Abstract
The NLRP3 inflammasome is involved in many obesity-associated diseases, such as type 2 diabetes, atherosclerosis, and gouty arthritis, through its ability to induce interleukin (IL)-1β release. The molecular link between obesity and inflammasome activation is still unclear, but free fatty acids have been proposed as one triggering event. Here we reported opposite effects of saturated fatty acids (SFAs) compared with unsaturated fatty acids (UFAs) on NLRP3 inflammasome in human monocytes/macrophages. Palmitate and stearate, both SFAs, triggered IL-1β secretion in a caspase-1/ASC/NLRP3-dependent pathway. Unlike SFAs, the UFAs oleate and linoleate did not lead to IL-1β secretion. In addition, they totally prevented the IL-1β release induced by SFAs and, with less efficiency, by a broad range of NLRP3 inducers, including nigericin, alum, and monosodium urate. UFAs did not affect the transcriptional effect of SFAs, suggesting a specific effect on the NLRP3 activation. These results provide a new anti-inflammatory mechanism of UFAs by preventing the activation of the NLRP3 inflammasome and, therefore, IL-1β processing. By this way, UFAs might play a protective role in NLRP3-associated diseases.
Keywords: obesity, inflammation, innate immunity, interleukin-1beta, palmitate, stearate, oleate, linoleate
Interleukin-1β (IL-1β) is a proinflammatory cytokine involved in many obesity-associated diseases, such as type 2 diabetes, atherosclerosis, and gouty arthritis (1). Its release is tightly regulated at both transcriptional and posttranscriptional levels at different steps, including synthesis, processing, and secretion (2). IL-1β is produced as an inactive precursor, the pro-IL-1β, which is cleaved by caspase-1 into the biologically functional form. Caspase-1-mediated IL-1β maturation occurs mostly through the assembly of multiprotein complexes called inflammasomes (3). Several NOD-like receptor (NLR) family members were identified to form inflammasomes, including NLRP1, NLRP3, NLRP7, and NLRC4, but non-NLR inflammasomes exist, such as the AIM2 inflammasome (3).
Recent studies suggest a role of NLRP3 in obesity and its complications, especially in type 2 diabetes (4, 5). NLRP3 interacts with the adaptor protein ASC [apoptosis-associated speck-like protein containing a CARD (caspase recruitment domain)], mediating recruitment and auto-activation of caspase-1. NLRP3 is mainly expressed in immune cells from the myeloid lineage, such as monocytes, dendritic cells, and macrophages (6). However, its expression is low compared with ASC and caspase-1 (7). The main caspase-1 substrate, the pro-IL-1β, is not expressed or is very weakly expressed in these cells. As NLRP3 and pro-IL-1β are the limiting factors of the NLRP3 inflammasome, a first signal commonly called the priming is necessary to increase their transcriptions (8). Next, a second stimulus is required to induce NLRP3 inflammasome assembly, caspase-1 activation, and, therefore, IL-1β maturation.
Macrophages, whose NLRP3 and pro-IL-1β expression are easily inducible (8), infiltrate the adipose tissue in correlation with fatness (9). In obese individuals, the adipose tissue macrophages surround the dead adipocytes (10) and have a proinflammatory phenotype called M1-polarization or “classically activated macrophages” (11). IL-1β, one of the various cytokines secreted by M1-polarized macrophages, is strongly incriminated in numerous obesity-associated diseases in a NLRP3-dependent way. Indeed, NLRP3-deficient mice are protected against obesity-induced insulin resistance (4, 5), atherosclerosis (12), and gouty arthritis (13).
The molecular link between obesity and NLRP3-mediated IL-1β release is not well established. Free fatty acids (FFAs), usually elevated in plasma of obese people, have been proposed as one triggering event (1). Recently, the saturated fatty acid (SFA) palmitate demonstrated its ability to activate the NLRP3 inflammasome in murine macrophages (14). In the present study, we tested the four most important FFAs present in blood (15) on human macrophages: two SFAs, palmitate and stearate, and two unsaturated fatty acids (UFAs), oleate and linoleate. We identified stearate as a new physiological NLRP3 inducer, acting with the same efficiency as palmitate on the caspase-1/ASC/NLRP3 pathway. UFAs, which did not activate the NLRP3 inflammasome, revealed an unexpected anti-inflammatory property through preventing NLRP3 inflammasome activation by SFAs and by various inducers including nigericin, alum, and monosodium urate (MSU).
MATERIALS AND METHODS
Preparation of FFA solutions
The palmitic acid (#P0500), stearic acid (#S4751), oleic acid (#O1008), and linoleic acid (#L1376) were purchased from Sigma. A 100 mM stock solution of sodium salt was prepared by dissolving fatty acids in 0.1 M NaOH at 30°C, 37°C, 65°C, and 75°C for linoleic acid, oleic acid, palmitic acid, and stearic acid, respectively. Stock solutions were aliquoted and stored at −20°C for less than one year. A 5% fatty acid free, low endotoxin BSA (Sigma, #A8806) solution was prepared in RPMI 1640. The FFA stock solution and the 5% BSA solution were mixed together to obtain a 2.5 mM working solution with FFA:BSA molar ratio at 3.4:1. After pH adjustment, the working solution was filtered through a 0.2 µm pore size membrane filter, aliquoted, and stored at −20°C for less than two months.
Cell culture and treatment
The THP-1 monocytic cell line (ATCC) (authentication in 2011 by DSMZ, Germany) and THP1-XBlue™ cells (InvivoGen) were cultured between 0.5 and 1.0 × 106 cells/ml in RPMI 1640 with glutamine supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, and 100 U/ml streptomycin and maintained at 37°C under 5% CO2 atmosphere. Medium and reagents were purchased from Lonza. THP-1 and THP1-XBlue™ cells were differentiated with 100 ng/ml of PMA (Sigma) for 24 h, washed with PBS, and kept one night at rest in fresh supplemented medium before stimulation. Primary monocytes were isolated from human peripheral blood mononuclear cells (PBMCs) by using CD14 MicroBeads (Miltenyi Biotec) according to the manufacturer's instructions. PBMCs were obtained by single-step density gradient centrifugation with Ficoll-Paque PLUS (GE Healthcare) from buffy coats (Red Cross, Belgium). Buffy coats were obtained from healthy donors after informed consent, and experiments were approved by the ethical committee of the University of Liege. Primary monocyte purity was evaluated by flow cytometry on a fluorescence-activated cell sorting (FACS) Canto II (BD Biosciences) by using a FITC-coupled anti-human CD14 antibody (BD Pharmingen), and it was greater than 92%. Freshly isolated monocytes were resuspended in supplemented RPMI 1640, plated, and kept one night at rest. The next day, primary cells were primed with 200 ng/ml of lipopolysaccharide (LPS) (Sigma) for 3 h before treatment. Monocyte-derived macrophages (MDMs) were obtained by culturing freshly isolated monocytes in RPMI 1640 with 20% FBS and 100 ng/ml of human M-CSF premium grade (Miltenyi Biotec) for six days, followed by additional 18 h of stimulation with 100 ng/ml LPS and 20 ng/ml interferon-γ (Miltenyi Biotec) to acquire M1-polarization. After priming or differentiation, monocytes/macrophages were treated with FFAs, LPS (Sigma), imject alum (Pierce), MSU (InvivoGen), nigericin (Enzo Life Sciences), muramyl dipeptide (MDP) (InvivoGen), doxorubicin (Sigma), anti-mycin A (Sigma), tunicamycin (Sigma), monensin (Sigma), ATP (InvivoGen), flagellin (InvivoGen), poly(dA:dT) (InvivoGen), or FSL-1 (InvivoGen). All these compounds were used at the times and concentrations indicated in figure legends. Flagellin and poly(dA:dT) were transfected using the DOTAP liposomal transfection reagent (Roche). Caspase inhibitor Z-VAD-FMK (Promega) or caspase-1 inhibitor Z-YVAD-FMK (BioVision), were added at 10 µM simultaneously to stimuli.
Generation of stable THP-1 cell line expressing shRNA
The THP-1 cells stably expressing shRNA were obtained by lentiviral transduction carried out by the GIGA-Viral Vectors Platform (GIGA, Liege, Belgium). In summary, shRNA and promoter sequences were amplified by PCR from commercial plasmids encoding a shRNA against human NLRP3 (Sigma, #TRCN0000427726) or a nontarget sequence (Sigma, #SHC002) and cloned into pHRSin-CSGW plasmid expressing GFP [a kind gift from Dr. Els Veroyen (University of Lyon)]. New lentiviral plasmids were cotransfected with pSPAX2 (Addgene, #12260) and a VSV-G-encoding plasmid (16) in Lenti-X 293T cells (Clontech, #632180). Viral supernatants were collected, filtered, and concentrated 100× by ultracentrifugation. Finally, THP-1 cells were transduced with these lentiviral vectors and GFP-positive cells were sorted by FACS.
esiRNA transfection
Cells were transfected by using the HiPerFect transfection reagent (Qiagen) according to the manufacturer's instructions for suspension cell lines, with minor modifications. THP-1 cells (0.2 × 105) were plated in a 24-well plate in 100 µl of supplemented medium. Transfection complexes were prepared in RPMI 1640 without serum by adding 3 µl of HiPerFect and 400 ng of esiRNA in a final volume of 100 µl and mixed with suspension cells. After 6 h, 400 µl of supplemented medium were added. The next day, differentiation was performed with PMA as previously described. The esiRNAs used were from Sigma: esiPYCARD (#EHU066851) for ASC, and esiNLRP3 (#EHU071121) and esiEGFP (#EHUEGFP) for negative control.
Immunoblot analysis
Cells were washed, scraped in lysis buffer [50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Igepal, 0.25% sodium deoxycholate, 1 mM PMSF, and complete protease inhibitor cocktail (Roche Applied Science)], and centrifuged to remove remnant pellets. Supernatants were stored at −20°C. For phosphorylation experiments, cells were lysed in total phospho lysis buffer (62.5 mM Tris-HCl at pH 6.8, 10% glycerol, 2% SDS, 3% β-mercaptoethanol, 0.03% bromophenol blue, 1 mM DTT, 1 mM sodium orthovanadate, 25 mM β-glycerophosphate, 15 mM sodium fluoride, 1 mM PMSF, and complete protease inhibitor cocktail), sonicated, and stored at −20°C. Culture supernatants were also collected at −20°C after a centrifugation to remove cells. Proteins (40 µg) or supernatant (20 µl) were subjected to SDS-PAGE. After electrophoresis, proteins were transferred from gel to a PVDF membrane (GE Healthcare). The primary antibodies used were as follows: anti-ASC (Adipogen, #AG-25B-0006), anti-cleaved IL-1β (Cell Signaling, #2021), anti-IL-1β propeptide (R & D Systems, #MAB6964), anti-IL-1β (R & D Systems, #AF-201-NA), anti-NLRP3 (Sigma, #HPA012878), anti-phospho-IκBα (Cell Signaling, #9246), and anti-phospho-p65 (Cell Signaling, #3031). The secondary antibodies conjugated to HRP were anti-rabbit (Cell Signaling, #7074), anti-goat (Dako, #P0160), and anti-mouse (Dako, #P0447). The detection was performed with ECL or ECL plus Western blotting substrate (Pierce) by using the digital imaging system ImageQuant LAS 4000 (GE Healthcare). Quantification was achieved with the ImageQuant TL software (version 7.0, GE Healthcare).
ELISA
Mature IL-1β was quantified in supernatants by ELISA with Quantikine for human IL-1β (R & D Systems) according to the manufacturer's recommendations.
qRT-PCR
Total RNAs were extracted with RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol, followed by DNase treatment (Roche). After quantification by spectrophotometer Nanodrop 1000 (Thermo scientific), 500 ng of RNA were reverse-transcribed by using the moloney murine leukemia virus reverse transcriptase (Invitrogen). Obtained cDNA was submitted to qRT-PCR on a LightCycler 480 (Roche Applied Science). Relative gene expression was calculated using the 2-delta delta CT method (17) with HPRT1 as housekeeping gene. Results are representative from three independent experiments quantified in triplicate. The following primers were used: IL-1β-Fw: 5′-CCTTGGGCCTCAAGGAAAA-3′ IL-1β-Rv: 5′-CTCCAGCTGTAGAGTGGGCTTA-3′ ASC-Fw: 5′-CGCGAGGGTCACAAACGT-3′ ASC-Rv: 5′-TGCTCATCCGTCAGGACCTT-3′ Caspase-1-Fw: 5′-CCGCAAGGTTCGATTTTCA-3′ Caspase-1-Rv: 5′-ACTCTTTCAGTGGTGGGCATCT-3′ NLRP3-Fw: 5′-CTGTAACATTCGGAGATTGTGGTT-3′ NLRP3-Rv: 5′-GACCAAGGAGATGTCGAAGCA-3′ HPRT1-Fw: 5′-TGACACTGGCAAAACAATGCA-3′ HPRT1-Rv: 5′-GCTTGCGACCTTGACCATCT-3′.
Measurement of caspase-1 activity
A 1 mM stock solution of fluorescence-quenching substrate for caspase-1 was prepared by dissolving the (7-methoxycoumarin-4-yl)acetyl- L-tyrosyl- L-valyl- L-alanyl- L-aspartyl- L-alanyl- L-prolyl- Nε- (2,4-dinitrophenyl)- L-lysine amide (MOCAc-YVADAPK(Dnp)-NH2) (PeptaNova, #3183-v) in DMSO, and then was aliquoted and stored at −20°C protected from light for one month. Cells were plated in 96-well ViewPlate (Perkin Elmer, #6005182) at 2 × 105 cells/well, primed with PMA, and stimulated as described before in a volume of 150 µl per well. After 6 h of treatment, 3 µl of caspase-1 substrate were added to obtain a 20 µM final concentration. After 90 min of incubation, fluorescence was measured by using the EnSpire 2300 Multilabel Reader (Perkin Elmer) at an excitation wavelength of 328 nm and an emission wavelength of 393 nm in well area scan mode.
ASC dimerization
PMA-differentiated THP-1 cells (107 cells/dish) were seeded in 60 cm2 culture dishes. After 4 h of treatment, cells were washed with PBS and oligoASC buffer [20 mM HEPES-KOH pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 320 mM sucrose, 0.1 mM PMSF, 1 mM DTT, 1 mM sodium orthovanadate, 25 mM β-glycerophosphate, 15 mM sodium fluoride, complete protease inhibitor cocktail, and 4 mM of disuccinimidyl suberate (DSS; Pierce)] was added for 15 min at room temperature under agitation. Cells were scraped and transferred to a microcentrifuge tube. After lysis by shearing 20× through a 21 gauge needle, cells were kept under agitation at room temperature for 20 min. Crosslinking was stopped by addition of Tris, and samples were kept on ice. To remove nuclei, samples were filtered through 5 µm pore size membrane filter. Crosslinked complexes were precipitated by centrifugation at full speed for 3 min at 4°C, and pellets were resuspended in total phospho lysis buffer before immunoblot analysis.
NF-κB/AP-1 activity assay
THP1-XBlue™ cells were differentiated and treated as describe above. After treatment, supernatants were mixed with QUANTI-Blue™ (InvivoGen) according to the manufacturer's instructions to detect alkaline phosphatase activity. Absorbance was measured at 630 nm on the EnSpire 2300 Multilabel Reader.
Statistical analyses
All statistical analyses were carried out using GraphPad Prism 5 for Windows (GraphPad Software, Inc.) and presented as means ± SEM. The Student t-test was performed for simple comparison (two groups) and the ANOVA test followed by Bonferroni post-test for multiple comparisons (more than two groups). Significance is indicated by a symbol.
RESULTS
SFA induce IL-1β secretion by activating the NLRP3 inflammasome in human monocytes/macrophages
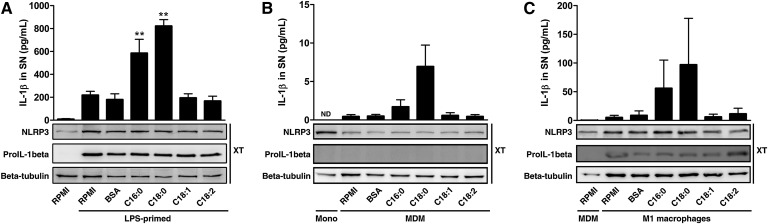
FFAs are poorly soluble compounds in aqueous solution, and most of them are bound to serum albumin in human physiology (18). For in vitro experiments, human serum albumin is often substituted by BSA as we did. Among FFAs, palmitate (C16:0) demonstrated its ability to induce IL-1β secretion in murine macrophages through a caspase-1/ASC/NLRP3-dependent pathway (14). In human primary monocytes primed with LPS to increase expression of both NLRP3 and pro-IL-1β (Fig. 1A, bottom), we showed that stearate (C18:0) was also able to induce IL-1β release with similar efficiency as palmitate when compared with vehicle alone (BSA) (Fig. 1A, top). On the other hand, oleate (C18:1) and linoleate (C18:2) had no effect on IL-1β secretion, showing opposite effects of SFAs compared with UFAs. Similar findings were observed in MDMs (Fig. 1B) and in M1-polarized macrophages (Fig. 1C), the predominant phenotype of macrophages present in the adipose tissue from obese. The release of IL-1β observed after LPS-priming (Fig. 1A) or M1-polarization (Fig. 1C) is linked to pro-IL-1β synthesis. Indeed, monocytes have a constitutive caspase-1 activity (19), resulting in a background of IL-1β release related to pro-IL-1β content.
Fig. 1.
SFAs, unlike UFAs, induce IL-1β release in human monocytes/macrophages. (A–C) ELISA for IL-1β in SNs (top) and immunoblotting for pro-IL-1β and NLRP3 in cell extracts (XT) (bottom) from (A) LPS-primed monocytes, (B) MDM, or (C) M1-polarized macrophages either treated with 200 µM of FFAs or vehicle (BSA) for 8 h. Results are presented as means ± SEM of three (A) or four (B, C) independent experiments. Beta-tubulin was blotted as a loading control in cell extracts. **P < 0.01 compared with BSA by one-way ANOVA followed by Bonferroni post-test. ND, not determined.
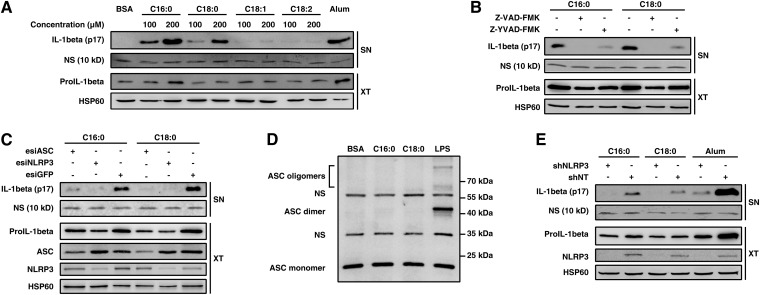
To explore the signaling pathway involved in SFA-induced IL-1β release, we differentiated the human monocytic cell line THP-1 into macrophage-like cells by using PMA. This classical (20, 21) and highly responsive model allowed us to easily detect large amounts of IL-1β in supernatant by Western blot and to focus on the maturation process. As in primary monocytes/macrophages, SFAs increased the IL-1β secretion in PMA-differentiated THP-1 cells in a dose-dependent manner (Fig. 2A). The 17 kDa form of IL-1β observed in Fig. 2A is known to be mainly produced after a proteolytic cleavage by caspase-1 (2, 3). To investigate caspase-1 involvement, the PMA-differentiated THP-1 cells were treated with SFAs alone or along with a pan-caspase inhibitor (Z-VAD) or with a caspase-1-specific inhibitor (Z-YVAD). Inhibition of caspases completely abolished SFA-induced IL-1β release, while caspase-1-specific inhibition repressed it almost completely (Fig. 2B). This result suggests a key role for caspase-1 in SFA-induced IL-1β processing, even if a minor contribution of a second caspase is not excluded.
Fig. 2.
SFAs activate the NLRP3 inflammasome in PMA-differentiated THP-1 cells. (A–C) IL-1β release in SNs from PMA-differentiated THP-1 cells determined by immunoblotting after 8 h of treatment (A) with various concentrations of FFAs, (B) with 200 µM of SFAs along with caspase inhibitor Z-VAD or caspase-1 inhibitor Z-YVAD, or (C) with 200 µM of SFAs after ASC or NLRP3 silencing by esiRNA. (D) PMA-differentiated THP-1 cells were treated with 200 µM of SFAs for 4 h. Cell extracts were crosslinked with DSS, and immunoblotting was performed using anti-ASC antibody. (E) IL-1β release in SNs from PMA-differentiated THP-1 cells stably expressing shRNA against NLRP3 (shNLRP3) or a nontarget shRNA (shNT) determined by immunoblotting after 8 h of treatment with 200 µM of SFAs. NLRP3 and ASC were blotted as a control for RNAi. Beta-tubulin or HSP60 was blotted as a loading control in cell extracts. LPS (1 µg/ml) was used as positive control in (D), and alum (400 µg/ml) in (A) and (E). A nonspecific band (NS) at 10 kDa was used as loading control in SNs. XT, cell extract. NS, nonspecific band.
ASC is an adaptor protein commonly involved in caspase-1 processing. It is composed of two domains mediating protein-protein interactions: the pyrin domain (PYD) and the CARD domain, which is essential for the homotypic interaction with caspase-1. When ASC was knocked down by esiRNA in PMA-differentiated THP-1 cells, a decrease in IL-1β secretion was observed after SFAs treatment (Fig. 2C). ASC can participate in caspase-1 activation through formation of two types of complexes: the pyroptosome and the inflammasome. The pyroptosome is produced after dimerization of ASC through its pyrin domain, allowing recruitment and activation of caspase-1 (7). To examine pyroptosome assembly, PMA-differentiated THP-1 cell extracts were crosslinked with DSS to detect ASC dimers. Compared with LPS at high concentration, a well-characterized pyroptosome inducer in PMA-differentiated THP-1 cells (7), neither palmitate nor stearate led to ASC dimerization (Fig. 2D). This indicates that ASC involvement in SFA-induced IL-1β secretion is not due to pyroptosome formation.
The second complex formed with ASC protein is the inflammasome. This complex appears by interaction between the PYD domain of ASC and the PYD domain of a second protein, such as NLRP3 or AIM2 (3). As for pyroptosome, ASC interaction leads to caspase-1 recruitment. Among the inflammasomes, the NLRP3 inflammasome is the most extensively studied, and a wide variety of compounds is known to activate it (3). The knockdown of NLRP3 in PMA-differentiated THP-1 cells by esiRNA led to a strong inhibition of SFA-induced IL-1β release (Fig. 2C). Similar results were obtained in THP-1 cells stably expressing shRNA against NLRP3 (Fig. 2E), confirming that both palmitate and stearate lead to a NLRP3 inflammasome-dependent IL-1β processing.
UFAs prevent SFA-induced IL-1β maturation
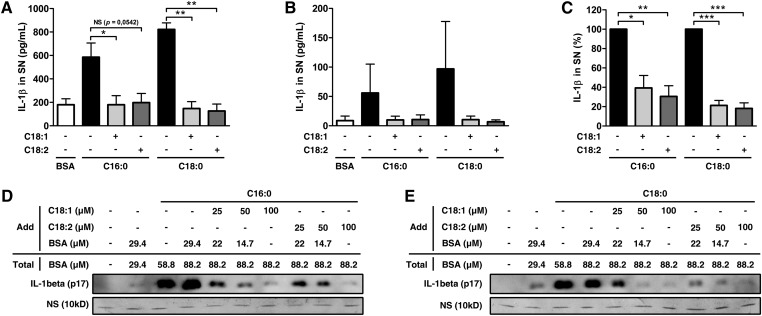
Unlike SFAs, UFAs possess double bounds that confer their particular properties. As described previously, they cannot activate the NLRP3 inflammasome compared with SFAs. To determine the impact of UFAs on SFA-induced IL-1β release, we treated LPS-primed primary monocytes with SFAs alone or in combination with UFAs. Interestingly, the presence of UFAs totally abolished the IL-1β secretion induced by SFAs, with equal efficiency for both oleate and linoleate (Fig. 3A). In M1-polarized macrophages, UFAs also decreased SFA-induced IL-1β release (Fig. 3B, C).
Fig. 3.
UFAs inhibit SFA-induced IL-1β release in monocytes/macrophages. (A, B) ELISA for IL-1β in SNs from (A) LPS-primed monocytes or (B) M1-polarized macrophages treated with 200 µM of SFAs alone or along with 200 µM of UFAs for 8 h. Results are presented as means ± SEM of three (A) or four (B) independent experiments. (C) Normalization of (B) to SFAs induction. (D, E) IL-1β release in SNs from PMA-differentiated THP-1 cells determined by immunoblotting after 8 h of treatment with 200 µM of (D) palmitate or (E) stearate in combination with UFAs and BSA at indicated concentrations. A nonspecific band (NS) at 10 kDa was used as loading control in SNs. *P < 0.05; **P < 0.01; ***P < 0.001; NS, nonsignificant by t-test.
The affinity of long-chain fatty acids for BSA weakly differs according to each FFA (22). Therefore, if SFAs have more affinity to BSA than do UFAs, the addition of BSA could mediate the inhibitory effect by decreasing the unbound fraction, which is the active form. To exclude this option, we normalized the BSA concentration to evaluate the specific impact of UFAs on palmitate-induced IL-1β secretion in PMA-differentiated THP-1 cells (Fig. 3D). With equal amounts of BSA, the inhibiting effect of UFAs persisted and was obviously dose dependent. Moreover, BSA alone had no effect. Similar results were obtained when cells were stimulated with stearate (Fig. 3E). UFAs were efficient in small concentrations, and the UFA:SFA ratio of 1:2 provided good inhibition. However, a ratio of 1:1 was used for all subsequent experiments.
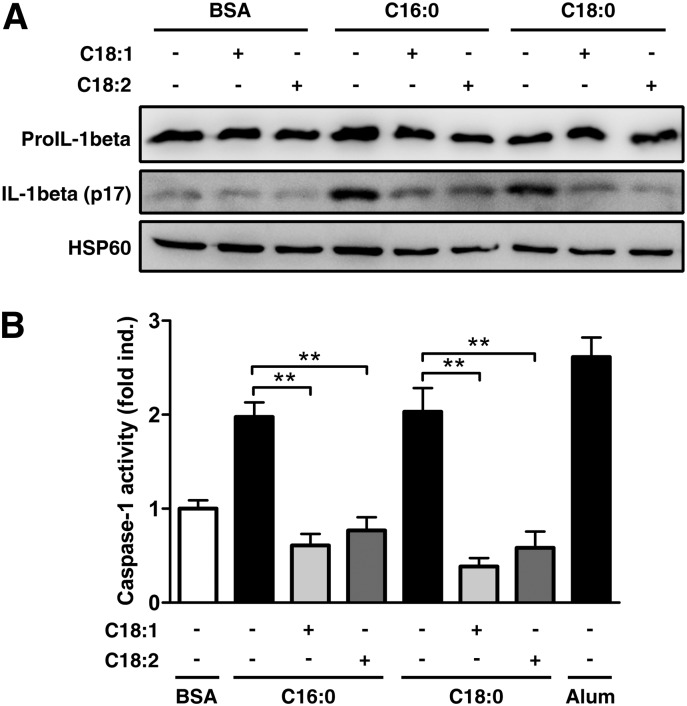
Processing and secretion are two important regulatory events for IL-1β release (2). To identify which step is targeted by UFAs, we assessed the maturation process in PMA-differentiated THP-1 cells. Cotreatment with UFAs strongly reduced the intracellular pro-IL-1β cleavage (Fig. 4A) and the caspase-1 activation (Fig. 4B) induced by SFAs. Considering that SFAs activate the NLRP3 inflammasome, the inhibiting effect of UFAs on IL-1β maturation probably operates by preventing SFA-induced NLRP3 inflammasome activation.
Fig. 4.
UFAs inhibit SFA-induced IL-1β maturation in PMA-differentiated THP-1 cells. (A, B) PMA-differentiated THP-1 cells were either treated with 200 µM of SFAs alone or along with 200 µM of UFAs. (A) IL-1β cleavage was determined by immunoblotting on cell extracts after 8 h. (B) Caspase-1 activity was measured by fluorescence. Results are presented as means ± SEM of three independent experiments. Alum (400 µg/ml) was used as positive control in (B). HSP60 was blotted as a loading control in cell extracts. **P < 0.01 by t-test.
UFAs decrease NLRP3 inflammasome activation by various inducers
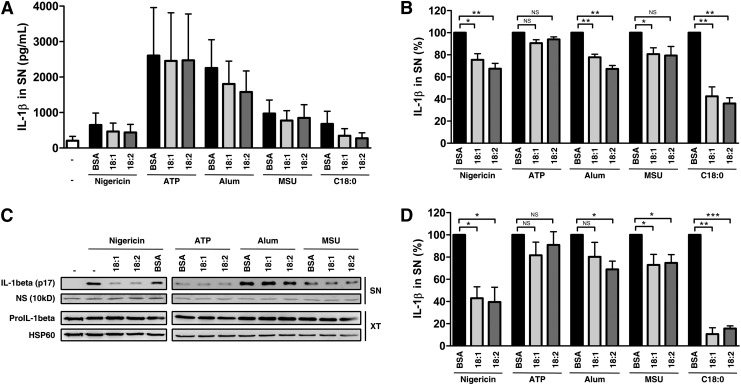
The hallmark of NLRP3 inflammasome formation is the interaction between NLRP3 and ASC. Unfortunately, the low amount of endogenous NLRP3 and the lack of good antibodies against the human form make this interaction difficult to observe without over-expressing a tagged NLRP3 protein. To overcome this problem, a different approach was used. As explained previously, the NLRP3 inflammasome is known to be activated by various compounds (3). Therefore, we tested UFAs outcomes on the most common NLRP3 activators such as nigericin, ATP, alum crystals and MSU in LPS-primed monocytes (Fig. 5A, B) and in PMA-differentiated THP-1 cells (Fig. 5C, D). Cotreatment with UFAs significantly inhibited the IL-1β release induced by SFAs but also by nigericin, alum, and MSU (Fig. 5B, D). On the other side, ATP seemed to be insensitive to UFAs after 8 h of cotreatment in both LPS-primed monocytes and PMA-differentiated THP-1 cells.
Fig. 5.
UFAs downregulate NLRP3 activation by classical inducers in primed-monocytes. (A) ELISA for IL-1β in SNs from LPS-primed monocytes after cotreatment with various NLRP3 inducers and 200 µM of UFAs or BSA. Cells were cotreated with nigericin (5 µM), ATP (5 mM), MSU (100 µg/ml) or alum (400 µg/ml) for 8 h. Results are presented as means ± SEM of four independent experiments. (B) Normalization of A to induction with each agent cotreated with BSA. (C) IL-1β release in SNs from PMA-differentiated THP-1 cells determined by immunoblotting after cotreatment with various NLRP3 inducers and 200 µM of UFAs or BSA. Cells were cotreated with nigericin (5 µM), ATP (10 mM), MSU (100 µg/ml), or alum (400 µg/ml) for 8 h. (D) Quantification of IL-1β in SN from C. Results are presented as means ± SEM of at least three independent experiments. A nonspecific band (NS) at 10 kDa was used as loading control in SN. HSP60 was blotted as a loading control in cell extracts. *P < 0.05; **P < 0.01; ***P < 0.001; NS, nonsignificant by t-test. XT, cell extract.
Several less frequently used compounds were described to activate the NLRP3 inflammasome, such as doxorubicin (23), tunicamycin (20), muramyl dipeptide (MDP) (24), antimycin A (21) and monensin (25). Cotreatment with UFAs significantly inhibited the IL-1β release induced by all these compounds in PMA-differentiated THP-1 cells (supplementary Fig. I-A, B). The NLRP3 inflammasome involvement was confirmed for all these compounds by using the shNLRP3 THP-1 cells (supplementary Fig. I-C).
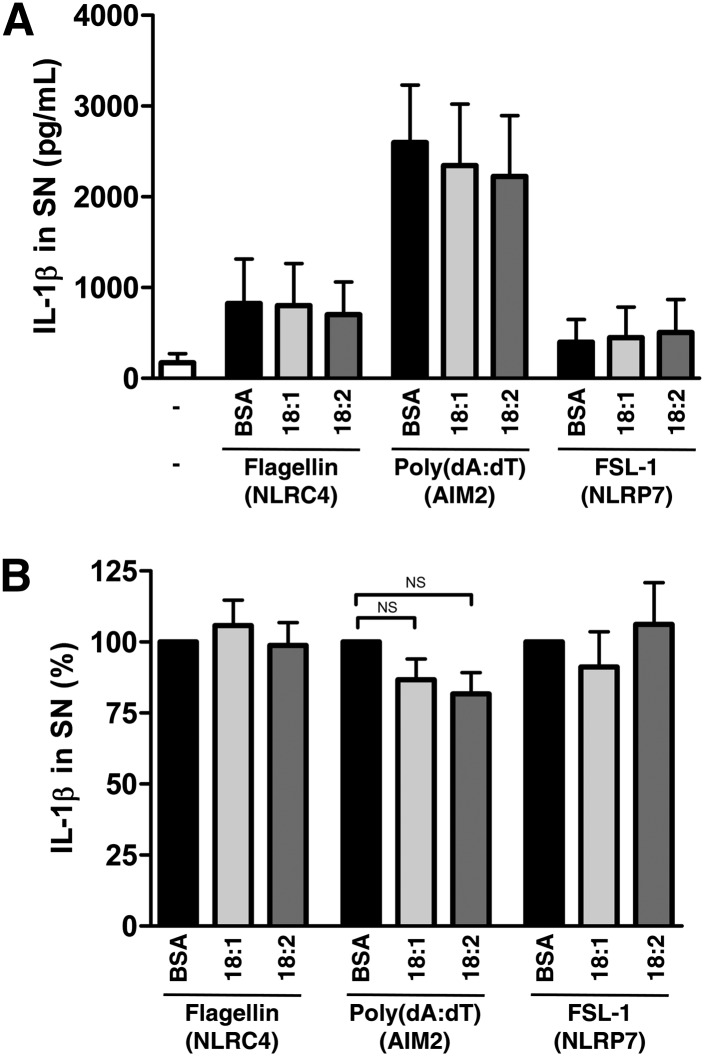
NLRP3 is one protein among many others leading to an inflammasome assembly. To explore other inflammasomes, we next treated LPS-primed monocytes with NLRC4, AIM2 or NLRP7 activators, respectively flagellin, poly(dA:dT) or FSL-1 (Fig. 6A). UFAs cotreatment had no significant effect on IL-1β release induced by these compounds (Fig. 6B). Taken together, these results suggest a specific action on the NLRP3 inflammasome, probably upstream of inflammasome assembly.
Fig. 6.
UFAs have no effect on NLRC4, NLRP7, or AIM2 inflammasomes. (A) ELISA for IL-1β in SNs from LPS-primed monocytes after cotreatment with various inflammasomes inducers and 200 µM of UFAs or BSA. Cells were treated with flagellin (2 µg), poly(dA:dT) (2 µg), or FSL-1 (0.2 µg/ml) for 8 h. Results are presented as means ± SEM of four independent experiments. (B) Normalization of A to induction with each agent cotreated with BSA. NS, nonsignificant by t-test.
UFAs reduce IL-1β processing but not transcription
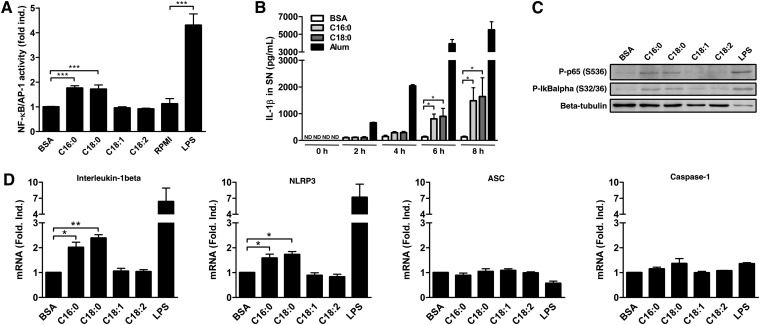
Nuclear factor-kappa B (NF-κB) family members are critical regulators of gene expression in mammals (26). These transcription factors bind as dimers to κB response elements in diverse gene promoters. Without stimulation, this dimer is sequestered in the cytoplasm by the inhibitory protein IκBα. Upon activation of the canonical pathway, IκBα is phosphorylated, ubiquitinated, and finally submitted to proteasomal degradation allowing the p50/p65 heterodimer translocation to nucleus. Various posttranslational modifications are known to change the transcriptional activity of p50/p65, such as the activating phosphorylation in the transactivation domain of p65 (26). SFAs are known to activate the canonical NF-κB pathway in nondifferentiated THP-1 cells (27). In PMA-differentiated THP1-XBlue cells, a THP-1 cell line stably expressing a reporter gene under the control of NF-κB and AP-1 transcription factors, an increase of the transcriptional activity was reported after SFA but not after UFA treatment (Fig. 7A). Since IL-1β also activates the NF-κB pathway and to exclude the artifact related to its release, subsequent experiments were performed before IL-1β secretion. IL-1β release occurred at around 4 h and became significant at 6 h post-treatment (Fig. 7B). After 3 h, SFAs increased phosphorylation of IκBα and p65 in PMA-differentiated THP-1 cells (Fig. 7C). No increase in phosphorylation was observed after UFA treatment.
Fig. 7.
SFAs increase the expression of NLRP3 and IL-1β in PMA-differentiated THP-1 cells. (A) NF-κB/AP-1 activity was assessed by colorimetric assay in SNs from PMA-differentiated THP1-XBlue™ cells after treatment with either 200 µM of FFAs or 250 ng/ml of LPS for 24 h. Results are presented as means ± SEM of three independent experiments. (B) ELISA for IL-1β in SNs from PMA-differentiated THP-1 cells after treatment with BSA, SFAs (200 µM), or alum (400 µg/ml) at different time points. Results are presented as means ± SEM of three independent experiments. (C) The phosphorylation of IκBα and p65 was analyzed by immunoblotting on total cell extracts after treatment with either 200 µM of FFAs for 3 h or 250 ng/ml of LPS for 1 h on PMA-differentiated THP-1 cells. Beta-tubulin was blotted as a loading control in cell extracts. (D) Relative mRNA expression measured by qRT-PCR for IL-1β, NLRP3, ASC, and caspase-1 after treatment with either 200 µM of FFAs for 4 h or 250 ng/ml of LPS for 2 h on PMA-differentiated THP-1 cells. Results are presented as means ± SEM of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 by t-test. ND, not determined.
Because NLRP3 and IL-1β are both NF-κB-dependent genes (8), SFAs could likely play a priming role as it was previously described in dendritic cells (28). Although IL-1β and NLRP3 genes were already induced by PMA in THP-1 cells, a further induction of NLRP3 and IL-1β mRNA was observed by qRT-PCR after treatment with SFAs but not with UFAs (Fig. 7D), suggesting a “second priming” concomitantly to NLRP3 inflammasome activation in PMA-differentiated THP-1 cells. ASC and caspase-1 mRNA did not change. This low induction in gene expression is specific of the PMA-differentiated THP-1 model, and no further induction of NF-κB activation was observed in LPS-primed monocytes (data not shown).
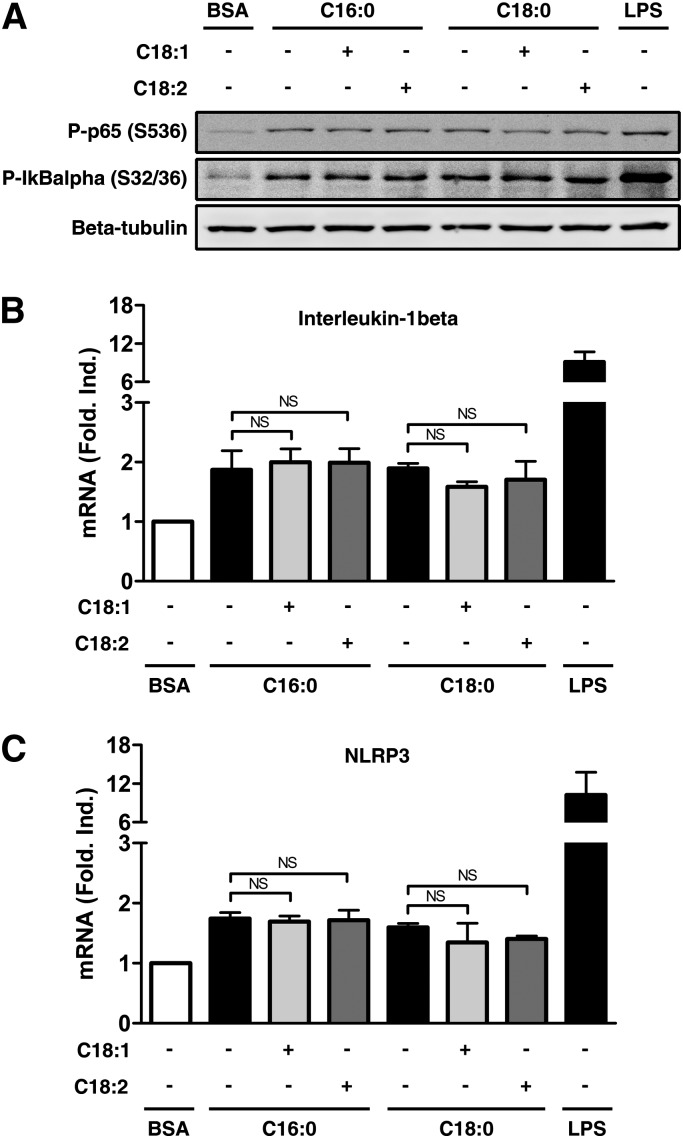
We next examined the impact of UFAs on this further induction of gene expression. Addition of UFAs did not change the phosphorylation of IκBα and p65 induced by SFAs in PMA-differentiated THP-1 cells (Fig. 8A). No significant effect was observed on IL-1β (Fig. 8B) or NLRP3 (Fig. 8C) mRNA levels when UFAs were present. Taken together, these results strongly suggest that i) SFAs increase little or no gene expression in primed cells, confirming that the IL-1β release observed is mainly due to an enhancement in processing; ii) the decrease in IL-1β secretion by cotreatment with UFAs is not linked to reduction of transcription but to reduction of IL-1β processing.
Fig. 8.
UFAs do not inhibit the expression of NLRP3 and IL-1β in PMA-differentiated THP-1 cells. (A–C) PMA-differentiated THP-1 cells were treated with 200 µM of SFAs alone or with 200 µM of UFAs. (A) The phosphorylation of IκBα and p65 was analyzed by immunoblotting on total cell extracts after 3 h. Beta-tubulin was blotted as a loading control. (B, C) Relative mRNA expression was measured by qRT-PCR for (B) IL-1β and (C) NLRP3 after 4 h of treatment. Results are presented as means ± SEM of three independent experiments. LPS (250 ng/ml) was used as positive control, and cells were treated for 1 h (A) or 2 h (B, C). NS, nonsignificant by t-test.
DISCUSSION
When the immune system is activated, all the resources are mobilized to eradicate the aggressor. At term, the inflammatory response ends and the immune system comes back to normal. In particular cases, the immune system is ineffective to resolve the trouble and a chronic inflammation appears. In obesity, an unexplained chronic, low-grade inflammation is present (29) in which SFAs could play a role. As reported here, both SFAs palmitate and stearate activated the NLRP3 inflammasome and led to IL-1β secretion. Unlike SFAs, the UFAs oleate and linoleate were unable to activate the NRLP3 inflammasome, showing opposite effects of SFAs versus UFAs. Our results confirm in human monocytes/macrophages previous work demonstrating the NLRP3 activation in mouse macrophages by palmitate (14) and identify stearate as a new inducer. Stearate is the second most important SFA in blood and represents with palmitate 90% of total SFAs (15), while other SFAs, such as laurate and myristate, are present in very low concentrations. This proinflammatory effect of SFAs in vitro corroborates interventional studies describing that SFA-rich diets increase IL-1β production (4, 30) as well as also other inflammatory processes that may be related to IL-1β (31–34).
In PMA-differentiated THP-1 cells, we demonstrated a weak induction of IL-1β and NLRP3 gene expression by SFAs. Even if it was low, this result means that the increase in IL-1β secretion after SFA treatment can also be, in part, of transcriptional origin. The lack of further induction reported in LPS-primed monocytes is likely inherent to the model used. Indeed, compared with PMA-differentiated THP-1 cells, there is no resting period between the priming and the stimulation with FFAs. This phenomenon is called “tolerance” and the NF-κB activation by priming agent leads to a transient unresponsive state in the second stimulation (35). Therefore, a priming action of SFAs is not excluded in unprimed monocytes, as it was clearly demonstrated in dendritic cells (28). In our study, primed monocytes were chosen to reduce to a minimum the transcriptional effect of SFAs and to focus on the second signal, the NLRP3 inflammasome activation.
IL-1β involvement in type 2 diabetes, gouty arthritis, or atherosclerosis is clear and well documented (1). It is strongly suggested that the NLRP3 inflammasome plays a crucial role in the transition between obesity and these diseases (36). Based on that, new treatments are developed in the hope of avoiding these obesity-associated complications. Treatment with IL-1β antagonists provided beneficial outcomes in many diseases, including type 2 diabetes (37) and gouty arthritis (38). Similar experiments are being conducted to prevent cardiovascular diseases (39), with very encouraging preliminary results (40). In our study, we report an unexpected effect of UFAs on NLRP3 inflammasome activation. Both oleate and linoleate prevent, with the same efficiency, the NLRP3-dependent IL-1β processing induced by SFAs but also by many activators, such as nigericin, alum, or MSU. However, no effect was observed on ATP-mediated NLRP3 inflammasome activation, which suggests an action upstream of the NLRP3 inflammasome assembly, probably by interfering with the activation pathways. Recently, Yan et al. reported that omega-3 fatty acids inhibit the NLRP3 inflammasome activation in macrophages (30). They also tested non-omega-3 fatty acids, such as oleate (but not linoleate), but they failed to observe an inhibition. This surprising result is likely due to their experimental conditions. Indeed, they used a very low concentration of oleate, lower than the physiological level, to work with a concentration corresponding to omega-3.
UFAs are known to exert a broad range of effects, such as changes in membrane composition (41), generation of anti-inflammatory compounds, such as resolvins (42), or activation of various receptors (43). G protein-coupled receptor (GPR)40 and GPR120 are both activated by long-chain fatty acids. Inhibition of the NLRP3 inflammasome by omega-3 was described to be dependent on these two receptors through initiation of β-arrestin-2 binding to NLRP3 (30). Therefore, one explanation could be that oleate and linoleate prevent NLRP3 inflammasome activation by binding these receptors but with less affinity than omega-3, explaining the requirement of a higher concentration. A second mechanism could involve the endoplasmic reticulum (ER). UFAs could prevent ER stress induction (44, 45), recently described to be involved in inflammasome activation (20, 46). In addition, SFAs or tunicamycin, both sensitive to UFA inhibition, are well-characterized ER stress inducers (20, 44–46).
An outstanding issue is the lack of response to UFAs when NLRP3 was activated by ATP. Numerous studies have described contradictory results depending on the activators tested (47–49). This wide heterogeneity in response to NLRP3 inducers clearly suggests that several pathways are able to trigger the NLRP3 inflammasome assembly. Currently, the ATP-activating pathway is commonly described as very simple with few places for UFA interference: ATP binds to P2X7 receptor leading to K+ efflux and NLRP3 inflammasome activation (50). An interesting finding reported that crystal activators, such as alum and MSU, trigger NLRP3 inflammasome activation indirectly through ATP release (51). If this new mechanism can be extended to all the NLRP3 inducers, one likely explanation could be that UFAs prevent ATP release but have no effect on exogenous ATP.
Although the mechanism is not yet fully elucidated, UFAs could play a protective role in diseases in which NLRP3 is involved, such as type 2 diabetes, atherosclerosis, or gouty arthritis. Unlike treatments with anakinra or other IL-1β antagonists requiring regular injections, simple dietary interventions could be helpful to obese people in decreasing Il-1β secretion and inflammation. Further investigation is required before concluding a protective effect of a high-UFA diet on inflammation, but several lines of evidence point in this direction. Numerous studies described a powerful anti-inflammatory effect for oleate and linoleate. Ex vivo macrophages from rats treated by gavage with oleate or linoleate showed a reduction of cytokine secretion, including IL-1β, compared with untreated rats (52), and intracerebroventricular injection of oleate reduced hypothalamic inflammation in rats (53). In most cases, intervention studies are performed in animals by changing the diet. Various groups reported an improvement in obesity-associated inflammation (32–34) and insulin sensitivity (34, 53) after a diet rich in UFAs, partially attributing these results to oleate- and linoleate-rich oils. In humans, there is an association between high-SFA and/or low-UFA concentrations and inflammation (54, 55) or metabolic syndrome (54). In some studies, these parameters can predict metabolic syndrome development (56, 57). Assays on small groups of people described a decrease of inflammatory gene expression (31) and an improvement of insulin sensitivity (58) after UFA-rich diet. Two large trials have been conducted to assess the benefit of a high-oleate diet on insulin sensitivity: the KANWU study (59) and, most recently, the LIPGENE study (60). In both studies, the oleate diet led to a significant increase in plasma oleate, but only the KANWU study reported an improvement of insulin sensitivity. Contradictory results are common in this type of study in which the design is crucial: choice of subjects (healthy or obese), ethnicity/genetics, initial diet, etc., may all affect the final outcome. In all the studies reporting an effect of UFAs, few mechanisms were proposed. Protection against inflammation and obesity-associated diseases reported after a high-UFA diet is not yet understood, but NLRP3 inflammasome inhibition might be a partial explanation.
In conclusion, we demonstrated that SFAs palmitate and stearate trigger IL-1β secretion in various models of human monocytes and macrophages in a caspase-1/ASC/NLRP3-dependent pathway. The UFAs oleate and linoleate, which did not activate the NLRP3 inflammasome, totally prevented the NLRP3 inflammasome activation induced by SFAs and, with less efficiency, by a broad range of NLRP3 activators. These results could be correlated with in vivo studies reporting anti-inflammatory properties of a UFA-rich diet on IL-1β production. By preventing NLRP3 inflammasome activation, UFAs might play a protective role in NLRP3-associated diseases, such as type 2 diabetes, atherosclerosis, and gouty arthritis.
Supplementary Material
Acknowledgments
The authors thank E. Di Valentin from the GIGA-Viral Vectors Platform (Liege, Belgium) for the stable cell lines generation and R. Stephan from the GIGA-Imaging and Flow Cytometry (Liege, Belgium) for the cell sorting of GFP-positive THP-1 cells.
Footnotes
Abbreviations:
- ASC
- apoptosis-associated speck-like protein containing a CARD
- CARD
- caspase recruitment domain
- DSS
- disuccinimidyl suberate
- FACS
- fluorescence-activated cell sorting
- IL
- interleukin
- LPS
- lipopolysaccharide
- MDM
- monocyte-derived macrophage
- MDP
- muramyl dipeptide
- MSU
- monosodium urate
- NF-κB
- nuclear factor-kappa B
- NLR
- NOD-like receptor
- PBMC
- peripheral blood mononuclear cell
- PYD
- pyrin domain
- SFA
- saturated fatty acid
- SN
- supernatant
- UFA
- unsaturated fatty acid
This work was supported by the Interuniversity Attraction Poles (IAP) program initiated by the Belgian Science Policy Office (BELSPO) (IAP Grant P7/32) and by the Belgian “Fonds National pour la Recherche Scientifique” (FNRS).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure.
REFERENCES
- 1.Wen H., Ting J. P., O'Neill L. A. 2012. A role for the NLRP3 inflammasome in metabolic diseases--did Warburg miss inflammation? Nat. Immunol. 13: 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eder C. 2009. Mechanisms of interleukin-1beta release. Immunobiology. 214: 543–553 [DOI] [PubMed] [Google Scholar]
- 3.Schroder K., Tschopp J. 2010. The inflammasomes. Cell. 140: 821–832 [DOI] [PubMed] [Google Scholar]
- 4.Vandanmagsar B., Youm Y. H., Ravussin A., Galgani J. E., Stadler K., Mynatt R. L., Ravussin E., Stephens J. M., Dixit V. D. 2011. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med. 17: 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stienstra R., van Diepen J. A., Tack C. J., Zaki M. H., van de Veerdonk F. L., Perera D., Neale G. A., Hooiveld G. J., Hijmans A., Vroegrijk I., et al. 2011. Inflammasome is a central player in the induction of obesity and insulin resistance. Proc. Natl. Acad. Sci. USA. 108: 15324–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guarda G., Zenger M., Yazdi A. S., Schroder K., Ferrero I., Menu P., Tardivel A., Mattmann C., Tschopp J. 2011. Differential expression of NLRP3 among hematopoietic cells. J. Immunol. 186: 2529–2534 [DOI] [PubMed] [Google Scholar]
- 7.Fernandes-Alnemri T., Wu J., Yu J. W., Datta P., Miller B., Jankowski W., Rosenberg S., Zhang J., Alnemri E. S. 2007. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 14: 1590–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauernfeind F. G., Horvath G., Stutz A., Alnemri E. S., MacDonald K., Speert D., Fernandes-Alnemri T., Wu J., Monks B. G., Fitzgerald K. A., et al. 2009. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183: 787–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curat C. A., Miranville A., Sengenes C., Diehl M., Tonus C., Busse R., Bouloumie A. 2004. From blood monocytes to adipose tissue-resident macrophages: induction of diapedesis by human mature adipocytes. Diabetes. 53: 1285–1292 [DOI] [PubMed] [Google Scholar]
- 10.Cinti S., Mitchell G., Barbatelli G., Murano I., Ceresi E., Faloia E., Wang S., Fortier M., Greenberg A. S., Obin M. S. 2005. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J. Lipid Res. 46: 2347–2355 [DOI] [PubMed] [Google Scholar]
- 11.Biswas S. K., Mantovani A. 2012. Orchestration of metabolism by macrophages. Cell Metab. 15: 432–437 [DOI] [PubMed] [Google Scholar]
- 12.Duewell P., Kono H., Rayner K. J., Sirois C. M., Vladimer G., Bauernfeind F. G., Abela G. S., Franchi L., Nunez G., Schnurr M., et al. 2010. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 464: 1357–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amaral F. A., Costa V. V., Tavares L. D., Sachs D., Coelho F. M., Fagundes C. T., Soriani F. M., Silveira T. N., Cunha L. D., Zamboni D. S., et al. 2012. NLRP3 inflammasome-mediated neutrophil recruitment and hypernociception depend on leukotriene B(4) in a murine model of gout. Arthritis Rheum. 64: 474–484 [DOI] [PubMed] [Google Scholar]
- 14.Wen H., Gris D., Lei Y., Jha S., Zhang L., Huang M. T., Brickey W. J., Ting J. P. 2011. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 12: 408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodson L., Skeaff C. M., Fielding B. A. 2008. Fatty acid composition of adipose tissue and blood in humans and its use as a biomarker of dietary intake. Prog. Lipid Res. 47: 348–380 [DOI] [PubMed] [Google Scholar]
- 16.Emi N., Friedmann T., Yee J. K. 1991. Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J. Virol. 65: 1202–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 18.Richieri G. V., Kleinfeld A. M. 1995. Unbound free fatty acid levels in human serum. J. Lipid Res. 36: 229–240 [PubMed] [Google Scholar]
- 19.Kahlenberg J. M., Dubyak G. R. 2004. Differing caspase-1 activation states in monocyte versus macrophage models of IL-1beta processing and release. J. Leukoc. Biol. 76: 676–684 [DOI] [PubMed] [Google Scholar]
- 20.Menu P., Mayor A., Zhou R., Tardivel A., Ichijo H., Mori K., Tschopp J. 2012. ER stress activates the NLRP3 inflammasome via an UPR-independent pathway. Cell Death Dis. 3: e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou R., Yazdi A. S., Menu P., Tschopp J. 2011. A role for mitochondria in NLRP3 inflammasome activation. Nature. 469: 221–225 [DOI] [PubMed] [Google Scholar]
- 22.Spector A. A., John K., Fletcher J. E. 1969. Binding of long-chain fatty acids to bovine serum albumin. J. Lipid Res. 10: 56–67 [PubMed] [Google Scholar]
- 23.Sauter K. A., Wood L. J., Wong J., Iordanov M., Magun B. E. 2011. Doxorubicin and daunorubicin induce processing and release of interleukin-1beta through activation of the NLRP3 inflammasome. Cancer Biol. Ther. 11: 1008–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinon F., Agostini L., Meylan E., Tschopp J. 2004. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr. Biol. 14: 1929–1934 [DOI] [PubMed] [Google Scholar]
- 25.Ichinohe T., Pang I. K., Iwasaki A. 2010. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 11: 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oeckinghaus A., Hayden M. S., Ghosh S. 2011. Crosstalk in NF-kappaB signaling pathways. Nat. Immunol. 12: 695–708 [DOI] [PubMed] [Google Scholar]
- 27.Huang S., Rutkowsky J. M., Snodgrass R. G., Ono-Moore K. D., Schneider D. A., Newman J. W., Adams S. H., Hwang D. H. 2012. Saturated fatty acids activate TLR-mediated proinflammatory signaling pathways. J. Lipid Res. 53: 2002–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds C. M., McGillicuddy F. C., Harford K. A., Finucane O. M., Mills K. H., Roche H. M. 2012. Dietary saturated fatty acids prime the NLRP3 inflammasome via TLR4 in dendritic cells-implications for diet-induced insulin resistance. Mol. Nutr. Food Res. 56: 1212–1222 [DOI] [PubMed] [Google Scholar]
- 29.Ye J., McGuinness O. P. 2013. Inflammation during obesity is not all bad: evidence from animal and human studies. Am. J. Physiol. Endocrinol. Metab. 304: E466–E477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan Y., Jiang W., Spinetti T., Tardivel A., Castillo R., Bourquin C., Guarda G., Tian Z., Tschopp J., Zhou R. 2013. Omega-3 Fatty Acids Prevent Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome Activation. Immunity. 38: 1154–1163 [DOI] [PubMed] [Google Scholar]
- 31.van Dijk S. J., Feskens E. J., Bos M. B., Hoelen D. W., Heijligenberg R., Bromhaar M. G., de Groot L. C., de Vries J. H., Muller M., Afman L. A. 2009. A saturated fatty acid-rich diet induces an obesity-linked proinflammatory gene expression profile in adipose tissue of subjects at risk of metabolic syndrome. Am. J. Clin. Nutr. 90: 1656–1664 [DOI] [PubMed] [Google Scholar]
- 32.Laugerette F., Furet J. P., Debard C., Daira P., Loizon E., Geloen A., Soulage C. O., Simonet C., Lefils-Lacourtablaise J., Bernoud-Hubac N., et al. 2012. Oil composition of high-fat diet affects metabolic inflammation differently in connection with endotoxin receptors in mice. Am. J. Physiol. Endocrinol. Metab. 302: E374–E386 [DOI] [PubMed] [Google Scholar]
- 33.Wang X., Cheng M., Zhao M., Ge A., Guo F., Zhang M., Yang Y., Liu L., Yang N. 2013. Differential effects of high-fat-diet rich in lard oil or soybean oil on osteopontin expression and inflammation of adipose tissue in diet-induced obese rats. Eur. J. Nutr. 52: 1181–1189 [DOI] [PubMed] [Google Scholar]
- 34.Tardif N., Salles J., Landrier J. F., Mothe-Satney I., Guillet C., Boue-Vaysse C., Combaret L., Giraudet C., Patrac V., Bertrand-Michel J., et al. 2011. Oleate-enriched diet improves insulin sensitivity and restores muscle protein synthesis in old rats. Clin. Nutr. 30: 799–806 [DOI] [PubMed] [Google Scholar]
- 35.Biswas S. K., Lopez-Collazo E. 2009. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 30: 475–487 [DOI] [PubMed] [Google Scholar]
- 36.Stienstra R., Tack C. J., Kanneganti T. D., Joosten L. A., Netea M. G. 2012. The inflammasome puts obesity in the danger zone. Cell Metab. 15: 10–18 [DOI] [PubMed] [Google Scholar]
- 37.Larsen C. M., Faulenbach M., Vaag A., Volund A., Ehses J. A., Seifert B., Mandrup-Poulsen T., Donath M. Y. 2007. Interleukin-1-receptor antagonist in type 2 diabetes mellitus. N. Engl. J. Med. 356: 1517–1526 [DOI] [PubMed] [Google Scholar]
- 38.Schumacher H. R., Jr, Sundy J. S., Terkeltaub R., Knapp H. R., Mellis S. J., Stahl N., Yancopoulos G. D., Soo Y., King-Davis S., Weinstein S. P., et al. 2012. Rilonacept (interleukin-1 trap) in the prevention of acute gout flares during initiation of urate-lowering therapy: results of a phase II randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 64: 876–884 [DOI] [PubMed] [Google Scholar]
- 39.Ridker P. M., Thuren T., Zalewski A., Libby P. 2011. Interleukin-1beta inhibition and the prevention of recurrent cardiovascular events: rationale and design of the Canakinumab Anti-inflammatory Thrombosis Outcomes Study (CANTOS). Am. Heart J. 162: 597–605 [DOI] [PubMed] [Google Scholar]
- 40.Abbate A., Van Tassell B. W., Biondi-Zoccai G., Kontos M. C., Grizzard J. D., Spillman D. W., Oddi C., Roberts C. S., Melchior R. D., Mueller G. H., et al. 2013. Effects of interleukin-1 blockade with anakinra on adverse cardiac remodeling and heart failure after acute myocardial infarction [from the Virginia Commonwealth University-Anakinra Remodeling Trial (2) (VCU-ART2) pilot study] Am. J. Cardiol. 111: 1394–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calder P. C. 2008. The relationship between the fatty acid composition of immune cells and their function. Prostaglandins Leukot. Essent. Fatty Acids. 79: 101–108 [DOI] [PubMed] [Google Scholar]
- 42.Zhang M. J., Spite M. 2012. Resolvins: anti-inflammatory and proresolving mediators derived from omega-3 polyunsaturated fatty acids. Annu. Rev. Nutr. 32: 203–227 [DOI] [PubMed] [Google Scholar]
- 43.Vinolo M. A., Hirabara S. M., Curi R. 2012. G-protein-coupled receptors as fat sensors. Curr. Opin. Clin. Nutr. Metab. Care. 15: 112–116 [DOI] [PubMed] [Google Scholar]
- 44.Nemcova-Furstova V., James R. F., Kovar J. 2011. Inhibitory effect of unsaturated fatty acids on saturated fatty acid-induced apoptosis in human pancreatic beta-cells: activation of caspases and ER stress induction. Cell. Physiol. Biochem. 27: 525–538 [DOI] [PubMed] [Google Scholar]
- 45.Ishiyama J., Taguchi R., Akasaka Y., Shibata S., Ito M., Nagasawa M., Murakami K. 2011. Unsaturated FAs prevent palmitate-induced LOX-1 induction via inhibition of ER stress in macrophages. J. Lipid Res. 52: 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oslowski C. M., Hara T., O'Sullivan-Murphy B., Kanekura K., Lu S., Hara M., Ishigaki S., Zhu L. J., Hayashi E., Hui S. T., et al. 2012. Thioredoxin-interacting protein mediates ER stress-induced beta cell death through initiation of the inflammasome. Cell Metab. 16: 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shenoy A. R., Wellington D. A., Kumar P., Kassa H., Booth C. J., Cresswell P., MacMicking J. D. 2012. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 336: 481–485 [DOI] [PubMed] [Google Scholar]
- 48.Subramanian N., Natarajan K., Clatworthy M. R., Wang Z., Germain R. N. 2013. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 153: 348–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hornung V., Bauernfeind F., Halle A., Samstad E. O., Kono H., Rock K. L., Fitzgerald K. A., Latz E. 2008. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Munoz-Planillo R., Kuffa P., Martinez-Colon G., Smith B. L., Rajendiran T. M., Nunez G. 2013. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 38: 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riteau N., Baron L., Villeret B., Guillou N., Savigny F., Ryffel B., Rassendren F., Le Bert M., Gombault A., Couillin I. 2012. ATP release and purinergic signaling: a common pathway for particle-mediated inflammasome activation. Cell Death Dis. 3: e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Magdalon J., Vinolo M. A., Rodrigues H. G., Paschoal V. A., Torres R. P., Mancini-Filho J., Calder P. C., Hatanaka E., Curi R. 2012. Oral administration of oleic or linoleic acids modulates the production of inflammatory mediators by rat macrophages. Lipids. 47: 803–812 [DOI] [PubMed] [Google Scholar]
- 53.Cintra D. E., Ropelle E. R., Moraes J. C., Pauli J. R., Morari J., Souza C. T., Grimaldi R., Stahl M., Carvalheira J. B., Saad M. J., et al. 2012. Unsaturated fatty acids revert diet-induced hypothalamic inflammation in obesity. PLoS ONE. 7: e30571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein-Platat C., Drai J., Oujaa M., Schlienger J. L., Simon C. 2005. Plasma fatty acid composition is associated with the metabolic syndrome and low-grade inflammation in overweight adolescents. Am. J. Clin. Nutr. 82: 1178–1184 [DOI] [PubMed] [Google Scholar]
- 55.Fernandez-Real J. M., Broch M., Vendrell J., Ricart W. 2003. Insulin resistance, inflammation, and serum fatty acid composition. Diabetes Care. 26: 1362–1368 [DOI] [PubMed] [Google Scholar]
- 56.Warensjo E., Riserus U., Vessby B. 2005. Fatty acid composition of serum lipids predicts the development of the metabolic syndrome in men. Diabetologia. 48: 1999–2005 [DOI] [PubMed] [Google Scholar]
- 57.Phillips C. M., Goumidi L., Bertrais S., Field M. R., McManus R., Hercberg S., Lairon D., Planells R., Roche H. M. 2012. Dietary saturated fat, gender and genetic variation at the TCF7L2 locus predict the development of metabolic syndrome. J. Nutr. Biochem. 23: 239–244 [DOI] [PubMed] [Google Scholar]
- 58.Summers L. K., Fielding B. A., Bradshaw H. A., Ilic V., Beysen C., Clark M. L., Moore N. R., Frayn K. N. 2002. Substituting dietary saturated fat with polyunsaturated fat changes abdominal fat distribution and improves insulin sensitivity. Diabetologia. 45: 369–377 [DOI] [PubMed] [Google Scholar]
- 59.Vessby B., Uusitupa M., Hermansen K., Riccardi G., Rivellese A. A., Tapsell L. C., Nalsen C., Berglund L., Louheranta A., Rasmussen B. M., et al. 2001. Substituting dietary saturated for monounsaturated fat impairs insulin sensitivity in healthy men and women: the KANWU study. Diabetologia. 44: 312–319 [DOI] [PubMed] [Google Scholar]
- 60.Tierney A. C., McMonagle J., Shaw D. I., Gulseth H. L., Helal O., Saris W. H., Paniagua J. A., Golabek-Leszczynska I., Defoort C., Williams C. M., et al. 2011. Effects of dietary fat modification on insulin sensitivity and on other risk factors of the metabolic syndrome--LIPGENE: a European randomized dietary intervention study. Int. J. Obes. (Lond.) 35: 800–809 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.