Abstract
Extracting high-quality RNA from hydrogels containing polysaccharide components is challenging, as traditional RNA isolation techniques designed for cells and tissues can have limited yields and purity due to physiochemical interactions between the nucleic acids and the biomaterials. In this study, a comparative analysis of several different RNA isolation methods was performed on human adipose-derived stem cells photo-encapsulated within methacrylated glycol chitosan hydrogels. The results demonstrated that RNA isolation methods with cetyl trimethylammonium bromide (CTAB) buffer followed by purification with an RNeasy® mini kit resulted in low yields of RNA, except when the samples were preminced directly within the buffer. In addition, genomic DNA contamination during reverse transcriptase–polymerase chain reaction (RT-PCR) analysis was observed in the hydrogels processed with the CTAB-based methods. Isolation methods using TRIzol® in combination with one of a Qiaex® gel extraction kit, an RNeasy® mini kit, or an extended solvent purification method extracted RNA suitable for gene amplification, with no evidence of genomic contamination. The latter two methods yielded the best results in terms of yield and amplification efficiency. Predigestion of the scaffolds with lysozyme was investigated as a possible means of enhancing RNA extraction from the polysaccharide gels, with no improvements observed in terms of the purity, yield, or amplification efficiency. Overall, this work highlights the application of a TRIzol®+extended solvent purification method for optimizing RNA extraction that can be applied to obtain reliable and accurate gene expression data in studies investigating cells seeded in chitosan-based scaffolds.
Introduction
Chitosan is a linear polysaccharide comprised of D-glucosamine and N-acetyl-D-glucosamine disaccharide repeating units that is readily chemically modifiable to facilitate crosslinking and formation of three-dimensional networks. Hydrogels derived from chitosan have been extensively studied as biomaterials for tissue engineering applications due to their favorable biocompatibility, biodegradability, and capacity for tailored bioactivity.1–4 These materials can be designed as cell delivery vehicles that crosslink in situ to encapsulate cell populations within target sites. In developing these regenerative approaches, gene expression analysis of the encapsulated cell populations by reverse transcriptase–polymerase chain reaction (RT-PCR) can provide useful information in characterizing the cellular response within the engineered microenvironments. However, the polysaccharide moieties within the scaffold materials can interfere with the RNA isolation process. Furthermore, RNA quality can be compromised during the cell lysis steps of the extraction in cationic scaffolds, such as those prepared from chitosan, as insoluble ionic complexes can form with existing soluble anions, including polysaccharides, glycosaminoglycans, and DNA fragments.5,6 Thus, conventional RNA extraction techniques used for two-dimensional cell cultures or whole tissues that rely on monophasic phenol and guanidine isothiocyanate solutions or the use of β-mercaptoethanol, N-laurosylsarcosine (sarkosyl), and density gradient centrifugation in cesium trifluoroacetate alone may not be adequate.7–9
In a recent publication, several methods were explored based on the premise that plant-based RNA extraction techniques could be applied to polysaccharide scaffolds due to their similarity in structure.10 In particular, a common approach with plant-derived tissues involves using a cationic extraction buffer, cetyl trimethylammonium bromide (CTAB), which is a strong detergent used to lyse plant cell walls and isolate the nucleic acid components from the polysaccharides.10 Commercially available RNA extraction kits such as the Qiagen RNeasy® Mini Plant kit or the RNeasy® Mini kit have also been used as methods for extracting RNA from agarose and gellan, as well as alginate-based scaffolds.11,12 In general, most of these methods initially involve a form of mechanical disruption to help separate the RNA. However, the use of enzymatic digestion to partially degrade the polysaccharide scaffolds has not yet been explored as a means of improving the efficiency of RNA extraction from the hydrogels. Chitosan is a partially N-deacetylated derivative of chitin and can be readily hydrolyzed at the amino and hydroxyl groups by either lysozyme or chitinase with similar broad substrate specificity.13 Moreover, in vivo studies have shown that chitosan scaffolds are degraded primarily by lysozyme to produce oligosaccharide products.14 Thus, a secondary objective of our study was to assess the effects of including a lysozyme predigestion step on the RNA extraction efficiency for chitosan-based scaffolds.
Overall, a comparative study was performed to evaluate the quality and yield of RNA extracted from human adipose-derived stem cells (ASCs) encapsulated in photo-crosslinkable N-methacrylated glycol chitosan (MGC) hydrogels using CTAB or TRIzol® with Qiagen kit isolation methods, as well as TRIzol® combined with an extended solvent purification technique. RNA purity (A260/A280 and A260/A230), concentration, and yield were assessed for each of the methods, both with and without lysozyme pretreatment. The isolated RNA samples were used in subsequent end-point RT-PCR analysis of gene expression of the common housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH), transferrin receptor (TfR), and 18S ribosomal RNA, as well as the adipogenic lineage marker peroxisome proliferator activated receptor-γ (PPARγ). The findings from this study provide important insight into the effectiveness of different techniques for RNA extraction from polysaccharide scaffolds such as those derived from chitosan.
Materials and Methods
Materials
Unless otherwise stated, all chemicals were purchased from Sigma-Aldrich Canada Ltd., and were used as received.
Preparation of cell-seeded MGC hydrogels
Human ASCs were isolated and expanded on tissue culture polystyrene in proliferation medium containing DMEM:Ham's F-12 medium supplemented with 10% fetal bovine serum (Thermo Scientific Hyclone, Cat. No. SH30396), and 1% pen-strep (100 U/mL penicillin and 0.1 mg/mL streptomycin) using previously established methods.15 Fresh proliferation medium was provided every 2–3 days and ASCs were passaged at 80% confluence. All prepared hydrogels were seeded with passage 2 (P2) ASCs. Human research ethics board approval for this study was obtained from Queen's University (REB No. CHEM-002-07).
MGC hydrogels were prepared using published methods.16 Briefly, purified glycol chitosan (80 kg/mol, ∼86% degree of deacetylation from Wako Chemical USA, Inc.) was dissolved in distilled water at 2.0% (w/v), and reacted with a 0.58:1.0 molar ratio of glycidyl methacrylate to free amine at pH 9 for 24 h. The solution was neutralized with 1.0 M hydrochloric acid, dialyzed twice against distilled water in 12 kg/mol dialysis tubing (Fisher Scientific) for 24 h, and lyophilized to form a white powder comprised of MGC. The degree of substitution (number of grafted methacrylate groups per 100 residues) was determined to be 20% by 1H NMR.
Dry MGC was dissolved in proliferation medium containing 0.1% (w/v) sterile filtered Irgacure 2959 (2-hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone) photoinitiator. A suspension of ASCs was incorporated to produce a mixture containing 1.5% (w/v) MGC and 1.0×107 cells/mL. Each gel was formed by photocrosslinking 0.1 mL of this mixture in a well of a 16-well chamber glass slide (Nalgene Nunc International) using UV light (320–390 nm, EXFO Lite; EFOS Corporation) at an intensity of 10.2 mW/cm2 for 3 min. All MGC hydrogels used for comparing RNA extraction methods were snap-frozen in liquid nitrogen immediately after fabrication and stored at −80°C until further use.
Enzymatic digestion with lysozyme
For each of the RNA extraction methods, a set of samples was subjected to enzymatic pretreatment with lysozyme. A 10 mg/mL lysozyme (protein ≥90%, ≥40,000 units/mg protein) digestion solution was prepared in 1×PBS (D-PBS; Thermo Scientific HyClone, Cat. No. SH30028; Fisher Scientific) at pH 7.4. Each hydrogel was suspended in 0.5 mL of the digest solution, finely minced with sharp scissors, and incubated for 48 h at 37°C in a dry heat block. After digestion, the samples were centrifuged for 10 min at 10,000 g and snap-frozen with liquid nitrogen to be stored at −80°C until further processing.
RNA extraction
Four different RNA extraction protocols were systematically investigated including a published method developed by Wang et al.10 involving CTAB in combination with the RNeasy® kit (Qiagen), TRIzol-based approaches with Qiagen kits, and an extended solvent purification protocol used in our lab. All extraction methods were tested in triplicate (n=3). A summary of the RNA extraction methods is outlined in Table 1.
Table 1.
RNA Isolation Methods
| CTAB+RNeasy® | TRIzol®+RNeasy® | TRIzol®+Qiaex® | TRIzol®+extended solvent purification |
|---|---|---|---|
| 1. Cryo-pulverize or mince samples as required | 1. Mince samples finely in 1 mL TRIzol® | 1. Mince samples finely in 1 mL of TRIzol® | 1. Mince samples finely in 1 mL of Trizol® |
| 2. Sonicate in 600 μL CTAB buffer | 2. Sonicate | 2. Sonicate | 2. Sonicate |
| 3. Incubate for 5 min at 65°C | 3. Incubate for 5 min at room temperature | 3. Incubate for 5 min at room temperature | 3. Incubate for 5 min at room temperature |
| 4. Add an equal volume of 24:1 chloroform-isoamyl alcohol (CHISAM) | 4. Add 200 μL of chloroform | 4. Add 200 μL of chloroform | 4. Add 200 μL of chloroform |
| 5. Vortex | 5. Incubate for 10 min at room temperature | 5. Incubate for 10 min at room temperature | 5. Vortex for 15 s |
| 6. Centrifuge at room temperature for 5 min at 15,000 g | 6. Centrifuge at 4°C for 10 min at 12,000 g | 6. Centrifuge at 4°C for 10 min at 12,000 g | 6. Incubate for 3 min at room temperature |
| 7. Mix upper aqueous phase with an equal volume of CHISAM (24:1) | 7. Add 250 μL of 70% ethanol to upper aqueous phase | 7. Collect upper aqueous phase and add 600 μL of QX1 buffer and 10 μL of Qiaex II bead suspension | 7. Centrifuge at 4°C for 15 min at 12,000 g |
| 8. Centrifuge at room temperature for 5 min at 15,000 g | 8. Centrifuge at room temperature for 15 min at 15,000 g | 8. Vortex sample for 30 s and incubate at 50°C for 10 min, with vortexing every 2 min | 8. Vortex for 15 s |
| 9. Add an equal volume of 99.9% isopropanol | 9. Resuspend pellet in 50 μL DEPC-treated water | 9. Centrifuge sample for 30 s at 15,000 g and carefully aspirate supernatant | 9. Incubate for 3 min at room temperature |
| 10. Centrifuge at room temperature for 5 min at 15,000 g | 10. Process sample with RNeasy® kit (Qiagen) according to the manufacturer's instructions | 10. Wash beads twice, centrifuging for 30 s at 15,000 g between washes, and air dry for 10 min | 10. Centrifuge at 4°C for 15 min at 12,000 g |
| 11. Discard supernatant and resuspend pellet in 50 μL of DEPC-treated water | 11. Elute with 20 μL of DEPC-treated water | 11. Collect upper aqueous phase and add 500 μL of phenol:chloroform:isoamyl (25:24:1) | |
| 12. Process sample with RNeasy® kit (Qiagen) according to the manufacturer's instructions | 12. Incubate at room temperature for 5 min | 12. Collect upper aqueous phase and add 500 μL of CHISAM (49:1) | |
| 13. Centrifuge for 30 s at 15,000 g | 13. Vortex for 15 s | ||
| 14. Repeat with an additional 20 μL of DEPC-treated water | 14. Incubate for 3 min at room temperature | ||
| 15. Centrifuge at 4°C for 15 min at 12,000 g | |||
| 16. Collect upper aqueous phase and add 500 μL of 100% isopropanol | |||
| 17. Mix by inversion | |||
| 18. Incubate at room temperature for 10 min | |||
| 19. Centrifuge 4°C for 10 min at 12,000 g | |||
| 20. Remove supernatant and wash pellet in 70% ethanol | |||
| 21. Centrifuge at 4°C for 5 min at 12,000 g | |||
| 22. Resuspend in DEPC-treated water |
CTAB, cetyl trimethylammonium bromide; DEPC, diethylpyrocarbonate.
The protocol developed by Wang et al. uses a combination of mechanical disruption, extraction with CTAB buffer and purification with the RNeasy® kit.10 To test the efficacy of this approach with our specific material and cell source, the hydrogels were either (1) cryo-pulverized with a mortar and pestle into a fine powder or (2) finely minced with sharp surgical scissors and then disrupted using an ultrasonic dismembrator (Fisher Scientific Model 100) with three 5-s bursts at a setting of 4 with intervals of cooling on ice in 600 μL of CTAB buffer [2% CTAB, 2% poly(vinylpyrrolidone) (PVP), 1.4 M NaCl, 100 mM Tris-HCl, 20 mM EDTA, and 1% beta-mercaptoethanol in diethylpyrocarbonate (DEPC)-treated water] prewarmed to 65°C. Following disruption, the samples were incubated for 5 min at room temperature. An equal volume of 24:1 chloroform-isoamyl alcohol (CHISAM) was added, and the samples were vortexed thoroughly and centrifuged at room temperature for 5 min at 15,000 g. After centrifugation the upper aqueous phase was mixed with an equal volume of CHISAM (24:1), followed by centrifugation at room temperature for 5 min at 15,000 g. An equal volume of 99.9% isopropanol was added to the upper aqueous phase and the samples were re-centrifuged at room temperature for 5 min at 15,000 g. The supernatant was discarded, the resulting pellet was washed once in 75% ethanol, and the sample was resuspended in 50 μL of DEPC-treated water. The sample was then treated according to the manufacturer's instructions for the RNeasy® kit (Qiagen) and the RNA pellet was analyzed immediately using a NanoDrop spectrophotometer, as described in the next section, and then stored at −80°C.
For the combined TRIzol® and RNeasy® kit (Qiagen) method, the hydrogels were finely minced with sharp scissors in 1 mL of TRIzol®, disrupted with the ultrasonic homogenizer as described previously, and incubated for 5 min at room temperature. Subsequently, 200 μL of chloroform was added and the samples were incubated at room temperature for 10 min before centrifuging at 4°C for 10 min at 12,000 g. The upper aqueous phase was collected and 250 μL of 70% ethanol was added to a maximum volume of 700 μL. The sample was centrifuged at room temperature for 15 min at 15,000 g and the pellet was resuspended in 50 μL of DEPC-treated water. Each sample was subsequently treated according to the manufacturer's instructions for the RNeasy® kit (Qiagen), analyzed using the NanoDrop spectrophotometer, and the RNA pellet was stored at −80°C.
For the combined TRIzol® and Qiaex® gel extraction kit (Qiagen) method, we investigated the possible adaption of a kit commonly used for extracting DNA from agarose hydrogels based on charge properties of the nucleic acids. The MGC hydrogels were finely minced and then homogenized in 1 mL of TRIzol® and incubated for 5 min at room temperature before adding 200 μL of chloroform. Following 10 min of incubation at room temperature, the samples were centrifuged at 4°C for 10 min at 12,000 g. The upper aqueous phase was collected and 600 μL of the QX1 buffer provided with the kit was added along with 10 μL of Qiaex II bead suspension, and the samples were vortexed thoroughly for 30 s. The sample was incubated at 50°C for 10 min to allow for the binding of the nucleic acids to the beads, with vortexing every 2 min to ensure that the beads were kept in suspension. Subsequently, the samples were centrifuged for 30 s at 15,000 g and the supernatant was carefully aspirated. The resulting pellet was washed with 500 μL of QX1 buffer and resuspended by vortexing. Centrifugation for 30 s at 15,000 g was repeated and the supernatant was carefully aspirated. The beads were washed twice with wash buffer, with centrifugation for 30 s at 15,000 g between each wash, and air-dried for 10 min. To elute the nucleic acids, 20 μL of DEPC-treated water was added and the solution was incubated at room temperature for 5 min followed by centrifugation for 30 s at 15,000 g. A second elution was performed with an additional 20 μL of DEPC-treated water. The extracted RNA was analyzed using the NanoDrop spectrophotometer and the RNA pellet was stored at −80°C.
For our lab protocol for RNA extraction with extended solvent purification,15 the hydrogels were finely minced and sonicated in 1 mL of TRIzol® and allowed to stand for 5 min at room temperature. Subsequently, 200 μL of chloroform was added and the mixture was vortexed for 15 s and incubated for 3 min at room temperature prior to centrifuging at 4°C for 15 min at 12,000 g. The upper aqueous phase was collected and 500 μL of phenol:chloroform:isoamyl (25:24:1) was added, followed by vortexing for 15 s, incubation at room temperature for 2 min, and centrifugation at 4°C for 15 min at 12,000 g. The upper aqueous phase was collected and 500 μL of CHISAM (49:1) was added to the sample followed by vortexing, incubation, and re-centrifugation at 12,000 g. The upper aqueous phase was combined with 500 μL of 100% isopropanol and mixed by inverting for 15 s, incubated at room temperature for 10 min, and centrifuged at 4°C for 10 min at 12,000 g. The supernatant was carefully removed without disturbing the RNA pellet, and the pellet was washed once in 500 μL of 70% ethanol and resuspended in 22 μL of DEPC-treated water. The samples were immediately analyzed using the NanoDrop before being stored at −80°C.
RNA assessment (purity, concentration, and yield)
The RNA purity and concentration were measured using a NanoDrop spectrophotometer (ND1000; Fisher Scientific) with a 1.25 μL sample volume. RNA purity was assessed according to the absorbance ratios, A260/A280 and A260/A230. Ideally, RNA samples should have absorbance readings between 1.9 and 2.1 for the A260/A280 ratio, and values over 2 for the A260/A230 ratio. The average RNA yield for the triplicate samples for each method was calculated using the RNA concentration and the total sample volume.
End-point RT-PCR gene expression analysis
Semi-quantitative RT-PCR analysis was conducted to compare the effects of the extraction protocols on the amplification of 3 common housekeeping genes (GAPDH, TfR, 18S), as well as one lineage marker (PPARγ) that would be expressed at low levels in all of the samples, using a Bio-Rad C1000™ thermal cycler. First-strand cDNA was synthesized from 1 μg of total RNA in a 20-μL reaction volume with first-strand buffer (50 mM Tris-HCl, 75 mM KCl, 3 mMMgCl2; Invitrogen), 10 mM dithiothreitol (DTT; Invitrogen), 0.09 OD260 units of random primers (Invitrogen, Cat. No. 48190-011), 0.5 mM of each dNTP (Invitrogen, Cat. No. 18427-013), and 200 units of SuperScript™ II RT (Invitrogen, Cat. No. 18064-104). Minus RT-controls were also prepared for each sample, replacing the RT enzyme with DEPC-treated water during cDNA synthesis. For the PCR step, the gene-specific primers (50 nM, desalted, Invitrogen) were designed using Primer3 software (Table 2) and had a melting temperature of 60°C. The PCRs were conducted using illustra™ puReTaq Ready-To-Go PCR beads (GE Healthcare, Cat. No. 27-95557-02) according to the manufacturer's instructions using 1 μL of cDNA per reaction in a 25-μL reaction volume. The PCR amplifications for PPARγ and TfR were performed under conditions of 95°C for 5 min, followed by 40 cycles of 30 s at 95°C (denaturation), 30 s at 58°C (annealing), and 30 s at 72°C (elongation). PCR amplifications for GAPDH and 18S were performed under similar conditions with a total of 25 and 30 cycles, respectively, to ensure amplification within the linear range. Minus-RT (to detect genomic contamination) and no-template (to detect reagent contamination) controls were included for all samples and all genes on every plate. The PCR products were separated via electrophoresis on 5% agarose gels (High Resolution Agarose; Sigma-Aldrich, Cat. No. A4718), stained with ethidium bromide, and detected under ultraviolet light (G:Box Chemi HR16, Syngene).
Table 2.
Primer Sequences
| Gene | Description | Accession No. | Primer | Fragment length (bp) | Genomic product length (bp) |
|---|---|---|---|---|---|
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | NM_002046 | F: ACAGTCAGCCGCATCTTCTT R: ACGACCAAATCCGTTGACTC |
94 | ≥324 |
| TfR | Transferrin receptor | NM_003234 | F:AGACTTTGGATCGGTTGGTG R:TTAAATGCAGGGACGAAAGG |
62 | 270 |
| 18S | 18S ribosomal RNA | X03205 | F:ACCGCGGTTCTATTTTGTTG R:CCCTCTTAATCATGGCCTCA |
51 | 51 |
| PPARγ | Peroxisome proliferator-activated receptor gamma | NM_138712 | F: TTCAGAAATGCCTTGCAGTG R: CCAACAGCTTCTCCTTCTCG |
84 | 13213 |
Densitometry analysis
The intensity of the bands for each of the genes was quantified using ImageJ analysis software (National Institute of Health, Bethesda, MD). The densitometry values were calculated based on the average pixel intensity in each band for the triplicate samples (n=3). The data presented are expressed as a mean±standard deviation (SD), and statistical analysis was performed by one-way ANOVA with a Tukey's post hoc comparison of means (p<0.05) using GraphPad Prism® software.
Results
RNA quality assessment
RNA recovered using each of the extraction methods from the ASCs photoencapsulated in the MGC hydrogels was evaluated in terms of the standard absorbance ratios, concentration (ng/μL), and total RNA (μg), as summarized in Table 3. In addition, the effects of predigestion of the MGC hydrogels with lysozyme were assessed to determine whether enzymatic treatment enhanced RNA liberation from the polysaccharide scaffolds. In general, the A260/A280 ratio was within the target range of 1.9–2.1 for all of the methods, suggesting that there were no high levels of sample contamination with protein, lipid, or DNA. However, the A260/A230 ratio values were all lower than 2, potentially indicative of organic solvent and/or polysaccharide contamination. Interestingly, lysozyme pretreatment did not enhance RNA recovery for any of the extraction methods with the exception of the TRIzol®+Qiaex® kit approach, although the results with enzymatic treatment in this group were variable. The lysozyme treatment was not observed to have a positive effect on the A260/A280 and A260/A230 ratios, indicating that including this enzymatic digestion step was not advantageous in terms of either the yield or purity. Overall, the highest concentration and total yield of RNA was isolated using the TRIzol®+extended solvent purification method without lysozyme digestion (9.17±1.76 μg), suggesting that this approach might be favorable, especially for scaffolds seeded at low cell densities.
Table 3.
RNA Extraction Results
| A260/A280 | A260/A230 | RNA concentration (ng/μL) | Total RNA (μg) | |
|---|---|---|---|---|
| Freeze grind+CTAB+RNeasy® | 2.01±0.04 | 0.32±0.28 | 59.8±18.4 | 1.80±0.55 |
| Mince+CTAB+RNeasy® | 2.05±0.01 | 1.44±0.68 | 266.4±42.1 | 7.99±1.26 |
| Lysozyme+CTAB+RNeasy® | 1.94±0.08 | 0.86±0.60 | 73.6±59.3 | 2.21±1.78 |
| TRIzol®+RNeasy® | 2.11±0.01 | 1.74±0.28 | 222.1±22.9 | 6.66±0.69 |
| Lysozyme+TRIzol®+RNeasy® | 2.10±0.03 | 1.47±0.20 | 129.8±43.2 | 3.90±1.30 |
| TRIzol®+Qiaex® | 1.88±0.02 | 0.65±0.08 | 76.9±11.6 | 3.08±0.47 |
| Lysozyme+TRIzol®+Qiaex® | 1.98±0.05 | 0.62±0.42 | 131.0±59.3 | 5.24±2.38 |
| TRIzol®+extended solvent purification | 1.96±0.02 | 0.72±0.36 | 416.9±80.1 | 9.17±1.76 |
| Lysozyme+TRIzol®+extended solvent purification | 1.94±0.03 | 0.46±0.19 | 192.90±33.31 | 4.23±0.73 |
Endpoint RT-PCR gene expression analysis
To further assess the quality of the RNA recovered, 1 μg of total RNA from each sample was analyzed by end-point RT-PCR to assess the amplification of common endogenous control genes (GAPDH and TfR) for ASCs, as well as the adipogenic marker PPARγ (Fig. 1). To accurately normalize the gene expression levels in the different samples, we conducted preliminary experiments to confirm that each of the genes was being analyzed within the linear range of the amplification curve. More specifically, for all of the genes, PCR amplification was conducted at 5-cycle intervals over 20 to 45 cycles and the changes in the agarose gel band intensity were semiquantitatively analyzed using ImageJ software. As expected, GAPDH was abundantly expressed in the undifferentiated ASC population, whereas TfR and PPARγ were generally expressed at lower levels. As such, for the subsequent studies, GAPDH was amplified for 25 cycles, and TfR and PPARγ were amplified for 40 cycles.
FIG. 1.
Representative end-point reverse transcriptase–polymerase chain reaction (RT-PCR) gene expression results for the housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and transferrin receptor (TfR), as well as the adipogenic marker peroxisome proliferator activated receptor-γ (PPARγ), for each of the RNA isolation methods investigated (n=3). The intensities of the bands were quantified by densitometry analysis, with the average and standard deviation values provided below each image. CTAB, cetyl trimethylammonium bromide.
The densitometry results indicated that the highest levels of housekeeping gene expression were detected when the (1) TRIzol®+RNeasy® and (2) TRIzol®+extended solvent purification methods were applied (Fig. 1). Notably, there was variability in the intensity of the bands for samples processed with and without lysozyme digestion, indicating that the enzymatic pretreatment might have affected the quality of the RNA to some extent. Bands indicative of PPARγ expression were also detected in the samples processed with each of these three methods, demonstrating that these techniques also enabled the amplification of genes expressed at low levels within the cell population. Overall, the CTAB+RNeasy® method had the lowest intensity bands for all of the genes studied in comparison to the other methods, indicative of less efficient amplification during the PCR in this group.
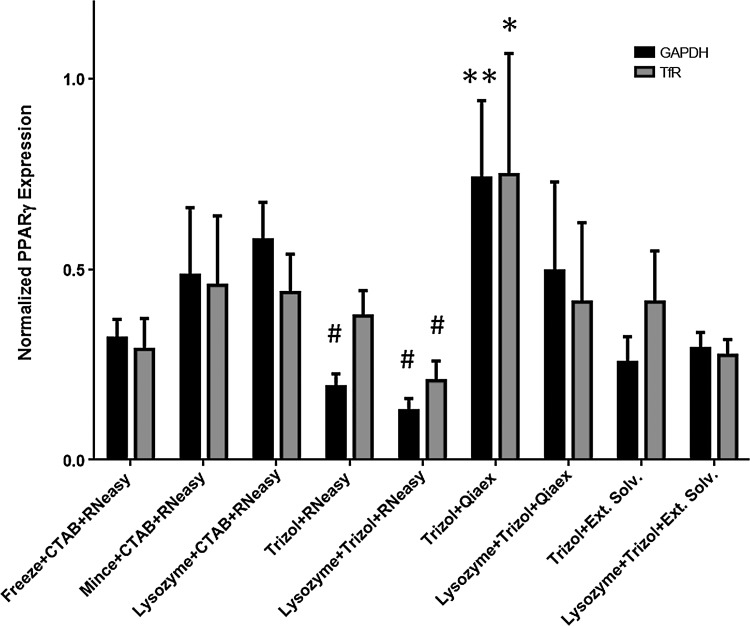
Based on the densitometry analysis, we normalized the levels of PPARγ expression to both GAPDH and TfR in each of the sample groups. In analyzing these results, while the intensity of the individual bands varied with the specific RNA isolation methods (Fig. 1), there were similar patterns observed in terms of the relative levels of PPARγ expression (Fig. 2). Positively, the normalized levels were on a similar order of magnitude for all of the sample conditions and for both of the housekeeping genes. In particular, consistent results were observed in the CTAB+RNeasy® samples and the TRIzol®+extended solvent purification samples, when either GAPDH or TfR was used as the endogenous control. In the TRIzol®+RNeasy® sample group, the trends suggest that the more abundant GAPDH was amplified with higher relative efficiency as compared to TfR. The results also suggest that the weakly expressed PPARγ was amplified with higher relative efficiency than either GAPDH or TfR in the TRIzol®+Qiaex® samples, potentially indicating that this kit might not be the most appropriate choice for RNA purification.
FIG. 2.
Relative PPARγ expression levels normalized to either GAPDH or TfR for each of the RNA isolation methods, based on densitometry analysis (n=3). Similar results were obtained for both housekeeping genes for all of the extraction methods studied. Statistical significance was determined using a one-way ANOVA with a Tukey's post hoc comparison of means (p<0.05). *Statistically different than all other groups. **Statistically different than the freeze+cetyl trimethylammonium bromide (CTAB)+RNeasy® method, the TRIzol®+RNeasy® methods, and the TRIzol®+extended solvent methods. #Statistically different than the mince+CTAB+RNeasy® method and the lysozyme+CTAB+RNeasy® methods.
Contamination with genomic DNA
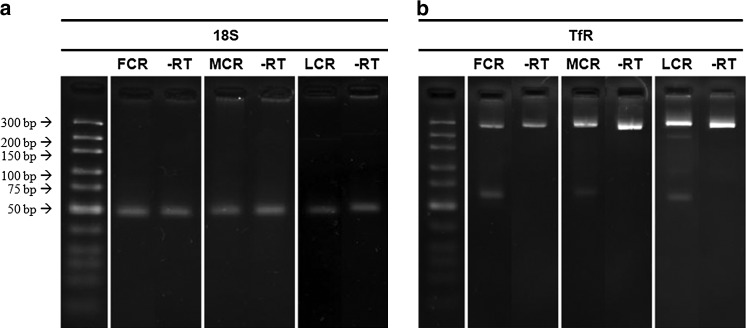
We assessed genomic contamination through PCR amplification of minus-RT controls for all of the samples. Based on these controls, there was evidence of DNA contamination only in the samples processed with the CTAB+RNeasy® methods. Genomic contamination in the CTAB groups can be readily elucidated by comparing the gel electrophoresis results for the 18S and TfR housekeeping genes (Fig. 3). When analyzing 18S, which contains no introns, single bands were detected in the CTAB samples and associated minus-RT controls (Fig. 3a). No bands were detected in the minus-RT controls for any of the samples processed with the TRIzol®-based methods. Importantly, we designed our primers for TfR to span an intron, so that genomic contamination in the CTAB groups was also distinguished from the desired RNA-derived product (62 bp) by the presence of a second, larger product on the gels (270 bp), as shown in Figure 3b, which was not observed in any of the other sample groups. The primers for GAPDH and PPARγ were similarly designed to span introns; however, the genomic product sizes for these genes were too large to resolve using the gel conditions in our study.
FIG. 3.
Representative end-point RT-PCR gene expression results for (a) 18S and (b) TfR from RNA isolated using the freeze grind+CTAB+RNeasy® (FCR) method, the mince+CTAB+RNeasy® (MCR) method, and the lysozyme+CTAB+RNeasy® (LCR) method. Genomic contamination was detected following agarose gel electrophoresis in all minus-RT controls. In addition, as shown in the TfR results, where the primers were designed to span an intron–exon boundary, two products were formed during the PCR, corresponding to a genomic product size of 270 bp and an mRNA product size of 62 bp.
Discussion
There is a need to establish standard methods for RNA isolation from polysaccharide scaffolds, as these biomaterials are being extensively studied as scaffolds for tissue engineering.17–20 In advancing these technologies, it is important to be able to accurately characterize the cell response using molecular biology techniques including RT-PCR analysis of gene expression. These types of studies will provide critical insight into how engineered cellular microenvironments can be tuned to optimize the cell response for each specific application. As such, we conducted a comparative analysis of four different RNA isolation protocols for RT-PCR studies of cells encapsulated within chitosan-based scaffolds.
In general, the method of choice should minimize polysaccharide and genomic contamination while preserving high-quality RNA for downstream RT-PCR processing, regardless of the specific levels of gene expression. Based on our analysis, an important observation is that while purity ratios and yields calculated from spectrophotometric data can provide an initial assessment of the RNA quality and can help to identify potential contaminants, there are cases when these ratios are within the expected ranges despite problems with the purity or integrity of the samples, which can only be detected during downstream processes such as PCR amplification.
Variations of the CTAB method have been used to extract RNA from plants and, in a recent study, from cells encapsulated within polysaccharide scaffolds including chitosan.10,21,22 In contrast to this previous work, our study demonstrated that the freeze grind+CTAB+RNeasy® combination resulted in the lowest overall A260/230 ratio and total RNA yield. The discrepancy in these results may be attributed to differences in the size of the constructs, the number of protonated amine groups present on the chitosan, and the degree of crosslinking. In addition, while freezing and grinding are common practices to disrupt cells in matrices, these steps may have contributed to the reduced yield in this method. An important consideration is that grinding of frozen samples can increase the risk of exposure to endogenous ribonucleases (RNases) if the sample is not immediately homogenized in an RNA extraction buffer that inactivates RNases.23 In general, the efficacy of cryo-pulverization may be dependent on the quantity of sample, the properties of the scaffold, and the availability of specialized equipment. For our samples, the small volumes and soft mechanical properties of the gels resulted in more effective RNA isolation using direct homogenization in the extraction solution, potentially because grinding with a mortar and pestle resulted in sample loss during transfer.
Our results demonstrate that the choice of buffer used in the initial steps of the RNA isolation is a critical factor in the separation process. The methods utilizing TRIzol® produced higher quality RNA with better PCR amplification, consistent with the findings of other published studies.12,24 The differences in the efficacy of the methods can be elucidated by analyzing the mechanisms involved in each of the separation processes. For example, the lower purities and yields, as well as the more variable amplification results observed with the samples processed with the Qiaex® kit, may be related to its design for processing solubilized agarose-based gels through nucleic acid adsorption onto QIAEX II silica-gel particles, under buffer conditions optimized for DNA isolation.
The CTAB+RNeasy® method removes polysaccharide fragments using a basic CTAB buffer solution (pH 8), rather than conventional acid guanidinium isothiocyanate solutions like in TRIzol®, to minimize complexation between positively charged chitosan fragments and negatively charged RNA.10 Although this may have advantages, isolating under basic conditions can impede the removal of contaminating DNA since polar DNA and RNA fragments will both partition into the aqueous phase during phase separation with CHISAM.25 DNA can be more efficiently separated in an acidic environment, typically at pH 4.8, since under these conditions the DNA backbone becomes neutralized by surrounding protons and dissolves into the organic phase.25 RNA fragments are not affected in the same manner since they are single stranded and the exposed nucleotides form hydrogen bonds with the water molecules and remain in the aqueous phase.
Following the initial extraction, subsequent liquid–liquid phase separations with phenol:chloroform-based solutions can remove a large amount of residual contamination that could potentially affect downstream procedures such as RT-PCR.26 The extended solvent purification protocol we investigated includes two additional phase extractions, using phenol:chloroform:isoamyl (25:24:1) and CHISAM (49:1), and resulted in the highest yield of RNA. The genomic contamination in the CTAB+RNeasy® samples may be related to the fact that treatment with phenol at low pH (optimally pH 4.8) was not incorporated in the isolation, as it would interfere with the PVP in the CTAB buffer.22,23 Soluble PVP acts as a purifying additive that binds to polyphenols and polysaccharides naturally found within the samples, which would otherwise form complexes with the RNA.27,28 The chloroform-based solutions used in the later purification steps in this protocol may have been less effective in removing residual proteins and DNA/RNA-protein aggregates as compared to treatment with phenol:chloroform solutions.26,29 Future modifications of this method could include replacing the PVP with polyvinylpolypyrrolidone, which is a highly crosslinked form of PVP that is insoluble and can be separated in subsequent treatment steps with phenol-based solutions. Alternatively, the PVP could be removed via ethanol precipitation of the RNA product before further phase extractions including phenol.26,30,31 The genomic contamination observed with the CTAB+RNeasy® methods highlights the importance of careful primer design and experimental controls. DNase treatment of the extracted RNA may be required if the CTAB+RNeasy® method is used, especially when analyzing genes that do not contain introns or if the primers designed do not span intron–exon boundaries.
Ultimately, selecting an appropriate RNA isolation technique depends on the properties of the sample. With the volume, mechanical properties, charge characteristics, and seeding density of our scaffolds, we obtained the most consistent results in terms of purity, yield, and gene amplification with the TRIzol®+extended solvent purification method. Overall, RNA isolation techniques for cells encapsulated in polysaccharide-rich hydrogels may be improved through direct homogenization of the sample in an acidic extraction buffer and by further purification using multiple phenol:chloroform-based phase separations. As economics may also be a factor, our estimated costs for the CTAB+RNeasy® method were between $9.00 and $10.00 USD/reaction, the TRIzol®+RNeasy® method were between $7.50 and $8.00 USD/reaction, and both the Qiaex® and extended solvent purification methods were between $3.50 and $4.50 USD/reaction.
Acknowledgments
Funding for this study was provided by the Canadian Institutes of Health Research (CIHR) Institute of Musculoskeletal Health and Arthritis (IMHA), with infrastructure support from the Natural Sciences and Engineering Research Council of Canada (NSERC) Research Tools and Instruments (RTI) program. The authors would like to thank Drs. J.F. Watkins, D. McKay, K. Meathrel, and J. Davidson and Mrs. K. Martin for clinical collaborations to support this work.
Disclosure Statement
No competing financial interests exist.
References
- 1.Fernandes S.C. Freire C.S. Silvestre A.J. Pascoal Neto C. Gandini A. Novel materials based on chitosan and cellulose. Polym Int. 2011;60:875. [Google Scholar]
- 2.Finkenstadt V.L. Natural polysaccharides as electroactive polymers. Appl Microbiol Biotechnol. 2005;67:735. doi: 10.1007/s00253-005-1931-4. [DOI] [PubMed] [Google Scholar]
- 3.Kumbar S.G. Toti U.S. Deng M. James R. Laurencin C.T. Aravamudhan A., et al. Novel mechanically competent polysaccharide scaffolds for bone tissue engineering. Biomed Mater. 2011;6:065005. doi: 10.1088/1748-6041/6/6/065005. [DOI] [PubMed] [Google Scholar]
- 4.Tan W. Krishnaraj R. Desai T.A. Evaluation of nanostructured composite collagen—chitosan matrices for tissue engineering. Tissue Eng. 2001;7:203. doi: 10.1089/107632701300062831. [DOI] [PubMed] [Google Scholar]
- 5.Kim I.-Y. Seo S.-J. Moon H.-S. Yoo M.-K. Park I.-Y. Kim B.-C., et al. Chitosan and its derivatives for tissue engineering applications. Biotechnol Adv. 2008;26:1. doi: 10.1016/j.biotechadv.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Suh J.K. Matthew H.W. Application of chitosan-based polysaccharide biomaterials in cartilage tissue engineering: a review. Biomaterials. 2000;21:2589. doi: 10.1016/s0142-9612(00)00126-5. [DOI] [PubMed] [Google Scholar]
- 7.Smale G. Sasse J. RNA isolation from cartilage using density gradient centrifugation in cesium trifluoroacetate: an RNA preparation technique effective in the presence of high proteoglycan content. Anal Biochem. 1992;203:352. doi: 10.1016/0003-2697(92)90324-z. [DOI] [PubMed] [Google Scholar]
- 8.Ullrich A. Shine J. Chirgwin J. Pictet R. Tischer E. Rutter W.J., et al. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977;196:243. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- 9.Blin N. Stafford D.W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976;3:2303. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L. Stegemann J.P. Extraction of high quality RNA from polysaccharide matrices using cetyltrimethylammonium bromide. Biomaterials. 2010;31:1612. doi: 10.1016/j.biomaterials.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S. Jia S. Liu G. Fang D. Zhang D. Osteogenic differentiation of muscle satellite cells induced by platelet-rich plasma encapsulated in three-dimensional alginate scaffold. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2012;114:32. doi: 10.1016/j.tripleo.2011.07.048. [DOI] [PubMed] [Google Scholar]
- 12.Wang C. Hao J. Zhang F. Su K. Wang D.-A. RNA extraction from polysaccharide-based cell-laden hydrogel scaffolds. Anal Biochem. 2008;380:333. doi: 10.1016/j.ab.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Hirano S. Yagi Y. The effects of N-substitution of chitosan and the physical form of the products on the rate of hydrolysis by chitinase from Streptomyces griseus. Carbohydr Res. 1980;83:103. doi: 10.1016/s0008-6215(00)85369-0. [DOI] [PubMed] [Google Scholar]
- 14.Hirano S. Tsuchida H. Nagao N. N-acetylation in chitosan and the rate of its enzymic hydrolysis. Biomaterials. 1989;10:574. doi: 10.1016/0142-9612(89)90066-5. [DOI] [PubMed] [Google Scholar]
- 15.Flynn L.E. The use of decellularized adipose tissue to provide an inductive microenvironment for the adipogenic differentiation of human adipose-derived stem cells. Biomaterials. 2010;31:4715. doi: 10.1016/j.biomaterials.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 16.Amsden B.G. Sukarto A. Knight D.K. Shapka S.N. Methacrylated glycol chitosan as a photopolymerizable biomaterial. Biomacromolecules. 2007;8:3758. doi: 10.1021/bm700691e. [DOI] [PubMed] [Google Scholar]
- 17.Pallela R. Venkatesan J. Janapala V.R. Kim S.-K. Biophysicochemical evaluation of chitosan-hydroxyapatite-marine sponge collagen composite for bone tissue engineering. J Biomed Mater Res A. 2011;100A:486. doi: 10.1002/jbm.a.33292. [DOI] [PubMed] [Google Scholar]
- 18.Guan S. Zhang X.-L. Lin X.-M. Liu T.-Q. Ma X.-H. Cui Z.-F. Chitosan/gelatin porous scaffolds containing hyaluronic acid and heparan sulfate for neural tissue engineering. J Biomater Sci Polym Ed. 2012 doi: 10.1080/09205063.2012.731374. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 19.Pok S. Myers J.D. Madihally S.V. Jacot J.G. A multi-layered scaffold of a chitosan and gelatin hydrogel supported by a Pcl core for cardiac tissue engineering. Acta Biomater. 2012;9:5630. doi: 10.1016/j.actbio.2012.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zakhem E. Raghavan S. Gilmont R.R. Bitar K.N. Chitosan-based scaffolds for the support of smooth muscle constructs in intestinal tissue engineering. Biomaterials. 2012;33:4810. doi: 10.1016/j.biomaterials.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang S. Puryear J. Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113. [Google Scholar]
- 22.Zeng Y. Yang T. RNA isolation from highly viscous samples rich in polyphenols and polysaccharides. Plant Mol Biol Rep. 2002;20:417. [Google Scholar]
- 23.Lader E.S. Methods and reagents for preserving RNA in cell and tissue samples. Patent US8178296. 2012 [Google Scholar]
- 24.Chomczynski P. Sacchi N. Single-step method of RNA isolation by acid guanidinium extraction. Anal Biochem. 1987;162:156. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 25.Brawerman G. Mendecki J. Lee S.Y. A procedure for the isolation of mammalian messenger ribonucleic acid. Biochemistry. 1972;11:637. doi: 10.1021/bi00754a027. [DOI] [PubMed] [Google Scholar]
- 26.Salzman R.A. Fujita T. Zhu-Salzman K. Hasegawa P.M. Bressan R.A. An improved RNA isolation method for plant tissues containing high levels of phenolic compounds or carbohydrates. Plant mol Biol Rep. 1999;17:11. [Google Scholar]
- 27.Claros M.G. Cánovas F.M. Rapid high quality RNA preparation from pine seedlings. Plant Mol Biol Rep. 1998;16:9. [Google Scholar]
- 28.Porebski S. Bailey L.G. Baum B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 1997;15:8. [Google Scholar]
- 29.Wang T. Zhang N. Du L. Isolation of RNA of high quality and yield from Ginkgo biloba leaves. Biotech Lett. 2005;27:629. doi: 10.1007/s10529-005-3629-1. [DOI] [PubMed] [Google Scholar]
- 30.Tong Z. Qu S. Zhang J. Wang F. Tao J. Gao Z., et al. A modified protocol for RNA extraction from different peach tissues suitable for gene isolation and real-time PCR analysis. Mol Biotech. 2012;50:229. doi: 10.1007/s12033-011-9433-3. [DOI] [PubMed] [Google Scholar]
- 31.Chen L. Sheng S.-J. Tan X.-M. Shen Y.-J. Li H.-Q. Zhao S.-J. An effective method of RNA isolation from Fallopia multiflora tuberous roots. Prep Biochem Biotech. 2012;42:87. doi: 10.1080/10826068.2011.566297. [DOI] [PubMed] [Google Scholar]