Abstract
Recent studies suggest that dysregulated innate immunity plays an important role in the pathogenesis of immunoglobulin A nephropathy (IgAN). The interleukin-20 subfamily and its receptor, interleukin-22 receptor alpha-1 (IL-22R1), were recently identified as immunomodulators in human diseases, acting as mediators of mucosal host defense. However, the potential role of IL-22R1 in the pathogenesis of IgAN has not been explored. In the current study, 194 patients with IgAN and 287 normal controls were genotyped for coding polymorphisms of the IL-22R1 gene and the association between the polymorphisms and IgAN was investigated. Local expression of IL-22R1 was examined in patients with IgAN and healthy controls using immunohistochemistry. Our case–control analysis showed that genotypes of rs3795299 were associated with childhood IgAN. Individuals with the CC genotype of rs3795299 had about 3-fold reduced risk of IgAN compared with those with the GG genotype in the codominant model (P=0.0028) and those with the genotypes containing the G allele (GG or GC) in the recessive model (P=0.002). After Bonferroni correction, the association between the rs3795299 CC genotype and reduced risk of developing IgAN remained significant. Furthermore, the renal expression of IL-22R1 was significantly higher in healthy controls compared with subjects with IgAN. Our data suggest that the CC genotype of rs3795299 polymorphism in the IL-22R1 gene is associated with the reduced risk of IgAN, and this genetic association was supported by the higher renal expression of IL-22R1 in healthy controls compared with patients with IgAN.
Introduction
Immunoglobulin A nephropathy (IgAN) is an immune complex-mediated glomerulonephritis, characterized by the deposition of IgA1 subclass in glomerular mesangium (Glassock 2011). It is the most common chronic glomerulonephritis in many parts of the world and is an important cause of end-stage renal disease. The pathogenesis of IgAN remains unclear. It is regarded as a complex disease that is initiated by more than one genetic factor combined with environmental factors (Lai 2012). Although previous genome-wide linkage studies of familial IgAN reported the associations of several genetic loci with the development of IgAN, the underlying genes remain elusive (Gharavi and others 2000; Bisceglia and others 2006; Paterson and others 2007). Two recent large-scale genome-wide association studies of IgAN in a Han Chinese population suggest that this disease is a heterogeneous disease having multiple susceptibility loci (Gharavi and others 2011; Yu and others 2012). One of these studies, Yu and others (2012), showed that the genes encoding α-defensin (DEFA) are implicated as susceptibility genes, suggesting dys-regulation of innate immunity in IgAN because α-defensins are innate immune molecules and involved in the inflammation response to infection. The onset or deterioration of IgAN is often associated with respiratory or gastrointestinal tract infection, which implies that IgAN may result from abnormal mucosal immunity. Several experimental and clinical data suggest that in the presence of underlying defects in the host immune system, exogenous antigens derived from various pathogens can trigger IgAN (Coppo and others 2010). The associations between IgAN and Toll-like receptors, the integral compositions of innate immunity, support this mechanism (Suzuki and others 2008; Park and others 2011).
The IL-10 cytokine family is an immunomodulator that elicits diverse host defense mechanisms. It can be divided into 3 groups of cytokines according to their biological functions—IL-10, IL-20 subfamily, and type III interferon. Among these 3 groups, IL-20 subfamily cytokines primarily act on epithelial cells and play an important role in innate immunity, especially against bacterial infection. They include 5 cytokines (IL-19, IL-20, IL-22, IL-24, and IL-26) that exert their biological effects via heterodimeric receptor complexes, composed of a type 1 receptor chain (R1) and a type 2 receptor chain (R2) (Ouyang and others 2011).
The interleukin-22 receptor alpha-1 (IL-22R1) is a shared receptor binding to 3 cytokines, IL-20, IL-22, and IL-24 (Wang and others 2002; Wegenka 2010; Ouyang and others 2011). It is a main receptor for IL-22, but also an alternative type 1 receptor chain for IL-20 and IL-24. Interestingly, IL-22R1 is not expressed in immune cells, but in a few nonimmune peripheral tissues such as skin, lung, liver, pancreas, and kidney (Wolk and others 2004). As a result, its expression determines the target site of its ligand cytokine (Wolk and Sabat 2006).
Recently, several studies showed that IL-20 subfamily cytokines act as mediators of mucosal host defense through IL-22R1. In skin, the GI tract, and trachea, IL-22 enhances innate immunity of mucosal cells against bacterial infection by producing a group of antimicrobial proteins, including β-defensin2 (BD2) and BD3, S100A8/9, RegIIIα, RegIIIβ, and lipocalin-2 (Wolk and others 2004; Wolk and others 2006; Aujla and others 2008). During hepatic inflammation, IL-22R1 mediates the protective role of its ligand cytokines to prevent and repair liver injury (Radaeva and others 2004; Hofmann and others 2012). Furthermore, recent studies suggest an association of IL-22R1 with chronic rhinosinusitis (CRS), an inflammatory disease of the nose and paranasal sinuses. Ramanathan and others (2007) demonstrated that mucosal expression of IL-22R1 was significantly decreased in patients with CRS with a nasal polyp (NP) who were resistant to medical and surgical therapy compared to normal controls and CRS without a NP. In addition, Endam and others (2009) reported that IL-22R1 polymorphisms were associated with severe CRS. All these data suggest that IL-22R1 and its cytokine complex may contribute to an important role in innate immunity. Therefore, we speculate that IL-22R1 may play a role in the pathogenesis of IgAN.
However, there has been no report regarding an association between IgAN and the IL-20 subfamily/IL-22R1although the kidney is one of the target sites in which IL-22R1 is expressed. Therefore, we conducted this study to evaluate whether coding the polymorphism of IL-22R1 may affect the susceptibility of IgAN and investigated the renal expression of IL-22R1 via immunohistochemistry (IHC) to confirm the association between IL-22R1 and IgAN.
Materials and Methods
Subjects for genotyping
One hundred ninety four patients with IgAN diagnosed by renal biopsy and 287 healthy control subjects were enrolled for this study. Demographic and clinical findings of subjects in this study are summarized in Table 1. A sex-matched control group was recruited from healthy subjects who came for routine health screening tests and volunteered to participate in this study. It was practically difficult to recruit age-matched controls because of several reasons. First, it is difficult to obtain blood samples from healthy children who do not need to perform laboratory tests. More importantly, IgAN is known to develop in young adults in their twenties, and healthy individuals aged more than 20 years are required as controls to avoid the possibility of subclinical IgAN. In addition, it is a genotyping, which is not significantly affected by age. The screening included the completion of a questionnaire that contained the following: symptoms, medical history, blood pressure, electrocardiography, abdominal sonography, laboratory tests, such as complete blood count, fasting glucose level, total cholesterol, triglyceride levels, high-density lipoprotein cholesterol levels, rheumatoid factor, hepatitis B and C viral markers, hemoglobin A1C, liver enzymes, blood urea nitrogen, creatinine, electrolytes, and a urinalysis. Controls without any abnormal results were selected. This study was approved by the Kyung Hee University Institutional Review Board, Seoul, Korea. Written informed consent was obtained from all of the subjects.
Table 1.
Demographic Characteristics of Subjects for Genotyping and Patient Subgroups
| Total subjects (n=481) | IgAN | Control | P value |
|---|---|---|---|
| Number of subjects (n) | 194 | 287 | |
| Male/Female | 114/80 | 156/131 | 0.339 |
| Age (mean±SD) | 12.55±5.14 | 37.51±13.55 | <0.001 |
| Patient subgroups (n=194) | n | ||
| 1) Gross hematuriaa | |||
| Positive/negative | 37/157 | ||
| 2) Proteinuriab (mg/m2/h) | |||
| >4/ ≤4 | 90/104 | ||
| 3) Nephrotic-range proteinuriac (mg/m2/h) | |||
| >40/ ≤40 | 14/180 | ||
| 4) Podocyte foot process effacement | |||
| Positive/negative | 72/122 | ||
| 5) Pathologic gradingd | |||
| Advanced/mild | 18/176 |
Gross hematuria developed as a first symptom of IgA nephropathy.
Means the proteinuria and the nephrotic-range proteinuria developed at the time of renal biopsy, respectively.
Patients were subgrouped into pathologically mild (Grade I–II) and advanced (III–IV) disease groups according to the modified H. S. Lee's histological grading system.
n, number of subjects; IgAN, immunoglobulin A nephropathy; SD, standard deviation.
Patient subgroups
IgAN patients were divided according to the presence or absence of the following clinical characteristics: (1) gross hematuria as a first symptom of IgAN; (2) proteinuria (>4 mg/m2/h) or (3) nephrotic-range proteinuria (>40 mg/m2/h) at the time of renal biopsy. Subgroups were also formed on the basis of the following histopathological findings; (4) the presence of podocyte foot process effacement on renal biopsy; and (5) the degree of pathological severity (pathologically mild [Grades I-II] or advanced [Grades III-IV]) assessed by the modified H. S. Lee's histological grading system (Lee and others 2005).
SNP selection and genotyping
We searched for coding SNPs (cSNPs) in the IL-22R1 gene based on the results of database searches (www.ncbi.nlm.nih.gov/SNP, dbSNP BUILD 132). On the IL-22R1 gene, there are 16 cSNPs. Among these SNPs, the SNPs without genotype frequency data (rs34967816, rs17852648, rs16829204, and rs61775472) and with a heterozygosity below 0.1 (rs34929192, rs117673187, rs35401673, rs34782294, rs34379702, rs35240302, rs34900099, and rs76898131) or a minor allele frequency below 0.1 (rs75716137 and rs17852649, particularly in the Asian population) were excluded. Finally, one missense SNP (rs3795299, Arg518Gly, C->G) and one synonymous SNP (rs10903022, Pro45Pro, C->T) were selected. The heterozygosities of SNPs were 0.475 for rs3795299 and 0.472 for rs10903022, respectively.
DNA was isolated from peripheral blood samples using a DNA Extraction kit (Roche), and SNP genotyping was performed by direct sequencing. Genomic DNA was amplified using primers specific for each SNP of the IL-22R1 gene (Table 2). The samples were sequenced using the ABI PRISM 3730XL analyzer (PE Applied Biosystems) and sequence data were analyzed using SeqManII software (DNASTAR).
Table 2.
Sequences of Primers Used for Each SNP in IL22R1 Gene
| SNP | Sequence (5′-3′) | Product size (bp) | Temperature (°C) | |
|---|---|---|---|---|
| rs3795299 | Sense | GACCCAAATGTGCTACACAGTG | 267 | 58 |
| Anti-sense | GCCTCTGAAAAGAGAATCCAGT | |||
| rs10903022 | Sense | TATATGGGGCAGGACTCTGACT | 304 | 58 |
| Anti-sense | AACATATGGAGAGGAGGGGAAT | |||
SNP, single-nucleotide polymorphism; bp, base pair.
Tissue samples for IL-22R1 expression analysis
IL-22R1 expression was evaluated in renal tissues from 35 patients with child-onset IgAN, compared with those from 14 controls. Controls samples were obtained from children or adolescents who performed renal biopsy because of abnormal urinary abnormalities occurring for at least 6 months, but had no abnormality in biopsied tissues. The use of renal tissues was approved by the Kyung Hee University Institutional Review Board. All cases were independently reviewed by 2 pathologists (A. Moon and Y.K. Park), and the diagnoses were confirmed in all instances.
IHC and scoring
IL-22R1expression was assessed by IHC using the Bond Polymer Intense Detection System (Vision BioSystems) according to the manufacturer's instructions with minor modifications. Briefly, 4-μm-thick sections of formalin-fixed, paraffin-embedded tissue were deparaffinized with Bond Dewax Solution (Vision BioSystems), and an antigen retrieval procedure was performed using Bond ER Solution (Vision BioSystems) for 30 min at 100°C. Endogenous peroxidases were quenched by incubation with hydrogen peroxide for 5 min. The sections were incubated for 15 min at an ambient temperature with the polyclonal rabbit antibody against Anti-IL-22R-1 (1:200; Millipore) using a biotin-free polymeric horseradish peroxidase–linker antibody conjugate system in the Bond-max autostainer (Vision BioSystems). Nuclei were counterstained with hematoxylin. The control sample was normal renal tubules, and the negative control was treated in an identical manner with an isotype rabbit IgG instead of the primary antibody. All slides were examined and scored by 2 independent pathologists (A. Moon and Y.K. Park), who were blinded to the clinicopathological data and patient identity. The results of IHC were scored by a modified semiquantitative method (Choi and others 2011). Staining intensity was scored as 0 (absent), 1 (weak), and 2 (intense). Disagreements between the 2 pathologists were resolved by consensus.
Statistical analysis
For the case–control association study, all SNPs in cases and controls were assessed for the Hardy–Weinberg equilibrium (HWE) using SNPstats software (Biostatistics and Bioinformatics Unit) (Sole and others 2006). Logistic regression analysis was used to calculate the odds ratios (OR), 95% confidence intervals (CIs), and P values. For multiple logistic regression analyses to determine associations of SNP with IgAN, we used multiple inheritance models, including a codominant model (the relative hazard differed between subjects with 1 minor allele and those with 2 minor alleles), a dominant model (subjects with 1 or 2 minor alleles had the same relative hazard for the disease), or a recessive model (only subjects with 2 minor alleles were at increased risk of the disease). The Bonferroni correction was applied by multiplying P values by the number of SNPs analyzed (n=2). For comparisons between patients with IgAN and healthy controls, the data were adjusted for gender only, and for comparisons among the IgAN subgroups, gender and age were controlled. A linkage disequilibrium (LD) block of polymorphisms was tested using Haploview version 4.1 (Broad Institute). SNPstats and HapAnalyzer Pro version 1.0 (www.hap.ngri.go.kr), and HelixTree (Golden Helix, Inc.) were used to analyze the association of SNPs and haplotypes. We calculated the power of the sample size to verify our data, using a genetic power calculator (http://pngu.mgh.harvard. Edu/∼purcell/gpc/cc2.html). In this study, sample powers of SNPs were 0.929 (rs3795299, number of effective sample for 80% power=133) and 0.928 (rs10903022, number of effective sample for 80% power=133), respectively (α=0.05, genotype relative risk=2-fold). Therefore, our data were thought to be acceptable.
For the comparison of clinicopathologic parameters according to IL-22R1 expressions, the chi-square test was used for discrete variables, and Pearson's correlation coefficient was used for continuous variables. Comparisons of scores between groups were evaluated by the Pearson's chi-square test. Statistical analyses were performed using the package of SPSS statistics 18.0 software (SPSS, Inc.), and a value of P<0.05 was considered statistically significant.
Results
Genetic analysis
The genotype distributions of all SNPs in this study were compatible with the HWE (control group, P>0.05). The frequencies of genotypes for IL-22R1 coding polymorphisms are summarized in Table 3. As shown in Table 3, rs3795299 showed a significant association between IgAN patients and control subjects in codominant and recessive models. In the codominant model, the frequencies of GG, GC, and CC genotypes were 51.6%, 38.3%, and 10.1% in the control group and 48.5%, 48.5%, and 3.1% in the IgAN group, respectively (P=0.0028, Pc=0.0056). Individuals with the CC genotype had about 3-fold reduced risk of IgAN compared with those with the GG genotype (OR=0.32, 95% CI=0.13–0.80, P=0.0028). In the recessive model, individuals with the CC genotype also had a 3-fold decreased risk of IgAN compared with those with the genotypes containing the G allele (GG or GC) (OR=0.28, 95% CI=0.11–0.69, P=0.002, Pc=0.004). Based on the results, the CC genotype of rs3795299 was shown to have a protective effect on the development of IgAN. However, the other SNP, rs10903022, was not significantly associated with IgAN. Two SNPs of the IL-22R1 gene were analyzed for LD and haplotypes using Haploview version 4.1 and HapAnalyzer. One LD block among 2 SNPs was identified by the Gabriel method (Gabriel and others 2002). However, the haplotypes in the LD block were not associated with IgAN (data not shown). We also evaluated the genetic associations between the 2 SNPs and subgroups of IgAN patients determined by clinical or histopathological characteristics as mentioned in the Method. However, no association was observed (data not shown).
Table 3.
Logistic Regression Analysis of IL22R1 Gene Polymorphisms Between IgAN Patients and Controls
| SNP | Model | Genotype | Control (287) n (%) | IgAN (194) n (%) | OR (95% CI) | P value | Pcvalue |
|---|---|---|---|---|---|---|---|
| rs3795299 | Codominant | G/G | 148 (51.6) | 94 (48.5) | Ref | 0.0028 | 0.0056 |
| Arg518Gly | G/C | 110 (38.3) | 94 (48.5) | 1.33 (0.91–1.95) | |||
| (C->G) | C/C | 29 (10.1) | 6 (3.1) | 0.32 (0.13–0.80) | |||
| Dominant | G/G | 148 (51.6) | 94 (48.5) | Ref | 0.54 | 1 | |
| G/C-C/C | 139 (48.4) | 100 (51.5) | 1.12 (0.78–1.62) | ||||
| Recessive | G/G-G/C | 258 (89.9) | 188 (96.9) | Ref | 0.002 | 0.004 | |
| C/C | 29 (10.1) | 6 (3.1) | 0.28 (0.11–0.69) | ||||
| rs10903022 | Codominant | T/T | 154 (53.7) | 99 (51.0) | Ref | 0.41 | 0.82 |
| Pro45Pro | T/C | 112 (39.0) | 85 (43.8) | 1.17 (0.80–1.71) | |||
| (C->T) | C/C | 21 (7.3) | 10 (5.2) | 0.71 (0.32–1.58) | |||
| Dominant | T/T | 154 (53.7) | 99 (51.0) | Ref | 0.62 | 1 | |
| T/C-C/C | 133 (46.3) | 95 (49.0) | 1.10 (0.76–1.58) | ||||
| Recessive | T/T-T/C | 266 (92.7) | 184 (94.8) | Ref | 0.29 | 0.58 | |
| C/C | 21 (7.3) | 10 (5.2) | 0.66 (0.30–1.44) |
Bold characters represent statistically significant values.
The P values were from logistic regression analysis with codominant, dominant, and recessive models controlling gender as covariate.
OR, odds ratio; CI, confidence interval; Ref, reference; n, number of subjects; Pc value, corrected by the Bonferroni method.
IL-22R1 expression in the normal and diseased kidneys
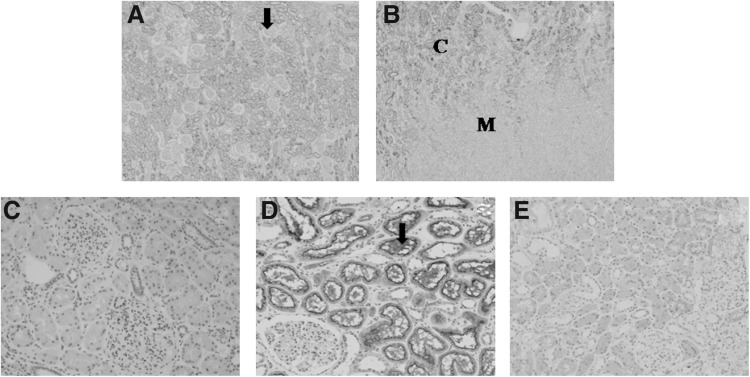
IL-22R1 staining was performed on biopsied samples from patients with IgAN (n=35) and normal controls (n=14). IL-22R1 expression was mainly present on cortical tubules, but not in the medullary portion of the kidneys (Fig. 1A, B). Furthermore, it appears to be more strongly expressed in the brush border of proximal tubules than in distal tubules (Fig. 1D). These patterns of IL-22R1 expression remained in IgAN kidneys. However, IL-22R1 expression was significantly decreased in patients with IgAN (Fig. 1E), as compared to normal controls (Fig. 1D). Figure 2 shows representative examples of IL-22R1 immunostaining according to the semiquantitative scores and relative degrees of IL-22R1 expression in the 2 groups. In the IgAN group (n=35), 13 patients (37%) had a score of 0, 15 patients (43%) a score of 1, and 7 patients (20%) a score of 2 (mean=0.83). In contrast, in the control group (n=14), none had a score of 0, 4 subjects (29%) a score of 1, and 10 subjects (71%) a score of 2 (mean=1.71). Based on the results, patients with IgAN had significantly lower scores than controls (0.83 vs. 1.71, P<0.001).
FIG. 1.
Immunostaining of the interleukin-22 receptor alpha-1 (IL-22R1) in normal controls (A–D) and a patient with IgA nephropathy (E). (A) Renal cortex: IL-22R1 is expressed in cortical tubules, not expressed in glomerulus (arrow, ×40). (B) Corticomedullary junction: IL-22R1 is not expressed in the medullar of kidney (×40) (C, cortex; M, medullar). (C) Control immunostaining of the renal cortex was performed with an isotype rabbit IgG (×200). (D) Renal cortex: proximal tubules show uniformly strong IL-22R1 expression, especially in brush borders (arrow). Distal tubules show weak IL-22R1 expression (×200). (E) Decreased IL-22R1 expression in a representative patient with IgA nephropathy (×200).
FIG. 2.
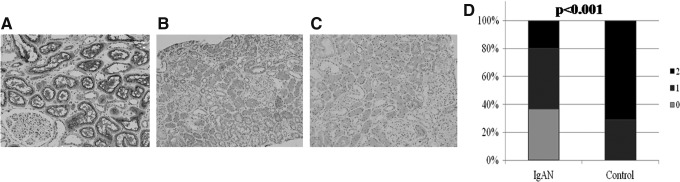
Representative examples of IL-22R1 immunostaining according to the semiquantitative scores (A; score of 2, B; score of 1, C; score of 0) and relative degrees (D) of IL-22R1 expression in patients with IgA nephropathy and normal controls, P<0.001).
Discussion
In this study, we demonstrate that a coding polymorphism in the IL22R1 gene is associated with the development of IgAN in Korean children. Our results showed that individuals with the CC genotype of rs3795299 had a significantly lower risk of developing IgAN, compared to those with other genotypes, suggesting that the CC genotype of rs3795299 in the IL-22R1 gene might be a protective genetic factor against the development of IgAN. In the comparison of IL-22R1 expression, the renal expression of IL-22R1 was also significantly higher in healthy controls compared with subjects with IgAN.
IL-22R1, the main receptor for IL-22 and alternative type 1 receptor chain for IL-20 and IL-24, is a recently identified immunomodulator. It is restrictively expressed in nonimmune peripheral tissues, subsequently determining the target of the IL-20 subfamily and receptor complex (Wolk and Sabat 2006). Although there has been no previous study for evaluating the role of the IL-20 subfamily and its receptor complex in IgAN, our results suggest that IL-22R1 may play a protective role for IgAN. Earlier studies investigating the association of IL-22R1 and CRS showed similar findings (Ramanathan and others 2007; Endam and others 2009). Ramanathan and others demonstrated that IL-22R1mRNA was decreased in patients with recalcitrant CRS with NP compared to patients with CRS without NP and normal controls. Endam and others also showed that IL-22R1 polymorphisms were correlated with severe CRS. As a mechanism for their results, they suggested that reduced expression of IL-22R1 leads to decreased production of proinflammatory Th1 cytokines, such as IL-6 and IL-8, as well as antimicrobial proteins, such as human BD2. This creates a mucosal Th1/Th2 imbalance and a decreased innate immune response in recalcitrant CRS with NP, favoring a Th2 response. We speculate similar mechanisms in the pathogenesis of IgAN regarding the role of IL-22R1. The decreased innate immune response in the pathogenesis of IgAN has been investigated (Lim and others 2001). In patients with IgAN, there is an impaired elimination of mucosal antigens derived from common microbes or foods. As a result, the IgA mucosal immune system is dealing with a continuous antigenic challenge, which triggers the production of nephrogenic IgA (Coppo and others 2010). When outer membrane antigens of Haemophilus parainfluenza were administered to mice, these antigens induced glomerular deposition of IgA and mesangial proliferation with increased IgA antibodies to the antigens (Yamamoto and others 2002). The missense SNP of the IL-22R1 gene (rs3795299, Arg518Gly, C->G) is a coding polymorphism that has potential to alter the expression of the IL-22R1 mRNA and/or protein. We speculate that this coding polymorphism may contribute to produce a dysfunctional IL-22R1, subsequently decreasing the innate immune response and increasing the susceptibility for the disease. However, we do not know how much the rs3795299 polymorphism can contribute to the alterations of protein expression. Interestingly, we did not find that individuals with the G allele had a higher risk of developing IgAN, although the subjects with the CC genotype have a lower risk of developing IgAN. This result suggests that there may be more important polymorphisms or epigenetic mechanisms in reducing the expression of the IL-22R1 mRNA or protein. To the best of our knowledge, there have also been no previous reports regarding the association of the rs3795299 polymorphism and IL-22R1 expression. In our unpublished IHC study, we found a trend of increased production of IL-22R1 in subjects with the CC genotype compared with those with other genotypes, although it was not statistically significant due to the small number of the subjects. Therefore, further studies are warranted to verify how the substitution from C to G in the IL-22R1 gene reduces IL-22R1 expression and modifies its biological functions in the kidney.
The IL-20 subfamily and its receptor complex are known to play either a protective or a pathogenic role in chronic inflammatory disease, depending on the nature of the affected tissue and the local cytokine milieu (Witte and others 2010). They exert broad biological functions, which are essential for epithelial integrity and innate host defense. However, uncontrolled responses from these cytokines can be harmful to tissues, subsequently leading to some autoimmune disease, such as psoriasis, rheumatoid arthritis, and inflammatory bowel disease (Radaeva and others 2004; Brand and others 2006; Sa and others 2007; Kragstrup and others 2008; Wolk and others 2009). Especially, with regard to renal disease, the associations with IL-20 and chronic kidney disease were reported (Li and others 2008; Wei and others 2008). These results are opposite to our results that suggest the protective role of IL-22R1 to kidney disease. And IL-22RA2 (known also as the IL-22 binding protein) participates in the IL-22-mediated processes. It acts as an IL-22 antagonist. It is known to be expressed in monocytes, activated B cells, and some tissues, including the placenta, skin, inflamed appendix, gastrointestinal tract, lymph node, thymus, and spleen (Xu and others 2001). However, there was no detectable level of IL-22RA2 gene expression in the kidney. Therefore, it is not clear whether IL-22RA2 plays a role in renal diseases. Further studies to investigate the role of the IL-20 subfamily and its receptor complex for renal diseases, including IgAN are needed.
In this study, we could not find significant associations between the G/C SNPs and patient subgroups based on clinical and histologic severity of IgAN. These results suggest that there is a significant association between a missense SNP of IL-22R1 and the development of IgAN, but it may not be the case between this SNP and severity of IgAN.
In summary, this is the first report of association between IL-22R1 and childhood IgAN. A coding polymorphism of IL-22R1 was associated with the prevalence of IgAN and the renal expression of IL-22R1 was reduced in patients with IgAN. This reduced expression of IL-22R1 may subsequently modify the IL-22-related immune response, such as diminished production of antimicrobial peptides, contributing to the development of IgAN.
Acknowledgments
We thank Jonathan Cho for English editing and participation in this research project. This study was partly supported by the program at the Kyung Hee University for the young researcher of medical science in 2009 (KHU-20091456).
Author Disclosure Statement
No competing financial interests exist.
References
- Aujla SJ. Chan YR. Zheng M. Fei M. Askew DJ. Pociask DA. Reinhart TA. McAllister F. Edeal J. Gaus K. Husain S. Kreindler JL. Dubin PJ. Pilewski JM. Myerburg MM. Mason CA. Iwakura Y. Kolls JK. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14(3):275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisceglia L. Cerullo G. Forabosco P. Torres DD. Scolari F. Di Perna M. Foramitti M. Amoroso A. Bertok S. Floege J. Mertens PR. Zerres K. Alexopoulos E. Kirmizis D. Ermelinda M. Zelante L. Schena FP. Genetic heterogeneity in Italian families with IgA nephropathy: suggestive linkage for two novel IgA nephropathy loci. Am J Hum Genet. 2006;79(6):1130–1134. doi: 10.1086/510135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand S. Beigel F. Olszak T. Zitzmann K. Eichhorst ST. Otte JM. Diepolder H. Marquardt A. Jagla W. Popp A. Leclair S. Herrmann K. Seiderer J. Ochsenkuhn T. Goke B. Auernhammer CJ. Dambacher J. IL-22 is increased in active Crohn's disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am J Physiol Gastrointest Liver Physiol. 2006;290(4):G827–G838. doi: 10.1152/ajpgi.00513.2005. [DOI] [PubMed] [Google Scholar]
- Choi JW. Lee JH. Park HS. Kim YS. PAI-1 expression and its regulation by promoter 4G/5G polymorphism in clear cell renal cell carcinoma. J Clin Pathol. 2011;64(10):893–897. doi: 10.1136/jclinpath-2011-200182. [DOI] [PubMed] [Google Scholar]
- Coppo R. Amore A. Peruzzi L. Vergano L. Camilla R. Innate immunity and IgA nephropathy. J Nephrol. 2010;23(6):626–632. [PubMed] [Google Scholar]
- Endam LM. Bosse Y. Filali-Mouhim A. Cormier C. Boisvert P. Boulet LP. Hudson TJ. Desrosiers M. Polymorphisms in the interleukin-22 receptor alpha-1 gene are associated with severe chronic rhinosinusitis. Otolaryngol Head Neck Surg. 2009;140(5):741–747. doi: 10.1016/j.otohns.2008.12.058. [DOI] [PubMed] [Google Scholar]
- Gabriel SB. Schaffner SF. Nguyen H. Moore JM. Roy J. Blumenstiel B. Higgins J. DeFelice M. Lochner A. Faggart M. Liu-Cordero SN. Rotimi C. Adeyemo A. Cooper R. Ward R. Lander ES. Daly MJ. Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gharavi AG. Kiryluk K. Choi M. Li Y. Hou P. Xie J. Sanna-Cherchi S. Men CJ. Julian BA. Wyatt RJ. Novak J. He JC. Wang H. Lv J. Zhu L. Wang W. Wang Z. Yasuno K. Gunel M. Mane S. Umlauf S. Tikhonova I. Beerman I. Savoldi S. Magistroni R. Ghiggeri GM. Bodria M. Lugani F. Ravani P. Ponticelli C. Allegri L. Boscutti G. Frasca G. Amore A. Peruzzi L. Coppo R. Izzi C. Viola BF. Prati E. Salvadori M. Mignani R. Gesualdo L. Bertinetto F. Mesiano P. Amoroso A. Scolari F. Chen N. Zhang H. Lifton RP. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet. 2011;43(4):321–327. doi: 10.1038/ng.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharavi AG. Yan Y. Scolari F. Schena FP. Frasca GM. Ghiggeri GM. Cooper K. Amoroso A. Viola BF. Battini G. Caridi G. Canova C. Farhi A. Subramanian V. Nelson-Williams C. Woodford S. Julian BA. Wyatt RJ. Lifton RP. IgA nephropathy, the most common cause of glomerulonephritis, is linked to 6q22-23. Nat Genet. 2000;26(3):354–357. doi: 10.1038/81677. [DOI] [PubMed] [Google Scholar]
- Glassock RJ. The pathogenesis of IgA nephropathy. Curr Opin Nephrol Hypertens. 2011;20(2):153–160. doi: 10.1097/MNH.0b013e3283436f5c. [DOI] [PubMed] [Google Scholar]
- Hofmann SR. Rosen-Wolff A. Tsokos GC. Hedrich CM. Biological properties and regulation of IL-10 related cytokines and their contribution to autoimmune disease and tissue injury. Clin Immunol. 2012;143(2):116–127. doi: 10.1016/j.clim.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Kragstrup TW. Otkjaer K. Holm C. Jorgensen A. Hokland M. Iversen L. Deleuran B. The expression of IL-20 and IL-24 and their shared receptors are increased in rheumatoid arthritis and spondyloarthropathy. Cytokine. 2008;41(1):16–23. doi: 10.1016/j.cyto.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Lai KN. Pathogenesis of IgA nephropathy. Nat Rev Nephrol. 2012;8(5):275–283. doi: 10.1038/nrneph.2012.58. [DOI] [PubMed] [Google Scholar]
- Lee HS. Lee MS. Lee SM. Lee SY. Lee ES. Lee EY. Park SY. Han JS. Kim S. Lee JS. Histological grading of IgA nephropathy predicting renal outcome: revisiting H. S. Lee's glomerular grading system. Nephrol Dial Transplant. 2005;20(2):342–348. doi: 10.1093/ndt/gfh633. [DOI] [PubMed] [Google Scholar]
- Li HH. Cheng HH. Sun KH. Wei CC. Li CF. Chen WC. Wu WM. Chang MS. Interleukin-20 targets renal mesangial cells and is associated with lupus nephritis. Clin Immunol. 2008;129(2):277–285. doi: 10.1016/j.clim.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Lim CS. Zheng S. Kim YS. Ahn C. Han JS. Kim S. Lee JS. Chae DW. Koo JR. Chun RW. Noh JW. Th1/Th2 predominance and proinflammatory cytokines determine the clinicopathological severity of IgA nephropathy. Nephrol Dial Transplant. 2001;16(2):269–275. doi: 10.1093/ndt/16.2.269. [DOI] [PubMed] [Google Scholar]
- Ouyang W. Rutz S. Crellin NK. Valdez PA. Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- Park HJ. Hahn WH. Suh JS. Kim MJ. Kang SW. Lee JS. Kim JW. Chung JH. Cho BS. Association between toll-like receptor 10 (TLR10) gene polymorphisms and childhood IgA nephropathy. Eur J Pediatr. 2011;170(4):503–509. doi: 10.1007/s00431-010-1325-1. [DOI] [PubMed] [Google Scholar]
- Paterson AD. Liu XQ. Wang K. Magistroni R. Song X. Kappel J. Klassen J. Cattran D. St George-Hyslop P. Pei Y. Genome-wide linkage scan of a large family with IgA nephropathy localizes a novel susceptibility locus to chromosome 2q36. J Am Soc Nephrol. 2007;18(8):2408–2415. doi: 10.1681/ASN.2007020241. [DOI] [PubMed] [Google Scholar]
- Radaeva S. Sun R. Pan HN. Hong F. Gao B. Interleukin 22 (IL-22) plays a protective role in T cell-mediated murine hepatitis: IL-22 is a survival factor for hepatocytes via STAT3 activation. Hepatology. 2004;39(5):1332–1342. doi: 10.1002/hep.20184. [DOI] [PubMed] [Google Scholar]
- Ramanathan M., Jr. Spannhake EW. Lane AP. Chronic rhinosinusitis with nasal polyps is associated with decreased expression of mucosal interleukin 22 receptor. Laryngoscope. 2007;117(10):1839–1843. doi: 10.1097/MLG.0b013e31811edd4f. [DOI] [PubMed] [Google Scholar]
- Sa SM. Valdez PA. Wu J. Jung K. Zhong F. Hall L. Kasman I. Winer J. Modrusan Z. Danilenko DM. Ouyang W. The effects of IL-20 subfamily cytokines on reconstituted human epidermis suggest potential roles in cutaneous innate defense and pathogenic adaptive immunity in psoriasis. J Immunol. 2007;178(4):2229–2240. doi: 10.4049/jimmunol.178.4.2229. [DOI] [PubMed] [Google Scholar]
- Sole X. Guino E. Valls J. Iniesta R. Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- Suzuki H. Suzuki Y. Narita I. Aizawa M. Kihara M. Yamanaka T. Kanou T. Tsukaguchi H. Novak J. Horikoshi S. Tomino Y. Toll-like receptor 9 affects severity of IgA nephropathy. J Am Soc Nephrol. 2008;19(12):2384–2395. doi: 10.1681/ASN.2007121311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. Tan Z. Zhang R. Kotenko SV. Liang P. Interleukin 24 (MDA-7/MOB-5) signals through two heterodimeric receptors, IL-22R1/IL-20R2 and IL-20R1/IL-20R2. J Biol Chem. 2002;277(9):7341–7347. doi: 10.1074/jbc.M106043200. [DOI] [PubMed] [Google Scholar]
- Wegenka UM. IL-20: biological functions mediated through two types of receptor complexes. Cytokine Growth Factor Rev. 2010;21(5):353–363. doi: 10.1016/j.cytogfr.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Wei CC. Li HH. Hsu YH. Hsing CH. Sung JM. Chang MS. Interleukin-20 targets renal cells and is associated with chronic kidney disease. Biochem Biophys Res Commun. 2008;374(3):448–453. doi: 10.1016/j.bbrc.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Witte E. Witte K. Warszawska K. Sabat R. Wolk K. Interleukin-22: a cytokine produced by T, NK and NKT cell subsets, with importance in the innate immune defense and tissue protection. Cytokine Growth Factor Rev. 2010;21(5):365–379. doi: 10.1016/j.cytogfr.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Wolk K. Haugen HS. Xu W. Witte E. Waggie K. Anderson M. Vom Baur E. Witte K. Warszawska K. Philipp S. Johnson-Leger C. Volk HD. Sterry W. Sabat R. IL-22 and IL-20 are key mediators of the epidermal alterations in psoriasis while IL-17 and IFN-gamma are not. J Mol Med (Berl) 2009;87(5):523–536. doi: 10.1007/s00109-009-0457-0. [DOI] [PubMed] [Google Scholar]
- Wolk K. Kunz S. Witte E. Friedrich M. Asadullah K. Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21(2):241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Wolk K. Sabat R. Interleukin-22: a novel T- and NK-cell derived cytokine that regulates the biology of tissue cells. Cytokine Growth Factor Rev. 2006;17(5):367–380. doi: 10.1016/j.cytogfr.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Wolk K. Witte E. Wallace E. Docke WD. Kunz S. Asadullah K. Volk HD. Sterry W. Sabat R. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur J Immunol. 2006;36(5):1309–1323. doi: 10.1002/eji.200535503. [DOI] [PubMed] [Google Scholar]
- Xu W. Presnell SR. Parrish-Novak J. Kindsvogel W. Jaspers S. Chen Z. Dillon SR. Gao Z. Gilbert T. Madden K. Schlutsmeyer S. Yao L. Whitmore TE. Chandrasekher Y. Grant FJ. Maurer M. Jelinek L. Storey H. Brender T. Hammond A. Topouzis S. Clegg CH. Foster DC. A soluble class II cytokine receptor, IL-22RA2, is a naturally occurring IL-22 antagonist. Proc Natl Acad Sci U S A. 2001;98(17):9511–9516. doi: 10.1073/pnas.171303198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto C. Suzuki S. Kimura H. Yoshida H. Gejyo F. Experimental nephropathy induced by Haemophilus parainfluenzae antigens. Nephron. 2002;90(3):320–327. doi: 10.1159/000049068. [DOI] [PubMed] [Google Scholar]
- Yu XQ. Li M. Zhang H. Low HQ. Wei X. Wang JQ. Sun LD. Sim KS. Li Y. Foo JN. Wang W. Li ZJ. Yin XY. Tang XQ. Fan L. Chen J. Li RS. Wan JX. Liu ZS. Lou TQ. Zhu L. Huang XJ. Zhang XJ. Liu ZH. Liu JJ. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet. 2012;44(2):178–182. doi: 10.1038/ng.1047. [DOI] [PubMed] [Google Scholar]