Abstract
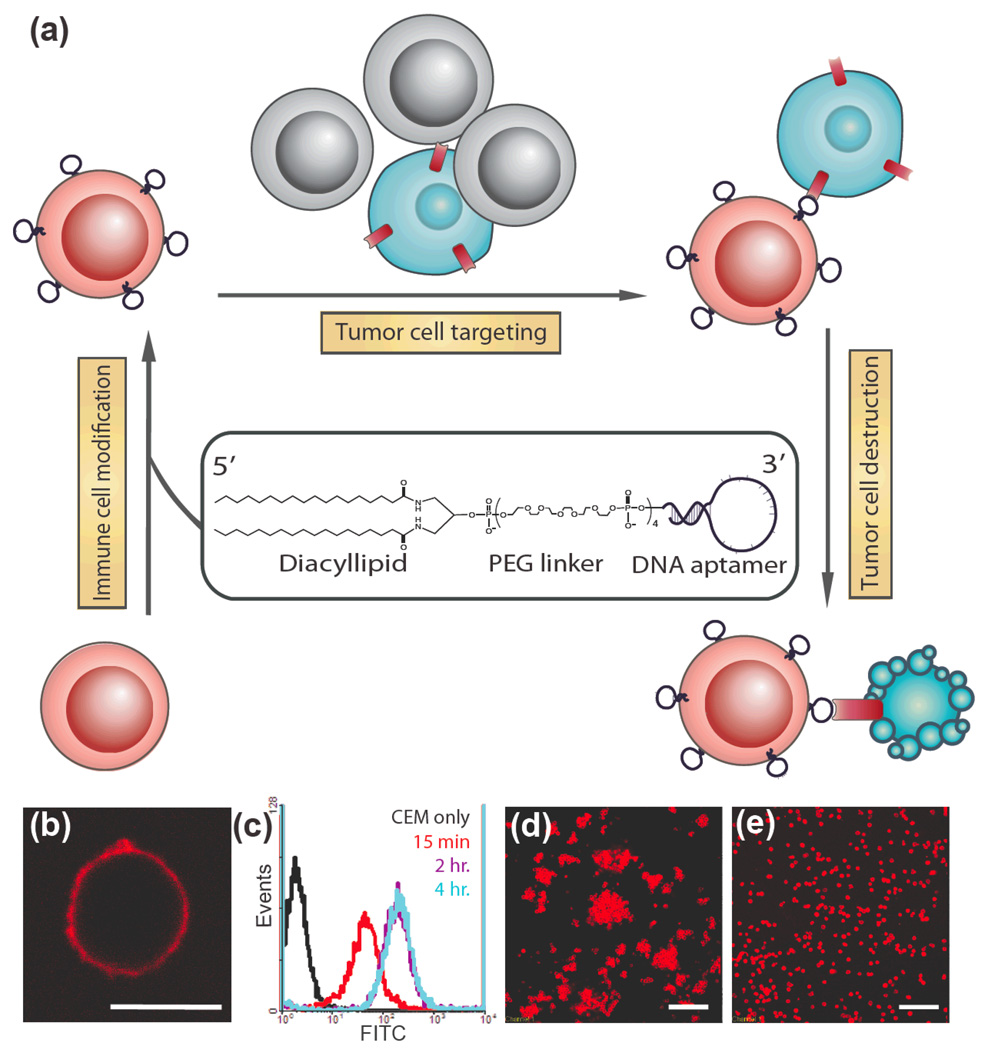
One important issue using cells as therapeutics is targeted delivery. Engineering cell surfaces to improve delivery efficiency is thus of great interest. Here we report a simple, efficient and effective way to modify the cell surface with target-specific ligands, i.e., DNA aptamers, while minimizing the effects on the modified cells. We demonstrated that after incubating with lipo-aptamer probes (shown in expansion), immune cells (red) recognize cancer cells (blue) in the cell mixture, and kill cancer cells.
Keywords: DNA aptamer, cell modification, cell targeting, cell-based immunotherapy
Cell-based therapy has shown considerable potential in treating diseases such as leukemia and osteoporosis. Certain types of cells, for instance killer lymphocytes that naturally attack cancer cells, are being studied for direct cancer treatment[1]. Other cells, like mesenchymal stem cells (MSC), can be genetically engineered to produce therapeutics in situ after delivery[2]. However, one important issue in cell-based therapy is the targeted delivery of cells in vivo, which not only improves therapeutic efficacy, but also minimizes side effects. In this regard, several approaches have been studied. A straightforward method involves physical delivery of cells to the site of interest, with the help of proper medical devices[3]. Other targeting strategies, including the use of native cell-homing machinery[4] and expression or coating of cells with targeting ligands, are under intensive study. Carbohydrates[5], short peptides[6] and extracellular domains of cell membrane receptors[7] have all been used as targeting moieties.
In recent years, oligonucleotide-based probes, termed aptamers, have been developed with the specificity and affinity required for diagnostic[8] and therapeutic applications[9]. Similar to antibodies, aptamers can specifically recognize a wide range of targets that vary from small molecules to cancer cells[8, 10–12]. However, they have additional properties that make them more attractive than antibodies. For example, aptamers are usually smaller, resulting in better tissue penetration ability. Furthermore, no immunogenic reactions have thus far been reported for any in vivo experiment with aptamers. Finally, since nucleic acids can be synthesized chemically, aptamers can be readily adapted for modifications to meet different needs.
Previously, our lab developed diacyl phospholipid-DNA conjugates[13], and in this paper, we report the use of this lipid-DNA probe to modify cell surfaces for specific cell targeting. We hypothesized that aptamers would induce cellular adhesion upon spontaneous receptor-ligand binding in a manner that mimics the natural process of cell-cell adhesion. We used leukemia cell lines to demonstrate that aptamers anchored on the cell surface could act as targeting ligands that specifically recognize their target cells. Further, we explored the potential of this probe in adoptive cell therapy. Immune effector cells modified by the probe showed improved affinity, while remaining cytotoxic to target cancer cells. Our method of aptamer-mediated cell targeting is illustrated schematically in Figure 1a.
Figure 1.
Modification of cell membranes with aptamers. (a) Schematic representation of targeting cancer cells (blue) with aptamer-modified immune cells (red). After incubating with lipo-aptamer probes (shown in expansion), immune cells recognize cancer cells in the cell mixture, and kill cancer cells. (b) Confocal microscope image of lipo-Lib-TMR-treated CEM cells. Red fluorescent probes were found only on the cell surface. Scale bar: 10 µm. (c) CEM cells were treated with lipo-Lib-FITC for different time intervals in cell culture medium. The maximum insertion was reached in 2 hours. (d) Ramos cells spontaneously aggregate after treatment with lipo-TD05-TMR. Scale bar: 100 µm. (e) Control experiments showed no assembly when Ramos cells were treated with lipo-lib-TMR. Scale bar: 100 µm.
To label the cell surface with aptamers, diacyl lipid-DNA aptamer conjugates were synthesized as previously described[13]. A membrane-anchored aptamer can be divided into three distinct segments (Figure 1a). The first segment is an aptamer sequence selected by a process called cell-SELEX (systematic evolution of ligands by exponential enrichment)[11, 12]. We have demonstrated in several cancer cell models that aptamers can recognize the molecular differences between target and control cell membranes by preferentially binding to target cells. In this study, two different aptamers, Sgc8, which targets protein tyrosine kinase 7 on CCRF-CEM cell membrane[11, 14], and TD05, which targets the immunoglobulin heavy mu chain on Ramos cells surface[12, 15], were used for testing. These aptamers exhibit high affinity (KdSgc8: 0.8nM, KdTD05: 74nM) and excellent selectivity towards their target leukemia cells, as required for mimicking native cell-surface ligand-receptor interactions. In addition, because multiple aptamers are presented on each cell surface, multivalent interaction with target proteins can greatly improve binding[16]. The second segment is a PEG linker, which allows DNA to extend out from the cell surface, thereby minimizing nonspecific and steric interactions between the cell-surface molecules and the aptamer. As a consequence, the PEG linker facilitates the conformational folding of the aptamer, which is important for aptamer-target binding. The third segment, a synthetic diacyllipid tail with two stearic acids, is conjugated at the 5’-end as the membrane anchor. By its hydrophobic nature, the diacyllipid tail could firmly insert into the cell membrane with excellent efficiency[13].
To demonstrate lipid insertion, a fluorescent dye molecule (TAMRA) was conjugated to the 3’-end of the oligonucleotides. After incubation with cells, the labeled lipid-DNA probes were detected on the cell membrane by confocal microscopy (Figure 1b). Aptamer density on the cell surface can be easily controlled by varying the incubation time or initial DNA probe concentration. As shown by flow cytometry, a higher initial concentration generally resulted in more aptamers anchored on the cell surface, and after 1µM concentration, the increment of probe concentration did not improve insertion much for CEM cells (Supporting Information, Figure S1a). Also, lipid insertion could be observed within 15 min and reached equilibrium after two hours in cell culture medium (Figure 1c). Similarly, immune effector cells such as natural killer (NK) cells and T cells can be modified with lipo-DNA probes as well (Supporting Information, Figure S1b-c).
To test whether the aptamers could fold properly to recognize their targets after anchoring on the cell membrane, we first designed a homotypic cell targeting experiment. We expected that cells modified with their targeting aptamer would form a cell-aptamer-cell assembly. As shown in Figure 1d, the TD05-treated Ramos cells spontaneously formed sequence-specific aggregates. In control experiments where Ramos cells were incubated with a random sequence (lipo-Lib-TMR), no aggregates were observed (Figure 1e). Similar homotypic assemblies were observed for CEM cells modified with Lipo-Sgc8-TMR (Supporting Information, Figure S2). The above experiments supported our hypothesis that membrane-anchored aptamers could induce cellular adhesion in a defined target-specific fashion.
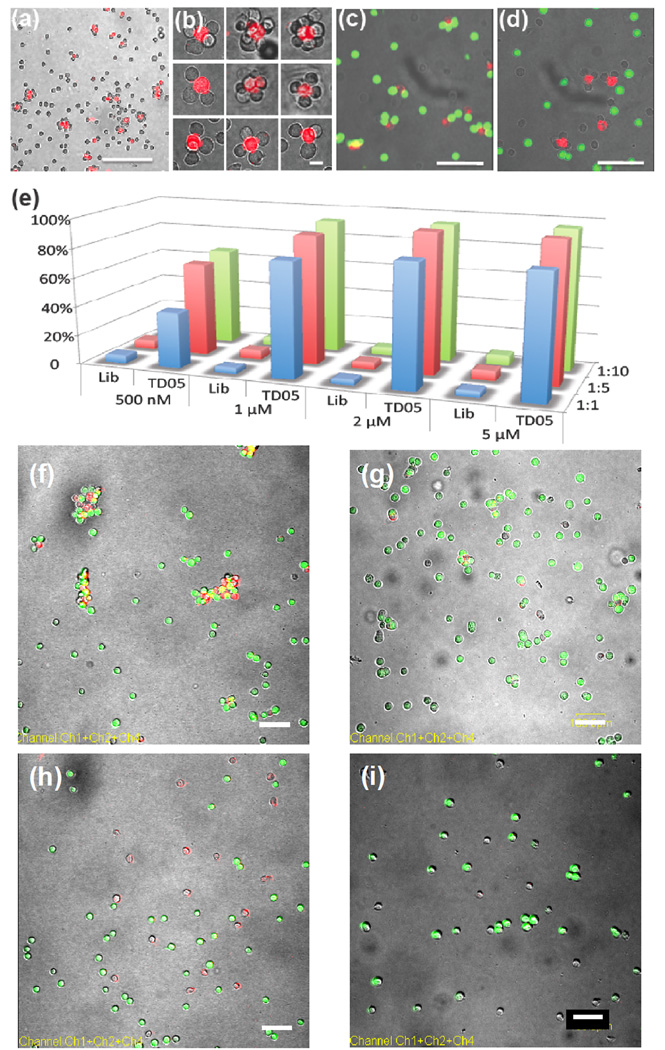
To further demonstrate aptamer specificity, we designed experiments to show different types of cell assemblies. Ramos cells were first treated with lipo-Sgc8-TMR (fluorescent), and mixed with unmodified CEM cells (nonfluorescent) at a 1:10 ratio. Cell aggregates with flower-like structure were observed (Figure 2a and 2b). Each cluster contained two types of cells: surface-modified fluorescent cells and nonfluorescent target cells. Heterotypic cell adhesions were also observed between CEM and Ramos cells using lipo-TD05-TMR (Supporting Information, Figure S3). The generality of this assembly strategy was demonstrated by crosslinking CEM/Ramos with other cell lines, such as Jurkat and K562, using similar procedures (Supporting Information, Figure S4, S5).
Figure 2.
Aptamer-directed assembly and disassembly of cell aggregates and quantitative analysis of aggregation. (a) 1:10 mixture of lipo-sgc8-TMR-modified Ramos and CEM cells. Scale bar: 100 µm. (b) Discrete cell clusters at higher magnification. Scale bar: 20 µm. (c) Confocal micrograph of selective cell assembly. Lipo-TD05-TMR-modified K562 cells (red fluorescence) were incubated with CellTracker Green-labeled Ramos cells (green fluorescence) and unlabeled CEM cells. No mismatched cell assembly (K562 and CEM cells or Ramos and CEM cells) was observed. Scale bar: 50 µm. (d) Confocal micrograph of selective cell assembly of lipo-Sgc8-TMR-modified K562 cells (red fluorescence) and unlabeled CEM cells. (e) Aggregation percentage of CEM cells in different sample sets. CEM-to-Ramos cell ratios are represented by blue (1:1), red (1:5) and green (1:10) bars. (f) 1:5 mixture of lipo-TD05-TMR-modified CEM (red) cells and Ramos (green) cells after 20 min incubation. CEM and Ramos cells formed aggregates. Scale bar: 100 µm. (g) Mixture of Lipo-TD05-TMR-modified CEM (red) cells and Ramos (green) cells from (f) treated with DNase I. Scale bar: 100 µm. (h) 1:5 mixture of lipo-Lib-TMR-modified CEM (red) cells and Ramos (green) cells after 25 min incubation. CEM and Ramos cells remained apart. Scale bar: 100 µm. (i) Mixture of Lipo-Lib-TMR-modified CEM (red) cells and Ramos (green) cells from (h) treated with DNase I. Scale bar: 100 µm.
Controlled cell aggregation could also be realized in mixed cell populations. For example, K562 cells were labeled with lipo-Sgc8-TMR or lipo-TD05-TMR, and then a mixture of CEM (nonfluorescent) and Ramos cells (CellTracker Green-labeled) was added. Depending on which aptamer was used, K562 specifically recognized CEM or Ramos in the mixture (Figure 2c and 2d), while no mismatched cell aggregates were observed. This indicated that aptamers anchored on the cell membranes were still very specific and could therefore be applied in a biological system.
Finally, we quantified the cell aggregation using flow cytometry. As presented in Figure 2e, 40–95% CEM cells formed aggregates with Ramos cells when modified with TD05, compared with less than 5% aggregate formation in DNA library-modified CEM cells (see also Supporting Information, Table S1). The percentage of aggregation can be controlled by the aptamer concentration and the modified-to-target cell ratio. An increase in probe concentration up to 1µM can significantly increase the aggregation percentage. From 1µM to 2 and 5µM, the increase is small because 1µM modification was sufficient to cause 80%–90% cell aggregation, and the number of lipo-DNA on cell surface was close to each other (Supporting Information, Figure S1a). On the other hand, since an increased number of target cells can increase the chance of interaction, more aggregates formed when an excess of Ramos cells was used.
To further confirm that the cell assembly was induced by DNA aptamers anchored on cell surfaces, the aggregates were treated with nuclease and protease that can destroy either aptamers or target proteins, respectively. First, CEM cells modified with lipo-Lib-TMR or lipo-TD05-TMR probes were mixed with 5 equivalents of Ramos cells (CellTracker Green-labeled). Images taken immediately after mixing showed that most CEM and Ramos cells remained apart with only a few small aggregates (Supporting Information, Figure S7). After 20 min incubation, as expected, almost all the TD05 aptamer-modified CEM cells were surrounded by Ramos cells, resulting in a yellow (combination of red and green) fluorescent signal (Figure 2f and Supporting Information, Figure S8b), while library-labeled CEM cells did not form any aggregates with Ramos cells (Figure 2h and Supporting Information, Figure S8a). Finally, these cell aggregates were treated with deoxyribonuclease I, which can cleave single- and double-stranded DNA. As shown in Figure 2g and Figure 2i, after treatment, the red fluorescent signal on CEM cells disappeared because the TMR dye molecule had been cleaved from the cell surface. Moreover, fewer aggregates can be seen in Figure 2g compared to Figure 2f, indicating that the aggregation caused by aptamer-target recognition had been disrupted by removal of DNA aptamers from the modified cell surfaces. It is noteworthy that some small aggregates remained, as shown in Figure 2g, and that the interface between these two cells remained yellow. One possible reason is that the binding of these two cells was so tight that DNAse was prevented from interacting with the aptamers by steric hindrance; consequently, the aggregates remained intact. We also treated cellular aggregates with proteinase K, a protein known to digest the target surface proteins of Ramos cells[15], resulting in disassembly of aggregated cells and uniform dispersion of individual cells (Supporting Information, Figure S9).
Since delivered cells are intended for therapy, modification of the cell surface should not affect cellular functions. The components of the probe, including oligonucleotides, PEG polymers and diacyl lipids, are not known to be cytotoxic. To test whether the insertion had any toxic effect on cells, cell necrosis and apoptosis were tested after modification, and minimal cell death was observed (Supporting Information, Figure S10a). The proliferation rate was monitored using a cell proliferation assay, and no significant change was observed in any cell lines with 1µM modification (Supporting Information, Figure S10b). These results suggested that the probe itself is not cytotoxic at 1µM concentration.
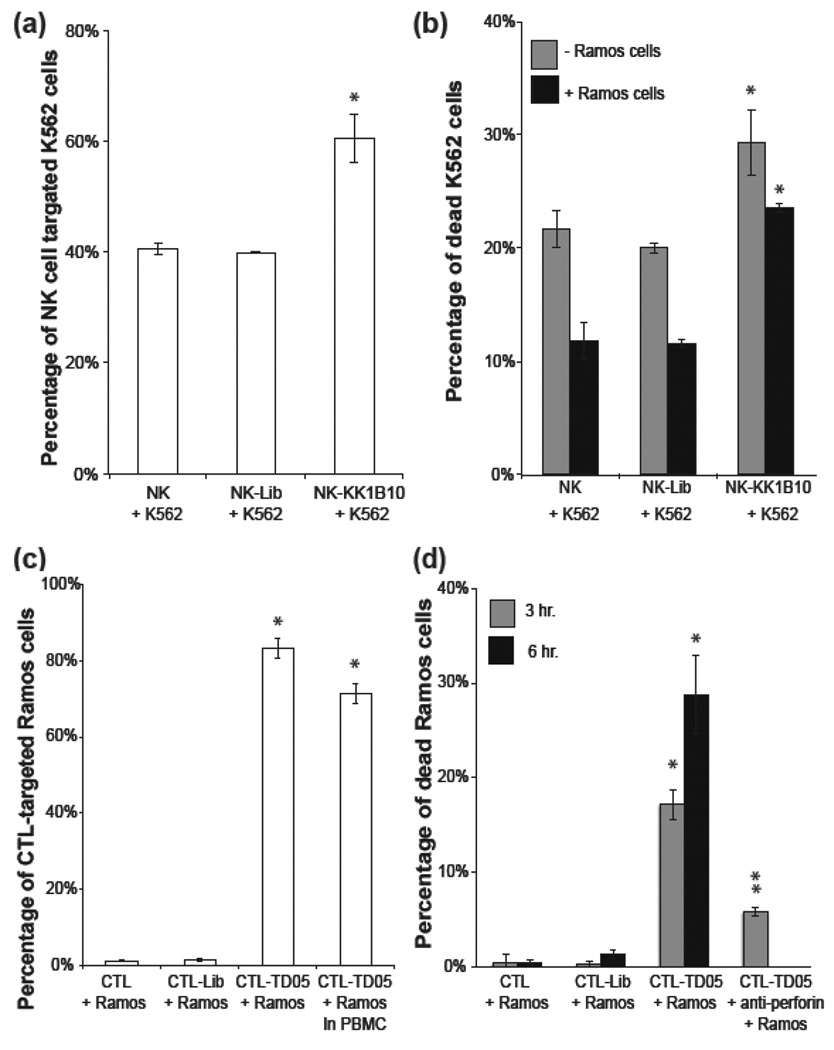
To further investigate the cellular effect of the lipid-DNA probe on immune effector cells, we used NK cells, a type of killer lymphocyte, as a model. NK cells recognize cells independent of major histocompatibility complex (MHC), and they kill cells that do not express MHC by releasing granule proteins[17]. K562, a type of chronic myelogenous leukemia cell line, is often used as a target for in vitro NK cell assays[18]. The aptamer used in this study was KK1B10, an aptamer that specifically recognizes K562 cells[19]. In the binding assay (Figure 3a), unmodified and library-modified NK groups showed a higher background binding (about 40%) compared with previously demonstrated CEM-Ramos binding assays. This is because NK cells recognize K562 cells spontaneously. Aptamer modification NK cells targeted 60% K562 cells, which improved NK cell targeting efficiency by 50%.
Figure 3.
Aptamer-assisted immune effector cell targeting and killing of leukemia cells (a) NK-K562 cell binding assay. NK cells recognize K562 cells spontaneously; however, aptamer modification improved the targeting efficiency by 50%. (b) NK-K562 cell killing assay. Aptamer-modified NK cells killed K562 cells more efficiently, especially in the presence of background cells (Ramos). (c) CTL-Ramos cell binding assay. Without presenting the specific MHC1:peptide on Ramos cell surfaces, CTL cannot recognize Ramos cells. On the other hand, aptamer-modified CTL recognized 80% Ramos cells. (d) CTL-Ramos cell killing assay. Assisted by membrane-anchored aptamers, CTL targeted and killed Ramos cells via cell-mediated immunity. CTL-mediated cytotoxicity was confirmed by blocking the perforin/granzyme pathway with antibody. Values are means with SD (n=3). The single asterisk indicates a significant difference between aptamer-modified and unmodified or Lib-modified groups determined by the one-tailed t-test at P<0.01. The double asterisks indicate a significant difference between aptamer-modified and anti-Perforin treated groups determined by the one-tailed t-test at P<0.01.
To determine if NK cells modified with the KK1B10 aptamer would affect their killing functions, we set up a NK cytolytic assay, as previously reported[20]. Compared with unmodified NK cells, control library-modified NK cells showed the same killing effect on K562 cells (around 21%, shown in Figure 3b), indicating that lipo-DNA would not interfere with the cell-mediated cytotoxicity. Moreover, NK cells modified with KK1B10 aptamer killed about 30% of K562 cells, which was 50% more than unmodified NK cells. It is worth mentioning that the incremental killing efficiency correlated well with targeting efficiency. The virtue of adding aptamers as extra targeting ligands was revealed in the presence of excess nontarget cells. In this experiment, K562 cells were mixed with 4 equivalents of nontarget Ramos cells first and then incubated with NK cells. Within the same amount of time, NK cells killed fewer K562 cells, but the decrease was considerably smaller in the aptamer-modified group, suggesting that aptamer modification can improve cell targeting without affecting immune effector function (Figure 3b).
These results suggested the feasibility of a novel T-cell killing model, in which the specific cytotoxicity was controlled by membrane-anchored aptamers. A primary immortalized cytomegalovirus (CMV)-specific CD8+ cytotoxic T lymphocyte (CTL) clone (HLA A0201) preactivated with 12-myristate 13-acetate (PMA) and ionomycin was used as the immune effector cell type. The target cells were Ramos cells, a B-cell Burkitt’s lymphoma cell line. Ionomycin synergizes with PMA to activate protein kinase C, resulting in perforin-granule release from CTL[21]. The perforin/granzyme cell death pathway requires direct contact between effector and target cells[22]. Therefore, without a CMV-specific peptide:MHC I complex on Ramos cell surfaces, the CMV-specific CTL cannot recognize Ramos cells, resulting in only background cytotoxic effect towards Ramos. Assisted by surface-anchored TD05 aptamers, however, the modified CTL have increased affinity toward target Ramos cells, thereby facilitating and prolonging interactions between effector and target cells. Thus, enhanced killing of Ramos cells should be observed.
To test this, we first performed a binding assay to study the binding between CTL and Ramos cells. Figure 3c shows that CTL did not recognize Ramos cells naturally, unless they were modified with the targeting aptamers. Within 20 minutes, 80% of Ramos cells were targeted by CTL in the cell culture. To test the targeting efficiency in a more biologically relevant condition, Ramos cells were spiked into peripheral blood mononuclear cells (PBMC) and then incubated with the CTL. The targeting efficiency was slightly influenced by the spiked PBMC, resulting in targeting 70% of Ramos cells.
We then designed the CTL-Ramos cell-killing assay in a manner similar to the previously described NK-K562 cell-killing assay (Supporting Information, Figure S11). As shown in Figure 3d, about 15% and 30% of Ramos cells were dead in the aptamer-modified CTL group in 3 hours and 6 hours, respectively, while < 5% dead Ramos cells were found in Lib-modified or unmodified CTL groups. To confirm that Ramos cell death was caused by CTL, an anti-perforin antibody that blocks the perforin/granzyme pathway[23] was added to the aptamer-modified group. The percentage of dead Ramos cells was greatly reduced, from 15% to 5%. This aptamer-modified CTL-killing assay demonstrated that aptamers could redirect specific CTL killing towards the desired target cells, which shed light on potential applications of the lipo-aptamer probes in adoptive immunotherapy of cancer. It is noteworthy that aptamers endowed CTL with new targeting specificity, and the recognition was independent of MHC, a major limitation of CTL therapy. An in vivo model to study the trafficking of aptamer-modified cells and immune effector killing is under investigation in our group.
In conclusion, we have successfully engineered a novel targeting ligand on cell membranes for specific cell targeting. We demonstrated that the noncovalent modification is simple, yet effective, with no short-term effect on modified cells. The selective assembly of multiple types of cells via aptamer-protein recognition is rapid and target-specific. Immune effector cells modified with aptamers can recognize leukemia cells via MHC nonrestricted structures, leading to elevated cancer cell targeting and killing. Using a CMV-specific CD8+ CTL clone as immune effector cells and Ramos as target cells, we demonstrated, for the first time, a redirected T cell-killing where the specificity was controlled by aptamers. Further study will focus on the metabolism of the synthetic lipid and in vivo cell trafficking. We believe that the diversity of aptamer targets and the facile modification make this strategy attractive in cell-based delivery and therapy.
Experimental Section
Materials
Unless otherwise stated, all solvents and chemicals were obtained from Sigma-Aldrich and used without further purification. DNA synthesis reagents were purchased from Glen Research. PEG phosphoramidite (DMT-Hexaethyloxy-Glycol phosphoramidite) was purchased from ChemGenes Corporation (Wilmington, MA). CellTracker™ Green CMFDA, CellTrace™ Far Red DDAO-SE, Annexin V/Dead Cell Apoptosis Kit and the CellTrace™ CFSE cell proliferation kit were purchased from Invitrogen. Interleukins were purchased from PeproTech. Purified Mouse Anti-Human Perforin was purchased from BD Biosciences.
Cell Surface Labeling
Cells (200µL, 1×106 cells/mL) were suspended in a 96-well plate and incubated with lipo-DNA probes (1µM lipid-DNA, with or without fluorescent dye) in cell culture medium at 37°C for 2 hours. Cells were then washed three times with PBS to remove free probes and resuspended in the desired buffer or cell culture medium.
Imaging of Lipo-DNA on Cell Surface
Ramos cells were incubated with TMR-labeled lipo-Lib probes, as described above. Images were taken and collected in the perpendicular lateral (x-y) plane by laser scanning confocal microscopy with a 488 nm argon laser and a 543/633 nm helium/neon laser.
Homotypic and Heterotypic Cell Assembly
For homotypic cell assembly, Ramos cells labeled with lipo-TD05-TMR or lipo-Lib-TMR were shaken at 300 rpm for 20–30 min at 25°C. For heterotypic cell assembly, Ramos cells were labeled with lipo-Sgc8-TMR or lipo-Lib-TMR first, and a proper ratio of CEM cells was then combined in binding buffer and shaken at 300 rpm for 30 min at 25°C. Aliquots were analyzed by laser scanning confocal microscopy. All experiments were repeated 5 times.
Selective Cell Assembly in Cell Mixture
Green-stained Ramos cells and untreated CEM cells were mixed together at a 1:1 ratio. K562 cells modified with either Lipo-TD05-TMR or lipo-Sgc8-TMR probes were co-incubated with 5 equivalents of the above cell mixtures, respectively, and shaken at 300 rpm for 30 min at 25°C. Aliquots were analyzed using laser scanning confocal microscopy.
Cell Assembly Treated with DNase I
After cell aggregates were formed, cells were centrifuged and resuspended in 1×DNaseI buffer with 20U/mL DNaseI and incubated at 37°C for 10 min. Aliquots were analyzed by laser scanning confocal microscopy.
Quantitative analysis of cell aggregates
CEM cells were stained with CellTrace Far Red DDAO-SE, and Ramos cells were stained with CellTracker Green CMFDA. After washing, CEM cells were incubated with various concentrations of lipo-Lib or lipo-TD05 (without any dye molecules), washed, and then incubated with different quantities of Ramos cells, as described above. The fluorescent signals from channels 1 (green) and 4 (far red) were determined by flow cytometry (Accuri C6 flow cytometer). The thresholds of channels 1 and 4 were set by comparing the fluorescent signals generated from unstained CEM and Ramos cells. The percentage of aggregation was calculated as 100 times the ratio of the double positive (green and far-red) population (upper right region in Supporting Information, Figure S6) to the total CEM cell (far-red) population (upper region in Supporting Information, Figure S6). Each set of samples was analyzed in triplicate.
Aptamer-assisted immune cell-killing assay
K562 or Ramos cells were washed with PBS buffer and labeled with 1 µM carboxyfluorescein succinimidyl ester (CFSE), suggested by the manufacturer, and then aliquotted to a 96-well microtiter plate at 1×104 cells/well. Immune effector cells were added to each well at the desired E:T ratio. The final reaction volume was adjusted to 200 µL. The plate was kept in a humidified atmosphere of 5% CO2 and 37 °C for 2–3 hours. Before flow cytometry (Accuri C6 flow cytometer) analysis, propidium iodide (PI) was added to each sample and incubated at RT in the dark for 30 min to label dead cells. The target cell death was calculated as the number of CFSE- and PI-positive cells over the total number of CFSE-positive cells.
Supplementary Material
Table 1.
Oligonucleotide sequences used in this work
| Probe Name | Sequence |
|---|---|
| Lipo-Sgc8 | 5′ Diacyllipid-(PEG)4-TTT TTT TAT CTA ACT GCT GCG CCG CCG GGA AAA TAC TGT ACG GTT AGA -3’ |
| Lipo-TD05 | 5’ Diacyllipid-(PEG)4-AAC ACC GGG AGG ATA GTT CGG TGG CTG TTC AGG GTC TCC TCC CGG TGA -3’ |
| Lipo-KK1B10 | 5’ Diacyllipid-(PEG)4-ACA GCA GAT CAG TCT ATC TTC TCC TGA TGG GTT CCT ATT TAT AGG TGA AGC TGT -3’ |
| Lipo-DNA | 5’ Diacyllipid-(PEG)4-NNN NNN NNN NNN NNN NNN NNN NNN NNN NNN NNN NNN NNN NNN NNN NNN -3’ |
Acknowledgments
This work is supported by grants awarded by the National Institutes of Health (GM066137, GM079359 and CA133086), by the National Key Scientific Program of China (2011CB911000) and China National Instrumentation Program 2011YQ03012412.
References
- 1.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. Journal of Immunology. 2007;178:2617–2621. [PubMed] [Google Scholar]; Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Sakaguchi K, Appella E, Yannelli JR, Adema GJ, Miki T, Rosenberg SA. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:6458–6462. doi: 10.1073/pnas.91.14.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]; Albertsson PA, Basse PH, Hokland M, Goldfarb RH, Nagelkerke JF, Nannmark U, Kuppen PJK. Trends in Immunology. 2003;24:603–609. doi: 10.1016/j.it.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Conrad C, Gupta R, Mohan H, Niess H, Bruns CJ, Kopp R, von Luettichau I, Guba M, Heeschen C, Jauch K-W, Huss R, Nelson PJ. Current Gene Therapy. 2007;7:249–260. doi: 10.2174/156652307781369119. [DOI] [PubMed] [Google Scholar]; Reiser J, Zhang XY, Hemenway CS, Mondal D, Pradhan L, La Russa VF. Expert Opinion on Biological Therapy. 2005;5:1571–1584. doi: 10.1517/14712598.5.12.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]; Elzaouk L, Moelling K, Pavlovic J. Experimental Dermatology. 2006;15:865–874. doi: 10.1111/j.1600-0625.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- 3.Parr AM, Kulbatski I, Tator CH. Journal of Neurotrauma. 2007;24:835–845. doi: 10.1089/neu.2006.3771. [DOI] [PubMed] [Google Scholar]; Poh K-K, Sperry E, Young RG, Freyman T, Barringhaus KG, Thompson CA. International Journal of Cardiology. 2007;117:360–364. doi: 10.1016/j.ijcard.2006.04.092. [DOI] [PubMed] [Google Scholar]
- 4.Millan CG, Marinero MLS, Castaneda AZ, Lanao JM. Journal of Controlled Release. 2004;95:27–49. doi: 10.1016/j.jconrel.2003.11.018. [DOI] [PubMed] [Google Scholar]; Freitas RA. Journal of Nanoscience and Nanotechnology. 2006;6:2769–2775. doi: 10.1166/jnn.2006.413. [DOI] [PubMed] [Google Scholar]
- 5.Foxall C, Watson SR, Dowbenko D, Fennie C, Lasky LA, Kiso M, Hasegawa A, Asa D, Brandley BK. Journal of Cell Biology. 1992;117:895–902. doi: 10.1083/jcb.117.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sarkar D, Spencer JA, Phillips JA, Zhao WA, Schafer S, Spelke DP, Mortensen LJ, Ruiz JP, Vemula PK, Sridharan R, Kumar S, Karnik R, Lin CP, Karp JM. Blood. 2011;118:E184–E191. doi: 10.1182/blood-2010-10-311464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandelboim O, Vadai E, Fridkin M, Katzhillel A, Feldman M, Berke G, Eisenbach L. Nature Medicine. 1995;1:1179–1183. doi: 10.1038/nm1195-1179. [DOI] [PubMed] [Google Scholar]; Nestle FO, Alijagic S, Gilliet M, Sun Y, Grabbe S, Dummer R, Burg G, Schadendorf D. Nat Med. 1998;4:328–332. doi: 10.1038/nm0398-328. [DOI] [PubMed] [Google Scholar]
- 7.Jena B, Dotti G, Cooper LJN. Blood. 2010;116:1035–1044. doi: 10.1182/blood-2010-01-043737. [DOI] [PMC free article] [PubMed] [Google Scholar]; Porter DL, Levine BL, Kalos M, Bagg A, June CH. New England Journal of Medicine. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]; Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 9.Keefe AD, Pai S, Ellington A. Nature Reviews Drug Discovery. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajendran M, Ellington AD. Combinatorial Chemistry & High Throughput Screening. 2002;5:263–270. doi: 10.2174/1386207023330246. [DOI] [PubMed] [Google Scholar]; Rajendran M, Ellington AD. Analytical and Bioanalytical Chemistry. 2008;390:1067–1075. doi: 10.1007/s00216-007-1735-8. [DOI] [PubMed] [Google Scholar]; Geiger A, Burgstaller P, vonderEltz H, Roeder A, Famulok M. Nucleic Acids Research. 1996;24:1029–1036. doi: 10.1093/nar/24.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shangguan D, Li Y, Tang ZW, Cao ZHC, Chen HW, Mallikaratchy P, Sefah K, Yang CYJ, Tan WH. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang ZW, Shangguan D, Wang KM, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan WH. Analytical Chemistry. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 13.Liu HP, Zhu Z, Kang HZ, Wu YR, Sefan K, Tan WH. Chemistry-a European Journal. 2010;16:3791–3797. doi: 10.1002/chem.200901546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. Journal of proteome research. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallikaratchy P, Tang ZW, Kwame S, Meng L, Shangguan DH, Tan WH. Molecular & Cellular Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y-F, Chang H-T, Tan W. Analytical Chemistry. 2008;80 doi: 10.1021/ac702322j. [DOI] [PubMed] [Google Scholar]
- 17.Ljunggren HG, Karre K. Immunology Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 18.Ortaldo JR, Oldham RK, Cannon GC, Herberman RB. Journal of the National Cancer Institute. 1977;59:77–82. doi: 10.1093/jnci/59.1.77. [DOI] [PubMed] [Google Scholar]
- 19.Sefah K, Tang ZW, Shangguan DH, Chen H, Lopez-Colon D, Li Y, Parekh P, Martin J, Meng L, Phillips JA, Kim YM, Tan WH. Leukemia. 2009;23:235–244. doi: 10.1038/leu.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcusson-Stahl M, Cederbrant K. Toxicology. 2003;193:269–279. doi: 10.1016/s0300-483x(03)00302-0. [DOI] [PubMed] [Google Scholar]
- 21.Chatila T, Silverman L, Miller R, Geha R. Journal of Immunology. 1989;143:1283–1289. [PubMed] [Google Scholar]
- 22.Trapani JA, Smyth MJ. Nature Reviews Immunology. 2002;2:735–747. doi: 10.1038/nri911. [DOI] [PubMed] [Google Scholar]
- 23.Otten HG, van Ginkel WGJ, Hagenbeek A, Petersen EJ. Leukemia. 2004;18:1401–1405. doi: 10.1038/sj.leu.2403414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.