Abstract
In the title compound, C14H17NO3, the nine-membered 1H-indole ring system is essentially planar [maximum deviation = 0.019 (1) Å]. In the crystal, molecules are linked via N—H⋯O hydrogen bonds, forming chains along [001]. These chains are linked via C—H⋯O hydrogen bonds and C—H⋯π interactions, forming a two-dimensional network lying parallel to the ac plane.
Related literature
For medicinal applications of the drug indomethacin (systematic name: 2-{1-[(4-chlorophenyl)carbonyl]-5-methoxy-2-methyl-1H-indol-3-yl}acetic acid), see: Paneth (1995 ▶); McIntyre et al. (2001 ▶); Abou-Ghannam et al. (2012 ▶). For the synthesis and reactions of indomethacin with other non-steroidal anti-inflammatory molecules, see: Mohamed et al. (2012 ▶).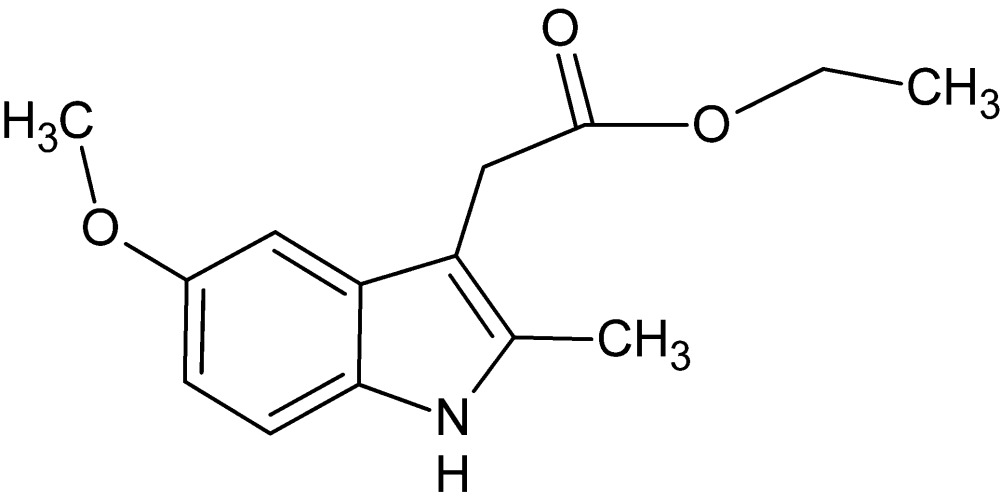
Experimental
Crystal data
C14H17NO3
M r = 247.29
Monoclinic,
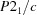
a = 7.8117 (5) Å
b = 17.1953 (12) Å
c = 9.9003 (7) Å
β = 106.756 (1)°
V = 1273.39 (15) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 150 K
0.23 × 0.21 × 0.06 mm
Data collection
Bruker SMART APEX CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2013 ▶) T min = 0.84, T max = 1.00
22947 measured reflections
3374 independent reflections
2889 reflections with I > 2σ(I)
R int = 0.043
Refinement
R[F 2 > 2σ(F 2)] = 0.044
wR(F 2) = 0.117
S = 1.08
3374 reflections
170 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.31 e Å−3
Δρmin = −0.24 e Å−3
Data collection: APEX2 (Bruker, 2013 ▶); cell refinement: SAINT (Bruker, 2013 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶) and DIAMOND (Brandenburg & Putz, 2012 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813018618/su2618sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813018618/su2618Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813018618/su2618Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 is the centroid of the C1–C6 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O2i | 0.914 (18) | 2.013 (18) | 2.8987 (14) | 162.7 (16) |
| C7—H7B⋯O2ii | 0.98 | 2.57 | 3.4271 (18) | 146 |
| C13—H13A⋯O1iii | 0.99 | 2.55 | 3.4236 (16) | 148 |
| C7—H7A⋯Cg1iv | 0.98 | 2.99 | 3.9550 (16) | 169 |
Symmetry codes: (i) 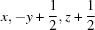 ; (ii)
; (ii) 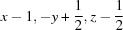 ; (iii)
; (iii) 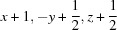 ; (iv)
; (iv) 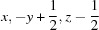 .
.
Acknowledgments
Manchester Metropolitan University, Tulane University and Erciyes University are gratefully acknowledged for supporting this study.
supplementary crystallographic information
Comment
Indomethacin, chemically named 2-{1-[(4-chlorophenyl)carbonyl]-5-methoxy-2-methyl-1H-indol-3-yl}acetic acid is a non-steroidal drug (NSAID) and is commonly used as an anti-inflammatory drug by inhibiting cyclooxygenase (COX) 1 and 2 enzymes. It also clinically used as a tocolytic agent to delay premature labor (preterm birth; PTB), reduce amniotic fluid in polyhydramnios, and to close patent ductus arteriosus (PDA). PTB is a major cause of neonatal morbidity and mortality worldwide (Paneth, 1995; McIntyre et al., 2001; Abou-Ghannam et al., 2012). In view of these facts and as part of our ongoing study incorporating NSAID's as a substructure in the synthesis of potential bio-active pharmacophors (Mohamed et al., 2012), indomethacin has been hydrolysed during its esterification in acidic medium with ethanol to afforded the title corresponding ethyl ester.
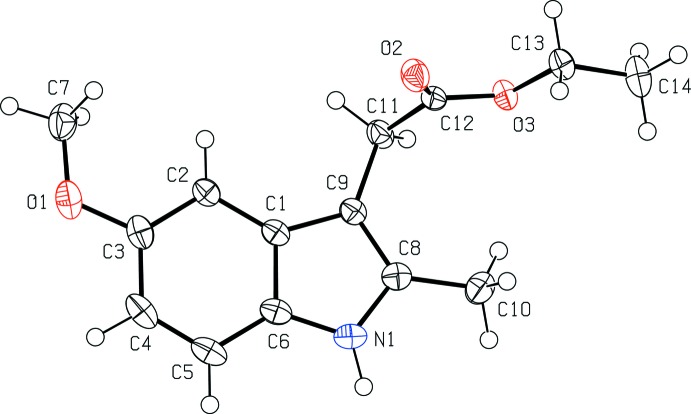
In the title compound, Fig. 1, the nine-membered 1H-indole ring system (N1/C1–C6/C8/C9) is essentially planar with a maximum deviation of 0.019 (1) Å for N1. The C2–C3–O1–C7, C1–C9–C8–C10, C1–C9–C11–C12, C9–C11–C12–O2, C11–C12–O3–C13 and C12–O3–C13–C14 torsion angles are -6.53 (18), -177.77 (12), 117.42 (13), -69.51 (15), 177.50 (9) and 171.98 (10)°, respectively.
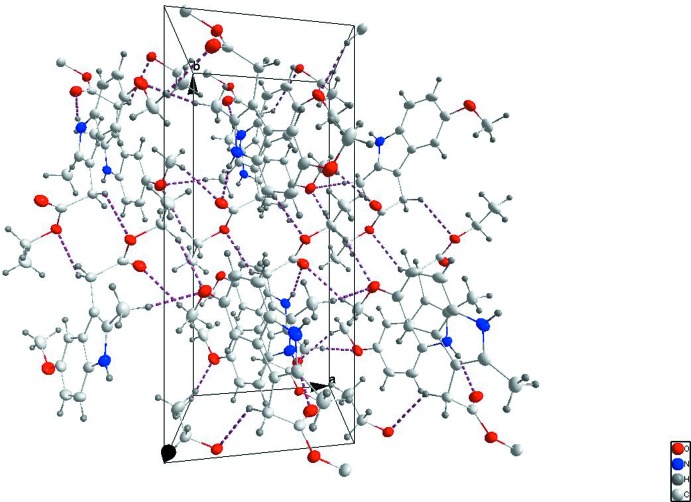
In the crystal, molecules are linked via N-H···O hydrogen bonds forming chains along [001]. These chains are linked via C-H···O hydrogen bonds and C-H···π interactions forming a two-dimensional network lying parallel to the ac plane (Fig. 2 and Table 1).
Experimental
A mixture of 0.03 mol indomethacin (10.57 g m) in 150 ml of absolute ethanol and 6 ml of concentrated H2SO4 was refluxed for 6 h. The mixture was cooled to room temperature and neutralized with NaHCO3 solution. The ester was separated as an organic layer, washed with water and extracted with diethyl ether (3 × 50mL). The combined ether layers were dried over MgSO4, filtered and left for 3–4 days until brown crystals formed. The solid was collected and recrystallized from cyclohexane to give the pure ester as silver-coloured crystals (m.p. 347–350 K) suitable for X-ray diffraction. Spectroscopic data for the title compound are available in the archived CIF.
Refinement
The C-bound H atoms were placed geometrically [C—H = 0.95 Å (aromatic H), C—H = 0.98 Å (methyl H) and 0.99 Å (methylene H)], and refined using a riding model with Uiso(H) = 1.5Ueq(C-methyl) and = 1.2Ueq(C) for other H atoms. The N-bound H atom was located in a difference Fourier synthesis and freely refined [N1—H1 = 0.914 (18) Å].
Figures
Fig. 1.
The molecular structure of the title molecule, with the atom numbering scheme. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
A perspective view along the c axis of the crystal packing of the title compound. The hydrogen bonds are shown as dashed lines - see Table 1 for details.
Crystal data
| C14H17NO3 | F(000) = 528 |
| Mr = 247.29 | Dx = 1.290 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 9989 reflections |
| a = 7.8117 (5) Å | θ = 2.4–29.1° |
| b = 17.1953 (12) Å | µ = 0.09 mm−1 |
| c = 9.9003 (7) Å | T = 150 K |
| β = 106.756 (1)° | Plate, colourless |
| V = 1273.39 (15) Å3 | 0.23 × 0.21 × 0.06 mm |
| Z = 4 |
Data collection
| Bruker SMART APEX CCD diffractometer | 3374 independent reflections |
| Radiation source: fine-focus sealed tube | 2889 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.043 |
| Detector resolution: 8.3660 pixels mm-1 | θmax = 29.1°, θmin = 2.4° |
| φ and ω scans | h = −10→10 |
| Absorption correction: multi-scan (SADABS; Bruker, 2013) | k = −23→23 |
| Tmin = 0.84, Tmax = 1.00 | l = −13→13 |
| 22947 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: difference Fourier map |
| Least-squares matrix: full | Secondary atom site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.044 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.117 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.08 | W = 1/[Σ2(Fo2) + (0.0539P)2 + 0.3155P] where P = (Fo2 + 2Fc2)/3 |
| 3374 reflections | (Δ/σ)max < 0.001 |
| 170 parameters | Δρmax = 0.31 e Å−3 |
| 0 restraints | Δρmin = −0.24 e Å−3 |
Special details
| Experimental. Spectroscopic data for the title compound: IR (KBr cm-1): (C=O ester 1728), (NH 3317), (C—H aliphatic, 2833–2924), (C—H, Ar, 2975–3002). 1H-NMR: (DMSO-D6) δ at 1.3(t, 3H, CH3 of ethyl group), 4.0(q, 2H, –CH2 aliphatic in ethyl group), 2.3(s, 3H, CH3), 3.4(s, –CH2), 3.7(s, 3H, –OCH3), 10.8(s, 1H, –NH), 6.8(s, 1H, Ar), 6.6(d, 1H, Ar), 7.2(d, 1H, Ar). 13C-NMR: 171 (C=O ester), 11(CH3 in indole), 14(CH3 of ethyl group), 29(–CH2), 55 (–OCH3), 59(–CH2 of ethyl group). 99, 103, 109, 110, 128, 129, 133,152 (8 C, aromatics). There are two signals at 29 and 59 p.p.m. oriented downward in the DEPT spectrum confirming the existence of two –CH2 groups. |
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement on F2 for ALL reflections except those flagged by the user for potential systematic errors. Weighted R-factors wR and all goodnesses of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The observed criterion of F2 > σ(F2) is used only for calculating -R-factor-obs etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.17116 (13) | 0.36046 (6) | −0.24184 (10) | 0.0376 (3) | |
| O2 | 0.75724 (12) | 0.08270 (5) | −0.02263 (9) | 0.0312 (3) | |
| O3 | 0.75501 (11) | 0.01325 (4) | 0.16773 (9) | 0.0251 (2) | |
| N1 | 0.65214 (13) | 0.29386 (6) | 0.27056 (11) | 0.0264 (3) | |
| C1 | 0.47230 (14) | 0.25588 (6) | 0.05968 (12) | 0.0216 (3) | |
| C2 | 0.34868 (15) | 0.26677 (6) | −0.07390 (13) | 0.0240 (3) | |
| C3 | 0.29070 (16) | 0.34165 (7) | −0.11378 (13) | 0.0282 (3) | |
| C4 | 0.35262 (18) | 0.40558 (7) | −0.02372 (15) | 0.0336 (4) | |
| C5 | 0.47322 (17) | 0.39589 (7) | 0.10729 (15) | 0.0309 (3) | |
| C6 | 0.53295 (15) | 0.32062 (6) | 0.14800 (13) | 0.0245 (3) | |
| C7 | 0.09045 (18) | 0.29785 (9) | −0.33106 (15) | 0.0367 (4) | |
| C8 | 0.66375 (15) | 0.21415 (7) | 0.26458 (12) | 0.0243 (3) | |
| C9 | 0.55670 (14) | 0.18869 (6) | 0.13595 (12) | 0.0220 (3) | |
| C10 | 0.77677 (16) | 0.16917 (8) | 0.38672 (13) | 0.0312 (4) | |
| C11 | 0.52395 (15) | 0.10556 (6) | 0.08854 (13) | 0.0258 (3) | |
| C12 | 0.68958 (14) | 0.06698 (6) | 0.06967 (12) | 0.0224 (3) | |
| C13 | 0.91945 (15) | −0.02529 (7) | 0.16271 (13) | 0.0272 (3) | |
| C14 | 0.98064 (19) | −0.07392 (8) | 0.29348 (16) | 0.0395 (4) | |
| H1 | 0.700 (2) | 0.3250 (10) | 0.347 (2) | 0.052 (5)* | |
| H2 | 0.30620 | 0.22390 | −0.13500 | 0.0290* | |
| H4 | 0.31030 | 0.45630 | −0.05410 | 0.0400* | |
| H5 | 0.51440 | 0.43900 | 0.16800 | 0.0370* | |
| H7A | 0.18300 | 0.26750 | −0.35610 | 0.0550* | |
| H7B | 0.00600 | 0.31840 | −0.41700 | 0.0550* | |
| H7C | 0.02660 | 0.26440 | −0.28160 | 0.0550* | |
| H10A | 0.72640 | 0.11700 | 0.38710 | 0.0470* | |
| H10B | 0.77960 | 0.19600 | 0.47470 | 0.0470* | |
| H10C | 0.89850 | 0.16500 | 0.37860 | 0.0470* | |
| H11A | 0.42780 | 0.10380 | −0.00200 | 0.0310* | |
| H11B | 0.48250 | 0.07610 | 0.15910 | 0.0310* | |
| H13A | 1.01170 | 0.01370 | 0.16000 | 0.0330* | |
| H13B | 0.89690 | −0.05840 | 0.07760 | 0.0330* | |
| H14A | 1.00740 | −0.04020 | 0.37670 | 0.0590* | |
| H14B | 1.08850 | −0.10270 | 0.29240 | 0.0590* | |
| H14C | 0.88610 | −0.11070 | 0.29690 | 0.0590* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0347 (5) | 0.0333 (5) | 0.0417 (5) | 0.0089 (4) | 0.0059 (4) | 0.0110 (4) |
| O2 | 0.0383 (5) | 0.0288 (4) | 0.0283 (4) | 0.0066 (4) | 0.0124 (4) | 0.0065 (3) |
| O3 | 0.0260 (4) | 0.0219 (4) | 0.0277 (4) | 0.0049 (3) | 0.0084 (3) | 0.0047 (3) |
| N1 | 0.0275 (5) | 0.0268 (5) | 0.0265 (5) | −0.0049 (4) | 0.0104 (4) | −0.0073 (4) |
| C1 | 0.0206 (5) | 0.0194 (5) | 0.0273 (5) | −0.0005 (4) | 0.0111 (4) | −0.0010 (4) |
| C2 | 0.0233 (5) | 0.0217 (5) | 0.0283 (6) | 0.0017 (4) | 0.0093 (4) | −0.0001 (4) |
| C3 | 0.0258 (5) | 0.0269 (6) | 0.0340 (6) | 0.0052 (4) | 0.0122 (5) | 0.0067 (5) |
| C4 | 0.0348 (6) | 0.0200 (5) | 0.0503 (8) | 0.0051 (5) | 0.0191 (6) | 0.0048 (5) |
| C5 | 0.0352 (6) | 0.0196 (5) | 0.0432 (7) | −0.0024 (5) | 0.0198 (6) | −0.0048 (5) |
| C6 | 0.0251 (5) | 0.0222 (5) | 0.0299 (6) | −0.0027 (4) | 0.0140 (4) | −0.0039 (4) |
| C7 | 0.0297 (6) | 0.0455 (8) | 0.0335 (7) | 0.0111 (5) | 0.0071 (5) | 0.0055 (6) |
| C8 | 0.0227 (5) | 0.0268 (5) | 0.0255 (5) | −0.0017 (4) | 0.0104 (4) | −0.0018 (4) |
| C9 | 0.0199 (5) | 0.0205 (5) | 0.0265 (5) | −0.0002 (4) | 0.0080 (4) | −0.0006 (4) |
| C10 | 0.0282 (6) | 0.0394 (7) | 0.0249 (6) | 0.0023 (5) | 0.0058 (5) | 0.0005 (5) |
| C11 | 0.0216 (5) | 0.0191 (5) | 0.0345 (6) | −0.0002 (4) | 0.0047 (4) | −0.0003 (4) |
| C12 | 0.0239 (5) | 0.0164 (5) | 0.0243 (5) | −0.0006 (4) | 0.0027 (4) | −0.0016 (4) |
| C13 | 0.0244 (5) | 0.0247 (5) | 0.0323 (6) | 0.0060 (4) | 0.0080 (5) | 0.0028 (5) |
| C14 | 0.0364 (7) | 0.0369 (7) | 0.0444 (8) | 0.0134 (6) | 0.0106 (6) | 0.0145 (6) |
Geometric parameters (Å, º)
| O1—C3 | 1.3789 (16) | C11—C12 | 1.5128 (16) |
| O1—C7 | 1.4198 (18) | C13—C14 | 1.4990 (19) |
| O2—C12 | 1.2108 (15) | C2—H2 | 0.9500 |
| O3—C12 | 1.3309 (13) | C4—H4 | 0.9500 |
| O3—C13 | 1.4590 (15) | C5—H5 | 0.9500 |
| N1—C6 | 1.3782 (16) | C7—H7A | 0.9800 |
| N1—C8 | 1.3760 (16) | C7—H7B | 0.9800 |
| N1—H1 | 0.914 (18) | C7—H7C | 0.9800 |
| C1—C2 | 1.4072 (17) | C10—H10A | 0.9800 |
| C1—C6 | 1.4105 (15) | C10—H10B | 0.9800 |
| C1—C9 | 1.4327 (15) | C10—H10C | 0.9800 |
| C2—C3 | 1.3841 (16) | C11—H11A | 0.9900 |
| C3—C4 | 1.4102 (18) | C11—H11B | 0.9900 |
| C4—C5 | 1.376 (2) | C13—H13A | 0.9900 |
| C5—C6 | 1.3953 (16) | C13—H13B | 0.9900 |
| C8—C9 | 1.3779 (16) | C14—H14A | 0.9800 |
| C8—C10 | 1.4914 (17) | C14—H14B | 0.9800 |
| C9—C11 | 1.5033 (15) | C14—H14C | 0.9800 |
| C3—O1—C7 | 117.11 (11) | C5—C4—H4 | 119.00 |
| C12—O3—C13 | 116.57 (9) | C4—C5—H5 | 121.00 |
| C6—N1—C8 | 109.32 (10) | C6—C5—H5 | 121.00 |
| C6—N1—H1 | 123.0 (11) | O1—C7—H7A | 109.00 |
| C8—N1—H1 | 127.1 (11) | O1—C7—H7B | 109.00 |
| C2—C1—C6 | 119.63 (10) | O1—C7—H7C | 109.00 |
| C6—C1—C9 | 106.77 (10) | H7A—C7—H7B | 109.00 |
| C2—C1—C9 | 133.59 (10) | H7A—C7—H7C | 109.00 |
| C1—C2—C3 | 118.12 (10) | H7B—C7—H7C | 109.00 |
| O1—C3—C2 | 124.03 (11) | C8—C10—H10A | 109.00 |
| O1—C3—C4 | 114.61 (11) | C8—C10—H10B | 109.00 |
| C2—C3—C4 | 121.36 (12) | C8—C10—H10C | 109.00 |
| C3—C4—C5 | 121.23 (11) | H10A—C10—H10B | 109.00 |
| C4—C5—C6 | 117.76 (12) | H10A—C10—H10C | 109.00 |
| C1—C6—C5 | 121.90 (11) | H10B—C10—H10C | 109.00 |
| N1—C6—C5 | 130.45 (11) | C9—C11—H11A | 109.00 |
| N1—C6—C1 | 107.65 (9) | C9—C11—H11B | 109.00 |
| N1—C8—C9 | 109.04 (10) | C12—C11—H11A | 109.00 |
| N1—C8—C10 | 120.84 (11) | C12—C11—H11B | 109.00 |
| C9—C8—C10 | 130.11 (11) | H11A—C11—H11B | 108.00 |
| C1—C9—C8 | 107.18 (10) | O3—C13—H13A | 110.00 |
| C8—C9—C11 | 126.44 (10) | O3—C13—H13B | 110.00 |
| C1—C9—C11 | 126.24 (10) | C14—C13—H13A | 110.00 |
| C9—C11—C12 | 112.39 (10) | C14—C13—H13B | 110.00 |
| O2—C12—O3 | 123.10 (11) | H13A—C13—H13B | 109.00 |
| O2—C12—C11 | 124.77 (10) | C13—C14—H14A | 110.00 |
| O3—C12—C11 | 112.13 (10) | C13—C14—H14B | 109.00 |
| O3—C13—C14 | 106.78 (10) | C13—C14—H14C | 110.00 |
| C1—C2—H2 | 121.00 | H14A—C14—H14B | 109.00 |
| C3—C2—H2 | 121.00 | H14A—C14—H14C | 109.00 |
| C3—C4—H4 | 119.00 | H14B—C14—H14C | 109.00 |
| C7—O1—C3—C2 | 6.53 (18) | C6—C1—C9—C8 | −0.25 (13) |
| C7—O1—C3—C4 | −173.81 (12) | C6—C1—C9—C11 | 175.68 (11) |
| C13—O3—C12—O2 | 2.08 (16) | C1—C2—C3—O1 | 179.49 (11) |
| C13—O3—C12—C11 | −177.50 (9) | C1—C2—C3—C4 | −0.15 (19) |
| C12—O3—C13—C14 | 171.98 (10) | O1—C3—C4—C5 | −179.64 (13) |
| C8—N1—C6—C1 | −2.16 (14) | C2—C3—C4—C5 | 0.0 (2) |
| C8—N1—C6—C5 | 177.77 (13) | C3—C4—C5—C6 | 0.3 (2) |
| C6—N1—C8—C9 | 2.03 (14) | C4—C5—C6—N1 | 179.64 (13) |
| C6—N1—C8—C10 | −176.93 (11) | C4—C5—C6—C1 | −0.4 (2) |
| C6—C1—C2—C3 | −0.02 (17) | N1—C8—C9—C1 | −1.07 (13) |
| C9—C1—C2—C3 | 178.39 (13) | N1—C8—C9—C11 | −176.98 (11) |
| C2—C1—C6—N1 | −179.74 (11) | C10—C8—C9—C1 | 177.77 (12) |
| C2—C1—C6—C5 | 0.33 (18) | C10—C8—C9—C11 | 1.9 (2) |
| C9—C1—C6—N1 | 1.47 (13) | C1—C9—C11—C12 | 117.42 (13) |
| C9—C1—C6—C5 | −178.47 (12) | C8—C9—C11—C12 | −67.42 (15) |
| C2—C1—C9—C8 | −178.81 (13) | C9—C11—C12—O2 | −69.51 (15) |
| C2—C1—C9—C11 | −2.9 (2) | C9—C11—C12—O3 | 110.07 (11) |
Hydrogen-bond geometry (Å, º)
Cg1 is the centroid of the C1–C6 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O2i | 0.914 (18) | 2.013 (18) | 2.8987 (14) | 162.7 (16) |
| C7—H7B···O2ii | 0.98 | 2.57 | 3.4271 (18) | 146 |
| C13—H13A···O1iii | 0.99 | 2.55 | 3.4236 (16) | 148 |
| C7—H7A···Cg1iv | 0.98 | 2.99 | 3.9550 (16) | 169 |
Symmetry codes: (i) x, −y+1/2, z+1/2; (ii) x−1, −y+1/2, z−1/2; (iii) x+1, −y+1/2, z+1/2; (iv) x, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2618).
References
- Abou-Ghannam, G. M. D., Ihab, M., Usta, M. D., Anwar, H. & Nassar, M. D. (2012). Am. J. Perinatol. 29, 175–186. [DOI] [PubMed]
- Brandenburg, K. & Putz, H. (2012). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2013). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- McIntyre, J., van Overmeire, B. & van den Anker, J. N. (2001). Paediatr. Perinat. Drug Ther. 4, 85–91.
- Mohamed, S. K., Albayati, M. R., Omara, W. A. M., Abdelhamid, A. A., Potgeiter, H., Hameed, A. S. & Al-Janabi, K. M. (2012). J. Chem. Pharm. Res. 4, 3505–3517.
- Paneth, N. S. (1995). Future Child, 5, 19–34. [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813018618/su2618sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813018618/su2618Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813018618/su2618Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report