Abstract
Although numerous genes are known to regulate serum lipid traits, identified variants explain only a small proportion of the expected heritability. We intended to identify further genetic variants associated with lipid phenotypes in a self-contained population of Sorbs in Germany. We performed a genome-wide association study (GWAS) on LDL-cholesterol, HDL-cholesterol (HDL-C), and triglyceride (TG) levels in 839 Sorbs. All single-nucleotide polymorphisms with a P value <0.01 were subjected to a meta-analysis, including an independent Swedish cohort (Diabetes Genetics Initiative; n = ∼3,100). Novel association signals with the strongest effects were subjected to replication studies in an additional German cohort (Berlin, n = 2,031). In the initial GWAS in the Sorbs, we identified 14 loci associated with lipid phenotypes reaching P values <10−5 and confirmed significant effects for 18 previously reported loci. The combined meta-analysis of the three study cohorts (n(HDL) = 6041; n(LDL) = 5,995; n(TG) = 6,087) revealed a novel association for a variant in THOC5 (rs8135828) with serum HDL-C levels (P = 1.78 × 10−7; Z-score = −5.221). Consistently, the variant was also associated with circulating APOA1 levels in Sorbs. The small interfering RNA-mediated mRNA silencing of THOC5 in HepG2 cells resulted in lower mRNA levels of APOA1, SCARB1, and ABCG8 (all P < 0.05). We propose THOC5 to be a novel gene involved in the regulation of serum HDL-C levels.
Keywords: single nucleotide polymorphism, genome-wide-association study, mRNA silencing
There is strong evidence for the heritability of lipid traits and the genetic risk of cardiovascular diseases (1–6). During the last two decades, genetic linkage studies have dominated efforts aimed at identifying genes and polymorphisms that contribute to susceptibility for common diseases (7). However, current advances in high-throughput genotyping technologies have resulted in the recent discovery of numerous loci potentially affecting variability in complex traits, including total cholesterol, LDL-cholesterol (LDL-C), HDL-cholesterol (HDL-C), and/or triglycerides (TGs). More than 100 loci-influencing lipid phenotypes have been identified in genome-wide association studies (GWASs) and replicated in independent cohorts (1–6, 8–11). Well-powered GWASs, combined with fine mapping of the susceptibility loci, were able to detect even variants with small effect size (11–14). Nevertheless, only 10–12% of the total variance for plasma lipid traits can be explained by these variants (5). Newly developed statistical approaches, family-based studies, or epigenetic analyses might facilitate the discovery of further novel variants affecting complex polygenic traits. Recently, fine mapping efforts in genes for LDL-C identified by GWAS revealed genetic variants doubling the explained heritability (15). Moreover, focusing on special populations with reduced genetic heterogeneity as well as phenotypic complexity and homogenous environmental background might help to detect new allelic or haplotypic associations (16, 17). Therefore, we attempted to identify novel variants associated with lipid phenotypes using a self-contained population of Sorbs from Eastern Germany.
We performed a GWAS on LDL-C, HDL-C, and TG levels in 839 Sorbs. All single-nucleotide polymorphisms (SNPs) with a P value <0.01 were subjected to a meta-analysis, including an independent Swedish cohort [Diabetes Genetics Initiative (DGI); n = ∼3,100]. Novel association signals with the strongest effects were subjected to replication studies in an additional German cohort (Berlin, n = 2,031). The most promising candidate gene, THOC5, was significantly associated with HDL-C in a combined meta-analysis including all three study populations and has been further investigated in in vitro analyses to elucidate its potential functional role in HDL-C metabolism.
MATERIALS AND METHODS
Subjects and phenotyping
Sorbs.
All subjects are part of a sample from an extensively clinically characterized population from Eastern Germany, the Sorbs (18–20). From the 1,000 Sorbs individuals who are enrolled in the cohort, 839 subjects without lipid-lowering drugs (499 women and 340 men) were included in the present study. The individuals had a mean age of 46 ± 16 years and a mean body mass index (BMI) of 26.6 ± 4.9 kg/m2 (data are given as arithmetic means ± SD). Further clinical characteristics of the study subjects are provided in Table 1.
TABLE 1.
Clinical characteristics of the Sorbs
| Trait | Minimum | Maximum | Mean ± SD |
| Age, yr | 18 | 88 | 46 ± 16 |
| BMI, kg/m2 | 15.4 | 50.3 | 26.6 ± 4.9 |
| Total cholesterol, mmol/l | 2.45 | 10.10 | 5.34 ± 1.04 |
| HDL-C, mmol/l | 0.70 | 3.47 | 1.64 ± 0.40 |
| LDL-C, mmol/l | 0.72 | 7.23 | 3.39 ± 0.98 |
| Triglycerides, mmol/l | 0.32 | 8.03 | 1.28 ± 0.83 |
| Waist, cm | 59.0 | 139.0 | 89.9 ± 13.8 |
| Hip, cm | 70.0 | 146.0 | 103.5 ± 9.1 |
The Sorbs cohort included 499 women and 340 men. BMI, body mass index; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; WHR, waist-to-hip ratio.
All parameters were measured in fasting blood samples. Total serum cholesterol and TG concentrations were measured by standard enzymatic methods (CHOD-PAP and GPO-PAP; Roche Diagnostics). Serum LDL-C and HDL-C concentrations were determined with commercial homogeneous direct measurement methods (Roche Diagnostics). All assays were performed in an automated clinical chemistry analyzer (Hitachi/Roche Diagnostics) at the Institute of Laboratory Medicine, University Hospital Leipzig.
The study was approved by the ethics committee of the University of Leipzig, Leipzig, Germany. All subjects provided written informed consent.
DGI.
The DGI is a collaboration of the Broad Institute of MIT and Harvard, Lund University, and Novartis Institutes for BioMedical Research. To identify inherited risk factors that predispose to type 2 diabetes (T2D), a genome-wide association study (GWAS) involving approximately 3,000 individuals from Scandinavia was performed using the 500K Affymetrix GeneChip. The initiative studied 1,464 patients with T2D and 1,467 control subjects from Finland and Sweden, each characterized for 18 clinical traits, among them anthropometric measures, lipids and apolipoproteins, and blood pressure (3,171 individuals for HDL-C, 3,125 for LDL-C, and 3,217 for triglycerides). The samples were population based (1,022 T2D cases and 1,075 euglycemic control subjects, matched on gender, age, BMI, and region of origin) and family based (326 sibships discordant for T2D; 442 cases and 392 euglycemic control subjects). Genotyping of 500,568 SNPs was attempted in each sample. The overall call rate for passing SNPs was 99.2%. After filtering rare and monomorphic variants and applying stringent quality-control filters, high-quality genotypes for 386,731 common SNPs were obtained (4).
In DGI, total cholesterol, HDL-C, and TG levels were measured in fasting blood samples drawn at the baseline examination for each study according to standard enzymatic methods (2). LDL-C levels were calculated according to Friedewald's formula, and missing values were assigned to individuals with triglycerides >400 mg/dl (2).
Berlin cohort (Metabolic Syndrome Berlin Potsdam).
This replication set was recruited as a cross-sectional study with focus on traits of the metabolic syndrome. Participants were recruited by paper advertisements and by the outpatient clinic of the German Institute of Human Nutrition and the Department of Endocrinology at the Charité-University Clinic, Berlin, Germany. All measurements and analyses were standardized across recruitment centers. Anthropometry included weight, height, waist and hip circumference, and skin fold measurements. Blood pressure was determined after at least 15 min of rest in a sitting position. Lipid parameters included total cholesterol, LDL-C, HDL-C, and triglycerides. The quantitative measurements of TG (ABX Pentra Triglycerides CP), cholesterol (ABX Pentra Cholesterol CP), and HDL-C (ABX Pentra HDL Direct CP) were done by colorimetry using a Pentra 400 Clinical Chemistry Analyzer (Horiba Instruments, Inc., CA). LDL-C concentrations were calculated according to Friedewald's formula. All parameters were measured in fasting blood samples.
A total of 2,172 Caucasian individuals from the MesyBepo (Metabolic Syndrome Berlin Potsdam) study population from the region of Berlin/Potsdam, Germany, were included in the present study. A total of 141 subjects who had been receiving lipid-lowering medication were excluded from further analyses. The remaining 2,031 subjects consisted of 651 men and 1,380 women (mean male age, 52 ± 14 years; mean female age 51 ± 13 years; mean male BMI, 29.1 ± 5.9 kg/m2; mean female BMI, 29.9 ± 6.9 kg/m2; data are given as arithmetic means ± SD), 312 subjects had type 2 diabetes (based on oral glucose tolerance test according to the WHO guidelines [21]). The study was approved by the ethics committees of the University of Potsdam and the Charité-University (Medical Department), Berlin, Germany. All subjects provided written informed consent.
DNA extraction, genotyping, and quality control in the Sorbs
Genomic DNA was extracted using a QIAmp DNA Blood Midi Kit (Qiagen Inc., Valencia, CA) according to the manufacturer's protocol. Genotyping was performed using the 500K Affymetrix GeneChip and the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Inc.) by the Microarray Core Facility of the Interdisciplinary Centre for Clinical Research, University of Leipzig, Germany, and by ATLAS Biolabs GmbH, Berlin, Germany. Genotypes were determined with GeneChip Genotyping Analysis Software (GTYPE) using the BRLMM algorithm for the 500K arrays and the Birdseed Algorithm for Genome-Wide Human SNP Array 6.0 (Affymetrix, Inc.). Data underwent quality control, and only SNPs fulfilling the following criteria were included: missing rate per SNP <5%, missingness per sample <7%, Hardy-Weinberg equilibrium P > 0.0001, minor allele frequency >0.05. The average genotyping rate was 99.0%. In all, 390,619 autosomal markers overlapping between the 500K Affymetrix GeneChip and the AffymetrixGenome-Wide Human SNP Array 6.0 were included in the analyses.
Statistical methods and software
The calculation of minor allele frequencies, Hardy-Weinberg equilibrium, and missing rates per SNP was performed with PLINK (22). Genome-wide association with lipid phenotypes was assessed by linear regression in PLINK. All non-normally distributed parameters were transformed to approximate normal distribution. We corrected the analyses for age, gender, and BMI and relatedness by using genomic control for Sorbs (λ = 1.31). Linkage disequilibrium (LD) metrics were calculated in Haploview 4.1 (23). A weighted meta-analysis was performed using METAL (24) Study-specific P values and effect directions were converted to Z statistics and weighted with sample size of each study. Two-sided P values <0.05 were considered to provide nominal evidence for association and are presented without Bonferroni correction for multiple testing. Only associations that would reach the P values adjusted for multiple testing (Bonferroni correction; P = 0.05 divided by the number of tested SNPs) were considered statistically significant.
Statistical analyses in replication studies were performed using SPSS version 18.0.2 (SPSS, Inc., Chicago, IL).
Genotyping for replication in the Berlin cohort
Genotyping of selected SNPs for replication in an independent cohort from Berlin was performed using the TaqMan allelic discrimination assay (Applied Biosystems, Inc.). Oligonucleotide sequences are available upon request. The TaqMan genotyping reaction was performed according to the manufacturer's protocol on an ABI PRISM 7500 sequence detector (Applied Biosystems Inc.).
Selection of THOC5 tagging SNPs and genotyping in the Sorbs
Five tagging SNPs (rs4823045, rs8140060, rs2283860, rs737975, and rs11704899) were selected from the HapMap database (r2 > 0.8; minor allele frequency >0.01) to cover all common genetic variants in THOC5. Genotyping in the Sorbs was performed using the TaqMan allelic discrimination assay (Applied Biosystems, Inc.). Oligonucleotide sequences are available upon request.
Small interfering RNA mediated knock-down of THOC5 in vitro
Briefly, human hepatic carcinoma (HepG2) cells were cultured in DMEM (high glucose: 4.5 g/l) (Invitrogen, Karlsruhe, Germany) supplemented with 10% calf serum (PAA Laboratories GmbH, Cölbe, Germany) and 2% penicillin-streptomycin (final concentration: 100 U/ml penicillin, 100 µg/ml streptomycin; abx) (Invitrogen) at 37°C and 5% CO2. Before the knock-down experiment, the cells were split into 24-well plates (6.0 × 104/well) and cultured in abx-free medium for 48 h to reach 50–70% confluence. Small interfering (siRNA) knock-down experiments were performed using the ON-TARGET plus SMARTpool siRNA (Thermo Scientific) for THOC5 (human) and a NonTargeting Pool (scrambled siRNA for human, mouse and rat) as control. DharmaFECT4 (Thermo Scientific) was used as transfection reagent. Four different transfection schemes were followed: untreated cells (100 µl calf serum- and abx-free growth medium [transfection medium]), mock-transfection (98 µl transfection medium and 2 µl transfection reagent), nontargeting pool (73 µl transfection medium, 2 µl transfection reagent, and 25 µl nontargeting siRNA pool [2 µM]), and THOC5 siRNA pool (73 µl transfection medium, 2 µl transfection reagent, and 25 µl THOC5 siRNA pool [2 µM]). The medium was removed from the cells, and, after washing with PBS (2×), 400 µl fresh abx-free culture medium were added to each well. Cells were treated with individual transfection mix (100 µl) (six wells/approach, three wells/time point [24/48 h]). All experiments were done in triplicate.
mRNA expression
The mRNA levels of THOC5 and of candidate genes related to HDL metabolism, apolipoprotein A1 (APOA1), scavenger receptor class B member 1 (SCARB1), ATP-binding cassette transporter A1 (ABCA1), and ATP-binding cassette subfamily G member 5 and 8 (ABCG5/ABCG8) were measured 24, 48, and 72 h after transfection. Taqman gene-expression assays (Applied Biosystems) were performed on ABI PRISM 7500 sequence detector (Applied Biosystems Inc.) according to the manufacturer's instructions.
The data were calculated using the ΔΔCt method with β-actin as a normalization gene and were corrected for individual PCR efficiency.
RESULTS
Genome-wide scan for lipid traits in the Sorbs
A genome-wide scan for association of SNPs with lipid parameters in the Sorbs revealed 14 novel loci reaching nominal P values <10−5 (supplementary Table I) (considering 390,619 SNPs, a P < 1.3 × 10−7 would be required to reach genome wide significance after Bonferroni correction for multiple testing). Several previously published variants were nominally associated with lipid traits in the Sorbs (supplementary Table II): six SNPs with HDL-C [in/near LIPC (8), CETP (8), PCIF1 (3), LPL (8), APOC1 (2, 6), and LIPC (8)], five SNPs with LDL-C [in/near APOC1 (2, 6), PSRC1 (6, 8), APOB (1, 2, 6, 8), CELSR2 (6), and TIMD4 (3)], and six SNPs with TG [in or near PCIF1 (3), LPL (two SNPs) (8), BUD13 (2), APOB (1, 2, 6, 8), and ZNF259 (8)]. The variant rs17145738 near MLXIPL previously published for association with HDL-C (5) was associated with LDL-C in the Sorbs (supplementary Table II).
Meta-analysis (Sorbs and DGI)
SNPs with a P value < 0.01 in the Sorbs (4,182 SNPs for HDL-C, 4,437 for LDL-C, and 4,395 for TG) were taken forward into stage 2 for a meta-analysis (herein referred to as second-stage meta-analysis) together with the DGI data (supplementary Fig. I). In the second-stage meta-analysis, five SNPs at four loci were associated with HDL-C, 13 SNPs at nine loci with LDL-C, and 26 SNPs at seven loci with serum TG concentrations with combined P values < 5 × 10−5 (supplementary Fig. I and supplementary Table III). Considering there were approximately 4,000 SNPs tested, associations of two SNPs with HDL-C, six SNPs with LDL-C, and 17 SNPs with TG would reach statistical significance after Bonferroni correction for multiple testing (P < 1.3 × 10−5). Nevertheless, after exclusion of previously reported variants (two SNPs for HDL-C, three SNPs for LDL-C, and eight SNPs for TG; supplementary Table III), SNPs with strong evidence for association and representative for their LD groups were taken forward to replication in an independent German cohort from Berlin. In summary, rs8135828 (THOC5) and rs12657936 (SPEF2) were selected for replication of associations with HDL-C, rs7251213 (APG4D) with LDL-C, and rs9444205 (TBX18) and rs17422816 (FAM79B) with TGs (supplementary Fig. I and supplementary Table III).
Replication of top association signals in the Berlin cohort and meta-analysis in three cohorts (Sorbs, DGI, Berlin)
HDL-C.
The rs8135828 in the intronic region of the THOC5 was significantly associated with HDL-C in the Berlin cohort (Table 2). Minor allele carriers (A-allele) showed lower serum HDL concentrations (P = 0.002 [Table 2]; genotype-specific means are provided in supplementary Table IV). The meta-analysis including Sorbs, the DGI sample, and the Berlin cohort revealed an association reaching P = 1.78 × 10−7 (Table 2).
TABLE 2.
Results of meta-analysis of replicated variants in all cohorts
| Nearby Gene | SNP | Chr | A1 Sorbs | MAF Sorbs | MAF DGI | MAF Berlin | P (Sorbs) | P (DGI) | P (Berlin) | Beta (SE) Sorbs | Beta (SE) DGI | Beta (SE) Berlin | Meta P (Sorbs, DGI) | Meta P (Sorbs, DGI, Berlin) |
| HDL-C | ||||||||||||||
| SPEF2 | rs12657936 | 5 | A | 0.10 | 0.11 | 0.12 | 1.50 × 10−3 | 3.90 × 10−3 | 0.92 | 0.06 (0.028) | 0.13 (0.044) | 2.00 × 10−3 (0.016) | 4.70 × 10−5 | 1.71 × 10−3 |
| THOC5 | rs8135828 | 22 | A | 0.14 | 0.15 | 0.15 | 1.62 × 10−4 | 6.18 × 10−3 | 2.00 × 10−3 | −0.06 (0.025) | −0.11 (0.039) | −0.05 (0.015) | 2.30 × 10−5 | 1.78 × 10−7 |
| LDL-C | ||||||||||||||
| ATG4D | rs7251213 | 19 | G | 0.41 | 0.39 | 0.41 | 9.47 × 10−3 | 7.69 × 10−5 | 0.34 | −0.11 (0.042) | −0.11 (0.028) | −0.02 (0.029) | 2.38 × 10−6 | 2.91 × 10−5 |
| Triglycerides | ||||||||||||||
| TBX18 | rs9444205 | 6 | A | 0.09 | 0.11 | 0.11 | 1.59 × 10−4 | 1.09 × 10−3 | 0.93 | 0.16 (0.042) | 0.14 (0.043) | −4.00 × 10−3 (0.04) | 2.70 × 10−6 | 5.04 × 10−4 |
| FAM79B | rs17422816 | 3 | T | 0.33 | 0.38 | 0.32 | 8.55 × 10−4 | 2.15 × 10−3 | 0.21 | −0.08 (0.039) | −0.09 (0.028) | −0.03 (0.027) | 1.67 × 10−5 | 4.70 × 10−5 |
SNPs replicated in the Berlin cohort (n = 2,031) and results of the combined P value of all three cohorts. Nearby gene is defined as the closest gene to the SNP in a 250 kbp window. A1, minor allele; Chr, chromosome; DGI, Diabetes Genetics Initiative; MAF, minor allele frequency; Meta, meta-analysis.
Despite consistent effect direction, the SPEF2 locus did not show a significant effect in the Berlin sample (P = 0.92) (Table 2). In the meta-analysis including Sorbs, the Berlin cohort, and the DGI sample, the A-allele was significantly associated with lower HDL serum levels (P = 1.71 × 10−3) (Table 2).
LDL-C.
Based on the second-stage meta-analysis of the Sorbs and DGI sample, we selected the rs7251213 in APG4D for further replication analyses in the Berlin cohort. Rs7251213 (APG4D) did not reach a nominal level of significance in the Berlin sample (Table 2), but in a combined analyses of all cohorts the G-allele was associated with reduced LDL-C concentrations (P = 2.91 × 10−5) (Table 2).
TG.
Based on the result of the second-stage meta-analysis, rs9444205 near TBX18 and rs17422816 in the intronic region of the FAM79B were selected for replication in the Berlin cohort. No significant effects on serum TG concentrations were detected (P = 0.93 and P = 0.21, respectively) (Table 2). However, meta-analysis of all three study populations showed an association of these loci with TG levels (P = 5.04 × 10−4 and 4.70 × 10−5, respectively) (Table 2).
Effects of additional THOC5 tagging SNPs on lipid traits in the Sorbs
Five tagging SNPs covering the common genetic variation within THOC5 were genotyped for fine mapping of the causal variant responsible for the initially identified association between rs8135828 and HDL-C. As expected, rs11704899, which was in complete LD with the rs8135828, showed significant association with HDL-C (supplementary Table V). Rs8140060 showed a nominal effect on HDL-C serum concentrations (P = 0.024, additive mode of inheritance). The A-allele was associated with reduced HDL levels in the Sorbs (supplementary Table V). Finally, rs11704899 showed nominal association with lower serum APOA1 levels (P = 0.028, β = −0.026; SE = 0.012; standardized for the minor allele; additive mode of inheritance).
Subjects on lipid-lowering drugs were excluded from statistical analyses because it may be assumed that the effects of individual genetic variants would be masked by the drug effects, whose contribution to the phenotype appears much stronger than that of the genetic polymorphism. Nevertheless, because excluding participants may lead to bias in the regression, we performed additional association analyses of all THOC5 polymorphisms with lipid parameters in the complete Sorbs cohort, which included subjects on lipid-lowering drugs. To correct for the effects of drugs, we estimated the effect of medication on lipid levels in regression analyses and used the obtained effect size to calculate expected proportional changes in the circulating lipid concentrations. Based on these calculations, factor 1.02 for total cholesterol, 1.08 for HDL-C, 1.03 for LDL-C, and 1.31 for TGs were used to adjust for the expected change in the phenotype of the treated patients. The results remained materially unchanged (supplementary Table V). Specifically, except for the rs11704899, none of the THOC5 SNPs provided statistically significant associations with HDL-C concentrations. However, even in our initial analyses using datasets without subjects on lipid-lowering drugs, the other SNPs showed only nominal associations with HDL-C, which would not withstand Bonferroni corrections for multiple testing (P < 0.01 required for statistical significance when considering five tested SNPs).
Effects of THOC5 on mRNA levels of genes related to HDL-C metabolism
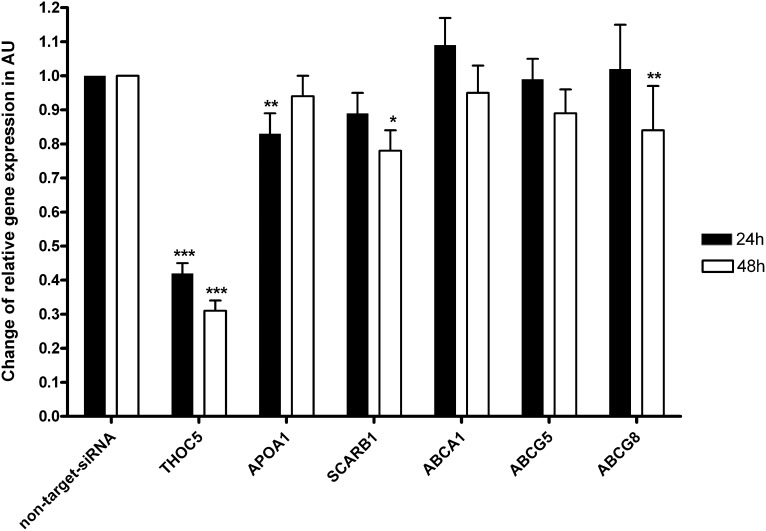
Because THOC5 had not been associated with HDL-C metabolism before this study, we performed THOC5 knock-down experiments in the human HepG2 cell line to assess the impact of THOC5 on mRNA levels of selected genes related to HDL-C metabolism (APOA1, SCARB1, and ABCG8) and to support the observed associations with HDL-C. The outcome of the THOC5 knock-down on mRNA levels of APOA1, SCARB1, and ABCG8 is shown in Fig. 1. The respective silencing of THOC5 by siRNA resulted in significantly reduced relative expression of THOC5, with a maximum of approximately 70% after 48 h (all P ≤ 4.14 × 10−13). This effective knock-down of the THOC5 mRNA expression led to diminished APOA1 mRNA levels after 24 h (P = 2.7 × 10−3) as well as SCARB1 (P = 1.3 × 10−2) and ABCG8 (P = 3.4 × 10−3) mRNA levels after 48 h (Fig. 1).
Fig. 1.
Effects of THOC5 knock-down in vitro. Change of mRNA levels in arbitrary units (AU) after 24 and 48 h using β-actin as a normalization gene and related to a nontarget-siRNA approach. THOC5 showed significant reduction in mRNA levels at each time point (*** both P values ≤ 4.14 × 10−13). Decreased mRNA levels have been shown for APOA1 (**P = 2.7 × 10−3) after 24 h and for SCARB1 (*P = 1.3 × 10−2) and ABCG8 (**P = 3.4 × 10−3) after 48 h.
DISCUSSION
Although numerous genetic determinants of serum lipid profiles have been recently identified using GWAS, only a small proportion of the estimated heritability can be explained (3). Focusing on variants with strong effects in genetically homogenous populations might help to identify loci of interest among SNPs not ranked in the top tier of a classical meta-analysis but still of potential physiological significance. Therefore, in the present study we followed a study design in which we preselected SNPs based on the GWAS in the self-contained population of Sorbs and proceeded with these SNPs in subsequent meta-analyses. In our GWAS in the Sorbs, we successfully replicated several of the previously shown associations with plasma lipid traits. This indicates that our cohort had sufficient power to detect signals found in larger meta-analyses and indirectly supports a potential physiological role of these variants in lipid metabolism.
In a meta-analysis including the Sorbs and the DGI sample followed by further replication in an independent cohort from Berlin, we identified a THOC5 variant (rs8135828) associated with reduced HDL-C levels. The association of other independent variants in the THOC5 locus with HDL levels in the Sorbs further minimizes the risk of a false-positive result. However, this locus did not show up in the published GWAS and meta-analyses that can be readily accessed, including imputed datasets in “The database of Genotypes and Phenotypes” (25). THOC5 also did not come up in the recent meta-analysis of the Global Lipids Genetics Consortium (5). Even though the effect direction from the Global Lipids Genetics Consortium was consistent with the results of our study, there was no significant association of rs8135828 with HDL-C concentrations (P = 0.30) (5). It remains to be clarified whether this is due to genetic heterogeneity between various study populations. Such heterogeneity of effect sizes in different populations could also be driven by gene-gene and gene-environment interactions. Another recent GWAS on genetic modulators of carotid atherosclerosis risk identified variants in the THOC5 region (26). Although this might indirectly support the potential role of THOC5 genetic variation in lipid metabolism, the two studies report different SNPs. The SNPs reported by Shrestha et al. (26) map ∼66 kb away from rs8135828 shown in our study. Moreover, according to the HapMap database (27), rs8135828 is not in LD with either of the SNPs (rs13053817 and rs5763254) studied by Shrestha et al. (26) (r2 = 0.07 for both SNPs). Therefore, even though fine mapping of the respective chromosomal region would be inevitable to draw further conclusions, it seems unlikely that the two studies point to the same region/variant affecting both the lipid traits and the risk of carotid atherosclerosis.
THOC5 is ubiquitously expressed in most mammalian tissues. THOC5 protein expression is elevated in blood cells, kidney, and liver (28). THOC5 is part of the RNA spliceosome complex and influences C/EBP expression in adipocyte differentiation (29, 30). It is also part of the TREX (transcription/export) complex, which is required for correct mRNA processing, in particular transcription elongation and nuclear export of mRNAs (especially heat shock mRNAs) (31, 32). Furthermore, the THOC5/FMIP complex is essential for hematopoietic primitive cell survival in vivo (33). Using a THOC5-depleted mouse fibroblast cell line, Guria et al. (31) identified several genes that were transcribed but not exported into the cytoplasm in absence of THOC5. However, little is known about the potential relationship between THOC5 and HDL-C metabolism. Therefore, we elucidated the effects of a targeted THOC5 mRNA knock-down on genes related to HDL-C metabolism. Our results showed that APOA1 expression was significantly reduced 24 h after THOC5 silencing. In line with this, rs11704899, which is in LD with the HDL-associated rs8135828 initially detected in the GWAS, showed nominal association with reduced circulating APOA1 levels. Considering the fact that APOA1 is the main protein of HDL particle formation, our data suggest that the effects of THOC5 variants on HDL-C concentrations might be mediated via effects on APOA1. Lower APOA1 levels are known to be associated with increased risk of coronary artery disease and related events (34). It has been demonstrated that variants in the APOA1 were associated with amyloidosis (35), which may lead to increased carotid intima-media thickness and to endothelial dysfunction (36). Knock-out of APOA1 in apoB-transgenic female mice, LDL receptor-deficient mice, and LDL receptor/apoE-deficient mice results in an increased risk of atherosclerosis (37, 38). Surprisingly, THOC5 knock-down performed in vitro also leads to reduced mRNA expression of SCARB1 and ABCG8 in HepG2 cells. These data were achieved in in vitro experiments using a cell culture system that, due to the substantially lower molecular complexity, cannot entirely resemble the in vivo situation. Therefore, we interpret these findings with caution and acknowledge that further functional studies are warranted to elucidate the precise molecular mechanisms by which THOC5 affects plasma HDL-C.
Our study has several limitations. First, the sample size of our initial GWAS might have been too small to reveal highly significant variants that might be truly causative or close to causal variation. Second, we cannot rule out that the heterogeneous genetic background in the different study populations might have affected the outcome of the meta-analyses. We are also aware that both the Sorbs and the Berlin study cohorts seem to have rather high BMIs (26.4 and 29 kg/m2, respectively). We therefore adjusted all analyses for BMI to avoid bias driven by BMI effects in the lipid profile. This does not necessarily preclude the possibility that some of the SNP associations with lipid phenotypes are affected by BMI. On the other hand, using the present study design with inclusion of obese subjects provides a broader distribution of the targeted phenotype. Nevertheless, we performed the replication analyses by stratifying the subjects based on BMI but found no differences between lean and obese subjects regarding genetic associations with lipid phenotypes (e.g., the rs8135828 did not show association with HDL levels in either the lean [<25 kg/m2] or the obese [>30 kg/m2] groups; P values of 0.19 and 0.31, respectively).
Running GWAS with imputed datasets might have resulted in additional association signals that would be worth being pursued. Nevertheless, even though by not including imputed datasets the study may have restricted the final outcome of the GWAS and may have missed additional association signals, combining genetic association studies with functional in vitro experiments provided clear evidence for the role of THOC5 in the lipid metabolism.
Taken together, our data suggest that THOC5 might be a novel player involved in the genetic control of serum lipid profiles.
Supplementary Material
Acknowledgments
The authors thank all those who participated in the studies, John Broxholm from the Bioinformatics Core Unit of the Wellcome Trust Centre for Human Genetics, and Andre Rothe from the Coordination Centre for Clinical Trials, University of Leipzig.
Footnotes
Abbreviations:
- DGI
- Diabetes Genetics Initiative
- GWAS
- genome-wide association study
- LD
- linkage disequilibrium
- siRNA
- small interfering RNA
- SNP
- single-nucleotide polymorphism
- TG
- triglyceride
This work was supported by grants from the DFG (KFO 152, SFB 1052/1, B03) and from the German Diabetes Association (Y.B., A.T., and P.K.), by a Travel Grant from BIF (A.T.), by IFB AdiposityDiseases Leipzig (K7-37 and MetaRot position to A.T.; K7-38 and K7-39 to Y.B.), and by the DHFD, Diabetes Hilfs- und Forschungsfonds Deutschland. J.S. was supported by a research group of the BMBF (Molecular Nutrition), a Heisenberg-Professorship (SP716/2-1) of the Deutsche Forschungsgemeinschaft (DFG), and a graduate school of the DFG (GK1208). Y.B. has been supported by a research group of the IFB AdiposityDiseases ADI-K50D. M.K. has been supported by K7-39. D.S. and B.E. are funded by Boehringer Ingelheim Foundation. The authors declare they have no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five tables and one figure.
REFERENCES
- 1.Aulchenko Y. S., Ripatti S., Lindqvist I., Boomsma D., Heid I. M., Pramstaller P. P., Penninx B. W., Janssens A. C., Wilson J. F., Spector T., et al. 2009. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 41: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., et al. 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40: 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxena R., Voight B. F., Lyssenko V., Burtt N. P., de Bakker P. I. W., Chen H., Roix J. J., Kathiresan S., Hirschhorn J. N., Daly M. J., et al. 2007. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 316: 1331–1336 [DOI] [PubMed] [Google Scholar]
- 5.Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 466: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandhu M. S., Waterworth D. M., Debenham S. L., Wheeler E., Papadakis K., Zhao J. H., Song K., Yuan X., Johnson T., Ashford S., et al. 2008. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 371: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson C. S., Eberle M. A., Kruglyak L., Nickerson D. A. 2004. Mapping complex disease loci in whole-genome association studies. Nature. 429: 446–452 [DOI] [PubMed] [Google Scholar]
- 8.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40: 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asselbergs F. W., Guo Y., van Iperen E. P., Sivapalaratnam S., Tragante V., Lanktree M. B., Lange L. A., Almoguera B., Appelman Y .E., Barnard J., et al. 2012. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am. J. Hum. Genet. 91: 823–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weissglas-Volkov D., Aguilar-Salinas C. A., Nikkola E., Deere K. A., Cruz-Bautista I., Arellano-Campos O., Muñoz-Hernandez L. L., Gomez-Munguia L., Ordoñez-Sánchez M. L., Reddy P. M., et al. 2013. Genomic study in Mexicans identifies a new locus for triglycerides and refines European lipid loci. J. Med. Genet. 50: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Surakka I., Whitfield J. B., Perola M., Visscher P. M., Montgomery G. W., Falchi M., Willemsen G., de Geus E. J., Magnusson P. K., Christensen K., et al. 2012. A genome-wide association study of monozygotic twin-pairs suggests a locus related to variability of serum high-density lipoprotein cholesterol. Twin Res. Hum. Genet. 15: 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansen C. T., Wang J., Lanktree M. B., Cao H., McIntyre A. D., Ban M. R., Martins R. A., Kennedy B. A., Hassell R. G., Visser M. E., et al. 2010. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 42: 684–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irvin M. R., Zhang Q., Kabagambe E. K., Perry R. T., Straka R. J., Tiwari H. K., Borecki I. B., Shimmin L. C., Stuart C., Zhong Y., et al. 2012. Rare PPARA variants and extreme response to fenofibrate in the Genetics of Lipid-Lowering Drugs and Diet Network Study. Pharmacogenet. Genomics. 22: 367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy M. V. P. L., Iatan I., Weissglas-Volkov D., Nikkola E., Haas B. E., Juvonen M., Ruel I., Sinsheimer J. S., Genest J., Pajukanta P., et al. 2012. Exome sequencing identifies 2 rare variants for low high-density lipoprotein cholesterol in an extended family. Circ Cardiovasc Genet. 5: 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanna S., Li B., Mulas A., Sidore C., Kang H. M., Jackson A. U., Piras M. G., Usala G., Maninchedda G., Sassu A., et al. 2011. Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet. 7: e1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kristiansson K., Naukkarinen J., Peltonen L. 2008. Isolated populations and complex disease gene identification. Genome Biol. 9: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., Hunter D. J., Ramos E. M., Cardon L. R., Chakravarti A., Cho J. H., et al. 2009. Finding the missing heritability of complex diseases. Nature. 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veeramah K. R., Tönjes A., Kovacs P., Gross A., Wegmann D., Geary P., Gasperikova D., Klimes I., Scholz M., Novembre J., et al. 2011. Genetic variation in the Sorbs of eastern Germany in the context of broader European genetic diversity. Eur. J. Hum. Genet. 19: 995–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tönjes A., Zeggini E., Kovacs P., Böttcher Y., Schleinitz D., Dietrich K., Morris A. P., Enigk B., Rayner N. W., Koriath M., et al. 2010. Association of FTO variants with BMI and fat mass in the self-contained population of Sorbs in Germany. Eur. J. Hum. Genet. 18: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tönjes A., Koriath M., Schleinitz D., Dietrich K., Böttcher Y., Rayner N. W., Almgren P., Enigk B., Richter O., Rohm S., et al. 2009. Genetic variation in GPR133 is associated with height: genome wide association study in the self-contained population of Sorbs. Hum. Mol. Genet. 18: 4662–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organization and International Diabetes Federation Definition, diagnosis and classification of diabetes mellitus and its complications. 1999. WHO Document Production Services, Geneva. [Google Scholar]
- 22.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., et al. 2007. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett J. C., Fry B., Maller J., Daly M. J. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 21: 263–265 [DOI] [PubMed] [Google Scholar]
- 24.Willer C. J., Li Y., Abecasis G. R. 2010. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 26: 2190–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Center for Biotechnology Information (NCBI) The database of Genotypes and Phenotypes (dbGaP); Available from: http://www.ncbi.nlm.nih.gov/gap
- 26.Shrestha S., Irvin M. R., Taylor K. D., Wiener H. W., Pajewski N. M., Haritunians T., Delaney J. A., Schambelan M., Polak J. F., Arnett D. K., et al. 2010. A genome-wide association study of carotid atherosclerosis in HIV-infected men. AIDS. 24: 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Center for Biotechnology Information (NCBI) International HapMap Project; Available from: http://hapmap.ncbi.nlm.nih.gov/
- 28.GeneCards Human Gene Database THOC5 Gene - GeneCards | THOC5 Protein | THOC5 antibody. Accessed July 19, 2013, at http://www.genecards.org/cgi-bin/carddisp.pl?gene=THOC5
- 29.Rehwinkel J., Herold A., Gari K., Köcher T., Rode M., Ciccarelli F. L., Wilm M., Izaurralde E. 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 11: 558–566 [DOI] [PubMed] [Google Scholar]
- 30.Mancini A., El Bounkari O., Norrenbrock A-F., Scherr M., Schaefer D., Eder M., Banham A. H., Pulford K., Lyne L., Whetton A. D., et al. 2007. FMIP controls the adipocyte lineage commitment of C2C12 cells by downmodulation of C/EBPalpha. Oncogene. 26: 1020–1027 [DOI] [PubMed] [Google Scholar]
- 31.Guria A., Tran D. D. H., Ramachandran S., Koch A., El Bounkari O., Dutta P., Hauser H., Tamura T. 2011. Identification of mRNAs that are spliced but not exported to the cytoplasm in the absence of THOC5 in mouse embryo fibroblasts. RNA. 17: 1048–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köhler A., Hurt E. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8: 761–773 [DOI] [PubMed] [Google Scholar]
- 33.Mancini A., Niemann-Seyde S. C., Pankow R., El Bounkari O., Klebba-Färber S., Koch A., Jaworska E., Spooncer E., Gruber A. D., Whetton A. D., et al. 2010. THOC5/FMIP, an mRNA export TREX complex protein, is essential for hematopoietic primitive cell survival in vivo. BMC Biol. 8: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Besler C., Lüscher T. F., Landmesser U. 2012. Molecular mechanisms of vascular effects of high-density lipoprotein: alterations in cardiovascular disease. EMBO Mol. Med. 4: 251–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaefer E. J., Santos R. D., Asztalos B. F. 2010. Marked HDL deficiency and premature coronary heart disease. Curr. Opin. Lipidol. 21: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modesto K. M., Dispenzieri A., Gertz M., Cauduro S. A., Khandheria B. K., Seward J. B., Kyle R., Wood C. M., Bailey K. R., Tajik A. J., et al. 2007. Vascular abnormalities in primary amyloidosis. Eur. Heart J. 28: 1019–1024 [DOI] [PubMed] [Google Scholar]
- 37.Moore R. E., Navab M., Millar J. S., Zimetti F., Hama S., Rothblat G. H., Rader D. J. 2005. Increased atherosclerosis in mice lacking apolipoprotein A-I attributable to both impaired reverse cholesterol transport and increased inflammation. Circ. Res. 97: 763–771 [DOI] [PubMed] [Google Scholar]
- 38.Voyiaziakis E., Godberg I. J., Plump A. S., Rubin E. M., Breslow J. L., Huang L-S. 1998. ApoA-I deficiency causes both hypertriglyceridemia and increased atherosclerosis in human apoB transgenic mice. J. Lipid Res. 39: 313–321 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.