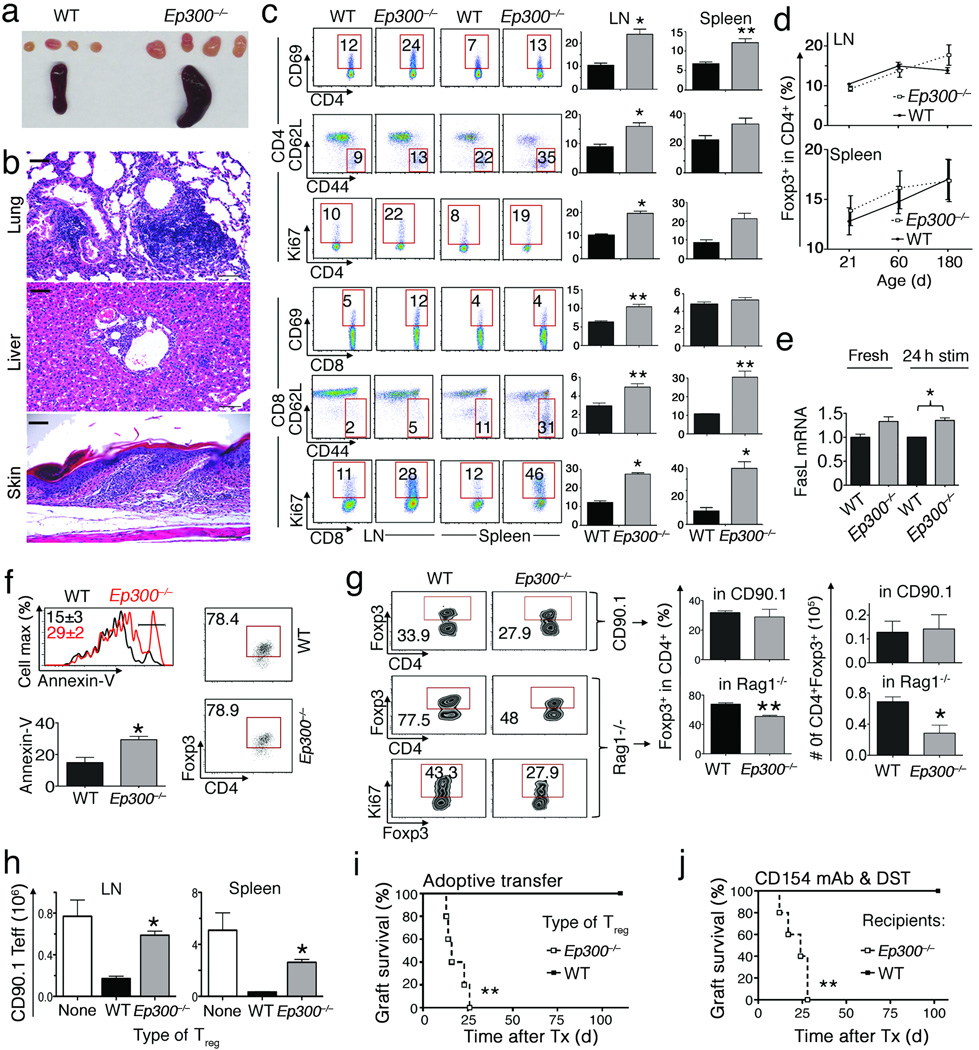
We conditionally deleted Ep300 in Tregs by crossing Ep300fl/fl and Foxp3YFP-cre mice (Supplementary Fig. 1a); Ep300−/− mice were born at expected Mendelian ratios, but from 10 weeks developed mild weight loss, dermatitis, lymphadenopathy and splenomegaly (Fig. 1a); focal mononuclear infiltrates in lung, liver and skin (Fig. 1b, Supplementary Table 1); and mildly decreased hematocrit and hemoglobin levels (Supplementary Fig. 1b). Compared to littermates, Ep300−/− mice showed increased T-cell activation and production of proinflammatory cytokines; though overall T-cell proportions were similar (Supplementary Fig. 1c,d). They also had increased serum IgG1 (Supplementary Fig. 1e), and ~20% developed autoantibodies (Supplementary Table 2). Microarray analysis showed that compared to wild-type (WT) Tregs, Ep300−/− Tregs overexpressed pro-apoptotic genes (Supplementary Fig. 2a,b). While Ep300 deletion did not affect CD4+Foxp3+ cell numbers or levels of Foxp3 protein under basal conditions (Fig. 1d, Supplementary Fig. 1 and 3a), upon activation Ep300−/− Treg had increased FasL mRNA (Fig. 1e, Supplementary Fig. 3b) and apoptosis (Fig. 1f). Likewise, adoptive transfer of Ep300−/− Tregs into immunodeficient, but not immunocompetent, mice led to decreased Treg survival compared to WT Tregs (Fig. 1g). Suppressive functions of Ep300−/− Tregs were modestly impaired and declined with age (Supplementary Fig. 4a). Hence, unlike scurfy mice, wherein Foxp3 mutation disrupts DNA binding and causes lethal autoimmunity by 3 weeks, Ep300 deletion has moderate effects on Treg biology, comparable to other mice with conditional Treg-targeting14–16.
Figure 1.
Effects of conditional deletion of Ep300 in Foxp3+ Tregs. Data in panels a–c are from 3-month old mice (4 mice/group). (a) Spleen and lymph nodes from Ep300−/− versus WT mice. (b) Focal mononuclear infiltrates in tissues from Ep300−/− mice; scale bars = 100 µ. (c) T cells activation markers in Ep300−/− versus WT mice; %-gated cells shown and representative of 4–6 experiments. (d) Analysis of CD4+Foxp3+ Treg percentage in Ep300−/− and WT mice. (e) FasL mRNA level in Ep300−/− versus WT mice upon activation with CD3/CD28 mAbs. (f) Analysis of CD4+Foxp3+ and Annexin–V level in Ep300−/− versus WT Tregs upon activation with CD3/CD28 mAbs. Cells were gated on CD4+7-AAD−. (g) Analysis of CD90.2+CD4+Foxp3+ and CD90.2+CD4+Foxp3+Ki67+ level of adoptively transferred Ep300−/− versus WT Tregs in CD90.1+ (upper panel) or Rag1−/− host mice (lower panel). (h) Suppression of homeostatic in vivo proliferation of CD90.1+CD4+CD25− Teff cells by Ep300−/− versus WT Tregs. (i–j) Cardiac allograft survival. (i) Rag1−/− mice (5/group) received BALB/c cardiac allografts and adoptive transfer of WT Teff cells plus Tregs from Ep300−/− or WT mice. (j) WT or Ep300−/− mice (5/group) received BALB/c cardiac allografts and tolerance was induced using CD154 mAb and donor splenocyte transfusion (DST). As indicated in respective panels, *p<0.05, and **p<0.01, and data from 2–3 independent experiments.
To assess Treg function in vivo, we transferred Ep300−/− or WT Tregs plus conventional Teff cells into immunodeficient mice. Compared to WT Tregs, Ep300−/− Tregs were poor suppressors of homeostatic Teff cell proliferation (Fig. 1h), and had reduced cell survival (Supplementary Fig. 4b). In a second in vivo test of Ep300 deletion in Tregs, we undertook cardiac allografts. Immunodeficient recipients adoptively transferred with Teff cells developed acute rejection by 14 d post-transplant, whereas mice receiving cotransfer of WT Teff and Treg cells maintained allografts long-term (>100 d). However, mice given WT Teff cells and Ep300−/−Tregs rejected their allografts by 25 d post-transplant (Fig. 1i). In a third in vivo test, we transplanted hearts into WT or Ep300−/− mice, and treated recipients with CD154 mAb and donor splenocytes (DST)17. Costimulation blockade induced allograft tolerance in WT but not Ep300−/− recipients (Fig. 1j). Collectively, data from conditional targeting show p300 is important for normal Treg survival and suppressive function.
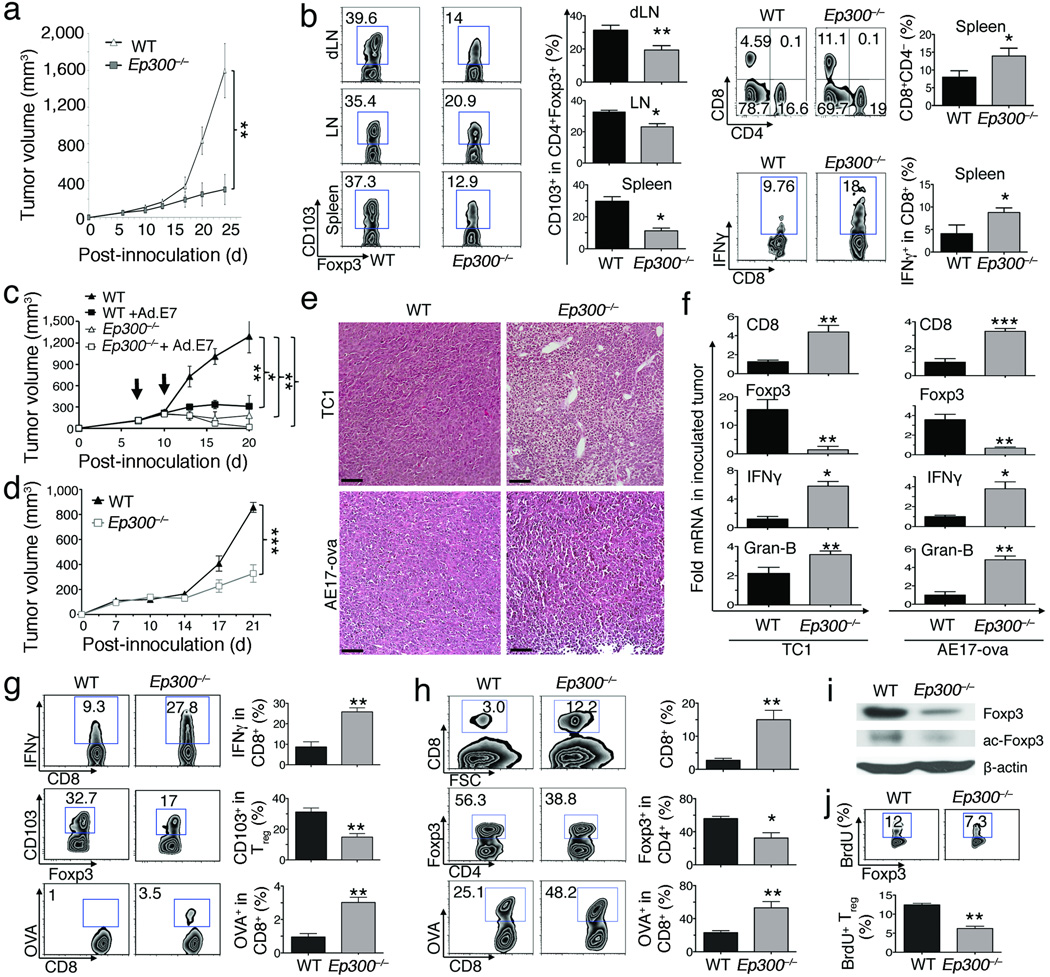
We next tested if Ep300 deletion in Tregs promoted anti-tumor immunity. Growth of TC1 lung adenocarcinomas, which express HPV-E7, was impaired in Ep300−/− versus WT tumor-bearing mice (Fig. 2a). Deletion of Ep300 did not affect lymphoid CD4+Foxp3+ Treg numbers (data not shown) but decreased their expression of CD103, important for Treg recruitment to tumor sites18,19, and increased lymphoid CD8+ Teff cell and IFN–γ production (Fig. 2b). Likewise, Ep300 deletion enhanced the effects of Ad.E7 vaccination on TC1 tumor growth (Fig. 2c), and was also effective in mice bearing AE17 mesotheliomas (Fig. 2d). Both tumors showed increased mononuclear cell infiltration (Fig. 2e). Impaired tumor growth in both models was accompanied by increased intratumoral CD8, granzyme–B and IFN–γ mRNA, but reduced Foxp3 mRNA (Fig. 2f), and immunohistology showed decreased Treg infiltration and increased intratumoral CD8+ Teff cells (Supplementary Fig. 5). AE17 tumor-bearing Ep300−/− mice had fewer lymphoid CD4+Foxp3+CD103+ Tregs, but more CD8+IFN–γ+ and OVA (tumor)-specific CD8+ Teff cells (Fig. 2g). AE17 tumor-bearing Ep300−/− mice also had reduced intratumoral Treg, and decreased levels of both total and acetylated Foxp3 protein, but increased CD8+OVA− specific Teff cells (Fig. 2h,i). Deletion of Ep300 reduced Treg proliferation (BrdU+, Ki67+) in tumor-bearing mice (Fig. 2i, Supplementary Fig. 6a). Hence, Ep300 conditional targeting can diminish Treg proliferation and accumulation within tumors, and enhance anti-tumor immunity.
Figure 2.
Treg-specific deletion of Ep300 enhances anti-tumor immunity. Ep300−/− or WT C57BL/6 mice (n=7–10/group) were inoculated with TC1 or AE17 tumor cells for 21 d (*p<0.05, **p<0.01, ***p<0.001). (a) TC1 tumor growth in Ep300−/− versus WT mice. (b) Analysis of CD4+Foxp3+CD103+, CD4+, CD8+, CD8+IFN–γ+ in lymphoid tissues from Ep300−/− or WT TC.1 tumor-bearing mice. (c) Ad.E7 vaccination of Ep300−/− versus WT mice, bearing TC1 tumors, at 7 and 10 d (arrows). (d) AE17 tumor growth in Ep300−/− versus WT mice (e) Mononuclear cell infiltration in Ep300−/− versus WT mice, bearing TC1 or AE17 tumor; scale bars: 100 µm. (f) Analysis of mRNA level of CD8, IFN–γ, granzyme–B and Foxp3 in tumor samples from Ep300−/− versus WT mice, bearing TC1 or AE.17 tumors (5/group). (g) Analysis of the percentage of CD4+Foxp3+CD103+, CD8+IFN–γ+, and OVA-specific CD8+ T cells in the spleens of Ep300−/− versus WT AE17 tumor-bearing mice. (h) Analysis of the frequency of CD4+Foxp3+, CD8+, OVA specific CD8+ T cells in tumor site of Ep300−/− versus WT mice, bearing AE17 tumor. (i) Western blot showing Foxp3 and acetyl–Foxp3 (K31) level in TC1 tumors of Ep300−/− versus WT tumor bearing mice (β–actin loading control). (j) Analysis of the percentage of Foxp3+BrdU+ in Ep300−/− versus WT mice, bearing AE17 tumors.
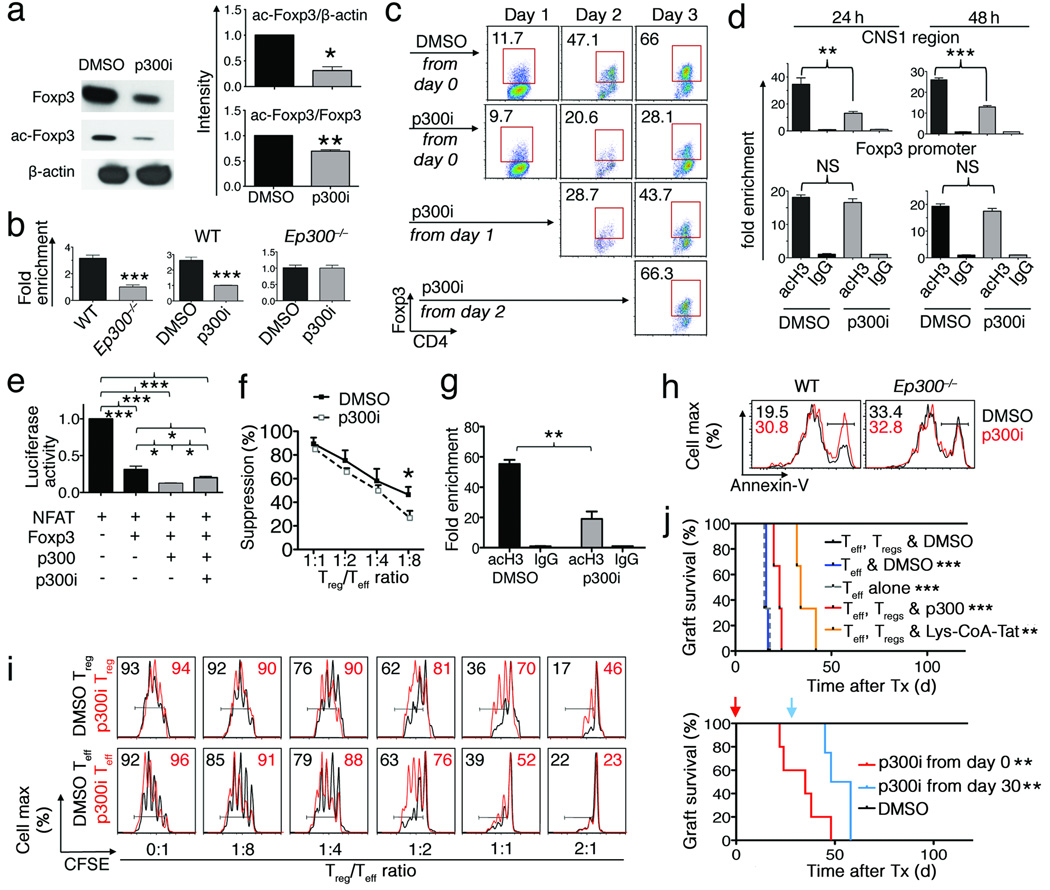
From a translational perspective, we wondered if a p300 small–molecule inhibitor (p300i, C646)20 would display similar utility. We began with in vitro studies of the effects of p300i on Foxp3 and Tregs, and found that p300i decreased acetylation of Foxp3 by p300 (Fig. 3a), and decreased Treg expression of Foxp3 mRNA and protein (Supplementary Fig. 6b). Tregs lacking Ep300 had lower levels of histone H3 acetylation at the Foxp3 promoter region compared to WT Tregs, and p300i decreased the levels of acetylation at the Foxp3 promoter in WT but not p300−/− Tregs, indicating the specificity of this approach (Fig. 3b). Use of p300i also decreased TGF–β-induced conversion of naïve CD4+CD25− T-cells into Foxp3+ Tregs, especially in the first 2 days of culture (Fig. 3c), and reduced acetylation at the CNS1 enhancer21,22, a critical region mediating peripheral conversion, but not at the Foxp3 promoter (Fig. 3d). Treatment with p300i inhibited Foxp3 transactivation (Fig. 3e), and decreased Treg suppression function (Fig. 3f). Analysis of cells from mice treated in vivo with p300i showed decreased acetyl-H3 accumulation at the Foxp3 promoter (Fig. 3g), increased Treg apoptosis in WT but not in Ep300−/− Treg (Fig. 3h), and decreased Treg suppressive function (Fig. 3i and Supplementary Fig. 6c), but did not promote immune activation or affect Treg numbers (Supplementary Figs. 7a,b).
Figure 3.
Use of p300i decreased Foxp3 acetylation and Treg function (*p<0.05, **p<0.01, ***p<0.005; data from 4 independent experiments). (a) Western blot showing Foxp3 and acetyl–Foxp3 level in 293T cells co-transfected with p300 and Foxp3 treated with DMSO or p300i. (b) ChIP-qPCR for acetyl-H3 binding to Foxp3 promoter. CD4+CD25+ Tregs from WT or Ep300−/− mice (left panel); from WT mice (middle panel) or Ep300−/− mice (right panel) treated with p300i or DMSO. (c) Flow cytometry analysis of CD4+Foxp3+ frequency. Sorted CD4+YFP− Teff cells cultured under Treg-inducing conditions and treated with p300i or DMSO. (d) ChIP-qPCR for acetyl-H3 binding to Foxp3 CNS1 and promoter regions. (e) NFAT-IL–2 luciferase reporter assay of 293T cells transfected with NFAT, Foxp3 and p300, treated with p300i. (f) In vitro Treg assay with p300i (5 µM). (g) ChIP-qPCR assay detecting acetyl–H3 at the Foxp3 promoter in Tregs treated with p300i in vivo. (h) Annexin–V in Tregs; C57BL/6 mice treated with p300i in vivo for 7 d; CD4+YFP+ cells were sorted from treated mice, stimulated with CD3/CD28 mAbs in vitro and stained for CD4, Annexin–V, 7–AAD. (i) Treg suppression assay comparing T-cell functions of p300i–treated mice versus control. (j) Cardiac allograft survival in Rag1−/− recipients treated with p300i, Lys–CoA–Tat peptide or DMSO (5/group, upper panel). Treg-dependent tolerance in WT recipients (4/group) was induced using DST and treated with p300i or DMSO (via 14 d Alzet pump delivery) begun at (red), or 30 days after (blue) transplantation (Tx).
We used a parent-to-F1 adoptive transfer model, involving allo-antigen-induced T-cell activation, proliferation and cytokine production23 to assess effects of p300i on conventional T-cell functions in vivo. T-cell activation and proliferation were comparable in groups receiving p300i versus DMSO control (Supplementary Fig. 8a). We next evaluated effects of p300i on T-cell responses in cardiac allograft recipients9. Administration of p300i, but not DMSO carrier, abrogated Treg-dependent allograft survival, as seen using C646 or a peptidic p300i (Lys-CoA-Tat)24 (Fig. 3j). To preclude p300i affecting graft survival by altering CD4+CD25− T–cell function, recipients were adoptively transferred with CD4+CD25− T–cells and treated with p300i or DMSO control. In both groups, acute rejection developed similarly (Supplementary Fig. 8b). We also tested if p300i affected grafts directly, by infusing p300i into isograft recipients; no effects on long-term graft survival were seen (>100 d survival, data not shown), supporting preferential effects of p300i on Tregs. Moreover, p300i can prevent, or abrogate established, Treg-dependent immune responses, as shown by the ability of p300i, given from the day of surgery or from 30 days post-transplant, to restore rejection in mice rendered tolerant by therapy with CD154 mAb/DST17 (Fig. 3j). Hence, p300i administration differentially affects Tregs vs. Teff cells in vivo; critically, p300i use can decrease Treg suppressive function while simultaneously leaving intact protective Teff cell responses.
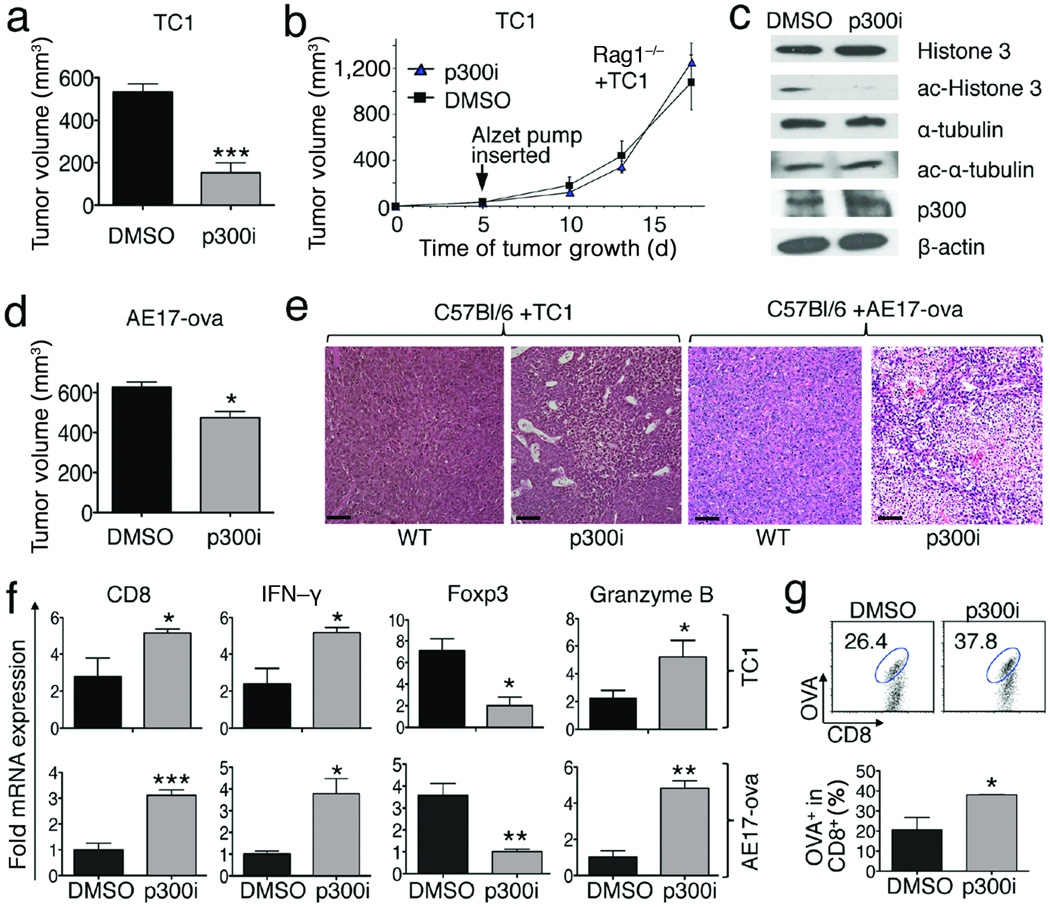
Lastly, we tested the effects of p300i on tumor growth. We found that p300i treatment significantly decreased TC1 tumor growth in normal (Fig. 4a) but not immunodeficient (Fig. 4b) mice. The compound reached tumor sites in the latter immunodeficient mice as shown by decreased acetylation of histone-3, whereas no effect was seen on the acetylation of p300-independent targets (e.g. α–tubulin) (Fig. 4c). The p300i also significantly inhibited growth of AE17 tumors in WT mice (Fig. 4d). In both models, p300i promoted tumor infiltration by host mononuclear cells (Fig. 4e), and decreased intratumoral Foxp3 but increased CD8, granzyme-B and IFN–γ expression (Fig. 4f). Therapy with p300i also increased antigen-specific CD8+OVA+ Teff cells within tumors (Fig. 4g).
Figure 4.
p300i therapy impaired tumor growth in immunocompetent mice (*p<0.05, **p<0.01, data representative of 4 independent experiments, 7–10 mice/group). (a–b) TC1 tumor growth in WT (day 18) (a) or Rag1−/−; (b) mice treated with p300i or DMSO. (c) Western blotting assays showed α-tubulin and H3-acetylation in TC1 tumors in immunodeficient hosts (day 18) treated with p300i or DMSO. (d) AE17 tumor growth in WT mice (day 18) treated with p300i versus DMSO. tumor volumes at end of study (day 18). (e) Mononuclear cell infiltration and tumor necrosis in TC1 and AE17 models in WT mice (day 18) treated with p300i or DMSO; scale bars = 100 µ. (f) qPCR analysis of Foxp3, CD8, granzyme-B and IFN-γ mRNA expression in TC1 and AE17 tumors treated with p300i versus DMSO. (g) p300i increased CD8+OVA+ T cells within AE17 tumors (Analyses of CD8+OVA+ percentage in mice bearing AE17 tumors treated with p300i or DMSO) (day 18).
Collectively, our studies show that p300 targeting impaired Treg homeostasis and function, and boosted anti-tumor immunity. In the steady state, Ep300 deletion in Tregs favored a pro-apoptotic phenotype, but without affecting actual Treg frequency or number until subject to immune activation. Given our evidence of the importance of p300 to iTreg development, and knowledge that tumors can promote iTreg production25, p300i may act not only by impairing the proliferation and function of existing Tregs, but by limiting the ability of cancers to avoid immune destruction by promoting iTreg conversion26. In contrast to Treg-depletional therapies, p300i use provided reasonable safety since Treg numbers were unaffected, even in p300−/− mice, and did not lead to severe autoimmunity. This ability of p300 targeting to modulate Tregs, likely as a result of affecting the acetylation and function of multiple transcription factors in Tregs and not just that of Foxp3, is quite unexpected given the ubiquitous expression of p300. However, p300i therapy had no effects on tumor growth in immunodeficient mice, and no untoward effects on conventional T-cells or other immune cells (e.g. parent-to-F1 and cardiac transplant experiments). As a result, we propose that p300 targeting has potential therapeutic applications in cancer, HIV and other conditions in which Foxp3+ Treg cells limit protective host immune responses.
ONLINE METHODS
Ethics statement
Studies were approved by the Institutional Animal Care and Use Committee of the Children’s Hospital of Philadelphia (#2008-7-746 and #2010-6-561).
Mice
We purchased standard CD90.2+ C57BL/6 (B6), B6/CD90.1+, BALB/c and B6/Rag1−/− mice (The Jackson Laboratory), and p300fl/fl mice27 and Foxp3YFP-cre mice28 were described previously. Mice were housed under pathogen-free conditions and used at 6–24 weeks of age.
Antibodies, flow cytometry and cell sorting
We purchased standard conjugated monoclonal antibodies (mAbs) for flow cytometry (BD Pharmingen), plus anti-Foxp3 mAb (FJK-16s, eBioscience), and rabbit antibodies to β–actin, α–tubulin and acetyl–α–tubulin (Cell Signaling), p300 (Santa Cruz), and histone-3 and acetyl-histone-3 (Upstate). Flow cytometry was performed using a Cyan flow cytometer (Beckman Coulter), and data were analyzed using FlowJo 8 software (Tree-Star). CD4+YFP+(Foxp3+) and CD4+YFP−(Foxp3−) cells were sorted from Foxp3YFP-cre or p300−/− mice using a FACS Aria cell sorter (BD Bioscience, UPenn Cell Sorting Facility).
Plasmids and p300i
A p300 expression vector was provided by Dr. Xiao-Jiao Yang (McGill University, Montreal, Canada), and we purchased NFAT expression vectors and NFAT–IL–2 promoter luciferase reporter (Addgene). Foxp3-MinR1 was described previously4, as were details of non-peptidic20 and peptidic24 p300i.
Treg suppression assay
CD4+CD25− T-cells and CD4+CD25+ Tregs were isolated from Foxp3YFP-cre or p300−/− mice using CD4+CD25+ Treg isolation kits (130-091-041, Miltenyi Biotec). Cell Trace Violet-labeled Teff cells (5×105) were stimulated with CD3 (5 µg/ml) in the presence of 5 × 105 irradiated syngeneic T-cell depleted splenocytes (130-049-101, Miltenyi Biotec) and varying ratios of Tregs9. After 72 h, proliferation of Teff cells was determined by flow cytometric analysis of Cell Trace Violet dilution.
Homeostatic proliferation assay
One million CD90.1+CD4+CD25− T-cells were mixed with half million CD4+YFP+ Tregs sorted from Foxp3YFP-cre or p300−/− mice (CD90.2+), and adoptively transferred to Rag1−/− mice9. Spleen and lymph nodes were isolated after 7 d, and total CD90.1+ CD4+ T-cells were determined by flow cytometry.
Quantitative PCR (qPCR)
RNA from Treg or Teff cells, fresh or activated with CD3/28 mAb-coated beads (Invitrogen), was isolated using RNeasy Kit (Qiagen, CA). RNA was isolated from tumor samples using STAT-60 (AmsbiP). cDNA was synthesized with TaqMan reverse transcription reagents (Applied Biosystems). qPCR was performed using TaqMan Universal PCR Master Mix (Applied Biosystems), and specific primers from Applied Biosystems. Gene expression data were normalized to 18S RNA.
Luminex
Serum immunoglobulin isotypes were assessed with a Multiplex mouse immunoglobulin assay kit (Millipore), using LiquiChip Luminex 100 (Qiagen).
Hematology and autoantibody detection
Citrated blood samples were tested using an automated hematology analyzer modified and calibrated for mouse blood samples. Pooled sera from male p300−/− or WT mice were diluted 1:5 and incubated with cryosections from normal male and female C57BL/6 mice, washed in PBS, followed by goat anti-mouse IgG FITC-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, 1:200). In addition to pooled WT healthy sera, controls included incubation with secondary antibody alone. Pooled sera from NZB mice with known autoantibodies served as positive controls. If any autoantibodies were detected, serum from each mouse was re-analyzed separately.
Luciferase assays
293T cells were maintained (37 °C, 5% CO2) in RPMI-1640 plus 10% heat-inactivated FBS, penicillin and streptomycin. Cells at 80–90% confluence were transfected with NFAT-IL-2 promoter luciferase reporter, plus NFAT, Foxp3, p300 expression vectors or empty vector, using Lipofectamine 2000 (Invitrogen). After 48 h, cells were treated with 6 ng mL−1 PMA and 1 µM ionomycin (Sigma) in the absence or presence of p300i for 5–6 h, and luciferase activities of whole-cell lysates analyzed with a dual-luciferase reporter assay kit (Promega).
Western blotting
Cell lysates were separated by SDS-PAGE, transferred to PVDF membranes and subjected to immunoblotting using the indicated Abs.
ChIP assays
For ChIP assays using EZ-Magna CHIP A Chromatin Immunoprecipitation Kit (17-408, Upstate), DNA-chromatin complexes were prepared from 2 × 106 Tregs, and genomic DNA precipitated using 10 µg of antibodies against p300 or acetylated histone H3, or using control rabbit IgG (Upstate). Genomic DNA in precipitates was probed by RT-PCR for Foxp3 promoter using forward primer 5’ TTCCCATTCACATGGCAGGCTTCA 3’, and reverse primer 5’ TGAGATAACAGGGCTCATGAGAAACCACA 3’; and for Foxp3 CNS1 region using forward primer 5’ TAAAGGAGACTGGAAGCCAACATGG 3’, and reverse primer 5’ ATAGAAGACATACACCACGGCG 3’.
Microarrays
RNA was extracted with RNeasy kits (Qiagen), and RNA integrity and quantity were analyzed by photometry (DU640; Beckman-Coulter). Microarray experiments were performed with whole-mouse-genome oligoarrays (Mouse430a 2.0; Affymetrix), and array data analyzed with MAYDAY 2.1229. Array data were subjected to robust multiarray average normalization. Normalized data were used for calculating fold changes of genes that were increased or decreased in expression with the Student’s t test, and data with >1.5–fold differential expression (p<0.05 with Storey’s FDR<0.3) were included in the analysis. Data underwent z-score transformation for display.
iTreg conversion assays
CD4+YFP− T-cells isolated by cell sorting from Foxp3YFP-cre mice were incubated with CD3/CD28 beads (Invitrogen), IL-2 (10 U mL−1) and TGF–β (2 ng mL−1) for 3 d and analyzed by flow cytometry.
Parent-to-F1 assay
CFSE-labeled single cells from C57BL/6 mice (H-2b) were injected into B6D2F1 mice (H-2d) (n=3/group), and recipients were treated with DMSO or p300i (C646, 8.9 mg kg−1 d−1) for 3 d. Thereafter, donor cells (H-2d negative) were analyzed for proliferation, cellular activation and cytokine production23. Allograft survival was monitored daily and rejection was defined as cessation of heartbeat. Data are representative of 2 independent experiments.
Cardiac transplantation
Heterotopic cardiac allografts were performed using BALB/c donors and B6/Rag1−/− recipients (n=5/group). In brief, B6/Rag1−/− mice engrafted with BALB/c hearts were adoptively transferred with 1 × 106 CD4+CD25− T-cells alone, or along with 5 × 105 Foxp3YFP-cre Tregs or p300−/− Tregs, and Alzet pumps were implanted subcutaneously to deliver p300i or DMSO infusions. In additional experiments, Treg-dependent allograft tolerance was induced by treatment of WT or Ep300−/− recipients at the time of engraftment with CD154 mAb plus 5 × 106 donor splenocytes17.
BrdU staining
WT or p300−/− AE17 tumor-bearing mice were injected twice per day with BrdU (1 mg, i.p.), for 3 d, sacrificed, and spleen cells were stained for CD4, Foxp3, and BrdU according to the manufacturer’s instructions (BrdU labeling kit, BD Bioscience), and analyzed by flow cytometry.
Cell lines and tumor models
TC1 cells, derived from mouse lung epithelial cells that were immortalized with HPV-16 E6 and E7, and transformed with the c-Ha-ras oncogene30, were provided by Dr. Yvonne Paterson (UPenn, Philadelphia, PA). The murine AE17.ova mesothelioma cell line (provided by Dr. Delia Nelson, University of Western Australia) was derived from mesothelioma cells developing in mice treated i.p. with asbestos, and then stably transduced with chicken ovalbumin31. Cells were grown in RPMI, 10% fetal bovine serum (FBS), 2 mM glutamine, and 5-µg ml−1 penicillin & streptomycin. Each mouse was shaved on their right flank and injected s.c. with 1.2 × 106 TC1 or 2 × 106 AE17 tumor cells. For p300i experiments, mice received p300i or DMSO via Alzet pumps inserted 7 d after initial tumor injection. Tumor volume was determined by the formula: (3.14 × long axis × short axis × short axis)/6. Tumors were paraffin-embedded and stained by H&E, or snap-frozen and stained by immunoperoxidase9.
Ad.E7 vaccination
1 × 106 TC1 (E7+) cells were injected subcutaneously into the right flanks of WT and Ep300−/− mice. One week later, mice bearing TC1 flank tumors (~100 mm3 in size) were either left untreated, or vaccinated subcutaneously in the left flank (contralateral to the tumor) with 1 × 109 plaque forming units (pfu) of Ad.E7 vector, as previously described32. Three days following initial vaccination, mice received a booster vaccine of 1 × 109 pfu of Ad.E7 in the left flank. Tumor sizes were monitored twice/week.
OVA-tetramer staining
Tumors and spleens from the experimental mice were harvested to determine OVA-specific CD8 T cells. Tumors were cut into small fragments and digested for 1 h at 37°C with a cocktail of collagenase type I (0.1 U mL−1, Worthington), collagenase type II (0.1 U mL−1, Worthington) collagenase type IV (0.1 U mL−1, Worthington), DNase I (0.05 U mL−1, Worthington), and elastase (5 U mL−1, Worthington), in L15-medium (Leibovitz). Single cell suspensions were blocked for 30 min with anti-Fc receptor antibody (eBioscience), washed (2% FBS in PBS), stained for 1 h with PE-conjugated H-2Kb tetramer loaded with ovalbumin peptide (SIINFEKEL) (iTag™ MHC Tetramer; Beckman Coulter), washed, and stained with APC-conjugated anti-CD8+ (Clone 53–6.7; BD Bioscience) for 30 min. A commercial kit (Fixable Aqua Dead Cell stain kit, Invitrogen) was used to exclude dead cells from data analyses. Cell acquisition was performed on a Cyan flow cytometer.
Statistics
Data were analyzed by GraphPad Prism 5.0d. All normally distributed data were displayed as mean ± standard error of the mean (SEM). Measurements between 2 groups were performed with a Student’s t test or Mann-Whitney U test. Groups of 3 or more were analyzed by one-way analysis of variance (ANOVA) or the Kruskal-Wallis test. Survival analysis was calculated using Log rank (Mantel Cox) test.
Supplementary Material
ACKNOWLEDGEMENTS
We thank E. Verdin (Gladstone Institute of Virology and Immunology, UCSF, USA) for anti-acetylated-Foxp3 antibody; Xiao-Jiao Yang (Department of Medicine, McGill University, Canada) for p300 expression vector; Yvonne Paterson (Department of Microbiology, UPenn, Philadelphia, PA) for TC1 cell lines; Delia Nelson (Department of Medicine, University of Western Australia) for AE17.ova mesothelioma cell line. Supported by grants from the National Institutes of Health (K08AI095353 to U.H.B., and P01AI073489 and 1R01CA158941 to W.W.H.).
Footnotes
ACCESSION CODES
The microarray data are deposited in Gene Expression Omnibus under accession code GSE47989.
SUPPLEMENTARY INFORMATION is available in the online version of this paper.
AUTHOR CONTRIBUTIONS
Y.L. and L.W. performed most studies, analyzed data and edited the manuscript. Y.L. wrote the manuscript. J.P., S.W., V.K., S.S. and S.M.A. performed tumor studies and analyzed tumor data. R.H. undertook mouse breeding and performed histology. U.H.B. analyzed microarray data, analyzed data and edited the manuscript. T.R.B. performed histology. T.A. performed autoantibody screening, analyzed data and edited the manuscript. P.K.B. and P.A.C. provided unique mice and reagents, respectively, and analyzed data. W.W.H. designed and directed this study, analyzed data and edited the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 2.Nishikawa H, Sakaguchi S. Regulatory T cells in tumor immunity. Int J Cancer. 2010;127:759–767. doi: 10.1002/ijc.25429. [DOI] [PubMed] [Google Scholar]
- 3.van Loosdregt J, et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115:965–974. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Wang L, Han R, Beier UH, Hancock WW. Two lysines in the forkhead domain of foxp3 are key to T regulatory cell function. PLoS ONE. 2012;7:e29035. doi: 10.1371/journal.pone.0029035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dranoff G. CTLA-4 blockade: unveiling immune regulation. J Clin Oncol. 2005;23:662–664. doi: 10.1200/JCO.2005.09.923. [DOI] [PubMed] [Google Scholar]
- 6.Powell DJ, Jr, et al. Administration of a CD25-directed immunotoxin, LMB-2, to patients with metastatic melanoma induces a selective partial reduction in regulatory T cells in vivo. J Immunol. 2007;179:4919–4928. doi: 10.4049/jimmunol.179.7.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fietta AM, et al. Systemic inflammatory response and downmodulation of peripheral CD25+Foxp3+ T-regulatory cells in patients undergoing radiofrequency thermal ablation for lung cancer. Hum Immunol. 2009;70:477–486. doi: 10.1016/j.humimm.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, de Zoeten EF, Greene MI, Hancock WW. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat Rev Drug Discov. 2009;8:969–981. doi: 10.1038/nrd3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tao R, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 10.de Zoeten EF, et al. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3(+) T-regulatory cells. Mol Cell Biol. 2011;31:2066–2078. doi: 10.1128/MCB.05155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beier UH, et al. Sirtuin-1 targeting promotes Foxp3+ T-regulatory cell function and prolongs allograft survival. Mol Cell Biol. 2011;31:1022–1029. doi: 10.1128/MCB.01206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beier UH, et al. Histone deacetylases 6 and 9 and sirtuin-1 control foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci Signal. 2012;5:ra45. doi: 10.1126/scisignal.2002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 14.Rudra D, et al. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2009;10:1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhry A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhry A, et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee I, et al. Recruitment of Foxp3+ T regulatory cells mediating allograft tolerance depends on the CCR4 chemokine receptor. J Exp Med. 2005;201:1037–1044. doi: 10.1084/jem.20041709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huehn J, et al. Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4+ regulatory T cells. J Exp Med. 2004;199:303–313. doi: 10.1084/jem.20031562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anz D, et al. CD103 is a hallmark of tumor-infiltrating regulatory T cells. Int J Cancer. 2011;129:2417–2426. doi: 10.1002/ijc.25902. [DOI] [PubMed] [Google Scholar]
- 20.Bowers EM, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: Identification of a selective small molecule inhibitor. Chem Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W, et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tone Y, et al. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 23.Ye Q, et al. BAFF binding to T cell-expressed BAFF-R costimulates T cell proliferation and alloresponses. Eur J Immunol. 2004;34:2750–2759. doi: 10.1002/eji.200425198. [DOI] [PubMed] [Google Scholar]
- 24.Lau OD, et al. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol Cell. 2000;5:589–595. doi: 10.1016/s1097-2765(00)80452-9. [DOI] [PubMed] [Google Scholar]
- 25.Zhou G, Levitsky HI. Natural regulatory T cells and de novo-induced regulatory T cells contribute independently to tumor-specific tolerance. J Immunol. 2007;178:2155–2162. doi: 10.4049/jimmunol.178.4.2155. [DOI] [PubMed] [Google Scholar]
- 26.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Kasper LH, et al. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol Cell Biol. 2006;26:789–809. doi: 10.1128/MCB.26.3.789-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Battke F, Symons S, Nieselt K. Mayday--integrative analytics for expression data. BMC Bioinformatics. 2010;11:121. doi: 10.1186/1471-2105-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin KY, et al. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 1996;56:21–26. [PubMed] [Google Scholar]
- 31.Jackaman C, et al. IL-2 intratumoral immunotherapy enhances CD8+ T cells that mediate destruction of tumor cells and tumor-associated vasculature: a novel mechanism for IL-2. J Immunol. 2003;171:5051–5063. doi: 10.4049/jimmunol.171.10.5051. [DOI] [PubMed] [Google Scholar]
- 32.Haas AR, et al. Cycloxygenase-2 inhibition augments the efficacy of a cancer vaccine. Clin Cancer Res. 2006;12:214–222. doi: 10.1158/1078-0432.CCR-05-1178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.