Abstract
Macrophages play a key role in atherogenesis in part through excessive uptake of oxidized LDL (OxLDL) via scavenger receptors. Binding of OxLDL to macrophages has traditionally been assessed using radiolabeled OxLDL. To allow more efficient and convenient measurements, we developed a nonradioactive binding assay in which biotinylated OxLDL (Bt-OxLDL) is added to macrophages in 96-well microtiter culture plates under various conditions and the extent of binding is determined using solid phase chemiluminescent immunoassay techniques. As examples, we show that Bt-OxLDL displayed high and saturable binding to macrophages in contrast to Bt-LDL, which showed very low binding. In competition assays, unlabeled OxLDL and the anti-OxLDL monoclonal antibody E06 inhibited Bt-OxLDL binding to macrophages in a dose-dependent manner. Specific binding of Bt-OxLDL to ApoE/SR-A/CD36 triple knockout macrophages was reduced by 80% as compared with binding to macrophages from ApoE knockout mice. Binding of Bt-OxLDL to CD36 transfected COS-7 cells showed enhanced saturable binding compared with mock-transfected cells. This assay avoids the use of radioactivity and uses small amounts of materials. It can be used to study binding of OxLDL to macrophages and factors that influence this binding. The techniques described should be readily adaptable to study of other ligands, receptors, and cell types.
Keywords: macrophages, oxidized LDL, CD36, SR-A, monoclonal antibody E06, atherosclerosis
Atherosclerosis is a chronic inflammatory disease that is a major cause of morbidity and mortality. Although its etiology is complex, oxidation of LDL trapped by the extracellular matrix in the intima, to generate oxidized LDL (OxLDL), is widely regarded as an important if not central event in atherogenesis (1). Intimal macrophages play a key role in the formation and progression of atherosclerosis by binding and taking up OxLDL via scavenger receptors, including CD36 and scavenger receptor A (SR-A) (2). Excessive and unregulated uptake of OxLDL by macrophages results in foam cell formation, which is the first stage of the developing atherosclerotic lesion (3).
Understanding the mechanisms responsible for binding and uptake of OxLDL by macrophages could lead to novel therapeutic approaches to inhibit foam cell formation and thus atherogenesis. For example, we have demonstrated that various oxidation specific monoclonal antibodies that bind to relevant epitopes on OxLDL can prevent OxLDL binding to macrophage scavenger receptors in culture conditions and when expressed in excess in vivo in murine models of atherosclerosis can inhibit progression of atherosclerosis (4, 5). The traditional in vitro techniques used to assess OxLDL binding to macrophages and the ability of a given antibody to block this binding requires radioactive labeling of the ligand, the use large culture plates and relatively large amounts of cells and reagents, and time-consuming processing of samples. In addition, the use of radioactivity is cumbersome and expensive and requires dedicated laboratory space and regulatory monitoring.
Our objective was to develop a nonradioactive binding assay that would allow for the rapid detection of binding of OxLDL to various macrophages that would be efficient and rapid, and that would use small amounts of reagents. Our laboratory is well versed in solid phase immunoassay techniques that use 96-well microtiter plates, the use of multichannel pipettes that facilitate reagent delivery, and the use of chemiluminescent plate readers that can detect chemiluminescence in each well of a 96-well plate in less than a minute. We adapted this basic technology to detect biotin-labeled OxLDL binding to macrophages cultured in sterile 96-well culture plates. The developed technique is fast, efficient, and practical and allows the possibility of testing many experimental parameters (e.g., various concentrations of ligands and competitors) under simultaneous and identical conditions. To demonstrate the potential of the method, we use this methodology to characterize the specific binding of OxLDL to macrophages and specifically to CD36 and the ability of the monoclonal antibody E06 to inhibit this binding.
Materials and Methods
Materials
J774 cells and COS-7 cells were from the American Type Culture Collection (Manassas, VA). Dulbecco's PBS (DPBS), Trypsin-EDTA, RPMI-1640, and DMEM with 4.5 g/l glucose, L-glutamine, and sodium pyruvate were from Mediatech Inc. (Manassas, VA). FBS was from Omega Scientific (Tarzana, CA), and BSA was from Sigma-Aldrich (St. Louis, MO). Versene and gentamicin (10 mg/ml) were from Invitrogen (Grand Island, NY). Hoechst reagent was from Calbiochem (Billerica, MA). FuGene6 was from Roche Applied Science (Indianapolis, IN). Levamisole solution was from Vector Laboratories (Burlingame, CA). EZ-Link Sulfo-NHS-Biotin, No-Weigh Format and alkaline phosphatase-conjugated NeutrAvidin were from Pierce Biotechnology (Rockford, IL). LumiPhos 530 was from Lumigen Inc. (Southfield, MI). Sterile 96-well microplates were from Greiner Bio-One (Cat#655083; Monroe, NC). The murine monoclonal IgM antibody E06 was generated and characterized in our laboratory (6).
Methods
Lipoprotein preparation and modification.
LDL (density = 1.019–1.063 g/ml) was isolated from normolipidemic human plasma collected in EDTA by ultracentrifugation (7). Throughout the report, all preparations of LDL (native, biotinylated, and oxidized) were handled as follows. For preparations that were to be used as native LDL, we added butylated hydroxytoluene (20 µM final concentration), and the sample was transferred to a dialysis tubing (MWCO 12–14 kDa) and dialyzed extensively five times against DPBS containing EDTA (0.1 mM) (at least 1:1,000; v/v) over >24 h at 4°C. LDL preparations were transferred to a sterile syringe and gently sterile filtered (0.45 μm; Millex-HV, Millipore) into sterile polypropylene cell culture tubes and stored at 4°C. The concentration of LDL was determined using the bicinchoninic acid assay reagent (8) and is expressed throughout this manuscript based on protein content. For LDL samples that were to be oxidized, we dialyzed the ultracentrifugal fraction against DPBS, and after oxidation (described below) we added butylated hydroxytoluene (20 μM final concentration) and then dialyzed against DPBS and EDTA and stored as described above.
Native LDL was biotinylated according to the manufacturer's protocol (Pierce Biotechnology) using the No-weigh EZ-Link Sulfo-NHS-Biotin reagent with the following modifications and considerations. The protocol describes that the biotinylation of IgG at 1–10 mg/ml using a 20-fold molar excess of the biotin reagent per mol IgG yields four to six biotin-tags per IgG molecule. Because LDL contains apoB and aminophospholipids, the biotinylation efficiency was expected to be different from IgG. It was therefore necessary to optimize the biotinylation reaction to detect bound LDL particles in the binding assay. Briefly, in our optimized protocol, the biotinylation reaction was carried out using 4 mg LDL-protein (7.27 nmol LDL, using 550 kDa as the molecular weight of LDL [apoB100]) in 1 ml DPBS, to which was added 60-fold molar excess of the Sulfo-NHS-Biotin reagent (38.6 μl of a 11.3 nmol biotin per µl solution of freshly dissolved 1 mg Sulfo-NHS-Biotin in 200 µl milli-Q H2O). The reaction mixture was incubated at room temperature (RT) for 1 h with occasional gentle mixing and swirling and handled as described above. In pilot experiments, where 6.7-, 20-, 60-, and 180-fold molar excess of the Sulfo-NHS-Biotin reagent were tested, the use of 60-fold molar excess of biotin reagent yielded the best combination of sensitivity and signal-to-noise ratio in the binding assay for various oxidation and aldehyde modifications of LDL (data not shown). The biotin-labeling may have to be optimized for different ligands and receptors and by each laboratory. For example, because this method of biotinylation uses derivative amines, including lysines, if one were to use this assay for assessing binding of LDL to fibroblasts, one may need to use an alternative labeling technique (e.g., one that binds carboxy groups) that would avoid modification of lysines that would impair binding of LDL to the LDL receptor.
Copper-induced oxidation of LDL (CuOxLDL) was prepared by incubating LDL at 4 mg/ml with 10 μM CuSO4 for 18 h at 37°C under sterile conditions (9). Biotinylated CuOxLDL (Bt-CuOxLDL) was prepared in parallel using biotinylated LDL (Bt-LDL). Oxidized LDL preparations were also handled as described above. The degree of oxidation was determined by measuring the amount of thiobarbituric acid-reactive substances (10) and by determining the binding of the antioxidized phospholipid IgM monoclonal antibody, E06, in ELISA. Although we have tested more than 6 months old native and oxidized Bt-LDL preparations without any detectable changes in binding characteristics, we have elected to prepare entirely new ligand preparations from freshly isolated LDL every 2 months.
Macrophage plating and culture conditions.
For each macrophage cell type used in these experiments, preliminary experiments were performed to determine the optimal plating conditions for a given cell type. All cell types were plated in sterile, opaque 96-well microplates (Greiner Bio-One) using 100 μl media per well. The following basal media were used. Media A contained DMEM (containing 2 mM L-glutamine and sodium pyruvate) containing 10% heat-inactivated FBS and Gentamicin (50 mg/l). Media B contained Media A conditioned on L929 cells for 4–5 days. Media C contained RPMI containing 10% heat-inactivated FBS and Gentamicin.
J774 macrophages were plated at 25,000 cells per well in a mixture of Media A and Media B (4:1; v/v) and incubated for 72 h at 37°C. Transfected COS-7 cells were plated at 20,000 cells per well in Media A and incubated 24 h at 37°C. Bone marrow-derived macrophages (BMDM) were prepared from BM cells removed aseptically from murine femurs and tibia and differentiated to macrophages using M-CSF in 10% FBS as described (11). These were plated at 100,000 cells per well in a mixture of Media C and Media B (7:3; v/v) and incubated for 24 h at 37°C. Thioglycollate-elicited peritoneal macrophages (TGEPM) were plated at 100,000 cells per well in Media A and incubated for 24 h at 37°C. BMDM and TGEPM were obtained from C57BL/6, SR-AI/II−/− (Sra−/−) (The Jackson Laboratory, Sacramento, CA; B6.Cg-Msr1tm1Csk/J, Stock #006096), Apoe−/−, or Cd36−/−/Sra−/−/ Apoe−/− triple knockout (ko) mice (a gift of Kathryn Moore) maintained in our vivarium. All animal experiments were performed according to National Institutes of Health guidelines and were approved by the University of California, San Diego Animal Subjects Committee.
Macrophage binding assay.
After incubations in the respective media described above, the following protocol was used for all cell types. The binding assay was carried out in the 96-well microplates without removing the cells. Cells were washed twice with 250 μl/well of PBS and placed on ice with 200 μl/well of ice-cold 1% BSA-PBS for 30 min to block nonspecific binding and to arrest cell growth. The media was then replaced with biotinylated ligands at varying concentrations in the absence or presence of competitors in 100 μl/well in 1% BSA-PBS (or DMEM) and incubated for 2 h on ice. Cells were washed twice with 250 μl of ice-cold PBS and fixed using 100 μl/well of 4% formaldehyde in ice-cold PBS for 15 min on ice, followed by 15 min at room temperature (RT). Macrophages were then washed twice with 250 μl of PBS at RT, and 100 μl of alkaline phosphatase-conjugated NeutrAvidin in 1% BSA-TBS (1:20,000) was added to each well. After 30 min of incubation at RT, macrophages were washed twice with 250 μl of TBS at RT. To detect bound ligands, macrophages were incubated with 50 μl/well of 50% LumiPhos 530 (a luminescent substrate) in double-distilled water for 90 min at RT in the dark. The light emissions were then measured as relative light units (RLU) per 100 ms (RLU/100 ms) using a Dynex Luminometer plate reader (DYNEX Technologies). All determinations were done in triplicate. At the end of the experiment, the number of cells present was quantified by use of Hoechst reagent and a standard curve as described (12). To determine specific binding of CuOxLDL, increasing concentrations of Bt-CuOxLDL were added to macrophages in the absence or presence of 20-fold excess unlabeled CuOxLDL. Specific binding was determined as the difference between total binding and binding in the presence of excess unlabeled OxLDL, as originally reported for acetyl LDL by Brown and Goldstein (13). Curve fitting and Kd values were determined using GraphPad Prism, Version 5.0c.
In competition binding experiments, a fixed and limiting dilution of Bt-CuOxLDL was added in each well in the absence and presence of increasing concentrations of various competitors, and the extent of binding was determined as described above. For these experiments, the data are expressed as the extent of binding in the presence of competitor (B) divided by binding in the absence of competitor (B0) to yield a B/B0 value, where binding in the absence of competitor equals 1.
Transfection of COS-7 cells with CD36.
COS-7 cells were transfected with CD36 DNA as previously described by Boullier et al. (9). COS-7 cells were maintained in Media A. For transfection, cells were cultured on a 6-well plate to approximately 70–80% confluence. They were then transfected with a mixture of 2 μg of human CD36 DNA subcloned into pSG5 (Stratagene, La Jolla, CA) and 6 μl of Fugene6 (Roche Applied Science). After 24 h, the cells were harvested with trypsin-EDTA and plated in sterile 96-well plates at 20,000 cells/well in Media A as described above. Cells transfected with empty vector pSG5 were used as a control. The binding and competition assays were performed 48 h after seeding of the transfected cells. Because of a high background chemiluminescence, Levamisole (1 drop/5 ml of substrate) was added to the LumiPhos 530 detection substrate in the final step of the binding assay to inhibit the endogenous alkaline phosphatase activity of the COS-7 cells.
Results
Macrophage binding assay
The macrophage binding assay was patterned after solid phase chemiluminescent immunoassays conducted in a 96-well plate format, enabling the use of high-throughput techniques, including multichannel pipettes and a chemiluminescent plate reader. Macrophages are cultured in sterile 96-well plates, which are then exposed to biotinylated ligands at varying concentrations in the presence and absence of competitors. The extent of bound ligand, such as biotinylated CuOxLDL (Bt-CuOxLDL), was determined by the use of alkaline phosphatase conjugated to avidin, which acts on the added LumiPhos 530 detection substrate to produce chemiluminescence, which was then measured by a plate reader. The extent of binding of a given ligand in each well is determined by the measured chemiluminescence, which is expressed as RLU/100 ms.
There were a number of important variables that needed to be optimized in developing the assay. Chief among these was finding the optimal number of cells to be plated per well that would lead to optimal receptor expression at the time of the binding assay, when plates were put on ice to inhibit internalization of bound ligand. This variable was dependent on the number of cells seeded per well initially, the media used, and the length of time in culture before actual performance of the binding assay. These conditions varied for the different macrophages studied and the optimal number of cells initially seeded per well, media used, and time in culture to achieve approximately 100% confluence at time of conduct of binding assay were established for J774 cells, BMDM, TGEPM, and CD36-transfected COS-7 cells. The conditions for each of these cell types are described in Materials and Methods. Because primary macrophages (BMDM and TGEPM) proliferate slowly in in vitro cultures, these cells were plated at high cell densities, ensuring fully confluent wells with optimally adhering macrophages 24 h later at the initiation of the binding assay. BMDM were cultured in 30% L929-conditioned media to ensure continued full differentiation to macrophages and reproducible and high expression of receptors, including scavenger receptors such as CD36. In contrast, TGEPM were fully differentiated in vivo and were therefore cultured in Media A without the need for growth factors. For J774 cells, we found that adherence and receptor-expression was not optimal 24 h after plating, which was probably due to the harvesting of cells cultured in flasks. Therefore, we plated 25,000 J774 cells per well, cultured them in 20% L929-conditioned media for 72 h, and found ∼100% confluent wells, which yielded ∼50% higher binding of Bt-CuOxLDL compared with J774 cells in Media A (without L929-conditioned media) (data not shown). Moreover, this improved binding applied to other biotinylated ligands, such as malondialdehyde-modified LDL (data not shown). Because transfected COS-7 cells were harvested with trypsin-EDTA, they were given 48 h in Media A because additional growth factors were not needed to reform surface receptors, adhere to wells, and proliferate to near confluency.
For each of these cell types, the conditions described resulted in appropriate cell density and scavenger receptor expression at the time of assay to yield the best CuOxLDL binding curves, as judged by apparent Kd and maximal binding determinations. In all assays, we added NeutrAvidin conjugated with alkaline phosphatase to detect the extent of binding of the Bt-CuOxLDL. For the macrophages tested (i.e., BMDM, TGEPM, and J774 cells), background chemiluminescence activity in the absence of added biotinylated ligands was low, showing little nonspecific signal in response to added NeutrAvidin-alkaline phosphatase. However, with COS-7 fibroblast cells there was considerable background activity, presumably reflecting endogenous alkaline phosphatase activity. We therefore added Levamisole to the final substrate solution, which inhibited endogenous alkaline phosphatase activity sufficiently to adequately reduce background. Preliminary experiments had demonstrated that Levamisole did not inhibit the alkaline phosphatase activity of the added NeutrAvidin-alkaline phosphatase complex, which is of a different isotype.
In initial experiments, we corrected the extent of binding of ligand in each well by the cell count in that well. The latter was determined by measuring the content of DNA in each well using Hoechst solution, measurement of fluorescence using SpectraMax Gemini EM (excitation 350 nm, emission 460 nm), and comparison to a simultaneously determined standard curve. The RLUs per well were divided by the DNA content to yield a corrected binding/cell value. An extensive set of preliminary studies indicated that for each cell type studied, when the reported conditions were used, the numbers of cells per well for a given plate at the end of the experiment were highly reproducible, and the corrected values did not differ significantly from uncorrected values. Therefore, in the data shown below, we report only the extent of binding per well, in RLU/100 ms. In all experiments, each value was determined in triplicate. Below we present examples of the application of this binding assay.
Specific binding of OxLDL to macrophages
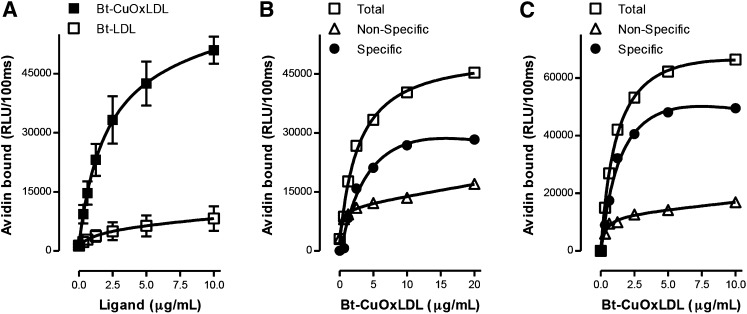
Figure 1A shows binding curves of Bt-CuOxLDL and Bt-LDL to J774 macrophages. Cells were cultured in 96-well microtiter plates for 24 h and then incubated with increasing concentrations of biotinylated ligands for 2 h. After appropriate washings, the extent of binding was determined. CuOxLDL displayed a high level of saturable binding, whereas that of native LDL revealed low but saturable binding, probably reflecting Bt-LDL recognition by the LDL receptor, which is sparsely expressed on J774 macrophages. We next measured the specific binding of CuOxLDL to J774 cells (Fig. 1B). Increasing concentrations of Bt-CuOxLDL were added to cells in the absence or presence of a 20-fold excess of unlabeled CuOxLDL. Data for total as well as nonspecific (binding in presence of excess unlabeled CuOxLDL) are graphed as are data for the calculated specific binding, which is the difference between total and nonspecific binding. The calculated apparent Kd for specific binding of CuOxLDL was 3.7 ± 1.0 μg/ml. Figure 1C shows the total, nonspecific, and specific binding curves of Bt-CuOxLDL to BMDM from Apoe−/− wild-type (wt) mice. For these data, the calculated apparent Kd for specific binding was 1.2 ± 0.2 μg/ml. These values agree well with similar Kd values reported for binding studies using iodinated OxLDL to various macrophage populations, which ranged from 1.0 to 7.4 μg/ml (14–18).
Fig. 1.
Total and specific binding of OxLDL to murine macrophages. In each panel, data are expressed as RLU/100 ms, and each point represents the average of triplicate determinations. A: Saturation binding curves of Bt-CuOxLDL and Bt-LDL to J774 macrophages. A total of 2.5 × 104 J774 macrophages/well were plated in 96-well culture plates and cultured for 72 h (see Materials and Methods). Cells were then washed and arrested on ice, and the media was replaced with ice-cold 1% BSA-PBS containing indicated amounts of Bt-CuOxLDL and Bt-LDL for 2 h. After extensive washing, the amount of biotinylated ligands bound to the plated cells was determined with the use of NeutrAvidin-alkaline phosphatase and a chemiluminescent ELISA technique as described in Materials and Methods. Data shown are mean ± SD of triplicate determinations from four independent experiments. B: Specific binding of CuOxLDL to J774 macrophages. Cells were plated and cultured in 96-well microtiter culture plates as in panel A. The cells were then washed and arrested on ice, and the media was replaced with 1% BSA-PBS containing indicated amounts of Bt-CuOxLDL in the absence or presence of a 20-fold excess of unlabeled CuOxLDL for 2 h. After extensive washing, the amount of Bt-CuOxLDL bound to the plated cells was determined as described in panel A. Specific binding was determined as the difference between total binding and binding in the presence of excess unlabeled CuOxLDL. Shown is one representative experiment of four. C: Specific binding of CuOxLDL to BMDM from Apoe−/− wt mice. After differentiation, 105 BMDM/well were plated in 96-well microtiter culture plates and incubated for 24 h (see Materials and Methods). Cells were washed and placed on ice, and then total and specific binding of OxLDL were determined as described in panel B. Data are from one representative experiment of two.
Role of scavenger receptors in binding of OxLDL
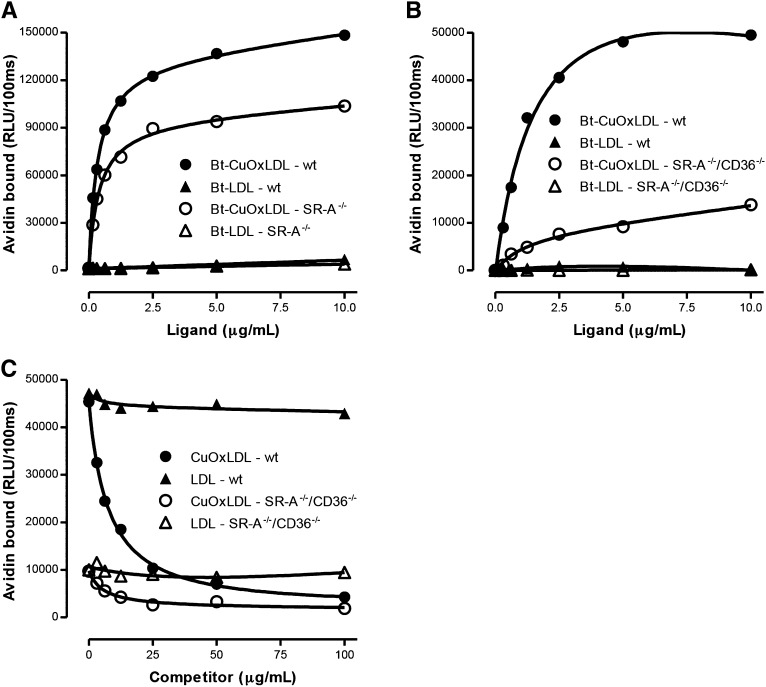
To demonstrate the role of scavenger receptors in binding OxLDL, we investigated the binding of CuOxLDL to macrophages deficient in SR-A or in SR-A and CD36. Figure 2A shows saturable binding of Bt-CuOxLDL and Bt-LDL to TGEPM from wt or scavenger receptor-AI/II knockout (Sra−/−) mice. Maximal binding of CuOxLDL was reduced approximately 30% in the Sra−/− macrophages compared with wt. Again, LDL showed minimal binding to wt or Sra−/− macrophages.
Fig. 2.
Binding of LDL and OxLDL to wt and scavenger receptor-deficient macrophages. A: Binding of Bt-OxLDL and Bt-LDL to thioglycollate-elicited peritoneal macrophages isolated from wt or SR-A−/− mice. Binding for each ligand was determined as described in the legend for Fig 1A. The apparent Kd for binding of CuOxLDL to wt macrophages was 0.42 ± 0.37 μg/ml and for binding to SR-A−/− macrophages was 0.46 ± 0.04 μg/ml. Maximal binding (Bmax) in ko macrophages was reduced ∼30% compared with wt. There was minimal binding of native LDL to wt or SR-A−/− macrophages. Data are representative of two experiments, with each point determined in triplicate. B: Specific binding of the same ligands to BMDM derived from Apoe−/− (wt) mice and from Cd36−/−/Sra−/−/ Apoe−/− triple ko mice (SR-A−/−/CD36−/−). Plating and culture of the BMDM and specific binding of CuOxLDL and LDL were performed as explained in the legend to Fig 1A. The apparent Kd for binding of CuOxLDL to Apoe−/− macrophages was 1.2 ± 0.2 μg/ml and to the triple ko macrophages was 3.5 ± 0.8 μg/ml. The Bmax to triple ko macrophages was reduced ∼80% compared with wt. Again, minimal binding of native LDL was noted to Apoe−/− or triple ko macrophages. Data are representative of two experiments, with each point determined in triplicate. Data for BMDM from Apoe−/− (wt) are the same as in Fig 1C. C: Competitive binding assay of Bt-CuOxLDL (2 μg/ml) to BMDM from Apoe−/− (wt) or Cd36−/−/Sra−/−/ Apoe−/− triple ko (SR-A−/−/CD36−/−) mice in the absence or presence of CuOxLDL or native LDL. Shown are the absolute levels of binding (in RLU/100 ms) for each ligand in the absence or presence of the indicated concentrations of competitors. The absolute binding of CuOxLDL to the triple ko macrophages is reduced about 80% compared with Apoe−/− macrophages. Unlabeled CuOxLDL was able to compete nearly all of the binding of Bt-CuOxLDL in Apoe−/− macrophages. In triple ko macrophages, unlabeled CuOxLDL was able to compete ∼75% of the residual Bt-CuOxLDL binding, consistent with the presence of other scavenger receptors capable of specific binding of OxLDL aside from CD36 and SR-A. Data are representative of two experiments, with each point determined in triplicate.
We next examined the binding of CuOxLDL to macrophages that were devoid of SR-A and CD36. Figure 2B shows specific binding of Bt-CuOxLDL and Bt-LDL to BMDM prepared from Apoe−/− or Cd36−/−/Sra−/−/Apoe−/− triple ko mice. Maximal binding of CuOxLDL to the triple ko macrophages was reduced approximately 80% compared with Apoe−/−. There was minimal binding of native LDL to Apoe−/− or triple ko macrophages.
We used a competitive binding assay to further examine the binding of native and modified LDL to different cell types. Figure 2C shows competitive binding of Bt-CuOxLDL (2 μg/ml) to BMDM prepared from Apoe−/− (wt) or Cd36−/−/Sra−/−/Apoe−/− ko mice in the absence or presence of CuOxLDL or native LDL. At the dosage of Bt-CuOxLDL used, the absolute binding of CuOxLDL to the triple ko macrophages was reduced approximately 80% compared with the Apoe−/− macrophages. Unlabeled Cu-OxLDL was able to compete nearly all of the binding of Bt-CuOxLDL in Apoe−/− macrophages, as expected, but was also able to compete approximately 75% of the residual Bt-CuOxLDL binding (i.e., nonSRA/non-CD36) in the triple ko macrophages. Thus, the absolute binding of CuOxLDL to SR-A/CD36 ko macrophages is decreased by ∼80%, consistent with prior reports (19, 20). However, the residual CuOxLDL binding also appears to be specific and likely accounted for by small numbers of other scavenger receptors that have been described to bind OxLDL (as reviewed in Ref. 2). As expected, native unlabeled LDL was not able to compete out the binding of Bt-CuOxLDL in Apoe−/− or triple ko macrophages.
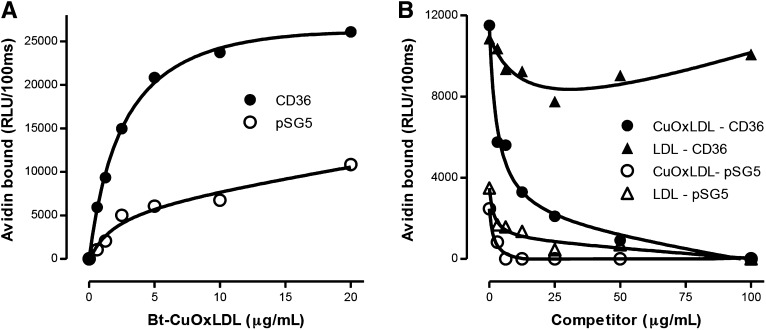
To further demonstrate the ability of CD36 to specifically bind OxLDL, we examined Bt-CuOxLDL binding to COS-7 fibroblast cells transfected with CD36. Figure 3A shows the specific binding of Bt-CuOxLDL to CD36 transfected or pSG5-mock-transfected COS-7 cells. Saturable and dose-dependent binding of CuOxLDL was observed for both cell types, but the apparent Kd for binding of CuOxLDL was 2.5 ± 0.2 μg/ml for the CD36 transfected cells and 5.8 ± 2.1 μg/ml for the mock-transfected cells. The apparent maximal binding value for the CD36 cells was ∼60% higher than for the mock-transfected cells. We also assessed specificity of CuOxLDL binding to the CD36 transfected cells. Figure 3B demonstrates that the absolute binding of Bt-CuOxLDL to mock-transfected cells was decreased approximately 60–70% compared with binding to CD36 transfected cells. Unlabeled CuOxLDL was able to fully compete out binding to CD36 transfected cells and could compete the low levels of binding to the pSG5 mock-transfected cells. Unlabeled LDL was able to compete to a minor degree the binding of OxLDL in these cells, possibly due to the presence of SR-B1, which is known to bind LDL and OxLDL (21).
Fig. 3.
OxLDL specifically binds to COS-7 cells transfected with CD36. A: Binding of Bt-CuOxLDL to CD36 transfected cells. COS-7 cells were transfected with a plasmid containing CD36 (CD36) or empty plasmid (pSG5) as described in Materials and Methods. A total of 20,000 cells/well were plated in 96-well microtiter culture plates and cultured 48 h as described in Materials and Methods. Cells were washed and specific binding of Bt-CuOxLDL was determined as described in the legend of Fig 1A. B: Competitive binding assay of Bt-CuOxLDL (2 μg/ml) to mock-transfected COS-7 cells (pSG5) or to CD36 transfected cells (CD36) in the absence or presence of CuOxLDL or native LDL. Shown are the absolute levels of binding (in RLU/100 ms) for each ligand in the absence or presence of the indicated concentrations of competitors. The absolute levels of binding of Bt-CuOxLDL to mock-transfected cells was decreased ∼60–70% compared with CD36-transfected cells. Unlabeled CuOxLDL was able to fully compete binding to CD36-transfected cells and could compete the low level of binding to the mock-transfected cells, suggesting a low level of endogenous scavenger receptor activity in the COS-7 cells. In these experiments, there was a significant level of background alkaline phosphatase activity, and, as explained in Materials and Methods, Levamisole was added to block this activity. In both panels, the data shown are representative of three experiments, with each point determined in triplicate.
Ability of monoclonal antibody E06 to block binding of CuOxLDL
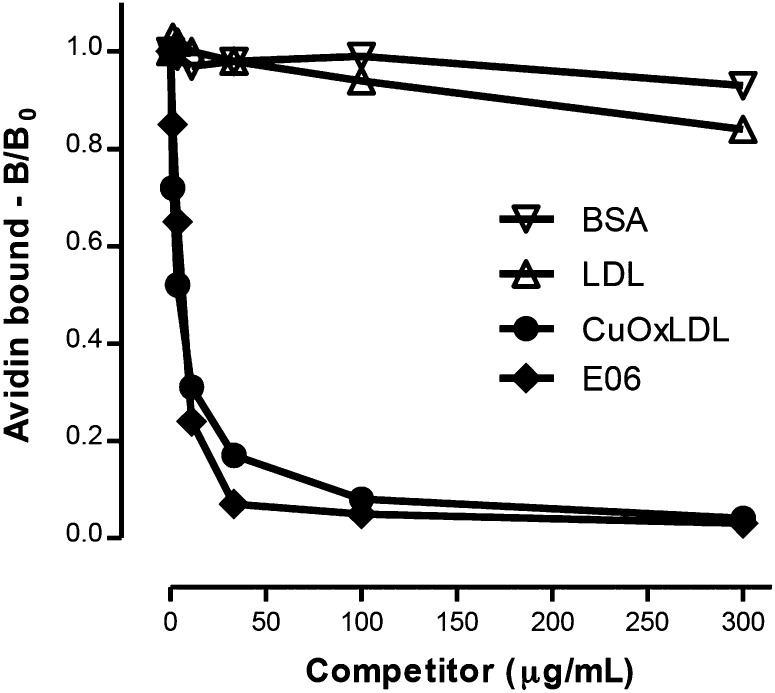
We previously used radioiodinated OxLDL binding assays to demonstrate that monoclonal antibody E06, directed to the phosphocholine head group of oxidized phospholipids, could inhibit the binding of OxLDL to macrophages, and specifically to CD36 and SR-B1 (9). In Fig. 4, we performed a similar assay using the current chemiluminescent technique. A fixed concentration of Bt-CuOxLDL was added to J774 cells in the absence or presence of increasing concentrations of E06 or indicated competitors. E06 exhibited high-affinity competition for binding, as did CuOxLDL, whereas native LDL and BSA were unable to compete (9).
Fig. 4.
Demonstration of the ability of monoclonal antibody E06 to inhibit binding of OxLDL to J774 macrophages. J774 cells were added to 96-well microtiter plates as described in Fig 1A. Bt-CuOxLDL (1 μg/ml) was added to each well in the absence or presence of the indicated concentrations of competitors: LDL, CuOxLDL, BSA, and E06 (a monoclonal IgM natural antibody that binds to the PC phosphocholine moiety of oxidized phospholipid). Only E06 affects the ability of CuOxLDL to bind to macrophage scavenger receptors. The experiment shown is the composite of two experiments, which were normalized by the use of B/B0 to describe the amount of ligand bound in the presence (B) or absence (B0) of competitor. Each value is the average of triplicate determinations.
Discussion
Brown and Goldstein (22, 23) made the seminal observation that native LDL could bind to specific receptors on the surface of fibroblasts, which led to the elucidation of the LDL receptor pathway, and how lipoprotein receptors regulated plasma and cellular lipid metabolism. Indeed, they extended this paradigm in another set of seminal studies by demonstrating that modified LDLs bound to a different set of receptors on macrophages that they termed “scavenger receptors” (13). These studies led to extensive investigations that are ongoing to understand the mechanisms by which native and modified lipoproteins regulate metabolism in health and disease.
The measurement of binding of OxLDL to various macrophage scavenger receptors continues to be an important component of studies elucidating mechanisms of inflammation and atherogenesis. The original binding studies of Brown and Goldstein used radioiodinated lipoproteins to define lipoprotein binding to cells. The protocols they described have been widely used by many laboratories, including our own (14, 15), and permit not only the ability to define binding parameters but to define rates of internalization and degradation of the lipoprotein. However, the use of radioactivity requires specialized training, a dedicated laboratory and equipment, and regulatory monitoring. Furthermore, we have previously shown that the iodination of LDL induces oxidation of LDL, which can continuously change the extent of LDL oxidation and change the binding characteristics of the “native” and oxidized LDL preparations (24). In addition, these assays require the use of relatively large culture plates and large amounts of cells and reagents. For many purposes, measurement of binding alone is sufficient. In this report, we describe the development and application of a nonradioactive binding assay of OxLDL to macrophage scavenger receptors that can be readily carried out in 96-well microtiter plates and that uses techniques originally developed for solid phase chemiluminescent immunoassays. The assay enables measurements of lipoprotein binding to cells of comparable sensitivity and specificity as achieved with iodinated lipoproteins. We have used this assay in prior reports (4, 5, 25, 26), but in this report we provide a more comprehensive description of the experimental variables, the use of different cells, and sufficient details whereby the assay could be duplicated in other laboratories. We provide the experimentally derived conditions to optimally conduct the assay with commonly used macrophage preparations, including TGEPM, BMDM, J774 macrophages, and COS-7 cells. We believe that the details provided should allow this assay to be readily adopted by most laboratories.
Previously, a nonradioactive “immunoreceptor” binding assay to study HDL binding to cells was described (27). An advantage of that assay is that it used an HDL ligand that was unlabeled. In that assay, HDL was incubated with cells in 0.7–1.0 ml of medium in 6-well plates for 48 h before the ligand incubation, after which the cells were washed, lysed, scraped, collected, and cleared by centrifugation, followed by protein quantification of the lysate. The lysate was then subjected to SDS-PAGE and Western blotting with an apoAI monoclonal antibody. Quantification was based on band intensities, standardized by known standards run in parallel. Although this assay offers an alternative technique, it appears to be more labor intensive and would be arduous to generate 96 determinations, whereas we routinely study three plates and thus 288 determinations in a typical binding study. Furthermore, such an immunoreceptor assay would not be amenable to detection of binding of OxLDL because during oxidation apoB is degraded (28) and results in multiple bands and even a smear on Western blotting with an apoB antibody (29). The use of E06, which binds oxidized phospholipid adducts with apoB, would also not be applicable because multiple apoB bands containing oxidized phospholipids are formed.
The binding assay described allows one to readily measure total and specific binding of a given ligand and to conduct competition assays with a variety of competitors to define specificity of binding.
A major advantage of this assay is the small volumes of reagents used. The examples shown used a total volume of 100 µl, but this could be reduced to 50 µl if needed. This makes it useful, for example, to assess the ability of reagents or antibodies to block binding of OxLDL to macrophages, a topic of interest to our laboratory. We show in Fig. 4 the ability of the innate natural antibody E06, which binds to the phosphocholine of oxidized phospholipids in OxLDL, to block binding of OxLDL to macrophage scavenger receptors. We have previously used this assay to measure the biological activity of such antibodies in murine plasma, using very small amounts of plasma (e.g., < 5 µl) (4, 5). Similarly, one could use this assay to screen a variety of small molecules, and in a dose-dependent manner, to inhibit OxLDL binding to macrophages or to identify drugs that regulate scavenger receptor expression.
The assay essentially uses techniques we have developed for solid phase chemiluminescent immunoassays (26). Because it uses the tools developed for such high-throughput assays, it is feasible to perform this assay on literally hundreds of samples at one time. However, several technical caveats about the conduct of the assay should be noted. First, in our routine chemiluminescent immunoassays, in which small antigens are directly plated in the wells, we use a semiautomated plate washer that simultaneously washes all 96 wells in the different washing steps required. For most cell types studied, we were able to use the plate washer after adjusting the setting to “cells,” which had a more gentle cycle. However, for the COS-7 cells, despite adjusting the conditions, use of the plate washer still led to an irreproducible removal of cells from wells of the plate. Therefore, in these assays, we added washing buffers “by hand” with multichannel pipettes and then, after allowing appropriate time for incubation, removed the buffer by inversion of the plate over a sink, followed by gentle tapping of the inverted plate on a paper towel to facilitate washing buffer removal.
Another technical point is that we routinely use biotinylated ligands and then NeutrAvidin-alkaline phosphatase for binding and detection of bound ligand. With the various macrophages studied in this report, including TGEM, BMDM, and the J774 cell line, we did not encounter endogenous alkaline phosphatase activity, which could add a high baseline value. However, it would be important to test for this activity in each new cell type studied by omission of the NeutrAvidin before addition of the luminescent substrate and measurement of the light emission. In the case of the COS-7 cells, we found a detectable but low level of such background activity, which was readily reduced by the use of Levamisole, which blocks the cellular alkaline phosphatase activity but does not interfere with the enzymatic activity of the NeutrAvidin alkaline phosphatase complex.
In summary, we have developed a nonradioactive binding assay of a ligand to plated cells. It uses techniques of solid phase chemiluminescent immunoassays and provides measurements of ligand binding and specificity and should be readily adaptable to other ligand-receptor interactions and other cell types.
Acknowledgments
The authors thank the Leducq Fondation for their support.
Footnotes
Abbreviations:
- BMDM
- bone marrow-derived macrophages
- Bt-CuOxLDL
- biotinylated copper-induced oxidation of low density lipoprotein
- Bt-OxLDL
- biotinylated oxidized low-density lipoprotein
- CuOxLDL
- copper-induced oxidation of low-density lipoprotein
- DPBS
- Dulbecco's phosphate-buffered saline
- ko
- knockout
- OxLDL
- oxidized low-density lipoprotein
- RLU
- relative light units
- RT
- room temperature
- SR-A
- scavenger receptor A
- TGEPM
- thioglycollate-elicited peritoneal macrophages
This work was supported by NHLBI grants HL086559 and HL088093 (J.W.), by a Post-Baccalaureate Supplement to Promote Diversity in Health-Related Research from these two grants (E.M.), and by an AHA Scientist Development Grant (0630228N) and research grants from The Lundbeck Foundation and The Danish Council for Independent Research-Medical Sciences (K.H.).
REFERENCES
- 1.Steinberg D., Witztum J. L. 2010. Oxidized low-density lipoprotein and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 30: 2311–2316 [DOI] [PubMed] [Google Scholar]
- 2.Kzhyshkowska J., Neyen C., Gordon S. 2012. Role of macrophage scavenger receptors in atherosclerosis. Immunobiology. 217: 492–502 [DOI] [PubMed] [Google Scholar]
- 3.Glass C. K., Witztum J. L. 2001. Atherosclerosis. the road ahead. Cell. 104: 503–516 [DOI] [PubMed] [Google Scholar]
- 4.Binder C. J., Horkko S., Dewan A., Chang M. K., Kieu E. P., Goodyear C. S., Shaw P. X., Palinski W., Witztum J. L., Silverman G. J. 2003. Pneumococcal vaccination decreases atherosclerotic lesion formation: molecular mimicry between Streptococcus pneumoniae and oxidized LDL. Nat. Med. 9: 736–743 [DOI] [PubMed] [Google Scholar]
- 5.Tsimikas S., Miyanohara A., Hartvigsen K., Merki E., Shaw P. X., Chou M. Y., Pattison J., Torzewski M., Sollors J., Friedmann T., et al. 2011. Human oxidation-specific antibodies reduce foam cell formation and atherosclerosis progression. J. Am. Coll. Cardiol. 58: 1715–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw P. X., Horkko S., Chang M. K., Curtiss L. K., Palinski W., Silverman G. J., Witztum J. L. 2000. Natural antibodies with the T15 idiotype may act in atherosclerosis, apoptotic clearance, and protective immunity. J. Clin. Invest. 105: 1731–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havel R. J., Eder H. A., Bragdon J. H. 1955. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34: 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150: 76–85 [DOI] [PubMed] [Google Scholar]
- 9.Boullier A., Friedman P., Harkewicz R., Hartvigsen K., Green S. R., Almazan F., Dennis E. A., Steinberg D., Witztum J. L., Quehenberger O. 2005. Phosphocholine as a pattern recognition ligand for CD36. J. Lipid Res. 46: 969–976 [DOI] [PubMed] [Google Scholar]
- 10.Yagi K. 1976. A simple fluorometric assay for lipoperoxide in blood plasma. Biochem. Med. 15: 212–216 [DOI] [PubMed] [Google Scholar]
- 11.Norris P. C., Reichart D., Dumlao D. S., Glass C. K., Dennis E. A. 2011. Specificity of eicosanoid production depends on the TLR-4-stimulated macrophage phenotype. J. Leukoc. Biol. 90: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papadimitriou E., Lelkes P. I. 1993. Measurement of cell numbers in microtiter culture plates using the fluorescent dye Hoechst 33258. J. Immunol. Methods. 162: 41–45 [DOI] [PubMed] [Google Scholar]
- 13.Goldstein J. L., Ho Y. K., Basu S. K., Brown M. S. 1979. Binding site on macrophages that mediates uptake and degradation of acetylated low density lipoprotein, producing massive cholesterol deposition. Proc. Natl. Acad. Sci. USA. 76: 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boullier A., Gillotte K. L., Horkko S., Green S. R., Friedman P., Dennis E. A., Witztum J. L., Steinberg D., Quehenberger O. 2000. The binding of oxidized low density lipoprotein to mouse CD36 is mediated in part by oxidized phospholipids that are associated with both the lipid and protein moieties of the lipoprotein. J. Biol. Chem. 275: 9163–9169 [DOI] [PubMed] [Google Scholar]
- 15.Gillotte-Taylor K., Boullier A., Witztum J. L., Steinberg D., Quehenberger O. 2001. Scavenger receptor class B type I as a receptor for oxidized low density lipoprotein. J. Lipid Res. 42: 1474–1482 [PubMed] [Google Scholar]
- 16.Endemann G., Stanton L. W., Madden K. S., Bryant C. M., White R. T., Protter A. A. 1993. CD36 is a receptor for oxidized low density lipoprotein. J. Biol. Chem. 268: 11811–11816 [PubMed] [Google Scholar]
- 17.De Rijke Y. B., Biessen E. A., Vogelezang C. J., van Berkel T. J. 1994. Binding characteristics of scavenger receptors on liver endothelial and Kupffer cells for modified low-density lipoproteins. Biochem. J. 304: 69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Kooij M. A., Morand O. H., Kempen H. J., van Berkel T. J. 1996. Decrease in scavenger receptor expression in human monocyte-derived macrophages treated with granulocyte macrophage colony-stimulating factor. Arterioscler. Thromb. Vasc. Biol. 16: 106–114 [DOI] [PubMed] [Google Scholar]
- 19.The Lipid Research Clinics Coronary Primary Prevention Trial results. II 1984. The relationship of reduction in incidence of coronary heart disease to cholesterol lowering. JAMA. 251: 365–374 [PubMed] [Google Scholar]
- 20.Kunjathoor V. V., Febbraio M., Podrez E. A., Moore K. J., Andersson L., Koehn S., Rhee J. S., Silverstein R., Hoff H. F., Freeman M. W. 2002. Scavenger receptors class A-I/II and CD36 are the principal receptors responsible for the uptake of modified low density lipoprotein leading to lipid loading in macrophages. J. Biol. Chem. 277: 49982–49988 [DOI] [PubMed] [Google Scholar]
- 21.Acton S. L., Scherer P. E., Lodish H. F., Krieger M. 1994. Expression cloning of SR-BI, a CD36-related class B scavenger receptor. J. Biol. Chem. 269: 21003–21009 [PubMed] [Google Scholar]
- 22.Brown M. S., Goldstein J. L. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science. 232: 34–47 [DOI] [PubMed] [Google Scholar]
- 23.Goldstein J. L., Brown M. S. 2009. The LDL receptor. Arterioscler. Thromb. Vasc. Biol. 29: 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khouw A. S., Parthasarathy S., Witztum J. L. 1993. Radioiodination of low density lipoprotein initiates lipid peroxidation: protection by use of antioxidants. J. Lipid Res. 34: 1483–1496 [PubMed] [Google Scholar]
- 25.Weismann D., Hartvigsen K., Lauer N., Bennett K. L., Scholl H. P., Charbel Issa P., Cano M., Brandstatter H., Tsimikas S., Skerka C., et al. 2011. Complement factor H binds malondialdehyde epitopes and protects from oxidative stress. Nature. 478: 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou M. Y., Fogelstrand L., Hartvigsen K., Hansen L. F., Woelkers D., Shaw P. X., Choi J., Perkmann T., Backhed F., Miller Y. I., et al. 2009. Oxidation-specific epitopes are dominant targets of innate natural antibodies in mice and humans. J. Clin. Invest. 119: 1335–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liadaki K. N., Liu T., Xu S., Ishida B. Y., Duchateaux P. N., Krieger J. P., Kane J., Krieger M., Zannis V. I. 2000. Binding of high density lipoprotein (HDL) and discoidal reconstituted HDL to the HDL receptor scavenger receptor class B type I. Effect of lipid association and APOA-I mutations on receptor binding. J. Biol. Chem. 275: 21262–21271 [DOI] [PubMed] [Google Scholar]
- 28.Fong L. G., Parthasarathy S., Witztum J. L., Steinberg D. 1987. Nonenzymatic oxidative cleavage of peptide bonds in apoprotein B-100. J. Lipid Res. 28: 1466–1477 [PubMed] [Google Scholar]
- 29.Palinski W., Rosenfeld M. E., Yla-Herttuala S., Gurtner G. C., Socher S. S., Butler S. W., Parthasarathy S., Carew T. E., Steinberg D., Witztum J. L. 1989. Low density lipoprotein undergoes oxidative modification in vivo. Proc. Natl. Acad. Sci. USA. 86: 1372–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]