Abstract
Blood levels of lipoprotein cholesterol and triglycerides (TGs) are highly heritable and are major risk factors for cardiovascular disease (CVD). Approximately 100 lipid-associated loci have been identified in populations of European ancestry. We performed a genome-wide association study of lipid traits in 1,782 Filipino women from the Cebu Longitudinal Health and Nutrition Survey, and tested for evidence of interactions with waist circumference. We conducted additional association and interaction analyses in 1,719 of their young adult offspring. Genome-wide significant associations (P < 5 × 10−8) were detected at APOE for low density lipoprotein cholesterol and total cholesterol, and at APOA5 for TGs. Suggestive associations (P < 10−6) were detected at GCKR for TGs, and at CETP and TOM1 for high density lipoprotein cholesterol. Our data also supported the existence of allelic heterogeneity at APOA5, CETP, LIPC, and APOE. The secondary signal (Gly185Cys) at APOA5 exhibited a single nucleotide polymorphism (SNP)-by-waist circumference interaction affecting TGs (Pinteraction = 1.6 × 10−4), manifested by stronger SNP effects as waist circumference increased. These findings provide the first evidence that central obesity may accentuate the effect of the TG-increasing allele of the APOA5 signal, emphasizing that CVD risk could be reduced by central obesity control.
Keywords: genome-wide association, lipid traits, interaction
Blood concentrations of lipoprotein cholesterol and triglycerides (TGs) are major risk factors for cardiovascular disease (CVD), the current leading cause of mortality worldwide (1). The most rapid increase in the prevalence of CVD is taking place in Asia, including the Philippines (1), although the obesity-related anthropometric measures remain lower in these Asian populations compared with those in populations of Europeans (2). Family-based studies have demonstrated that 40–50% of the total variation in lipid traits is genetically determined (3). Genome-wide association (GWA) studies have identified ∼100 loci associated with fasting levels of total cholesterol (TC), high density lipoprotein cholesterol (HDL-C), low density lipoprotein cholesterol (LDL-C), and TGs in Europeans (4–9). Many of these lipid-associated loci, including APOA5, LPL, GCKR, TRIB1, MLXIPL, CETP, LIPC, and ABCA1, were further confirmed in individuals of non-European ancestries (10–12). More recently, a large-scale GWA study conducted in East Asians identified additional HDL-C signals near MYL2 and HECTD4 (C12orf51) (11). The full gene names corresponding to the locus symbols are listed in supplementary Table I.
The GWA signals identified to date have only explained ∼10% phenotypic variation in each lipid trait, corresponding to ∼25% of the trait heritability (8). Additional heritability may be explained by the incomplete coverage of functional variants and by allelic heterogeneity (13, 14). Gene-by-environment interactions may also account for some heritability (15) because the interactive effects of environmental modifiers may hinder the detection of genetic associations (16). Earlier studies have demonstrated that diet, alcohol, smoking, and physical activity can modulate genetic effects on lipid levels (12, 17–19). In addition, evidence exists that anthropometric traits, including body mass index (BMI), waist-hip ratio, and body composition, modify the association between genetic variants and lipid traits (20).
In recent decades, the rising prevalence of CVD shows a positive link with the increasing prevalence of central obesity (21, 22). Waist circumference is believed to be a stronger predictor of CVD than BMI, particularly in Asians, who tend to have a higher risk of CVD at any given level of BMI compared with Europeans (23). Intra-abdominal visceral fat accumulation has been shown to affect the lipid levels either directly or via insulin resistance (24, 25). Despite considerable evidence that genetic variants and obesity traits independently and jointly influence lipid levels, few studies have explicitly examined the modifying effect of waist circumference on lipid associations identified by GWA studies.
To test the hypothesis that central obesity would influence the association between genetic variation and lipid levels, we performed a GWA study to investigate single nucleotide polymorphism (SNP) associations with lipoprotein cholesterol and TG levels in 1,782 Filipino mothers from the Cebu Longitudinal Health and Nutrition Survey (CLHNS) and tested for interactions between lipid-associated variants and waist circumference affecting lipid levels. We also conducted analyses in 1,719 young adult offspring of the CLHNS mothers using Metabochip genotypes to confirm SNP associations and interactions with waist circumference.
METHODS AND PROCEDURES
Study subjects and phenotypes
The CLHNS is an ongoing population-based birth cohort study that began in 1983. The original study population, study design, and recruitment protocols have been described in detail previously (26). Briefly, the baseline survey randomly recruited 3,327 pregnant women from the Metropolitan Cebu area of the Philippines in 1983–1984 (3,080 singleton live births), and since followed them and their offspring to the present. Trained field staff conducted in-home interviews and collected anthropometric measurements at each visit. Overnight fasting plasma samples for DNA extraction and biomarker measurements were obtained in 2005. Written informed consent was obtained from all participants, and study protocols were approved by the University of North Carolina Institute Review Board for the Protection of Human Subjects.
The current study used cross-sectional data from the 2005 survey available for 1,782 mothers and 1,719 male and female young adult offspring. Three measurements of waist circumference were taken to the nearest 0.1 cm, placing a plastic tape about two inches above the navel, after normal exhalation (27). The three measurements of waist circumference were taken at the same time and their average was used for analyses. TC was measured using enzymatic methods on the Beckman Diagnostics CX5 chemistry analyzer (Beckman Coulter Diagnostics, Fullerton, CA). HDL-C and LDL-C were determined using the homogenous assays Direct HDL-C and Direct LDL-C (Equal Diagnostics, Exton, PA). TGs were measured with a glycerol blank as a two-step reaction (28). Only one of the CLHNS mothers and one of the young adult offspring were taking lipid-altering medication.
Genotyping and imputation
SNP genotyping of the CLHNS mothers was performed with the Affymetrix Genomewide Human SNP Array 5.0 at the Vanderbilt Microarray Shared Resource at Vanderbilt University Medical Center, Nashville, TN, using the standard protocol recommended by the manufacturer. The quality control procedures have been previously described (29). As the imputation using the 1000 Genomes CEU+CHB+JPT reference panel (error rate 3.6% and MACH r2 0.86) led to a better imputation quality compared with that using the CHB+JPT panel (error rate 3.3% and MACH r2 0.81), we applied MACH to impute genotypes based on phased reference haplotypes from the 1000 Genomes Project CEU+CHB+JPT samples (June 2010 release). After exclusion of SNPs with poor imputation quality (MACH r2 < 0.3), ∼3.7 million imputed SNPs were tested for association with TC, HDL-C, LDL-C, and TGs. The candidate SNP APOE rs7412 was not available in the genome-wide data and was genotyped using TaqMan allelic discrimination (Applied Biosystems, Foster City, CA).
The 1,719 CLHNS offspring were genotyped by the University of North Carolina Mammalian Genotyping using the Metabochip (Illumina, San Diego, CA), a custom high-density genotyping array of SNPs designed to provide a high-density coverage for both overall SNPs and low frequency variants concentrated around GWA loci and/or signals for cardiovascular and metabolic traits (30). The genotyping and quality control of Metabochip SNPs in the CLHNS offspring were described previously (31).
In the CLHNS mothers, ∼350,000 markers were directly genotyped, and imputation was performed for all markers based on the 1000 Genomes Project; we used the imputed genotypes for all markers (29). In the offspring, all variants for analyses were directly genotyped by Metabochip.
Statistical analysis
Values of TC and TGs were natural log-transformed (ln) to approximate the normal distributions of the traits. We applied multiple linear regression models and assumed an additive mode of inheritance to test for the association between genotypes and HDL-C, LDL-C, ln TC, and ln TGs.
We constructed 10 principal components (PCs) of population substructure using EIGENSOFT (32). In the CLHNS mothers, we applied MACH2QTL to perform the GWA analyses, adjusting for age, age2, household assets (a score that counts the possession of land and house and household amenities, such as a TV or refrigerator) (33), natural ln household income, and the first seven PCs. To control for the effects of other potential factors that could confound the SNP-lipid associations, we also conducted additional analyses with additional covariates, including menopausal status, current smoking, and type 2 diabetes, in the CLHNS mothers. In the CLHNS offspring, we used SAS version 9.2 (SAS Institute, Cary, NC) to test the association between lipid traits and selected SNPs that were reported in previous GWA studies. As the ages of all offspring were within two years of each other, only sex, household assets, ln household income, and the first seven PCs were included as covariates. To examine the association in the combined cohort of mothers and offspring, we applied a general linear mixed model that accounted for the correlation of the outcome trait between mother-child pairs due to shared genetic and environmental exposures. Age, age2, sex, household assets, ln household income, the first seven PCs, and generation (mother/offspring) were used as covariates.
For each SNP that showed evidence of a main effect association with a lipid trait in the CLHNS (P < 0.1 in both mothers and offspring analyzed separately), we further tested for a genotype interaction with waist circumference by including a SNP-by-waist circumference interaction term in the linear regression model or in the linear mixed model. The same sets of covariates used in the main effect analyses were included in the interaction analyses. For each SNP that showed significant evidence of interaction (Pinteraction < 0.05 in both mother and offspring cohorts), we conducted a stratified analysis by categorizing the study samples based on quartiles of waist circumference and testing for the main effects of the SNP on lipids within quartiles. All interaction analyses were conducted in SAS version 9.2 (SAS Institute, Cary, NC).
RESULTS
The general characteristics of the 1,782 mothers and 1,719 young adult offspring from the CLHNS studied in these genetic analyses are shown in Table 1. Association analyses revealed genome-wide significant associations (P < 5 × 10−8) at APOE for both LDL-C (rs7412, P = 1.6 × 10−27) and TC (rs7412, P = 1.8 × 10−14), and at APOA5 for TGs (rs662799, P = 3.1 × 10−18) in the CLHNS mothers (Table 2). Suggestive evidence of association at a less stringent threshold of P < 10−6 was detected at the TG locus GCKR (rs780092, P = 1.6 × 10−7) and the HDL-C locus CETP (rs1800775, P = 6.3 × 10−7). In addition, we observed that variants at TOM1, a new HDL-C locus recently identified in a large-scale meta-analysis in individuals of European ancestry (9), also exhibited suggestive evidence of association in the mothers (rs138777, P = 5.0 × 10−7).
TABLE 1.
General characteristics of CLHNS mothers and young adult offspring
| Characteristic | Mothers (n = 1,782) | Offspring (n = 1,719) |
| Female (%) | 100 | 47.6 |
| Age in 2005 (years) | 48.4 ± 6.1 | 21.5 ± 0.3 |
| Household assets in 2005 (0 to 11) | 5.2 ± 2.0 | 5.2 ± 2.0 |
| Household income in 2005 (pesos/week) | 396.4 (244.6, 623.9) | 357.7 (213.6, 586.7) |
| TC (mmol/l) | 4.79 (4.09, 5.44) | 3.94 (3.44, 4.58) |
| HDL-C (mmol/l) | 1.06 ± 0.27 | 1.09 ± 0.29 |
| LDL-C (mmol/l) | 3.10 ± 0.87 | 2.44 ± 0.75 |
| TGs (mmol/l) | 1.25 (0.91, 1.78) | 0.99 (0.74, 1.41) |
| Waist circumference (cm) | 80.0 (73.5, 88.5) | 69.0 (65.0, 74.1) |
Data are mean ± SD, median (25th percentile, 75th percentile), or percent. Only one of the CLHNS mothers and one of the young adult offspring were taking lipid-altering medication.
TABLE 2.
Loci associated with levels of lipoprotein cholesterol and TGs in the CLHNS
| Locus | SNP | Trait | Effect Allele | Other Allele | Mothers (n = 1,782)a | Offspring (n = 1,719)b | Combinedc | |||||
| EAF | β (SE) | P | EAF | β (SE) | P | β (SE) | P | |||||
| APOE | rs7412 | LDL-C | T | C | 0.12 | −0.48 (0.04) | 1.6E-27 | 0.11 | −0.49 (0.04) | 1.0E-35 | −0.48 (0.03) | 2.7E-53 |
| APOE | rs7412 | TC | T | C | 0.12 | −0.08 (0.01) | 1.8E-14 | 0.11 | −0.11 (0.01) | 1.7E-20 | −0.09 (0.01) | 1.5E-30 |
| APOA5 | rs662799 | TGs | A | G | 0.76 | −0.18 (0.02) | 3.1E-18 | 0.73 | −0.12 (0.02) | 2.6E-10 | −0.14 (0.01) | 5.7E-24 |
| GCKR | rs780092 | TGs | A | G | 0.68 | 0.09 (0.02) | 1.6E-07 | 0.69 | 0.06 (0.02) | 3.5E-04 | 0.08 (0.01) | 1.8E-09 |
| CETP | rs1800775 | HDL-C | A | C | 0.40 | 0.05 (0.01) | 6.3E-07 | 0.43 | 0.04 (0.01) | 1.7E-04 | 0.04 (0.01) | 3.4E-09 |
| TOM1 | rs138777 | HDL-C | G | A | 0.59 | 0.05 (0.01) | 5.0E-07 | 0.61 | 0.02 (0.01) | 0.058 | 0.03 (0.01) | 4.0E-05 |
Results are shown if P < 10−6 in the CLHNS mothers. EAF, effect allele frequency.
Associations in the CLHNS mothers were adjusted for age, age2, household assets, ln household income, and the first seven PCs of population substructure.
Associations in the CLHNS offspring were adjusted for sex, household assets, ln household income, and the first seven PCs.
The mixed model analysis of combined samples was adjusted for age, age2, sex, household assets, ln household income, the first seven PCs, and generation (mothers/offspring).
We further evaluated these SNP associations in the CLHNS young adult offspring genotyped using the Metabochip (Table 2). We observed significant associations (P < 5 × 10−8) for the variants at APOE (P = 1.0 × 10−35 for LDL-C and 1.7 × 10−20 for TC) and APOA5 (P = 2.6 × 10−10), and nominal association for SNPs at GCKR (P = 3.5 × 10−4) and CETP (P = 1.7 × 10−4). For the SNP rs138777 at TOM1, we found marginal association with HDL-C in the offspring (P = 0.058). In the combined analysis including both mothers and offspring, all of these loci, except TOM1 (P = 4.0 × 10−5), reached significance at P < 5 × 10−8 (Table 2).
We next assessed whether additional SNP-lipid associations identified in previous GWA studies could be extended to Filipino middle-aged and young adults, groups that have not been widely studied. As all the loci were previously reported, we defined the evidence of association as P < 0.1 for the reported GWA index SNPs or their proxies in both mothers and offspring with a consistent direction of effect. Among the 95 previously reported lipid loci (8), 14 reached this threshold in the CLHNS. In addition to the four loci (APOE, APOA5, GCKR, and CETP) described in Table 2, the additional loci included three for TGs (LPL, MLXIPL, and ANGPTL3), three for HDL-C (LIPC, MMAB-MVK, and LIPG), two for LDL-C (ABO and APOB), and two for TC (TIMD4 and DNAH11) (Table 3). Analyses including both mothers and offspring showed that the variant rs588136 at LIPC also exhibited genome-wide significant association with HDL-C levels in the combined samples (P = 1.5 × 10−12).
TABLE 3.
Additional previously reported SNPs that exhibited evidence of association with lipoprotein cholesterol and TGs in the CLHNS
| Locus | Trait | SNP Reported Previouslya | SNP Analyzed in CLHNS | LD r2b | Mothers (n = 1,782) | Offspring (n = 1,719) | Combined | |||||
| EAF | β (SE) | P | EAF | β (SE) | P | β (SE) | P | |||||
| LPL | TGs | rs12678919 | rs328 | 0.99 | 0.95 | 0.11 (0.04) | 7.9E-03 | 0.95 | 0.07 (0.04) | 0.10 | 0.09 (0.03) | 1.6E-03 |
| MLXIPL | TGs | rs17145738 | rs17145738 | 1.00 | 0.10 | −0.05 (0.03) | 0.077 | 0.10 | −0.06 (0.03) | 0.040 | −0.06 (0.02) | 4.2E-03 |
| ANGPTL3 | TGs | rs2131925 | rs2131925 | 1.00 | 0.71 | 0.04 (0.02) | 0.036 | 0.69 | 0.04 (0.02) | 0.034 | 0.04 (0.01) | 5.1E-03 |
| LIPC | HDL-C | rs2070895 | rs588136 | 0.87 | 0.69 | −0.05 (0.01) | 5.1E-06 | 0.63 | −0.06 (0.01) | 1.1E-09 | −0.05 (0.01) | 1.5E-12 |
| LIPG | HDL-C | rs7241918 | rs2156552 | 0.94 | 0.14 | −0.02 (0.01) | 0.075 | 0.15 | −0.03 (0.01) | 0.031 | −0.03 (0.01) | 4.6E-03 |
| MMAB-MVK | HDL-C | rs2338104 | rs10774708 | 1.00 | 0.62 | −0.02 (0.01) | 0.076 | 0.61 | −0.03 (0.01) | 3.6E-03 | −0.02 (0.01) | 0.011 |
| ABO | LDL-C | rs9411489 | rs2519093 | 0.95 | 0.85 | −0.10 (0.05) | 0.033 | 0.83 | −0.13 (0.03) | 1.6E-04 | −0.12 (0.03) | 3.0E-05 |
| APOB | LDL-C | rs1367117 | rs1367117 | 1.00 | 0.88 | −0.09 (0.05) | 0.047 | 0.87 | −0.09 (0.04) | 0.023 | −0.08 (0.03) | 7.4E-03 |
| TIMD4 | TC | rs6882076 | rs6882076 | 1.00 | 0.65 | 0.01 (0.01) | 0.060 | 0.65 | 0.02 (0.01) | 0.035 | 0.02 (0.01) | 4.0E-03 |
| DNAH11 | TC | rs12670798 | rs5008148 | 0.94 | 0.55 | 0.02 (0.01) | 0.072 | 0.58 | 0.01 (0.01) | 0.052 | 0.02 (0.01) | 0.012 |
Results are shown if P < 0.1 in both CLHNS mothers and offspring with a consistent direction of effect. EAF, effect allele frequency.
LD r2 between the SNP pairs was estimated based on the 1000 Genomes Project phase I EUR data (November 2010 release).
Given prior evidence that up to 10 lipid-associated loci exhibited two or more signals (7, 8, 14), and in particular, that at least four lipid loci have been reported to contain multiple signals in East Asians (APOA5 for TG, CETP for HDL-C, and APOE and ABO for LDL-C) (14), we next tested for the association with these additional variants in the CLHNS samples. We conditioned on the SNP genotype of the initial signal and defined a secondary signal as the most strongly associated SNP with a Pconditional < 5 × 10−4 in combined mothers and offspring. After accounting for the effects of the most strongly associated SNPs at each of the five loci that reached GWA significance in the combined mothers and offspring, we confirmed the presence of secondary signals at four loci including the TG locus APOA5, the HDL-C loci CETP and LIPC, and the LDL-C locus APOE (Table 4). Except at the LDL-C locus APOE, the evidence of association of the second signals was attenuated when we conditioned on the first signals at the corresponding loci [|Δ(−log10P)| > 2], suggesting that these additional signals at APOA5, CETP, and LIPC were not completely independent of the first signals (Table 4). In further analyses adjusting for the additional covariates of menopausal status, current smoking, and type 2 diabetes status (supplementary Table II), we observed similar results for the SNPs described above [all |Δ(−log10P)| < 1, supplementary Table III].
TABLE 4.
Lipid loci showing evidence of two signals in the CLHNS
| Locus | Trait | SNP | Mothers (n = 1,782) | Offspring (n = 1,719) | Combined | LD (D′/r2)c | |||||
| EAF | Pinitiala | Pconditionalb | EAF | Pinitiala | Pconditionalb | Pinitiala | Pconditionalb | ||||
| APOA5 | TGs | rs662799 | 0.76 | 3.1E-18 | 2.5E-15 | 0.73 | 2.6E-10 | 7.7E-07 | 5.7E-24 | 1.1E-17 | 1.00/0.09 |
| rs3741297 | 0.98 | 3.1E-06 | 7.4E-03 | 0.97 | 3.5E-08 | 1.1E-04 | 3.2E-12 | 1.1E-05 | |||
| CETP | HDL-C | rs1800775 | 0.40 | 6.3E-07 | 1.1E-04 | 0.43 | 1.7E-04 | 0.013 | 3.4E-09 | 1.2E-05 | 0.64/0.06 |
| rs7499892 | 0.84 | 1.9E-06 | 3.1E-04 | 0.84 | 4.1E-09 | 2.4E-07 | 3.5E-12 | 1.1E-08 | |||
| LIPC | HDL-C | rs588136 | 0.69 | 5.1E-06 | 1.0E-04 | 0.63 | 1.1E-09 | 2.0E-07 | 1.5E-12 | 1.6E-09 | 0.34/0.06 |
| rs1532085 | 0.43 | 2.6E-04 | 6.1E-03 | 0.46 | 4.1E-05 | 6.0E-03 | 5.8E-07 | 4.5E-04 | |||
| APOE | LDL-C | rs7412 | 0.12 | 1.6E-27 | 5.9E-28 | 0.11 | 1.0E-35 | 2.6E-33 | 2.7E-53 | 5.4E-52 | 1.00/0.01 |
| rs769449 | 0.91 | 0.032 | 0.015 | 0.91 | 1.5E-08 | 4.4E-06 | 5.0E-07 | 1.5E-05 | |||
Results are shown for SNPs with Pinitial < 10−6 in the CLHNS mother and offspring combined cohort. EAF, effect allele frequency.
Pinitial: P values for main effect association.
Pconditional: P values produced by reciprocal conditional analyses, in which both the primary and secondary signals were included in the model.
As shown in supplementary Table IV, individuals with larger waist circumference have more atherogenic lipoprotein profiles. To investigate whether the associations in Tables 2–4 were affected by measures of waist circumference, a known predictor of CVD risk, we tested the SNP-lipid associations again while also adjusting for waist circumference in addition to the covariates included in the initial model. Results from analyses in the CLHNS mothers showed that the association evidence did not change substantially for most of the loci [|Δ(−log10P)| < 2] (supplementary Table III). However, adjusting for waist circumference appeared to influence the association of the APOA5 variant rs662799 with TGs (unadjusted P = 3.1 × 10−18; adjusted P = 2.8 × 10−21) (supplementary Table III).
We next assessed whether the lipid-associated SNPs exhibited evidence of interaction with waist circumference on lipid levels (supplementary Table V). Among the 20 signals at 15 loci with main effect associations shown in Tables 2–4, we observed SNP-by-waist circumference interactions (Pinteraction < 0.05) affecting TGs for APOA5 variants rs3741297 (Pinteraction = 9.6 × 10−3) and rs662799 (Pinteraction = 0.032), and affecting TC level for the APOE variant rs7412 (Pinteraction = 0.034) in CLHNS mothers. Additional analyses in the offspring confirmed the interaction for APOA5 rs3741297 (Pinteraction = 7.4 × 10−3). In a mixed model that combined the mothers and offspring together, the interaction remained significant (Pinteraction = 1.6 × 10−4), even after a conservative Bonferroni correction for multiple testing (P < 8.3 × 10−4, 0.05/60 tests).
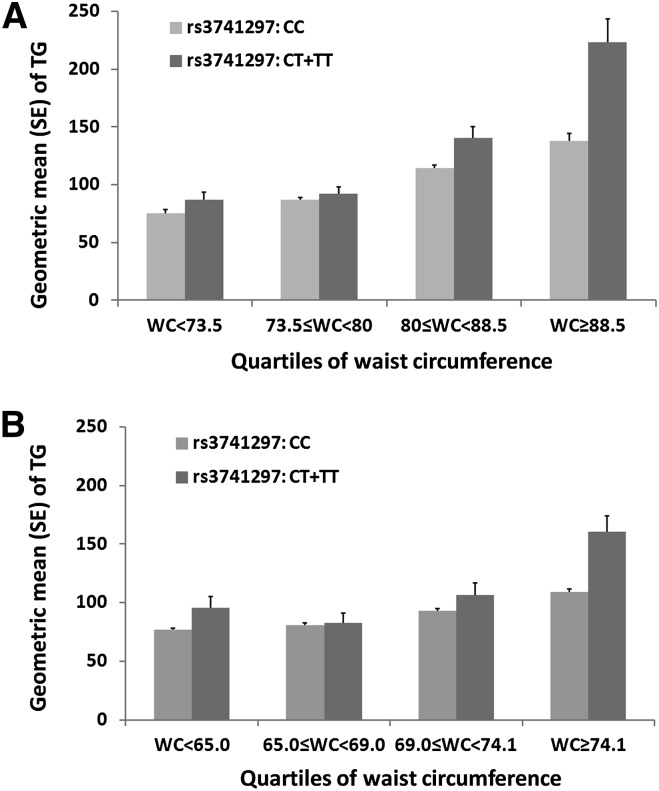
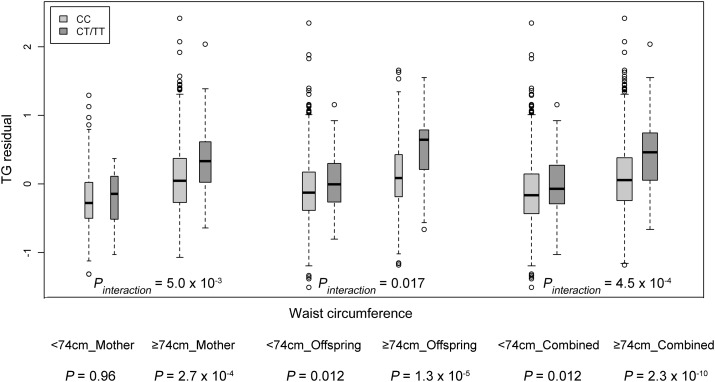
Further analyses stratified by quartiles of waist circumference supported the interaction between APOA5 variant rs3741297 and waist circumference affecting TG levels. In the mothers, the SNP had no effect on TGs in the lowest quartile of waist circumference <73.5 cm (quartile (Q)1: geometric mean of TG level in individuals carrying CC = 0.85 mmol/l and CT+TT = 0.98 mmol/l, P = 0.93, n = 440), but the minor allele was significantly associated with higher TG level in higher quartiles of waist circumference (Q2: CC = 0.98 mmol/l and CT+TT = 1.04 mmol/l, P = 0.047, n = 442; Q3: CC = 1.29 mmol/l and CT+TT = 1.59 mmol/l, P = 2.6 × 10−3, n = 444; Q4: CC = 1.56 mmol/l and CT+TT = 2.52 mmol/l, P = 4.5 × 10−3, n = 443; Fig. 1A). The magnitudes of the association became stronger as waist circumference increased (Q1: β = −0.01, Q2: β = 0.24, Q3: β = 0.51, and Q4: β = 0.52). Notably, we observed a consistent pattern of interaction in the offspring, with stronger SNP effects in individuals with higher waist circumference (Q1: β = 0.21, n = 408; Q2: β = 0.04, n = 411; Q3: β = 0.14, n = 415 and Q4: β = 0.37, n = 415; Fig. 1B). The mothers had a higher median waist circumference than the offspring; we next stratified the samples using a threshold of 74.0 cm, the median of waist circumference in the combined cohort. In the combined mothers and offspring, individuals carrying the T allele of rs3741297 had significantly higher TG levels compared with the CC carriers only in the larger waist circumference group (≥74.0 cm; β = 0.41, 0.38, and 0.38 in mothers, offspring, and combined cohorts, respectively), but not in the smaller waist group (<74.0 cm; β = −0.01, 0.14, and 0.12 in mothers, offspring, and combined cohorts, respectively) (Fig. 2).
Fig. 1.
Differences of TG levels between APOA5 variant rs3741297 CC and CT+TT carriers stratified by quartiles of waist circumference (WC). A: in CLHNS mothers; B: in CLHNS offspring. The T allele of rs3741297 is in complete LD (r2 = 1.0) with the C allele (185Cys) of rs2075291.
Fig. 2.
Differences of TG residual between rs3741297 CC and CT+TT carriers stratified by a waist circumference of 74.0 cm, the median of waist circumference in mother and offspring combined cohort, in the CLHNS mothers, offspring, and combined cohort. The TG residual was generated by adjusting for age, age2, household assets, ln household income, and first seven PCs in the CLHNS mothers; for sex, household assets, ln household income, and the first seven PCs in the CLHNS offspring; and for age, age2, sex, household assets, ln household income, the first seven PCs, and generation (mothers/offspring) in combined cohort.
As sex may influence the SNP association with lipid traits, we further tested the evidence of sex differences among the SNPs with main effect association. As shown in supplementary Table VI, all P values for SNP-by-sex interaction were >0.05, except for a variant rs7499892 at CETP, which exhibited suggestive evidence of sexual dimorphic association for HDL-C level in CLHNS offspring (Pinteraction = 0.045, without Bonferroni correction); a sex-stratified analysis showed a stronger genetic effect in women (β = 0.098, P = 1.4 × 10−6) than in men (β = 0.051, P = 5.5 × 10−4).
DISCUSSION
In this study of genetic associations with lipoprotein and TG levels in Filipinos from the CLHNS, we observed the associations at 15 loci, which accounted for 3.1, 4.7, 6.8, and 6.2% of the phenotypic variation of TC, HDL-C, LDL-C, and TGs, respectively, in the mothers from the CLHNS. Consistent with previous findings (7, 8), in particular those from populations of East Asians (14), our data also supported the existence of allelic heterogeneity at APOA5, CETP, LIPC, and APOE; the four secondary signals observed at these loci led to an average 23% increase in the amount of phenotypic variance compared with that explained by the most strongly associated variants alone. In addition, the findings of these consistent SNP-lipid associations in a young adult population aged ∼20 years, who had healthier lipid levels compared with middle-aged or older individuals, suggests a shared genetic influence on lipid levels across adulthood despite differing durations of environmental exposures. While we did not confirm the other previously reported loci, this might reflect our modest power to detect the association, differences in linkage disequilibrium (LD) between analyzed variants and underlying functional variants, or population-specific loci. Furthermore, the modifying effects of environmental factors could have impeded the detection of some associations (16).
A novel and important finding of this study is the evidence of a significant interaction between an APOA5 (apolipoprotein A-V) variant and waist circumference affecting TG levels in both mother and offspring cohorts in the CLHNS. Waist circumference, a measure of abdominal adiposity, may predict CVD risk better than BMI (34). Waist circumference is strongly associated with CVD and its risk factors including TGs (35). The increasing difference in TG levels between rs3741297 CC and CT+TT carriers with increasing waist circumference (Fig. 1) suggests that a larger waist circumference augments the genetic effect of the TG-elevating minor allele T. The range of the trait differed in the CLHNS mothers and offspring (Table 1), reflecting the trend toward increasing waist circumference with age (36). We observed that the associations were significant in the three higher quartiles of waist circumference in mothers (≥73.5 cm) and appeared stronger in the highest quartile in offspring (≥74.1 cm). When we stratified the individuals using a threshold of 74.0 cm, the median of waist circumference in the mothers and offspring combined cohort, the pattern of interaction was consistent across analyses in the mothers, the offspring, and the combined cohort. As the offspring were ∼20 years younger and had significantly lower TG levels and smaller waist circumference compared with those in their mothers, the consistent pattern of interaction suggested that the modifying effect of waist circumference influencing the APOA5-TG association was not affected by adult age. While the pattern of interaction was consistent in both cohorts, a combined sample size of ∼3,400 is still small, and further larger studies are needed to confirm these findings and evaluate the effect of duration of larger waist circumference.
Notably, the APOA5 rs3741297 is in complete LD (D′ = 1.0, r2 = 1.0 based on CLHNS offspring genotype data) with a nonsynonymous variant rs2075291 (Gly185Cys), which has been shown to regulate the activation of lipoprotein lipase (LPL) (37). Evidence from that study showed that the 185Cys substitution of APOA5 could cause reduced LPL-mediated hydrolysis leading to delayed TG clearance and elevated TG levels in the bloodstream (37). The in vitro identification of a tertiary structure change due to multimers of the 185Cys APOA5 protein further supports that residue 185 is critical in LPL activation (38).
The association between Gly185Cys and TGs was initially identified in a Chinese population (39) and subsequently replicated in Japanese (40). The very low frequency of Gly185Cys in other populations (minor allele frequency < 0.005 in Europeans and African Americans) compared with Asians (minor allele frequency ∼0.03) may explain why its association with TGs has only been detected in Asians to date (14). As Gly185Cys was not present on the GWA genotyping array or the 1000 Genomes Project reference panel (June 2010 release), the variant was not directly genotyped or imputed in the CLHNS mothers; however, our results based on the Metabochip genotyped data in the offspring showed the same significant main effect association on TG level and interaction with waist circumference as for the tested proxy variant rs3741297. In the CLHNS mothers, the ∼4% of individuals carrying the TG-increasing T allele of the proxy SNP rs3741297 (LD r2 = 1.0 with 185Cys), had significantly higher TG levels (geometric mean ± SE: 1.63 ± 0.09 mmol/l, approximately to the upper limit of 1.70 mmol/l for normal TG levels) compared with nonrisk allele carriers (1.28 ± 0.01 mmol/l).
The modifying effect of waist circumference on the APOA5 association with TGs is biologically plausible. The APOA5 185Cys variant's role in reducing LPL activation may be exacerbated by the effect of abdominal adiposity on LPL mass and activity. Intra-abdominal visceral fat accumulation has been shown to affect LPL mass and activity either directly or via insulin resistance, thereby impairing the hydrolysis of TGs (24, 25). Similarly, earlier studies demonstrated an interaction of the LPL-HindIII genotype with abdominal obesity on TG levels (41, 42) and hypothesized that the presence of central obesity exacerbates the alterations in LPL activity caused by genetic variation, thus preventing the possible compensatory effects that may occur in nonobese individuals (42). Our identification of a SNP-by-waist circumference interaction suggests that future studies should analyze genetic associations in different strata of anthropometric, life style, and other environmental factors.
In addition to the two APOA5 variants that we described in this study, many other variants at APOA5 have been shown to exhibit significant association with TG levels in diverse populations (4–8, 12, 14). Among these reported SNPs, several variants including rs964184 (7, 8), rs3741298 (14), and rs651821 (12, 14) also showed genome-wide significant association (all P < 1 × 10−11) in the CLHNS mothers; however, the significant level of these associations were substantially attenuated by conditioning on the most strongly associated variant rs662799 (all Pconditional > 0.45), suggesting that these variants and rs662799 would represent a same TG signal. Other variants, including rs12272004 (6), rs28927680 (5), rs12286037 (4), and rs3135506 (14), which were common and significantly associated with TGs in European or African ancestry populations, were found not to be variable in the 1000 Genomes Project ASN samples.
A previous study in the CLHNS reported that compared with women from the USA, the Filipino women had significantly lower levels of HDL-C at all BMI levels and exhibited a higher prevalence of isolated HDL-C phenotype, which is defined as a combination of low HDL-C (<35 mg/dl) but normal TGs (<200 mg/dl) (2). Although these findings suggested potentially different effects of genetic variants across populations, we did not identify any additional loci/variants that were specific to Filipinos or had substantially larger genetic effects in this population compared with those in Europeans (8, 14). Our study may not have sufficient power to detect such association, but the unusually low HDL-C level in the Filipinos is possibly due to the different lifestyle and/or environmental exposures across populations.
In conclusion, our findings emphasize the importance of central obesity prevention in public health efforts to reduce CVD risk, particularly in individuals who are genetically susceptible to the disease.
Supplementary Material
Acknowledgments
The authors thank the University of San Carlos-Office of Population Studies Foundation research and data collection teams and the study participants who generously provided their time for this study.
Footnotes
Abbreviations:
- BMI
- body mass index
- CLHNS
- Cebu Longitudinal Health and Nutrition Survey
- CVD
- cardiovascular disease
- GWA
- genome-wide association
- HDL-C
- high density lipoprotein cholesterol
- LD
- linkage disequilibrium
- LDL-C
- low density lipoprotein cholesterol
- ln
- log-transformed
- PC
- principal component
- Q1(2, 3, 4)
- quartile 1(2, 3, 4)
- SNP
- single nucleotide polymorphism
- TC
- total cholesterol
- TG
- triglyceride
This work was supported by National Institutes of Health Grants DK-078150, TW-05596, HL085144; and Pilot Funds RR-20649, ES-10126, DK-56350. A.F.M. was supported by an Integrative Vascular Biology Fellowship, National Institutes of Health Grant HL-69768. The authors have no competing interests to declare.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of six tables.
REFERENCES
- 1.World Health Organization 2011. Cardiovascular Diseases (CVDs): Fact Sheet No. 317. World Health Organization (WHO), Geneva. [Google Scholar]
- 2.Rutherford J. N., McDade T. W., Feranil A. B., Adair L. S., Kuzawa C. W. 2010. High prevalence of low HDL-c in the Philippines compared to the US: population differences in associations with diet and BMI. Asia Pac. J. Clin. Nutr. 19: 57–67 [PMC free article] [PubMed] [Google Scholar]
- 3.Namboodiri K. K., Kaplan E. B., Heuch I., Elston R. C., Green P. P., Rao D. C., Laskarzewski P., Glueck C. J., Rifkind B. M. 1985. The Collaborative Lipid Research Clinics Family Study: biological and cultural determinants of familial resemblance for plasma lipids and lipoproteins. Genet. Epidemiol. 2: 227–254 [DOI] [PubMed] [Google Scholar]
- 4.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40: 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., et al. 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40: 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aulchenko Y. S., Ripatti S., Lindqvist I., Boomsma D., Heid I. M., Pramstaller P. P., Penninx B. W., Janssens A. C., Wilson J. F., Spector T., et al. 2009. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 41: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 466: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M. L., Mora S., et al. 2013. Discovery and refinement of loci associated with lipid levels. Nat. Genet. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keebler M. E., Deo R. C., Surti A., Konieczkowski D., Guiducci C., Burtt N., Buxbaum S. G., Sarpong D. F., Steffes M. W., Wilson J. G., et al. 2010. Fine-mapping in African Americans of 8 recently discovered genetic loci for plasma lipids: the Jackson Heart Study. Circ Cardiovasc Genet. 3: 358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y. J., Go M. J., Hu C., Hong C. B., Kim Y. K., Lee J. Y., Hwang J. Y., Oh J. H., Kim D. J., Kim N. H., et al. 2011. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat. Genet. 43: 990–995 [DOI] [PubMed] [Google Scholar]
- 12.Tan A., Sun J., Xia N., Qin X., Hu Y., Zhang S., Tao S., Gao Y., Yang X., Zhang H., et al. 2012. A genome-wide association and gene-environment interaction study for serum triglycerides levels in a healthy Chinese male population. Hum. Mol. Genet. 21: 1658–1664 [DOI] [PubMed] [Google Scholar]
- 13.Sanna S., Li B., Mulas A., Sidore C., Kang H. M., Jackson A. U., Piras M. G., Usala G., Maninchedda G., Sassu A., et al. 2011. Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet. 7: e1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y., Waite L. L., Jackson A. U., Sheu W. H., Buyske S., Absher D., Arnett D. K., Boerwinkle E., Bonnycastle L. L., Carty C. L., et al. 2013. Trans-ethnic fine-mapping of lipid loci identifies population-specific signals and allelic heterogeneity that increases the trait variance explained. PLoS Genet. 9: e1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumitrescu L., Goodloe R., Brown-Gentry K., Mayo P., Allen M., Jin H., Gillani N. B., Schnetz-Boutaud N., Dilks H. H., Crawford D. C. 2012. Serum vitamins A and E as modifiers of lipid trait genetics in the National Health and Nutrition Examination Surveys as part of the Population Architecture using Genomics and Epidemiology (PAGE) study. Hum. Genet. 131: 1699–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreasen C. H., Andersen G. 2009. Gene-environment interactions and obesity–further aspects of genomewide association studies. Nutrition. 25: 998–1003 [DOI] [PubMed] [Google Scholar]
- 17.Hellstrand S., Sonestedt E., Ericson U., Gullberg B., Wirfalt E., Hedblad B., Orho-Melander M. 2012. Intake levels of dietary long-chain PUFAs modify the association between genetic variation in FADS and LDL-C. J. Lipid Res. 53: 1183–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Junyent M., Tucker K. L., Smith C. E., Garcia-Rios A., Mattei J., Lai C. Q., Parnell L. D., Ordovas J. M. 2009. The effects of ABCG5/G8 polymorphisms on plasma HDL cholesterol concentrations depend on smoking habit in the Boston Puerto Rican Health Study. J. Lipid Res. 50: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmad T., Chasman D. I., Buring J. E., Lee I. M., Ridker P. M., Everett B. M. 2011. Physical activity modifies the effect of LPL, LIPC, and CETP polymorphisms on HDL-C levels and the risk of myocardial infarction in women of European ancestry. Circ Cardiovasc Genet. 4: 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Surakka I., Isaacs A., Karssen L. C., Laurila P. P., Middelberg R. P., Tikkanen E., Ried J. S., Lamina C., Mangino M., Igl W., et al. 2011. A genome-wide screen for interactions reveals a new locus on 4p15 modifying the effect of waist-to-hip ratio on total cholesterol. PLoS Genet. 7: e1002333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deurenberg-Yap M., Chew S. K., Deurenberg P. 2002. Elevated body fat percentage and cardiovascular risks at low body mass index levels among Singaporean Chinese, Malays and Indians. Obes. Rev. 3: 209–215 [DOI] [PubMed] [Google Scholar]
- 22.Adair L. S. 2004. Dramatic rise in overweight and obesity in adult Filipino women and risk of hypertension. Obes. Res. 12: 1335–1341 [DOI] [PubMed] [Google Scholar]
- 23.Huxley R., James W. P., Barzi F., Patel J. V., Lear S. A., Suriyawongpaisal P., Janus E., Caterson I., Zimmet P., Prabhakaran D., et al. 2008. Ethnic comparisons of the cross-sectional relationships between measures of body size with diabetes and hypertension. Obes. Rev. 9(Suppl 1): 53–61 [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi J., Tashiro J., Murano S., Morisaki N., Saito Y. 1998. Lipoprotein lipase mass and activity in post-heparin plasma from subjects with intra-abdominal visceral fat accumulation. Clin. Endocrinol. (Oxf.). 48: 515–520 [DOI] [PubMed] [Google Scholar]
- 25.Rodriguéz-Lee M., Bondjers G., Camejo G. 2007. Fatty acid-induced atherogenic changes in extracellular matrix proteoglycans. Curr. Opin. Lipidol. 18: 546–553 [DOI] [PubMed] [Google Scholar]
- 26.Adair L. S., Popkin B. M., Akin J. S., Guilkey D. K., Gultiano S., Borja J., Perez L., Kuzawa C. W., McDade T., Hindin M. J. 2011. Cohort profile: the Cebu longitudinal health and nutrition survey. Int. J. Epidemiol. 40: 619–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carba D. B., Bas I. N., Gultiano S. A., Lee N. R., Adair L. S. 2013. Waist circumference and the risk of hypertension and prediabetes among Filipino women. Eur. J. Nutr. 52: 825–832 [DOI] [PubMed] [Google Scholar]
- 28.Kuzawa C. W., Adair L. S., Avila J. L., Cadungog J. H., Le N. A. 2003. Atherogenic lipid profiles in Filipino adolescents with low body mass index and low dietary fat intake. Am. J. Hum. Biol. 15: 688–696 [DOI] [PubMed] [Google Scholar]
- 29.Lange L. A., Croteau-Chonka D. C., Marvelle A. F., Qin L., Gaulton K. J., Kuzawa C. W., McDade T. W., Wang Y., Li Y., Levy S., et al. 2010. Genome-wide association study of homocysteine levels in Filipinos provides evidence for CPS1 in women and a stronger MTHFR effect in young adults. Hum. Mol. Genet. 19: 2050–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voight B. F., Kang H. M., Ding J., Palmer C. D., Sidore C., Chines P. S., Burtt N. P., Fuchsberger C., Li Y., Erdmann J., et al. 2012. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 8: e1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Croteau-Chonka D. C., Wu Y., Li Y., Fogarty M. P., Lange L. A., Kuzawa C. W., McDade T. W., Borja J. B., Luo J., AbdelBaky O., et al. 2012. Population-specific coding variant underlies genome-wide association with adiponectin level. Hum. Mol. Genet. 21: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909 [DOI] [PubMed] [Google Scholar]
- 33.Adair L. S. 2001. Size at birth predicts age at menarche. Pediatrics. 107: E59. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization 2008. Waist Circumference and Waist-Hip Ratio: Report of a WHO Expert Consultation (Geneva, Switzerland, December 8–11, 2008). [Google Scholar]
- 35.Shen W., Punyanitya M., Chen J., Gallagher D., Albu J., Pi-Sunyer X., Lewis C. E., Grunfeld C., Heshka S., Heymsfield S. B. 2006. Waist circumference correlates with metabolic syndrome indicators better than percentage fat. Obesity (Silver Spring). 14: 727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens J., Katz E. G., Huxley R. R. 2010. Associations between gender, age and waist circumference. Eur. J. Clin. Nutr. 64: 6–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y. J., Lin Y. L., Chiang C. I., Yen C. T., Lin S. W., Kao J. T. 2012. Functional importance of apolipoprotein A5 185G in the activation of lipoprotein lipase. Clin. Chim. Acta. 413: 246–250 [DOI] [PubMed] [Google Scholar]
- 38.Dorfmeister B., Zeng W. W., Dichlberger A., Nilsson S. K., Schaap F. G., Hubacek J. A., Merkel M., Cooper J. A., Lookene A., Putt W., et al. 2008. Effects of six APOA5 variants, identified in patients with severe hypertriglyceridemia, on in vitro lipoprotein lipase activity and receptor binding. Arterioscler. Thromb. Vasc. Biol. 28: 1866–1871 [DOI] [PubMed] [Google Scholar]
- 39.Kao J. T., Wen H. C., Chien K. L., Hsu H. C., Lin S. W. 2003. A novel genetic variant in the apolipoprotein A5 gene is associated with hypertriglyceridemia. Hum. Mol. Genet. 12: 2533–2539 [DOI] [PubMed] [Google Scholar]
- 40.Yamada Y., Ichihara S., Kato K., Yoshida T., Yokoi K., Matsuo H., Watanabe S., Metoki N., Yoshida H., Satoh K., et al. 2008. Genetic risk for metabolic syndrome: examination of candidate gene polymorphisms related to lipid metabolism in Japanese people. J. Med. Genet. 45: 22–28 [DOI] [PubMed] [Google Scholar]
- 41.Vohl M. C., Lamarche B., Moorjani S., Prud'homme D., Nadeau A., Bouchard C., Lupien P. J., Despres J. P. 1995. The lipoprotein lipase HindIII polymorphism modulates plasma triglyceride levels in visceral obesity. Arterioscler. Thromb. Vasc. Biol. 15: 714–720 [DOI] [PubMed] [Google Scholar]
- 42.Sentí M., Bosch M., Aubó C., Elosua R., Masià R., Marrugat J. 2000. Relationship of abdominal adiposity and dyslipemic status in women with a common mutation in the lipoprotein lipase gene. The REGICOR investigators. Atherosclerosis. 150: 135–141 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.