Abstract
To understand the role of N-glycosylation of lysosomal phospholipase A2 (LPLA2), four potential N-glycosylation sites in human LPLA2 (hLPLA2) were individually modified replacing asparagine (Asn) with alanine by site-direct mutagenesis. COS-7 cells transiently transfected with wild-type (WT) hLPLA2 gene produced catalytically active LPLA2. A single mutation at 273-, 289-, or 398-Asn partially reduced production of active LPLA2. A single mutation at 99-Asn and quadruple mutations at all four Asn sites resulted in a marked reduction of active LPLA2 and loss of active LPLA2, respectively. Western blot analysis using anti-hLPLA2 antibody showed that the LPLA2 expression level was similar between all transfectants. N-glycosidase F digestion revealed that multiple forms of LPLA2 found in individual transfectants are due to different N-glycans linked to the core protein. The LPLA2 activity in individual transfectants was mostly recovered in the soluble fraction and correlated to the quantity of LPLA2 detected in the soluble fraction. LPLA2 mutated at 99-Asn was mostly retained in the membrane fraction. The WT transfectants treated with tunicamycin markedly lost LPLA2 activity. These data indicate that the 99-Asn is the most critical N-glycosylation site for formation of native hLPLA2 in vivo and that the N-glycosylation of LPLA2 is crucial for biosynthesis of catalytically active hLPLA2.
Keywords: site-direct mutagenesis, lysosomal phospholipase A2, N-glycosylation
More than a decade ago, we identified a phospholipase A2 in MDCK cell homogenate with specificity toward glycerophospholipids such as phosphatidylcholine and phosphatidylethanolamine under acidic conditions (1). The phospholipase is calcium-independent, localized to lysosomes, has an acidic pH optimum, and transacylates short chain ceramides. The enzyme purified from bovine brain is a water-soluble glycoprotein consisting of a single peptide chain with a molecular mass of 45 kDa (2). The primary structure deduced from the DNA sequences is highly preserved between mammals (3). Later, the enzyme was classified as phospholipase A2, group XV (4). Lysosomal phospholipase A2 (LPLA2) has 49% of amino acid sequence identity to LCAT and belongs to the αβ-hydrolase superfamily (3). LPLA2 is highly expressed in alveolar macrophages (5). A marked accumulation of glycerophospholipids and extensive lamellar inclusion bodies, a hallmark of cellular phospholipidosis, are observed in alveolar macrophages in LPLA2−/− mice (6). In addition, older LPLA2−/− mice present similar symptoms to human autoimmune disease (7). Like other lysosomal enzymes, LPLA2 is secreted to extracellular space (8, 9). Recently, LPLA2 activity was found in the aqueous humor as well as ocular tissues in pig eye (10). The LPLA2 found in pig aqueous humor seems to be originated from ocular tissues such as the trabecular meshwork surrounding the anterior chamber and to be associated with clearance of the aqueous humor (10). These observations indicate that LPLA2 may play an important role for the host defense system in body (7) as well as for intracellular phospholipid homeostasis (6).
LPLA2 undergoes posttranslational modifications including a signal peptide cleavage and N-linked glycosylations (3). There are four potential N-glycosylation sites in human lysosomal phospholipase A2 (hLPLA2) molecules. Deglycosylation of enzymatically active LPLA2 by treatment with endoglycosidase F1, which releases high mannose and hybrid oligosaccharides but not complex oligosaccharides from N-linked glycoproteins (11), resulted in no change of the enzyme activity (3). Recombinant mouse LPLA2 is taken up into LPLA2-deficient alveolar macrophages via mannose receptor(s) and translocated to intracellular acidic compartments such as endosome and lysosome (8). This information indicates that N-linked glycosylations of LPLA2 may play a crucial role in the sorting and/or folding of LPLA2 molecules in the cell.
Recently, we observed that gene expression of LPLA2 in Escherichia coli or Sf-9 insect cells results in production of enzymatically inactive LPLA2 (unpublished data). LPLA2 is a glycoprotein containing N-linked high mannose type oligosaccharides (2, 3). Proteins produced in prokaryotic cells such E. coli are not glycosylated in the same way as proteins produced in mammalian cells (12). Unlike E. coli, the insect cell line, Sf-9 cells, can make proteins with posttranslational modifications similar to other eukaryotic cells. However, the N-glycans of mammalian glycoproteins attached in the Sf-9 cells are different from those attached in mammalian cells (13). In some cases, such a difference results in formation of functionally inactive mammalian glycoproteins in the insect cells (13), indicating that abnormal N-linked oligosaccharides of LPLA2 may evoke a partial or complete loss of the activity of physiologically functional LPLA2. Therefore, we have considered that the N-linked glycosylation of LPLA2 is a critical posttranslational modification in the formation of catalytically active LPLA2.
In the present study, to understand the role of N-linked glycosylations of LPLA2, we replaced the four potential N-glycosylation sites of hLPLA2 with an alanine (Ala) residue one by one and expressed mutated or nonmutated hLPLA2 protein in COS-7 cells. The whole cell extracts were tested for LPLA2 activity and immunoreactivity against anti-hLPLA2 antibody.
MATERIALS AND METHODS
Reagents
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), sulfatide, N-acetylsphingosine (NAS), and N-oleoyl-sphingosine were obtained from Avanti Polar Lipids Corp. (Alabaster, AL); high performance thin layer chromatography silica gel plates, 10 × 20 cm, were from Merck (Darmstadt, Germany); N-glycosidase F (PNGase F) and POD immunostain set were from Wako (Tokyo, Japan); anti-rabbit IgG goat polyclonal antibody, HRP-conjugate, was from MP Biomedicals (Solon, OH); anti-lamp1 mouse monoclonal antibody and anti-calnexin rabbit polyclonal antibody were from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antibody against human LPLA2 was generously gifted from Takeda Pharmaceutical Company, Limited.
Transacylase activity of LPLA2
Preparation of liposomes.
DOPC, sulfatide, and NAS were taken in a glass tube and dried down under a stream of nitrogen gas. The dried lipid mixture was dispersed into 50 mM sodium citrate (pH 4.5) by a probe-type sonicator for 8 min in an icy water bath.
Transacylase assay.
The reaction mixture consisted of 48 mM sodium citrate (pH 4.5), 10 μg/ml BSA, 38 μM NAS incorporated into phospholipid liposomes (containing 12.7 μM sulfatide and 127 μM DOPC), and the homogenate or soluble fraction obtained from each transfectant in a total volume of 500 μl. Individual transfected cell soluble fractions were obtained by centrifugation at 150,000 g of individual transfected cell homogenates (1 mg/ml of protein concentration). The reaction was initiated by adding the enzyme source, incubated for 0.5 to 20 min at 37°C, and terminated by adding 3 ml of chloroform/methanol (2:1, v/v) plus 0.3 ml of 0.9% (w/v) NaCl. The mixture was centrifuged at 800 g for 5 min at room temperature. The resultant lower organic layer was transferred into another glass tube and dried down under a stream of nitrogen gas. The dried lipid was dissolved in 40 μl of chloroform/methanol (2:1), applied on a high performance thin layer chromatography plate, and developed in a solvent system consisting of chloroform/acetic acid (90:10, v/v). The plate was dried and soaked in 8% (w/v) CuSO4, 5H2O, 6.8% (v/v) H3PO4, and 32% (v/v) methanol. The uniformly wet plate was briefly dried by a hair dryer and charred for 15 min in a 150°C oven. The plate was scanned and the content of the product (1-O-acyl-NAS) was estimated by National Institutes of Health-ImageJ 1.37v with N-oleoyl-sphingosine as a standard.
Construction of LPLA2 expression plasmids
The plasmid containing subcloned human LPLA2 (hLPLA2) cDNA was generously provided by Takeda Pharmaceutical Company, Limited. The entire open reading frame of hLPLA2 was obtained by polymerase chain reaction (PCR) of the plasmid in the presence of a primer pair, which consisted of the N-terminal primer (forward) having EcoRI site at the N terminus (EcoRI-hLPLA2) and the C-terminal primer (reverse) having XhoI site at the C terminus (hLPLA2-XhoI) (Table 1). The coding sequence of human LPLA2 was excised at EcoRI and XhoI sites from the PCR product. It then was subcloned into the EcoRI and XhoI sites of pcDNA3.
TABLE 1.
Oligonucleotides used for the site-directed mutagenesis of lysosomal phospholipase A2
| Oligonucleotide | Sequence |
| EcoRI-hLPLA2-F | 5′-GTGGAATTCATGGGCCTCCACCTCCGCCCCTAC-3′ |
| hLPLA2-XhoI-R | 5′-TTTATTCTCGAGTCAGGGCCCAAGGAGCAC-3′ |
| N1A-F | 5′-CTGGTTTACGCGAAAACATCC-3′ |
| N1A-R | 5′-GGATGTTTTCGCGTAAACCAG-3′ |
| N2A-F | 5′-CTGCCCTACGCTTACACATGG-3′ |
| N2A-R | 5′-CCATGTGTAAGCGTAGGGCAG-3′ |
| N3A-F | 5′-ACCCACAATCGCTTACACACTGTGGGA-3′ |
| N3A-R | 5′-GCAGTGTGTAAGCGATTGTGGGTGTCT-3′ |
| hLPLA2-N4A-XhoI-R | 5′-TATCTCGAGTCAGGGCCCAAGGAGCACACGTTTCAGATAGGCCAGGGTGGTGGCTGCGGCCAGCAT-3′ |
F and R indicate forward and reverse, respectively. Underlined sequences indicate the restriction endonuclease cleavage sites. Bold sequences mismatch with the template.
Site-directed mutation of the amino acid residues in four N-linked glycosylation sites of hLPLA2 was generated by the overlap extension method (14, 15). The asparagine (Asn) residues in the N-glycosylation sites in hLPLA2 were replaced with an Ala. In brief, the method requires a pair of PCRs to amplify the overlapping fragments and another PCR to fuse the fragments. To obtain the overlapping fragments, amplification primers (Table 1) and the template DNA consisting of the plasmid containing the hLPLA2 gene between EcoRI and XhoI sites were applied to PCR reaction. In the single mutation, 99-Asn to 99-Ala (N1A), 273-Asn to 273-Ala (N2A), 289-Asn to 289-Ala (N3A), and 398-Asn to 398-Ala (N4A), a primer pair consisting of one mutagenic primer (forward) and hLPLA2-XhoI primer or the same mutagenic primer (reverse) and EcoRI-hLPLA2 primer was used with hLPLA2 cDNA plasmid in the first PCR reaction. The single mutated hLPLA2 was obtained by the second PCR reaction using two overlapping fragments and EcoRI-hLPLA2 and hLPLA2-XhoI primers. To construct the single mutation of 398-Asn to 398-Ala (N4A), the PCR was employed with a primer pair of EcoRI-hLPLA2 and one mutagenic primer hLPLA2-XhoI containing N4A (hLPLA2-N4A-XhoI).
The quadruple mutations (Nt), 99-Asn, 273-Asn, 289-Asn, and 398-Asn replaced with alanines, were generated by the PCR reaction of two fragments, obtained from the first PCR reaction of EcoRI-hLPLA2 and N2A reverse primer pair with pcDNA3-hLPLA2-N1A plasmid as template, and N2A forward and hLPLA2-N4A-XhoI primer pair with pcDNA3-hLPLA2-N3A plasmid as template. And the second PCR was employed with a primer pair of EcoRI-hLPLA2 and hLPLA2-N4A-XhoI. Each PCR product was excised at EcoRI and XhoI sites and subcloned into the EcoRI and XhoI sites of pcDNA3. For all constructs, the DNA sequence was confirmed by sequencing in both directions.
Cell culture and transfection
To obtain wild-type (WT) and mutated hLPLA2-overexpressed cells, COS-7 cells were transiently transfected with pcDNA3 containing the entire open reading frame of individual hLPLA2.
COS-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco BRL) supplemented with 10% fetal bovine serum. For transient expression, COS-7 cells were cultured in 6-well plates. When the cells reached 80% of confluence, the cells were transfected with 4 μg purified plasmid using Lipofectamine 2000 (Invitrogen) in 1 ml Opti-MEM medium (Gibco BRL). One milliliter DMEM containing 20% fetal bovine serum was added after 4 h incubation at 37°C, 5% CO2. Twenty-four hours after transfection, the cells were washed three times with 2 ml of PBS, scraped with 2 ml 0.25 M sucrose, 10 mM HEPES (pH 7.4), and 1 mM EDTA and transferred into an plastic tube. The following procedures were carried out at 4°C. The cells were collected by centrifuge at 800 g for 10 min. Each cell pellet was suspended into 0.5 ml of 0.25 M sucrose, 10 mM HEPES, and 1 mM EDTA (pH 7.4) and dispersed by a probe-type sonicator for 10 s × 4 at 0°C. In the present study, the protein concentration of each homogenate was adjusted to 1 mg/ml with the same sucrose buffer.
Immunoblotting
The cell homogenate and soluble fraction were precipitated by the method of Bensadoun and Weinstein (16). The resultant pellet was dissolved with 30 μl of loading buffer plus 1.5 μl of 2 M Tris for SDS polyacrylamide gel electrophoresis. Proteins were separated using a 12% SDS polyacrylamide gel and transferred to a polyvinylidene difluoride membrane using the iBlot Gel Transfer System (Invitrogen). The membrane was incubated with an anti-human LPLA2 rabbit antibody. The antigen-antibody complex on the membrane was visualized with an anti-rabbit IgG HRP-conjugated goat antibody using the POD immunostain set including NADH, nitrotetrazolium blue, and hydrogen peroxide.
RESULTS
N-glycosylation sites and mutation
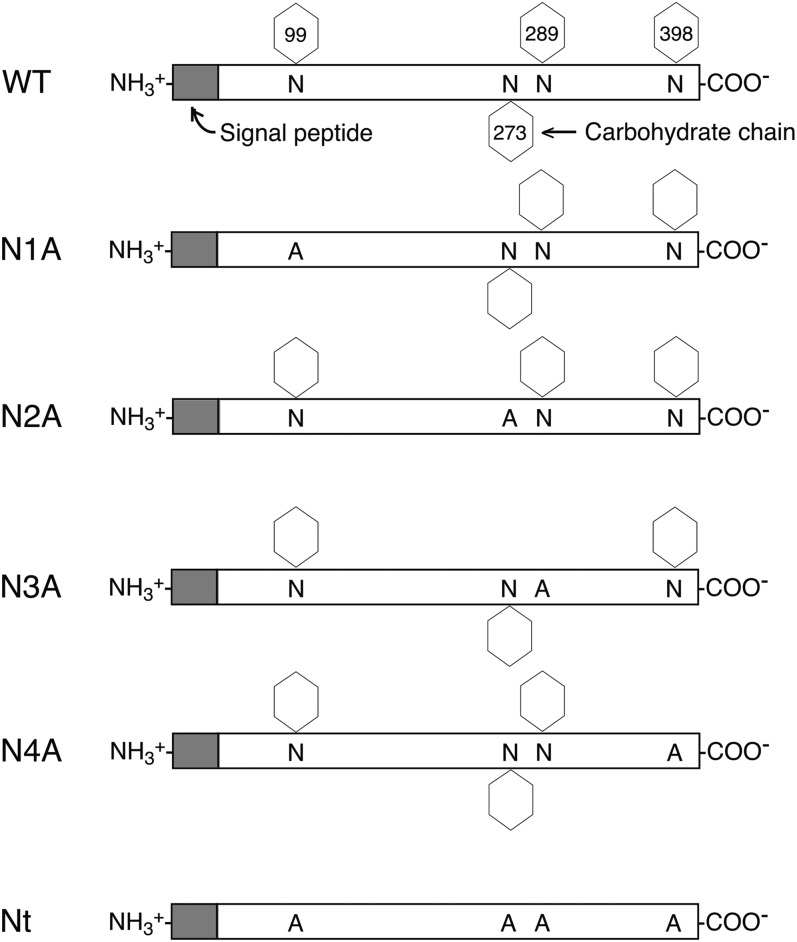
hLPLA2 has four potential N-glycosylation sites, at 99-Asn, 273-Asn, 289-Asn, and 398-Asn (Fig. 1). In the present study, these four Asn residues were individually replaced with an Ala residue by site-directed mutagenesis. In addition, all four N-glycosylation sites were modified with Ala residues. WT and mutated hLPLA2s were transiently expressed in COS-7 cells, which were used to examine the LPLA2 activity and cellular expression. Here, we use abbreviations, N1A, N2A, N3A, N4A, Nt, and WT as 99-Asn, 273-Asn, 289-Asn, 398-Asn, all four-Asn mutated hLPLA2s, and WT hLPLA2, respectively.
Fig. 1.
Schematic diagram of the human LPLA2 showing N-glycosylation sites and mutations. hLPLA2 has four potential N-glycosylation sites, Asn-99, -273, -289, and -398. Site-directed mutations of the Asn residues in the glycosylation sites of hLPLA2 were generated by the overlap extension method. COS-7 cells were transfected with purified plasmid using Lipofectamine PlusTM in Opti-MEM medium. The mutants, each missing a different N-glycosylation site, are numbed from the N terminus as N1A–N4A. Nt denotes the four glycosylation site mutant. WT shows hLPLA2, including a signal peptide and N-glycosylation sites, predicted from the cDNA of hLPLA2.
LPLA2 activity and expression of mutated hLPLA2 in COS-7 cells
LPLA2 has dual enzyme activities, transacylase and phospholipase A2 activities (1–3). The transacylation of short chain ceramide, NAS, by LPLA2 in the presence of glycerophospholipid as acyl group donor is a specific reaction under acidic conditions (3, 6, 9). Therefore, such a transacylation reaction has been used to detect the LPLA2 activity in the cell, tissue, and extracellular fluid (3, 6, 9, 10). In the present study, to measure the transacylase and phospholipase A2 activity of LPLA2 in the transfectants, DOPC/sulfatide/NAS liposomes were incubated with cell extracts obtained from individual transfectants under acidic conditions. The transacylase and phospholipase A2 activities were determined by the amounts of 1-O-acyl-NAS formed and fatty acid released, respectively, over time. LPLA2 expression was evaluated by Western blotting of the cell extracts using anti-hLPLA2-rabbit polyclonal antibody.
LPLA2 activity (transacylase and phospholipase A2 activities).
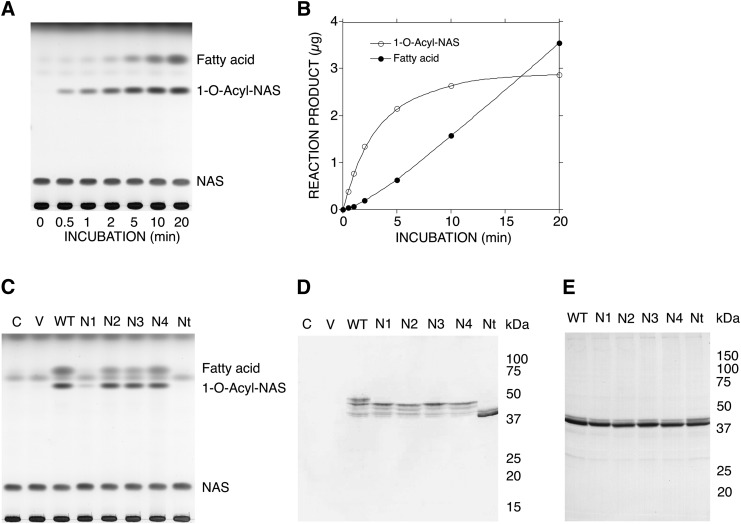
Figure 2A, B shows a representative sample of the kinetics of the formation of 1-O-acyl-NAS and release of fatty acid by the homogenate (2.5 μg of protein/assay) obtained from WT gene transfectants. As reported previously (1), in this reaction, the formation of 1-O-acyl-NAS preceded the release of fatty acid, and reached a plateau. In the reaction conditions shown in Fig. 2C, 20 μg protein of each homogenate was used for the assay. There was no significant formation of 1-O-acyl-NAS or release of fatty acid by the cell homogenate of COS-7 cells transfected with or without pcDNA expression vector and was considered the background control. About 3 μg of 1-O-acyl-NAS was produced when the cell homogenates obtained from WT, N2A, N3A, and N4A gene transfectants were used (Fig. 2C). According to the results shown in Fig. 2A, B, the formation of 1-O-acyl-NAS by those homogenates in Fig. 2C must reach a plateau. However, the fatty acid released in the same reaction increased in the order of N3A < N2A < N4A < WT gene transfectants. By contrast, the cell homogenate obtained from N1A gene transfectants showed marked reduction of both 1-O-acyl-NAS formation and fatty acid release as compared with those obtained from WT transfectants. In addition, the cell homogenate obtained from Nt gene transfectants showed no induction of either enzyme activity.
Fig. 2.
LPLA2 activity and LPLA2 protein of mutated hLPLA2. A: In the time course study of LPLA2 activity of WT, 2.5 μg protein of the cell homogenate prepared from WT gene transfectants was incubated for 0 to 20 min at 37°C with liposomes containing NAS as described in the Materials and methods. B: Reaction products, 1-O-acyl-NAS and fatty acid, produced by WT in (A) were plotted against the incubation time. C: In LPLA2 assay, 20 μg protein of each cell homogenate (1 mg protein/ml) was incubated for 20 min at 37°C with the same liposomes as described in (A). D: In Western blotting, 40 μg of protein in each cell homogenate was separated by SDS polyacrylamide gel electrophoresis and subjected to immunoblotting with an anti-hLPLA2 rabbit antibody, and WT and mutated hLPLA2s were visualized as described in the Material and Methods. C and V denote control and expression vector, respectively. E: Forty micrograms of protein of each cell homogenate were treated with PNGase F according to the manufacture's information. The positive proteins against anti-LPLA2 antibody were visualized as described in the Material and Methods.
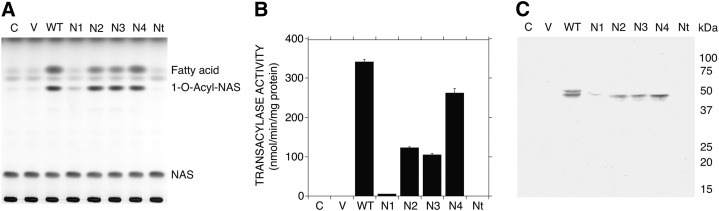
Generally, more than 85% of the total LPLA2 activity in cell homogenate is recovered in the soluble fraction (1). In this study, the LPLA2 activity of the soluble fractions from cells transfected with vectors containing various N-glycosylation site mutations was investigated. The soluble fraction of individual transfectants was obtained by centrifuging at 150,000 g the cell homogenates with the protein concentration of 1 mg/ml. The soluble fraction of individual transfectants showed similar transacylase and phospholipase A2 activities as observed in its homogenate (Fig. 2C and Fig. 3A). In addition, the initial velocity of the transacylase activity in a specific volume of each soluble fraction was measured to determine the specific activity of LPLA2 for each soluble fraction (Fig. 3B). The specific activities of soluble fractions obtained from N2A, N3A, and N4A gene transfectants showed 37, 32, and 82%, respectively, of that obtained from WT gene transfectants. By contrast, the specific activity of soluble fraction obtained from N1A gene transfectants was much lower (2% of WT) than that obtained from WT gene transfectants. In addition, the soluble fraction of Nt gene transfectants showed similar specific activity to the soluble fractions obtained from the control cells and the cells transfected with a control vector.
Fig. 3.
LPLA2 activity and LPLA2 protein of the soluble fraction obtained from WT and mutated hLPLA2s gene transfectants. The soluble fractions of individual transfectants were obtained by centrifugation for 1 h at 150,000 g of their cell homogenates (1 mg of protein/ml) at 4°C. A: In LPLA2 assay, 20 μl of each soluble fraction was incubated for 20 min at 37°C with liposomes containing NAS as described in the Material and Methods. B: To obtain the initial velocity of each fraction, the reaction time was adjusted in each assay. Error bars indicate SD (n = 3). C: In Western blotting, 40 μl of each soluble fraction was separated by SDS polyacrylamide gel electrophoresis and subjected to immunoblotting with an anti-hLPLA2 rabbit antibody, and WT and mutated hLPLA2s were visualized as described in the Material and Methods. C and V denote control and expression vector, respectively.
Immunoblotting.
The hLPLA2 expression was confirmed for all transfectants by immunoblotting using anti-LPLA2 antibody (Fig. 2D). In all transfectants, multiple bands immunoreactive against the antibody were observed. The degree of the expression of the total immunoreactive protein produced in COS-7 cells transfected with hLPLA2 or mutated hLPLA2 gene was similar between transfectants. The cell homogenates obtained from N1A, N2A, N3A, and N4A gene transfectants showed similar multiple bands with molecular mass between 42 and 45 kDa. In addition, the WT gene transfectant had an additional higher molecular band at about 48 kDa. By contrast, the Nt transfectant provided a clear dense band and a second faint band that corresponded to molecular sizes of 40 and 42 kDa, respectively. Furthermore, PNGase F treatment of cell homogenates from the single mutation transfectants or Nt transfectant resulted in formation of identical LPLA2 proteins with molecular sizes between 40 and 42 kDa (Fig. 2E), a size similar to the untreated cell homogenate obtained from Nt gene transfectants (Fig. 2D).
The immunoblotting pattern obtained from the soluble fractions prepared in Fig. 3 was quite different from that obtained from the homogenates in Fig. 2D (Fig. 3C). The molecular size of the soluble form of N1A, N2A, N3A, and N4A was 45 kDa. The soluble forms of WT were 45 and 48 kDa. Some multiple protein bands immunoreactive against anti-LPLA2 antibody with smaller molecular mass found in the homogenates obtained from WT, N1A, N2A, N3A, and N4A gene transfectants were hardly detected in their soluble fractions. The intensity of the 45 kDa band in the soluble fraction obtained from N1A gene transfected cell homogenate was greatly reduced (Fig. 3C). In addition, the quadruple mutation resulted in complete loss of the immunoreactive protein band against anti-LPLA2 antibody in the soluble fraction (Fig. 3C, Nt).
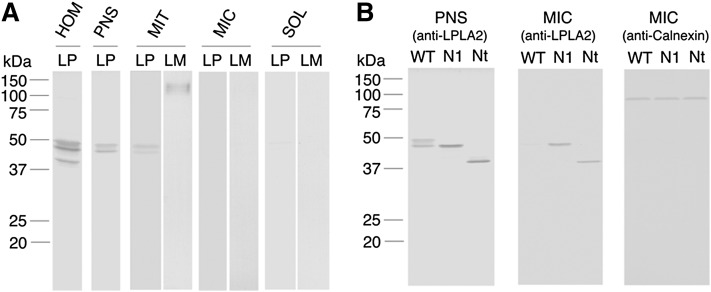
A subcellular fractionation study was carried out using differential centrifugation. As shown in Fig. 4A, LPLA2 proteins recovered in the post nuclear supernatant (PNS) prepared from WT gene transfectants were high-molecular forms (45 and 48 kDa) and distributed in the crude mitochondrial fraction (Fig. 4A), but not in the microsomal or soluble fractions (Fig. 4B). The mitochondrial fraction was positive against anti-lamp1 (lysosomal marker) antibody (Fig. 4A). By contrast, a substantial amount of LPLA2 was found in the microsomal fraction prepared from N1A and Nt gene transfectants, which was a positive fraction against anti-calnexin [endoplasmic reticulum (ER) marker] antibody (Fig. 4B).
Fig. 4.
Subcellular fractionation of WT and mutated hLPLA2 gene transfectants by differential centrifugation. All manipulations in the subcellular fractionation were carried out at 4°C. One milliliter of cell homogenate (1 mg protein/ml) obtained from WT, N1A, or Nt gene transfectants was centrifuged for 10 min at 600 g. The resultant supernatant and pellet were collected as post nuclear supernatant (PNS) and nuclear fraction, respectively. The PNS was centrifuged for 10 min at 15,000 g. The resultant pellets were collected as crude mitochondrial fraction (MIT). The supernatant obtained at 15,000 g was centrifuged for 1.5 h at 160,000 g. The resultant supernatant and pellet were collected as soluble fraction (SOL) and microsomal fraction (MIC), respectively. The pellet of MIT and MIC was dispersed with 1 ml of 0.25 M sucrose, 10 mM HEPES (pH 7.4), and 1 mM EDTA. Twenty microliters of the cell homogenate (HOM), PNS, MIT, and MIC suspensions, and SOL was used to assure the distribution of LPLA2 in the cell. SDS polyacrylamide gel electrophoresis and Western blotting were carried out as described in the Materials and Methods. In (A), distribution of WT in the cell was examined by treatment of the membrane with anti-hLPLA2 antibody (LP) or anti-lamp1 antibody (LM). In (B), N1A and Nt were found in MIC. The left and middle membranes were treated with anti-hLPLA2 antibody. The right membrane was treated with anti-calnexin antibody.
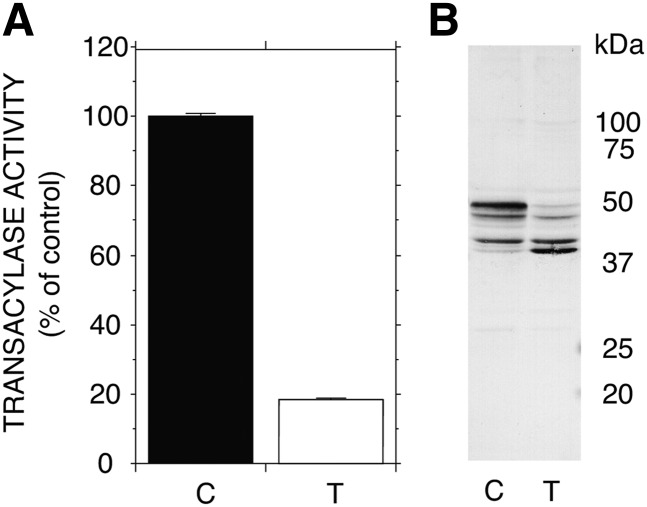
Tunicamycin treatment of WT LPLA2-transfected cells
Tunicamycin is known to inhibit the synthesis of N-linked glycoproteins (17). The COS-7 cells transiently transfected with WT gene were treated with or without 5 μM tunicamycin for 24 h. After the treatment, the cell homogenate was prepared as described in the Materials and Methods. The transacylase activity of the cell homogenate was markedly reduced by the treatment with tunicamycin (Fig. 5A). The immunoblot analysis showed that the cell homogenate obtained from the transfectants treated with tunicamycin had a greatly reduced amount of 45 and 48 kDa forms of hLPLA2 and gained 40 and 42 kDa forms of hLPLA2 (Fig. 5B).
Fig. 5.
Treatment of COS-7 cells transiently transfected with hLPLA2 gene with tunicamycin. COS-7 cells were transiently transfected with hLPLA2 gene for 48 h at 37°C. The transfectants were treated with or without 5 μM of tunicamycin for 24 h at 37°C. A: In LPLA2 assay, 10 μg protein of each cell homogenate was incubated for 1.5–5 min at 37°C with liposomes containing NAS as described in the Material and Methods. Error bars indicate SD (n = 3). C and T denote control (without tunicamycin) and tunicamycin, respectively. B: In Western blotting, 30 μg of protein of each homogenate was separated by SDS polyacrylamide gel electrophoresis, subjected to immunoblotting with an anti-hLPLA2 rabbit antibody, and visualized as described in the Material and Methods.
DISCUSSION
In the present study, N-glycosylation sites of hLPLA2 were modified by site-directed mutagenesis. Four individual hLPLA2 mutants in which N-linked Asn residues were individually replaced with an Ala residue, and one hLPLA2 mutant where all four Asn residues were replaced with Ala residues, were examined (Fig. 1). As shown in Fig. 2C, the formation of 1-O-acyl-NAS and release of fatty acid by the homogenate obtained from COS-7 cells transfected with empty vector was extremely low, which was similar to the homogenate obtained from COS-7 cells. As noted above, LPLA2 has dual enzyme activity, i.e., phospholipase A2 and transacylase (1, 2). As expected, the cell homogenate obtained from WT gene transfectants showed both transacylase and phospholipase A2 activities (Fig. 2A, C). In addition, the cell homogenates obtained from N2A, N3A, and N4A gene transfectants showed the transacylase and phospholipase A2 activities similar to WT. For WT, N2A, N3A, and N4A, the formation of 1-O-acyl-NAS by the cell homogenates containing N2A, N3A, N4A, and WT reached a plateau in the tested conditions (Fig. 2C), but the release of fatty acid was different between the homogenates. It was found that the formed 1-O-acyl-NAS is converted to NAS and fatty acid by the hydrolysis via LPLA2 (18). Thus, a certain equilibrium between formation and degradation of 1-O-acyl-NAS could be attained during the reaction. As shown in our previous studies (1, 2), the transacylation reaction by LPLA2 was found to reach the plateau much faster than the phospholipase A2 reaction by LPLA2 (Fig. 2A, B). The phospholipase A2 activity resulting from the fatty acid released by cell homogenates containing N2A and N3A was lower than that of WT (Fig. 2C), indicating that the LPLA2 activity of the cell homogenates containing N2A and N3A is lower than that of WT. By contrast, the apparent LPLA2 activities of the homogenates obtained from N1A and Nt gene transfectants were markedly reduced and lost, respectively (Fig. 2C). Thus the phospholipase A2 activity found in those homogenates is shown in order of WT, N4A, N2A, N3A, N1A, Nt from the highest to the lowest. In addition, immunoblotting revealed that the degree of the total LPLA2 expression in individual transfectants is similar among all transfectants (Fig. 2D). These results indicate that the formation of catalytically active LPLA2 is affected by N-glycosylation of the enzyme.
The N1, N2, N3, and N4 proteins contained at least three N-glycosylation sites and showed similar multiple bands of molecular size between 42 and 45 kDa (Fig. 2D). WT showed an extra band with molecular size of 48 kDa, which may be due to the presence of four potential N-linked glycosylation sites. Nt that lacked all four of N-glycosylation sites showed several LPLA2 bands between 40 and 42 kDa (Fig. 2D), which are nonglycosylated forms. To further evaluate the influence of glycosylation on protein molecular mass, cell homogenates were treated with PNGase F. PNGase F cleaves carbohydrate chains linked to Asn residues between the innermost GlcNAc and Asn of high mannose, hybrid, and complex oligosaccharides from N-linked glycoproteins (19). Treating with PNGase resulted in LPLA2 formation of the same molecular size as Nt (Fig. 2D, E). In addition, the treatment of the homogenate containing WT with endo-glycosidase F1 led to the formation of multiple LPLA2 protein molecules compared with PGNase F (data not shown). Endo-glycosidase F1 cleaves carbohydrate chains linked to Asn residues between the innermost GlcNAc and Asn of high mannose and hybrid oligosaccharides but not complex oligosaccharides (11). These results indicate that the multiple molecular forms found in the cell homogenate obtained from WT, N1A, N2A, N3A, and N4A gene transfectants are due to the difference of N-linked oligosaccharides as evidenced by the pattern of PNGase F and endo-glycosidase F1 treatments. The cell extracts obtained from Nt transfectants and single point mutations treated with PGNase F contained the same multiple forms of LPLA2 in spite of a lack of N-linked glycosylation sites (Fig. 2D, E). The main protein form in those extracts has apparent molecular size of 40 kDa (Fig. 2D, E). According to the primary amino acid sequence of hLPLA2, the molecular size of hLPLA2 protein core is 43 kDa without a signal peptide or any posttranslational modifications. This implies that hLPLA2 takes other posttranslational modifications such as proteolytic cleavage.
Interestingly, the majority of the LPLA2 activity is found in the soluble fractions of whole cell extracts obtained from cells transfected with the various vectors (Figs. 2C, 3A). The immunoblot pattern of individual transfected cell soluble fractions provided a strong correlation with the specific activity pattern of the transacylase activity in the soluble fractions (Fig. 3B, C). Namely, the LPLA2 proteins recovered in the soluble fraction are catalytically active forms as well as soluble glycosylated forms. By contrast, the insoluble glycosylated and nonglycosylated LPLA2 proteins have lower molecular sizes than the soluble glycosylated LPLA2 proteins (Figs. 2D, 3C) and appear to aggregate and bind to the membrane, irreversibly. Unlike N2A, N3A, N4A, and WT, the N1A protein is almost catalytically inactive and is mostly recovered in the membrane fraction. This means that the N-glycosylation at 99-Asn of hLPLA2 is critical for the production of catalytically active LPLA2. In addition, the subcellular fractionation study of WT, N1A, and Nt gene transfectants (Fig. 4) indicates that WT is steadily targeted to lysosome in the cell, and the hLPLA2s mutated at the 99-Asn are steadily retained in the ER but WT is not. There is a hydrophobic domain (72-tyrosin to 94-isoleucine) in the N-terminal region of LPLA2 molecule (15). N-glycans are hydrophilic and affect physicochemical properties of glycoproteins such as molecular size, solubility, and electrical charge (20). The 99-Asn is in the proximity of the N-terminal hydrophobic domain of the LPLA2 molecule. Thus, a lack of the carbohydrate chain at the 99-Asn probably increases the hydrophobicity of the N-terminal hydrophobic domain region, and evokes aggregation of LPLA2 and the nonspecific binding of LPLA2 to the membrane via the hydrophobic interaction. Thus, the N-glycan linked to 99-Asn of hLPLA2 may have a crucial role in protecting hLPLA2 from irreversible aggregation and providing hLPLA2 a proper folding and sorting pathway to form native conformation in the ER. As expected, the treatment of WT-transfected cells with tunicamycin, which is a known inhibitor of glycoprotein synthesis (17), resulted in a marked loss of the catalytically active forms (45 and 48 kDa forms), and accumulation of the catalytically inactive nonglycosylated forms (40 and 42 kDa forms) of hLPLA2 in the cells (Fig. 5). These results further support the idea that the N-linked glycosylation of LPLA2 is indispensable for the formation of enzymatically active LPLA2 in the cell.
In mammalian cells, N-glycosylation is a posttranslational modification proceeded in the ER and Golgi apparatus. The quality control of glycoprotein occurs in the ER and is carried out by the control of glycoprotein folding via chaperons, ER-associated degradation of misfolded glycoproteins linked to the ubiquitin-proteasome system and ER-derived quality control compartment responsible for the trimming of oligosaccharides (21). It is necessary for newly synthesized glycoproteins to acquire their native protein conformations. Improperly modified carbohydrate chain(s) of glycoprotein results in failure of the folding and sorting of the protein, in which the proteins misfolded in the ER cannot be translocated from the ER to the Golgi apparatus because of formation of insoluble aggregates in the ER (21). N-linked glycosylation of lysosomal lipolytic enzymes such as acid ceramidase and lysosomal acid lipase (LAL) has been reported (22, 23). Human acid ceramidase has the six individual potential N-glycosylation sites. The individual elimination of sites 1, 3, and 5 resulted in abolishing the catalytic activity. Human LAL also has the six individual potential N-glycosylation sites. The single mutation of two sites resulted in inactive LAL but not that of other sites. These results suggest that these lysosomal lipolytic enzymes have a crucial N-linked glycosylation site(s) to form the functional enzyme in the cell. There are four potential N-glycosylation sites in hLPLA2. Although the physiological role of N-glycosylation of LPLA2 still remains unknown, the present findings indicate that the N-glycan at the 99-Asn may function as a determinant in the folding, sorting, and quality control of hLPLA2 in the ER.
In conclusion, the present study showed that the N-glycan at 99-Asn of hLPLA2 is more important than other N-glycans of hLPLA2 for production and localization of catalytically active LPLA2 in the cell, suggesting that individual N-linked oligosaccharide chains of LPLA2 have a different function or role. The mutation of any N-linked glycosylation sites of LPLA2 reduces producing enzymatically active LPLA2 in the cell. The recent study revealed that older LPLA2-null mice show lymphoid hypertrophy, glomerulonephritis, and abnormal serologies including anti-dsDNA antibody, positive anti-nuclear antibodies, and high circulation of immunoglobulin levels, findings that resemble systemic lupus erythematosus (6). Thus, a chronic reduction of LPLA2 activity by the mutation of N-glycosylation sites of LPLA2 in vivo may induce the same or a similar phenotype as occurred in LPLA2-deficient mice.
Acknowledgments
The authors would like to thank Dr. Nishino for his valuable comments and encouragement of this study, and Dr. Robert Kelly for his kindly reading and careful advice of the manuscript.
Footnotes
Abbreviations:
- Ala
- alanine
- Asn
- asparagine
- hLPLA2
- human lysosomal phospholipase A2
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- ER
- endoplasmic reticulum
- LAL
- lysosomal acid lipase
- LPLA2
- lysosomal phospholipase A2
- N1A
- 99-Asn to 99-Ala single mutation
- N2A
- 273-Asn to 273-Ala single mutation
- N3A
- 289-Asn to 289-Ala single mutation
- N4A
- 398-Asn to 398-Ala single mutation
- NAS
- N-acetylsphingosine
- Nt
- quadruple mutation (99-Asn, 273-Asn, 289-Asn, and 398-Asn replaced with alanines)
- PNGase F
- N-glycosidase F
- WT
- wild-type
REFERENCES
- 1.Abe A., Shayman J. A., Radin N. S. 1996. A novel enzyme that catalyzes the esterification of N-acetylsphingosine. Metabolism of C2-ceramides. J. Biol. Chem. 271: 14383–14389 [DOI] [PubMed] [Google Scholar]
- 2.Abe A., Shayman J. A. 1998. Purification and characterization of 1-O-acylceramide synthase, a novel phospholipase A2 with transacylase activity. J. Biol. Chem. 273: 8467–8474 [DOI] [PubMed] [Google Scholar]
- 3.Hiraoka M., Abe A., Shayman J. A. 2002. Cloning and characterization of a lysosomal phospholipase A2, 1-O-acylceramide synthase. J. Biol. Chem. 277: 10090–10099 [DOI] [PubMed] [Google Scholar]
- 4.Schaloske R. H., Dennis E. A. 2006. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta. 1761: 1246–1259 [DOI] [PubMed] [Google Scholar]
- 5.Abe A., Hiraoka M., Wild S., Wilcoxen S. E., Paine R., 3rd, Shayman J. A. 2004. Lysosomal phospholipase A2 is selectively expressed in alveolar macrophages. J. Biol. Chem. 279: 42605–42611 [DOI] [PubMed] [Google Scholar]
- 6.Hiraoka M., Abe A., Lu Y., Yang K., Han X., Gross R. W., Shayman J. A. 2006. Lysosomal phospholipase A2 and phospholipidosis. Mol. Cell. Biol. 26: 6139–6148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shayman J. A. 2009. Lysosomal phospholipase A2 and phospholipidosis. Abstract in 50th International Conference on the Bioscience of Lipids. Elsevier, Regensburg, Germany.
- 8.Abe A., Kelly R., Kollmeyer J., Hiraoka M., Lu Y., Shayman J. A. 2008. The secretion and uptake of lysosomal phospholipase A2 by alveolar macrophages. J. Immunol. 181: 7873–7881 [DOI] [PubMed] [Google Scholar]
- 9.Abe A., Kelly R., Shayman J. A. 2010. The measurement of lysosomal phospholipase A2 activity in plasma. J. Lipid Res. 51: 2464–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abe A., Hiraoka M., Inatomi S., Ohguro I., Ohguro H. 2012. Lysosomal phospholipase A2 activity in pig aqueous humor. Invest. Ophthalmol. Vis. Sci. 53: 152–156 [DOI] [PubMed] [Google Scholar]
- 11.Trimble R. B., Tarentino A. L. 1991. Identification of distinct endoglycosidase (endo) activities in Flavobacterium meningosepticum: endo F1, endo F2, and endo F3. Endo F1 and endo H hydrolyze only high mannose and hybrid glycans. J. Biol. Chem. 266: 1646–1651 [PubMed] [Google Scholar]
- 12.Moens S., Vanderleyden J. 1997. Glycoproteins in prokaryotes. Arch. Microbiol. 168: 169–175 [DOI] [PubMed] [Google Scholar]
- 13.Altmann F., Staudacher E., Wilson I. B., Marz L. 1999. Insect cells as hosts for the expression of recombinant glycoproteins. Glycoconj. J. 16: 109–123 [DOI] [PubMed] [Google Scholar]
- 14.Horton R. M., Ho S. N., Pullen J. K., Hunt H. D., Cai Z., Pease L. R. 1993. Gene splicing by overlap extension. Methods Enzymol. 217: 270–279 [DOI] [PubMed] [Google Scholar]
- 15.Hiraoka M., Abe A., Shayman J. A. 2005. Structure and function of lysosomal phospholipase A2: identification of the catalytic triad and the role of cysteine residues. J. Lipid Res. 46: 2441–2447 [DOI] [PubMed] [Google Scholar]
- 16.Bensadoun A., Weinstein D. 1976. Assay of proteins in the presence of interfering materials. Anal. Biochem. 70: 241–250 [DOI] [PubMed] [Google Scholar]
- 17.Duksin D., Mahoney W. C. 1982. Relationship of the structure and biological activity of the natural homologues of tunicamycin. J. Biol. Chem. 257: 3105–3109 [PubMed] [Google Scholar]
- 18.Abe A., Hiraoka M., Shayman J. A. 2007. The acylation of lipophilic alcohols by lysosomal phospholipase A2. J. Lipid Res. 48: 2255–2263 [DOI] [PubMed] [Google Scholar]
- 19.Maley F., Trimble R. B., Tarentino A. L., Plummer T. H., Jr 1989. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180: 195–204 [DOI] [PubMed] [Google Scholar]
- 20.Sola R. J., Griebenow K. 2009. Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 98: 1223–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellgaard L., Helenius A. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4: 181–191 [DOI] [PubMed] [Google Scholar]
- 22.Ferlinz K., Kopal G., Bernardo K., Linke T., Bar J., Breiden B., Neumann U., Lang F., Schuchman E. H., Sandhoff K. 2001. Human acid ceramidase: processing, glycosylation, and lysosomal targeting. J. Biol. Chem. 276: 35352–35360 [DOI] [PubMed] [Google Scholar]
- 23.Zschenker O., Bahr C., Hess U. F., Ameis D. 2005. Systematic mutagenesis of potential glycosylation sites of lysosomal acid lipase. J. Biochem. 137: 387–394 [DOI] [PubMed] [Google Scholar]