Abstract
Chimeric mice are versatile model systems for the study of mammalian circadian biology. In chimeras, genetically different cells are combined within single animals, making them useful for assessing how normal cells interact with genetically altered cells in intact biological systems. In particular, the primary circadian pacemaker in the suprachiasmatic nucleus is amenable to analysis using series of chimeras that incorporate cells carrying mutations in circadian genes. The study of chimeras carrying circadian mutations can contribute to a better understanding of the function of the altered genes and of the fundamental physiology of circadian timing. Chimera analysis is a valuable approach for studying network properties in complex, integrated biological systems like that, which controls circadian behavior in mammals.
Introduction
The study of aggregation chimeric mice is a historically embryological technique that we have adopted to study circadian rhythmicity in mammals (Low-Zeddies and Takahashi, 2001). A mouse chimera is a genetically composite animal whose cell populations are derived from two different embryos. Aggregation chimeric mice are made by bringing two embryos together in vitro and allowing them to fuse together and then allowing the composite embryo to complete development. Chimeras can be made to contain a combination of wild-type and mutant cells (whether chemically induced, spontaneous, or the result of genetic engineering). Mutants are useful resources for studying physiology in that genetic perturbations enable one to better understand normal function (Benzer, 1973). A series of chimeric mice that incorporates a given mutant line enhances its utility and extends the information that can be obtained from the mutation. By studying chimeras in which normal cells are combined with cells carrying a mutation that affects circadian function, one potentially addresses two fundamental questions: what is the function of the altered gene and how does altering the function of the gene affect the physiology of the circadian system? Using chimeric mice, these two basic questions can be addressed at multiple levels of biological organization: spatially, from the cell to the tissue to whole animal behavior, and temporally, from the embryo to the adult. The physiological function of chimeric tissue can be studied in vivo in the intact organism or cultured in vitro.
It has become clear that most, if not all, cells in mammals are competent circadian oscillators. It is not yet well understood, however, how multi-oscillator networks are set up as tissues and organs that sustain and adapt their timing to each other and to the environment. In particular, in the suprachiasmatic nucleus (SCN), which functions as the mammalian central circadian pacemaker, the oscillations of thousands of individual cells are transformed into a coherent signal that controls the circadian rhythm of locomotor activity. Incorporating mutant cells into chimeras is a means to genetically dissect the system in vivo by effectively creating genetic lesions at the level of individual cells. By combining mutant with wild-type cells in the SCN, the experimenter alters the components of the pacemaking ensemble, potentially altering the properties of how the cells interact to produce circadian behavior. At the same time, because the genetically different cells are combined early—at embryonic day 2.5—a practically “seamless” coupling of cellular pacemakers is achieved in chimeric tissue. The intricate intercellular connectivity is preserved, and this is critical to the extraordinary complexity of this neural structure. In short, the study of a series of mutant chimeras, which we refer to as chimera analysis, is a versatile tool for studying the nature of network properties in integrated biological systems such as the circadian system.
Properties of Aggregation Chimeric Mice
Aggregation chimeric mice were first produced by Tarkowski (1961) and by Mintz (1962) as a means to study cell regulation during mammalian development. Each chimeric mouse must be generated experimentally— chimeras cannot be produced through breeding alone. When a normal embryo is combined with one carrying a particular mutation, the chimeric individual that results will contain a unique proportion and distribution of cells from its two component cell genotypes in all of its tissues. That is, across a population of chimeric mice, no two are the same. Distribution of the component cell types in chimeras is essentially random. Furthermore, an experimental population, or series, of aggregation chimeras will cover a 0–100% range of relative cell genotypic proportions (or, put another way, the dose of mutant cells varies), with all proportions being equally represented (Falconer and Avery, 1978). By using a reliable cell marker, one ensures that the genotype of each cell can be unambiguously identified in all tissues. A single set of chimeras can be used to study the effects of chimerism in tissues throughout the body. The proportions of the two cellular genotypes among different tissues and organs are highly correlated within each chimeric individual (Musci and Mullen, 1992). This characteristic, though, tends to interfere with efforts to determine a tissue focus of the mutation based on correlations between mutant phenotypes and composition of various tissues, a strategy used in studies of mosaic fruitflies, for example (Benzer, 1973). The two cellular genotypes are finely mixed in chimeric tissue in general (Dewey et al., 1976; Oster-Granite and Gearhart, 1981), including the SCN (Low-Zeddies and Takahashi, 2001). The close apposition of the two cellular genotypes in chimeric tissue enhances the potential for their functional interaction.
Chimeras can be made using any mutant mouse strain, whether spontaneous, induced, gene targeted, or transgenic. Studying an experimental series of chimeric mice can extend the utility of any given mutant line: in contrast to studying animals that are wholly homozygous mutant, heterozygote, or wild type, each chimeric mouse is a new experimental system. The variety of configurations and proportions of mutant versus normal cells represented in a series of chimeric mice potentially reveals all of the possible structural and functional outcomes of the interactions between these different cellular genotypes.
A Study of Clock Chimeras Principles for Circadian Studies
We used a series of mouse chimeras to study the effects of allowing Clock mutant cells to interact with wild-type cells on the circadian rhythm of locomotor behavior (Low-Zeddies and Takahashi, 2001). The Clock gene regulates the intrinsic circadian period and the persistence of circadian rhythmicity (or circadian amplitude) in constant conditions (Vitaterna et al., 1994). These two phenotypic traits are fundamental properties of the circadian clock system. We generated a population of 130 homozygous Clock aggregation chimeric mice that carried from 0 to 100% of Clock mutant versus wild-type cells. For each of these chimeric individuals, we analyzed the cellular composition of SCN tissue and assayed their circadian behavior (Low-Zeddies and Takahashi, 2001).
Across the varied population of Clock chimeric mice, we observed patterns of circadian behavior that spanned a range from normal to Clock mutant like. In general, the comprehensive range in relative mutant contribution from 0 to 100% across a series of chimeras is advantageous in that the widest potential range of phenotypes may be seen. Furthermore, this continuous distribution is advantageous for correlational analyses. In some cases, we observed phenotypic traits that were intermediate between those characteristic of wild type and mutant. For example, we documented stable circadian period lengths that fell between the 23.5-and the 28-h periods characteristic of the wild-type and Clock mutant strains (Vitaterna et al., 1994). In general, we found that intermediate and novel behavioral profiles that were observed in some Clock mutant chimeras served as valuable analytical tools. The phenotypic gradient across the chimeric series tracked a progression of mutant severity, revealing how the circadian pacemaker mechanism can be broken down incrementally as the mutant cell dose is increased. Unexpectedly, we found that some of these Clock chimeras that contained only homozygous Clock mutant and wild-type cells could behave as though they were Clock heterozygotes. This result indicated that the interaction between wild-type and Clock mutant alleles at an intercellular level in chimeras can resemble allelic interaction at an intracellular level. Finally, our study of Clock chimeras demonstrated that the effect of the Clock mutation on circadian period and its effect on circadian amplitude are likely to arise from physiological processes that involve different subsets of cells because they did not always covary in individual chimeric mice. As a general principle, how mutant versus wild-type phenotypic traits covary across a series of chimeras is informative as to their underlying mechanism and can be used to discriminate separable phenotypic traits of a mutation. The use of Clock chimeras, then, allowed us to reveal new aspects of how the Clock gene affects circadian function at the cellular level and how individual cellular oscillators within the SCN functionally interact with one another to produce an integrated circadian rhythm of behavior.
Using Chimeras to Study Intercellular Interactions in Circadian Systems
We have found that chimera analysis can be a generally useful approach to manipulate and then probe intercellular interaction in circadian physiology. Chimeras permit investigation into the interactions between genetically different cell populations and the effect of these interactions on organismal behavior. Chimeric mice also have the potential to be a powerful model system for the in vitro study of cell and tissue organization of the mammalian circadian system. Numerous lines of mice already exist in which genes implicated in circadian rhythms [e.g., the mouse Cryptochrome genes 1 and 2 (van der Horst et al., 1999; Vitaterna et al., 1999), the mouse Period genes 1, 2, and 3 (Bae et al., 2001; Zheng et al., 2001), BMAL 1/Mop3 (Bunger et al., 2000), and Clock (Vitaterna et al., 1994)] have been altered or knocked out and that show robust circadian behavioral phenotypes. All of these clock gene mutations, and those that continue to be produced, can be incorporated into experimentally informative chimeras. Our results outlined previously demonstrate how subtle aspects of the functional organization of the SCN can become apparent in chimeras.
Chimeras Can Be Used to Study Cell Interaction in the SCN
Chimeric mice can be used to study the intercellular interactions that mediate the coupling among cellular oscillators and confer a specialized pacemaking function to the SCN. It is now well established that circadian periodicity is an intrinsic property of individual SCN cells (Herzog et al., 1998; Liu et al., 1997). The chimeric SCN is a potentially useful system for recording the electrical activity of single cells in slice culture preparations, as a direct method of determining whether the rhythmicity of wild-type SCN cells can be disrupted by the presence of mutant cells and whether the two can synchronize one another. If desired, chimeric combinations of two different circadian mutations could be used to study interactions between cells with different oscillatory properties, e.g., short and long circadian period combinations. By studying the interaction between mutant and wild-type cells in chimeric tissue, one might ask which genes, when mutated, affect coupling properties among SCN neurons or their entrainment by light stimuli. For example, using chimeras, one might test the role of genes that encode neurochemicals or receptors that are thought to mediate signaling by light and/or interneuronal communication within the SCN, such as the VPAC2 receptor, or gastrin-releasing peptide (Aida et al., 2002; Harmar et al., 2002). An alternative assay to electrical recording of cellular activity in chimeric SCN preparations is to incorporate cells from one of the lines of mice now available that carry circadian reporter trans-genes to visually observe the dynamic interactions between cells of contrasting genotype. Mice have been genetically engineered to produce green fluorescent protein (GFP) or luciferase (luc) that are driven to oscillate with the circadian rhythm of the molecular clockwork in individual cells (Kuhlman et al., 2000; Wilsbacher et al., 2002; Yoo et al., 2004). These constructs that utilize mouse Period gene promoters are ubiquitously expressed and can be imaged in living tissue in real time.
Chimeras Can Be Used to Study the Effects of Cell Interaction on Behavior
An important question in circadian biology is, what are the mechanisms by which the output signals from a multiplicity of individual oscillators interact to form a functional pacemaker at the tissue and whole-animal levels? The properties of the multiple levels of cell and tissue organization that underlie circadian rhythms in behavior are not likely to be resolved by studying the behavior of single-cell oscillators. Using chimeras, one can manipulate the genetic composition of the network of oscillators in the SCN and assess how different cellular circadian phenotypes are ultimately integrated into a coherent behavioral output (Low-Zeddies and Takahashi, 2001). The expression of a complex behavior such as circadian locomotor activity relies on an intricate network of connections and feedback; by using chimeras, the behavior can be studied in the intact nervous system.
Chimeras Can Be Used to Study the Control of Circadian Rhythms in Peripheral Tissues
Similarly, chimeras may be used to study intercellular processes involved in conveying circadian timing signals from the SCN to other tissues and organs in the body (the periphery). The routes of communication from the SCN to the sites that generate locomotor activity behavior are thought to be complex and multimodal. Both neural mechanisms and diffusible signaling molecules have been implicated. Chimeras may be used to test the role of candidate molecules involved in output signaling from the SCN. For example, transforming growth factor-a has been identified as a candidate factor that mediates SCN output controlling the timing of locomotor behavior (Kramer et al., 2001). Chimeric models carrying various proportions of cells in which molecules such as this are deficient may help define their role in the circadian control of behavior.
Finally, chimera analysis can help explore how cellular circadian phenotypes are integrated in peripheral tissues. It is known that fundamental properties of circadian rhythms in tissues throughout the body differ from those of the circadian oscillators in the master pacemaker in the SCN. For example, the phases of the liver, lung, and skeletal muscle tissues lag the SCN rhythm by 7–11 h and respond differently from the SCN to shifts in the light cycle (Yamazaki et al., 2000). Isolated explants from peripheral tissues express a circadian rhythm in Period2-driven bioluminescence, suggesting that the cellular oscillators in these peripheral tissues are also coupled, although by what mechanism is not known (Yoo et al., 2004). Chimeras can be used to study how the interactions between individual cellular oscillators in nonneural tissues in the periphery compared with intercellular interactions in the neural pacemaker in the SCN. Can network properties be identified that differentiate central from peripheral tissues and account for the uniqueness of the SCN master pacemaker?
General Applications for Chimeras in Circadian Studies
Chimeras Can Reveal Gene Function at a Cellular Level
Chimeras have been conventionally employed to determine whether a mutant cell can affect the phenotype of neighboring wild-type cells, i.e., whether a cellular phenotype is cell autonomous. This remains an important use of chimeras that can help define in which cells the expression of a circadian gene is functionally important, particularly when the gene is widely expressed. Moreover, effects of mutations can be mediated directly or indirectly. For example, molecules such as receptors, signal transducers, or transcription factors, although expressed within a cell, may perform regulatory functions that affect extracellular molecules or neural communication, which in turn influence fates or functions of other cells. Thus, in the postgenomic age, even though function is often predicted from gene sequence, chimera analysis is valuable for assessing the role of specific genes in circadian physiology at the cellular level. Chimeras have also traditionally been a tool for tracing cell lineages during development and for addressing how various mutations affect the ability of cells to participate in normal development.
Chimeras Can Rescue Cells Carrying Mutations that Cause Early Developmental Lethality
Chimera analysis has taken on a new relevance with the popularity of gene knockout mice. In chimeras, the effects of phenomena that interfere with interpretation of the phenotype in a pure knockout animal may be alleviated. For example, the products of many genes are essential such that inactivating them results in lethality early in development, preventing further study of discrete behavioral effects. Given the pervasiveness of circadian rhythms in physiology and the importance of timing to developmental processes, one might expect that some proportion of genes that affect the circadian timing system will also have functions during development that are essential for viability. Cells carrying genetic alterations that cause early organismal lethality can often be rescued in a chimeric setting (e.g., Barsh et al., 1990; Campbell and Peterson, 1992). That is, disabling gene function in only a subset of cells within the organism may permit survival to a later developmental stage or adulthood, allowing examination of later roles for the gene in question. It can also be determined what proportion of cells and where they must be located to rescue normal function.
From a practical standpoint, when a mutation causes early lethality, the embryos used to produce homozygous mutant chimeras must come from heterozygous mutant crosses. It then becomes necessary to adopt a strategy to be able to determine the genotype of the mutant portion of the resulting chimeras because both homozygous mutant and heterozygous chimeric tissues will contain both mutant and wild-type alleles. For example, one may incorporate either two different wild-type alleles or two distinct mutant alleles in the component heterozygous embryos that can be distinguished by DNA genotyping of the resulting chimeras. Alternatively, it is possible, though challenging, to microdissect out single mutant cells from tissue in the chimera (identified by cell marker) to determine whether the cells are homozygous or heterozygous for the mutant allele. Another route would be to generate embryonic stem (ES) cell chimeras using known homozygous ES cells.
Chimeras Can Clarify the Mechanisms behind Pleiotropic Effects of a Mutation
In some cases, knocking out certain genes in mice results in no detectable phenotype due to gene redundancy mechanisms, or the genetic manipulation produces multiple phenotypic effects that make it difficult to discriminate separate roles for the gene product. Chimera analysis can provide valuable perspective to the study of a mutant phenotype. It is now well known that observed effects of a targeted mutation may not be directly related to the function of the gene per se due to redundancy of function among members of a gene family, alterations in the regulation of other genes, and other indirect effects. Combining mutant cells with wild-type cells in chimeras may help tease out phenotypic effects resulting from compensatory processes that obscure interpretation of the normal function of the gene. Chimera analysis can potentially clarify and separate the effects of a given genetic manipulation. If based on different underlying cell physiological processes, the pleiotropic effects of the mutation may be dissociated across a series of mutant-normal chimeras. Conversely, by assessing their covariance across a series of chimeras, it can be determined whether different traits share a common controlling cell population.
Chimeras Can Serve as Models for Circadian Dysfunction
Finally, chimeras that incorporate clock gene mutations may be used to model circadian dysfunction and how the loss of circadian control contributes to disease states. A gradient of chimeras may be able to mimic the breakdown in circadian organization that occurs with aging, for example.
Technique
Chimera Production
The production of mouse chimeras is a straightforward technique that reliably yields viable animals. Methods of aggregating two eight-cell embryos (morulae) are standard (Hogan et al., 1994)(Fig. 1). Superovulation is induced hormonally in female mice of both wild-type and mutant strains. Each female is then mated with a stud male of a matching genotype. On embryonic day 2.5 (vaginal plug == embryonic day 0.5), morulae are flushed from dissected oviducts into M2 mouse medium and then incubated briefly in acidic Tyrode’s solution to remove the zonae pellucidae. Pairs of embryos are then pushed together and cocultured overnight in CZB+ or KSOM medium at 37°,5%CO2. The following day, the aggregated embryos are transferred surgically into the uterine homs of 2.5-day pseudopregnant foster mothers where they finish development. Pseudopregnant recipient females are produced by mating naturally cycling females with vasectomized males. Chimeric pups are identifiable by the presence of variegated coat and eye pigmentation when pigmentation is used as a marker to differentiate wild-type from mutant parental strains. Coat color is a reliable indicator of the presence of chimerism in the central nervous system (CNS) because both CNS and coat pigment cells arise from the neural crest (Rawles, 1947). Nonchimeric littermates serve as useful controls to verify the lack of effect of the marker strain and to control for embryo manipulation.
Figure 1.
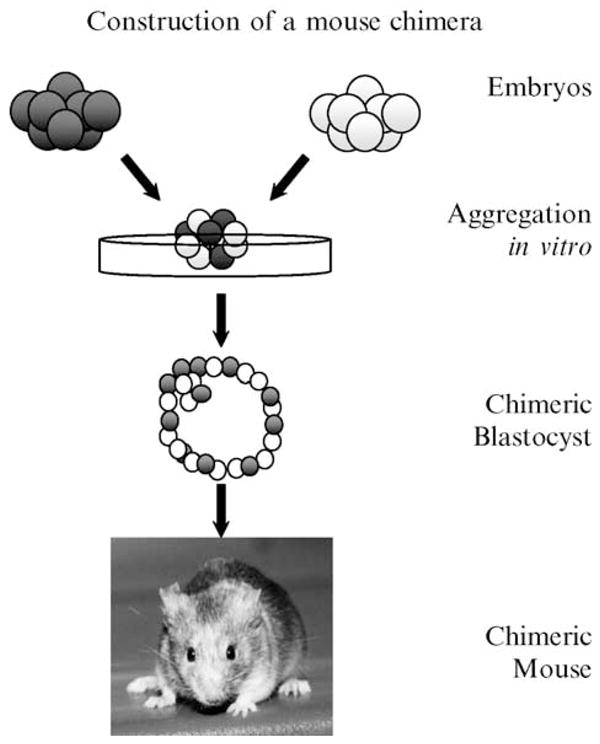
A schematic diagram of the method used to create a chimeric mouse. Two genetically different 2.5-day embryos are denuded and then aggregated together in a dish. The embryos fuse together in culture overnight. The resulting chimeric blastocyst is transferred into a foster mother, where it completes development into a chimeric mouse.
Cell Markers
To analyze the tissues of chimeric mice, it is essential to have a reliable, independent means of distinguishing mutant from wild-type cells in situ— a cell marker. The applicability of chimera analysis has been enhanced by the advent of strains of transgenic mice that express genes such as LacZ or GFP that are detected easily at the single-cell level and can serve as markers for identifying cell genotypes (Hadjantonakis and Nagy, 2001; Zambrowicz et al., 1997). For example, the ROSA26 strain is ideal for chimera analysis studies. ROSA26 constitutively expresses the, β-galactosidase enzyme in all of its cells (Zambrowicz et al., 1997)so that ROSA26 cells can be used as the marked wild-type component or ROSA26 can be crossed with the mutant strain to produce marked mutant cells. For LacZ detection in ROSA26 brain tissue, sections are incubated for 24 h at 37° in an X-gal staining solution containing 1 mg/ml X-gal (5-bromo-4-chloro-3-indolyl-,-d-galactoside dissolved in dimethyl sulfoxide), 5 mM K3Fe(CN)6,and 5 mM K4Fe(CN)6 in a phosphate-buffered saline-based wash buffer. Alternatively, living chimeric tissues can be imaged that incorporate strains genetically engineered to produce fluorescent proteins or luciferase (Hadjantonakis and Nagy, 2001; Kuhlman et al., 2000; Wilsbacher et al., 2002; Yoo et al., 2004). Using powerful software applications now available, it is possible to reconstruct serial sections into three-dimensional images that can help in visualizing the distribution of mutant versus wild-type cells in chimeric tissue.
Statistical Analysis of Chimera Data
Analysis of a series of chimeric mice yields data amenable to statistical analyses. With the potential for a phenotypic gradient across a population of chimeras, it is important to use a sensitive, quantitative scoring method, such as period length or a Fourier analysis measure of circadian amplitude. For a chimera analysis to be informative requires studying a sufficient number of chimeras to cover a comprehensive phenotypic range. Statistically significant numbers of animals necessary for a given analysis will depend on the complexity of the phenotype and its genetic control, which should be determined after analyzing an initial number of chimeras. In the analysis of complex behavioral data, statistical power will increase with the number of chimeric individuals studied.
Complex biological phenomena such as circadian behavior are described most effectively by multiple quantitative measures. Multivariate statistical analyses can simplify, organize, and reveal structure in large behavioral and anatomical data sets, as are generated by large populations of chimeras. These tools have allowed us to test hypotheses about how the relationships between variables reveal SCN functional organization (Low-Zeddies and Takahashi, 2001). We found principal components and cluster analyses useful for evaluating relationships among the period, amplitude, and SCN scores in Clock chimeras and to facilitate comparison of the multidimensional behavior of chimeras with that of the control genotypic groups. A principal components analysis yields a unique solution of weighted linear composites of the observed variables that can reduce a multivariate data set to fewer components, making data easier to visualize and understand. Cluster analysis is a procedure for detecting natural groupings in data. The method is based on measures of dissimilarity between objects, expressed as distances in a multidimensional space (defined by the number of variables taken into account). We have found that clustering algorithms can define inherent structure in complex behavioral data and be of heuristic value for comparing multidimensional behavioral profiles.
Other Chimeric or Mosaic Mouse Models
There are other methods of generating chimeric (carrying cells that differ in genomic content) or mosaic (all cells carry the same genomic content, but genes are activated differentially in different cell populations) mice. Technically, the preimplantation stage is the only feasible time to routinely manipulate mammalian embryos. Preimplantation chimeric mice can also be generated by injecting cultured ES cells into blastocysts. Unlike aggregation chimeras, however, which show a comprehensive range of mutant contribution to all tissues, the composition of blastocyst injection chimeras may be biased systematically according to characteristics of the ES cell line (Berger et al., 1995; Ioffe et al., 1995). The outcome of producing chimeras by morula aggregation compared with ES cell injection is that, across a series of chimeras, those made by aggregation show a more variable mutant contribution to all tissues. For chimeras made using ES cells, the hard work is primarily in the production and maintenance of the ES cell cultures. For morula aggregation chimeras, because the embryo “culture” occurs in the living mouse, the bulk of the effort goes toward mouse husbandry.
It is possible to create genetically composite mice by delivering genes to a limited proportion of cells in vivo using retroviruses (Soriano and Jaenisch, 1986), stem cell transplantation (Brustle et al., 1995), or even tissue transplantation (Ralph and Lehman, 1991). These methods have appropriate applications, but also disrupt the normal development of tissues, which does not occur (after the eight-cell stage) in aggregation chimeras. Methods of generating various kinds of mosaic mice include X inactivation (Nesbitt, 1971), laacZ/lacZ (Bonnerot and Nicolas, 1993), and Cre-loxP-mediated mosaicking strains (Betz et al., 1996; Dietrich et al., 2000; Guo et al., 2002). In contrast to aggregation chimeras, these methods do not have general applicability to all genes, they result in different patterns of juxtaposition between the component cell genotypes compared with aggregation chimeras, and they produce systematic biases in the proportions and distributions of the two contrasting cell genotypes. Finally, conditionally mutant lines of mice can be created in which particular genes are altered or deleted with spatial specificity. To create a conditional mutant requires a suitable gene promoter that reliably confers the required cell-type specificity. Production of these lines is technically demanding, requiring a substantial amount of work up front, and risks being subject to the regulatory peculiarities of each construct. The use of aggregation chimeras is a straightforward, generally applicable approach that complements more high-investment conditional genetic engineering manipulations. Indeed, a chimera analysis can be used as a first-line tool to indicate whether undertaking a more precise targeting strategy will be worthwhile and informative. We suggest that aggregation chimeras are the most efficient and straightforward chimera/mosaic model to use. With a viable, reasonably breeding mutant in hand, morula aggregation can be performed immediately and is guaranteed to produce viable experimental chimeras.
Summary
An important goal for the future in circadian rhythms research is to determine how individual cellular oscillators are integrated into higher-order structures to produce this complex behavior. Chimera analysis using clock mutants can help achieve this goal and builds on the progress that has been made in the dissection of circadian molecular components. Any clock mutant or knockout strain can be used to produce chimeric mice, each of which is a unique and novel experimental system that can potentially exhibit new circadian biological properties.
The analysis of chimeric mice can help reveal how multicellular circadian oscillatory structures are assembled from a functional standpoint. At the cellular level, chimera experiments with genetically differing oscillator populations demonstrate how multiple oscillators interact with one another and are a potentially rich source of data for mathematical modeling studies. At the tissue level, chimeras provide a way to probe the fundamental mechanisms by which signals from the individual cellular clocks in the SCN are integrated to produce a coherent timing signal that is broadcast to the rest of the body. At the organismal level, analyzing the wheel-running activity rhythms of chimeras allows the investigator to probe the functional organization of the intact circadian system.
Chimera analysis should be considered an essential part of a growing set of tools for manipulating gene activity in vivo, the results of which will be used to converge upon an understanding of complex biological processes such as circadian behavior.
References
- Aida R, Moriya T, Araki M, Akiyama M, Wada K, Wada E, Shibata S. Gastrin releasing peptide mediates photic entrainable signals to dorsal subsets of suprachiasmatic nucleus via induction of Period gene in mice. Mol Pharmacol. 2002;61:26–34. doi: 10.1124/mol.61.1.26. [DOI] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- Barsh GS, Lovett M, Epstein CJ. Effects of the lethal yellow (Ay) mutation in mouse aggregation chimeras. Development. 1990;109:683–690. doi: 10.1242/dev.109.3.683. [DOI] [PubMed] [Google Scholar]
- Benzer S. Genetic dissection of behavior. Sci Am. 1973;229:24–37. doi: 10.1038/scientificamerican1273-24. [DOI] [PubMed] [Google Scholar]
- Berger CN, Tam PP, Sturm KS. The development of haematopoietic cells is biased in embryonic stem cell chimaeras. Dev Biol. 1995;170:651–663. doi: 10.1006/dbio.1995.1244. [DOI] [PubMed] [Google Scholar]
- Betz UA, Vosshenrich CA, Rajewsky K, Muller W. Bypass of lethality with mosaic mice generated by Cre-loxP-mediated recombination. Curr Biol. 1996;6:1307–1316. doi: 10.1016/s0960-9822(02)70717-3. [DOI] [PubMed] [Google Scholar]
- Bonnerot C, Nicolas JF. Clonal analysis in the intact mouse embryo by intragenic homologous recombination. C R Acad Sci III. 1993;316:1207–1217. [PubMed] [Google Scholar]
- Brustle O, Maskos U, McKay RDG. Host-guided migration allows targeted introduction of neurons into the embryonic brain. Neuron. 1995;15:1275–1285. doi: 10.1016/0896-6273(95)90007-1. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hoganesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RM, Peterson A. An intrinsic neuronal defect operates in dystonia musculorum: A study of dt/dt$+/+ chimeras. Neuron. 1992;9:693–703. doi: 10.1016/0896-6273(92)90032-9. [DOI] [PubMed] [Google Scholar]
- Dewey MJ, Gervais AG, Mintz B. Brain and ganglion development from two genotypic classes of cells in allophenic mice. Dev Biol. 1976;50:68–81. doi: 10.1016/0012-1606(76)90068-3. [DOI] [PubMed] [Google Scholar]
- Dietrich P, Dragatsis I, Xuan S, Zeitlin S, Efstratiadis A. Conditional mutagenesis in mice with heat shock promoter-driven cre transgenes. Mamm Genome. 2000;11:196–205. doi: 10.1007/s003350010037. [DOI] [PubMed] [Google Scholar]
- Falconer DS, Avery PJ. Variability of chimeras and mosaics. J Emb Exp Morph. 1978;43:195–219. [PubMed] [Google Scholar]
- Guo C, Yang W, Lobe CG. A Cre recombinase transgene with mosaic, widespread tamoxifen-inducible action. Genesis. 2002;32:8–18. doi: 10.1002/gene.10021. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis AK, Nagy A. The color of mice: In the light of GFP-variant reporters. Histochem Cell Biol. 2001;115:49–58. doi: 10.1007/s004180000233. [DOI] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH. The VPAC2 receptor is essential for circadian function in the mouse suprachiasmatic nucleus. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Takahashi JS, Block GD. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nature Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E, editors. Manipulating the Mouse Embryo: A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. [Google Scholar]
- Ioffe E, Liu Y, Bhaumik M, Poirier F, Factor SM, Stanley P. WW6: An embryonic stem cell line with an inert genetic marker that can be traced in chimeras. Proc Natl Acad Sci USA. 1995;92:7357–7361. doi: 10.1073/pnas.92.16.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, Weitz CJ. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511–2515. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- Kuhlman SJ, Quintero JE, McMahon DG. GFP fluorescence reports Period1 circadian gene regulation in the mammalian biological clock. Neuroreport. 2000;11:1479–1482. [PubMed] [Google Scholar]
- Liu C, Weaver DR, Strogatz SH, Reppert SM. Cellular construction of a circadian clock: Period determination in the suprachiasmatic nucleus. Cell. 1997;91:855–860. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001;105:25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz B. Formation of genotypically mosaic mouse embryos. Am J Zool. 1962;4:432. [Google Scholar]
- Musci TS, Mullen RJ. Cell mixing in the spinal cords of mouse chimeras. Dev Biol. 1992;152:133–144. doi: 10.1016/0012-1606(92)90163-b. [DOI] [PubMed] [Google Scholar]
- Nesbitt MN. X chromosome inactivation mosaicism in the mouse. Dev Biol. 1971;26:252–263. doi: 10.1016/0012-1606(71)90125-4. [DOI] [PubMed] [Google Scholar]
- Oster-Granite ML, Gearhart J. Cell lineage analysis of cerebellar Purkinje cells in mouse chimeras. Dev Biol. 1981;85:199–208. doi: 10.1016/0012-1606(81)90251-7. [DOI] [PubMed] [Google Scholar]
- Ralph MR, Lehman MN. Transplantation: A new tool in the analysis of the mammalian hypothalamic circadian pacemaker. Trends Neurosci. 1991;14:362–366. doi: 10.1016/0166-2236(91)90164-p. [DOI] [PubMed] [Google Scholar]
- Rawles ME. Origin of pignment cells from the neural crest in the mouse embryo. Physiol Zool. 1947;20:248–266. doi: 10.1086/physzool.20.3.30151958. [DOI] [PubMed] [Google Scholar]
- Soriano P, Jaenisch R. Retroviruses as probes for mammalian development: Allocation of cells to the somatic and germ cell lineages. Cell. 1986;46:19–29. doi: 10.1016/0092-8674(86)90856-1. [DOI] [PubMed] [Google Scholar]
- Tarkowski AK. Mouse chimaeras developed from fused eggs. Nature. 1961;190:857–860. doi: 10.1038/190857a0. [DOI] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Komhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbacher LD, Yamazaki S, Herzog ED, Song EJ, Radcliffe LA, Abe M, Block G, Spitznagel E, Menaker M, Takahashi JS. Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice in vivo. Proc Natl Acad Sci USA. 2002;99:489–494. doi: 10.1073/pnas.012248599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi RI, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA ßgeo 26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells. Dev Biol. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]


