Abstract
The asymmetric unit of the title compound, C10H6N2S2, contains two crystallographically independent but conformationally similar molecules. The fused thiophene ring cores are almost planar [maximum deviation = 0.027 (3) Å] with the thiophene rings forming dihedral angles of 0.5 (4)° in one molecule and 1.91 (4)° in the other. The crystal packing is stabilized only by van der Waals interactions.
Related literature
For the biological activity of thiophene derivatives, see: Mabkhot et al. (2013 ▶); Mishra et al. (2011 ▶). For the synthesis of fused heterocyclic compounds, see: Cornel & Kirsch (2001 ▶); Mashraqui et al. (1999 ▶). For crystal data for related thiophene compounds, see: Gunasekaran et al. (2009 ▶); Mashraqui et al. (2004 ▶).
Experimental
Crystal data
C10H6N2S2
M r = 218.31
Triclinic,
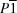
a = 7.2573 (11) Å
b = 10.1538 (15) Å
c = 13.665 (2) Å
α = 94.467 (3)°
β = 99.120 (4)°
γ = 95.850 (4)°
V = 984.5 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.50 mm−1
T = 273 K
0.37 × 0.15 × 0.11 mm
Data collection
Bruker SMART APEX CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2000 ▶) T min = 0.838, T max = 0.947
13821 measured reflections
4912 independent reflections
3074 reflections with I > 2σ(I)
R int = 0.053
Refinement
R[F 2 > 2σ(F 2)] = 0.055
wR(F 2) = 0.132
S = 0.99
4912 reflections
257 parameters
H-atom parameters constrained
Δρmax = 0.37 e Å−3
Δρmin = −0.24 e Å−3
Data collection: SMART (Bruker, 2000 ▶); cell refinement: SAINT (Bruker, 2000 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL, PARST (Nardelli, 1995 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813017960/rz5077sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813017960/rz5077Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813017960/rz5077Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at the King Saud University (Riyadh) for funding this study through the research grant No. RGP-VPP-007.
supplementary crystallographic information
Comment
Thiophene moieties containing heterocyclic compounds are known to have a number of biological activities, including anti-inflammatory, anti-oxidant and anti-glycation properties etc. (Mabkhot et al., 2013; Mishra et al. 2011). The title compound is a thiophene derivative, composed of two fused thiophene rings. It was synthesized as part of our ongoing research towards the synthesis of novel chemical entities with diverse biological activities.
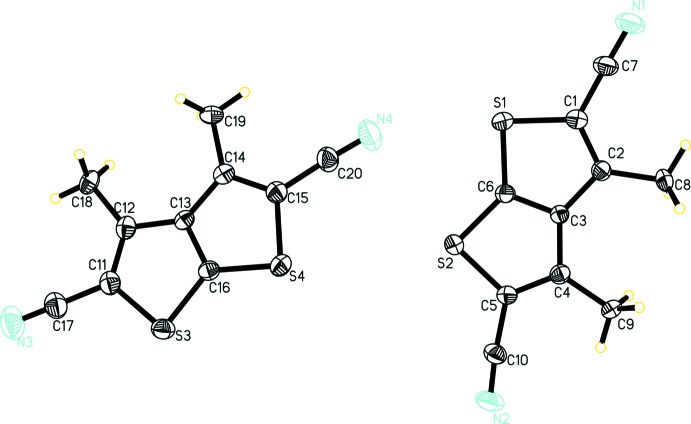
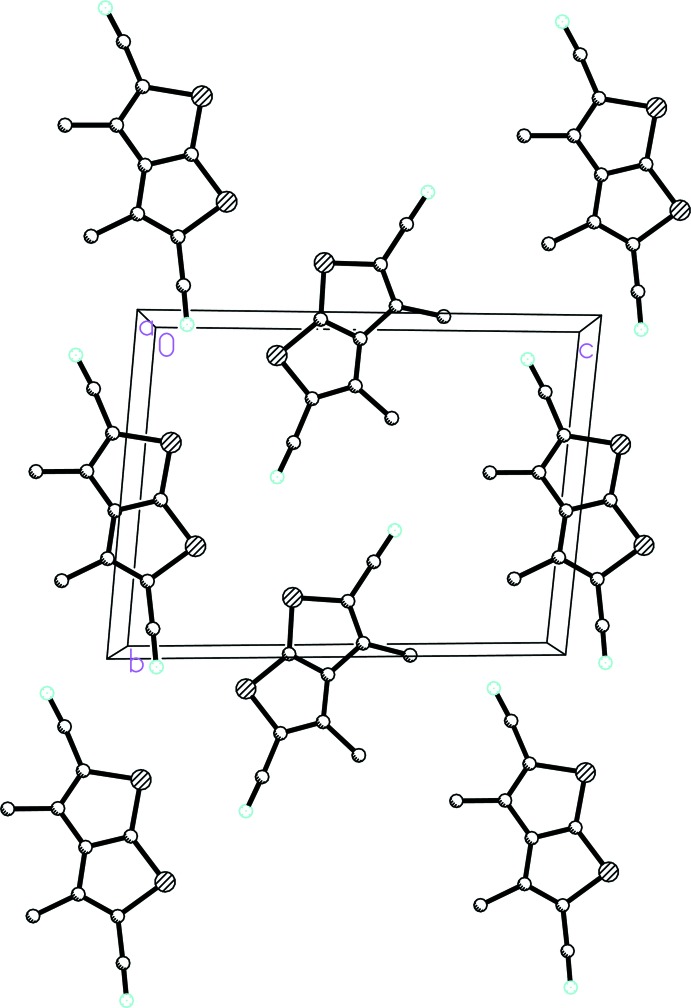
The title, compound C10H6N2S2, contains two independent (S1–S2/C1–C6 and S3–S4/C11–C16) in the asymmetric unit (Fig. 1). Structurally it is similar to the previous reported compound diethyl 3,4-bis(acetoxymethyl)thieno-[2,3-b]thiophene-2,5-dicarboxylate (Gunasekaran et al., 2009) with the difference that four acetoxy methyl substituents has been replaced by two nitrile and two methyl substituents. The fused thiophene ring cores are almost planar [maximum deviation 0.027 (3) Å for atom C16] as indicated by the dihedral angles formed by the thiophene rings of 0.5 (4)° in one molecule and 1.91 (4)° in the other. The crystal packing (Fig. 2) is stabilized only by van der Waals interactions.
Experimental
The title compound was synthesized by following the procedure described in the literature (Mashraqui et al., 1999; Cornel et al., 2001). The compound was crystallized by using a mixture of dimethyl formamide (DMF) and dichloromethane (CH2Cl2) (1:1 v/v) at room temperature to obtain light brown crystals. M. p. 498 K. Anal. calcd. for C10H6N2S2: C, 55.04; H, 2.75; N, 12.84. Found: C, 54.82; H, 2.92; N, 12.97.
Spectral Data: IR (KBr, cm-1): 2964, 2213 cm-1; 1H-NMR (400 MHz, CDCl3): δ 2.69 (s, 6H, CH3); 13C-NMR (100 MHz, CDCl3): δ 14.8 (2 CH3), 108.5 (2 CAr), 113.3 (2 CN), 134.1 (CAr), 143.1 (2 CAr), 150.8 p.p.m. (CAr).
Refinement
H Atoms were positioned geometrically and refined as riding, with C—H = 0.96 Å and with Uiso(H) = 1.5Ueq(C).
Figures
Fig. 1.
The molecular structure of the title compound with displacement ellipsoids drawn at the 30% probability level.
Fig. 2.
Partial packing diagram of the title compound. Hydrogen atoms are omitted for clarity.
Crystal data
| C10H6N2S2 | Z = 4 |
| Mr = 218.31 | F(000) = 448 |
| Triclinic, P1 | Dx = 1.473 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.2573 (11) Å | Cell parameters from 1631 reflections |
| b = 10.1538 (15) Å | θ = 1.5–28.4° |
| c = 13.665 (2) Å | µ = 0.50 mm−1 |
| α = 94.467 (3)° | T = 273 K |
| β = 99.120 (4)° | Block, brown |
| γ = 95.850 (4)° | 0.37 × 0.15 × 0.11 mm |
| V = 984.5 (3) Å3 |
Data collection
| Bruker SMART APEX CCD area-detector diffractometer | 4912 independent reflections |
| Radiation source: fine-focus sealed tube | 3074 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.053 |
| ω scan | θmax = 28.4°, θmin = 1.5° |
| Absorption correction: multi-scan (SADABS; Bruker, 2000) | h = −9→9 |
| Tmin = 0.838, Tmax = 0.947 | k = −13→13 |
| 13821 measured reflections | l = −18→18 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.055 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.132 | H-atom parameters constrained |
| S = 0.99 | w = 1/[σ2(Fo2) + (0.0565P)2] where P = (Fo2 + 2Fc2)/3 |
| 4912 reflections | (Δ/σ)max = 0.001 |
| 257 parameters | Δρmax = 0.37 e Å−3 |
| 0 restraints | Δρmin = −0.24 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.18703 (11) | 0.32325 (7) | 0.83846 (5) | 0.0460 (2) | |
| S2 | 0.24419 (10) | 0.62763 (7) | 0.91346 (5) | 0.0435 (2) | |
| S3 | 0.38935 (11) | 1.16617 (7) | 0.61752 (6) | 0.0477 (2) | |
| S4 | 0.37946 (11) | 0.89010 (8) | 0.70279 (5) | 0.0473 (2) | |
| N1 | 0.1667 (4) | −0.0317 (3) | 0.8989 (2) | 0.0718 (9) | |
| N2 | 0.3550 (4) | 0.8848 (3) | 1.1405 (2) | 0.0706 (9) | |
| N3 | 0.2705 (5) | 1.3690 (3) | 0.4034 (3) | 0.0902 (11) | |
| N4 | 0.2415 (4) | 0.5277 (3) | 0.6753 (2) | 0.0827 (10) | |
| C1 | 0.1990 (4) | 0.2211 (3) | 0.9361 (2) | 0.0424 (7) | |
| C2 | 0.2290 (4) | 0.2874 (3) | 1.0280 (2) | 0.0381 (6) | |
| C3 | 0.2441 (3) | 0.4276 (2) | 1.02172 (19) | 0.0336 (6) | |
| C4 | 0.2755 (4) | 0.5432 (3) | 1.0910 (2) | 0.0384 (6) | |
| C5 | 0.2804 (4) | 0.6552 (3) | 1.0431 (2) | 0.0396 (7) | |
| C6 | 0.2251 (4) | 0.4597 (3) | 0.9243 (2) | 0.0371 (6) | |
| C7 | 0.1799 (4) | 0.0807 (3) | 0.9144 (2) | 0.0520 (8) | |
| C8 | 0.2489 (4) | 0.2224 (3) | 1.1236 (2) | 0.0471 (7) | |
| H8A | 0.2582 | 0.1294 | 1.1097 | 0.071* | |
| H8B | 0.1410 | 0.2332 | 1.1547 | 0.071* | |
| H8C | 0.3601 | 0.2632 | 1.1674 | 0.071* | |
| C9 | 0.3013 (4) | 0.5424 (3) | 1.20166 (19) | 0.0447 (7) | |
| H9A | 0.3111 | 0.6320 | 1.2319 | 0.067* | |
| H9B | 0.4138 | 0.5039 | 1.2246 | 0.067* | |
| H9C | 0.1954 | 0.4909 | 1.2196 | 0.067* | |
| C10 | 0.3195 (4) | 0.7857 (3) | 1.0941 (2) | 0.0474 (7) | |
| C11 | 0.2885 (4) | 1.1564 (3) | 0.4924 (2) | 0.0457 (7) | |
| C12 | 0.2220 (4) | 1.0317 (3) | 0.4497 (2) | 0.0410 (7) | |
| C13 | 0.2546 (3) | 0.9366 (3) | 0.52151 (18) | 0.0341 (6) | |
| C14 | 0.2139 (4) | 0.7963 (3) | 0.5223 (2) | 0.0389 (6) | |
| C15 | 0.2732 (4) | 0.7597 (3) | 0.6151 (2) | 0.0441 (7) | |
| C16 | 0.3434 (4) | 0.9965 (3) | 0.6131 (2) | 0.0378 (6) | |
| C17 | 0.2795 (5) | 1.2748 (3) | 0.4434 (3) | 0.0590 (9) | |
| C18 | 0.1286 (4) | 0.9979 (3) | 0.34337 (19) | 0.0452 (7) | |
| H18A | 0.1082 | 1.0783 | 0.3128 | 0.068* | |
| H18B | 0.0102 | 0.9451 | 0.3412 | 0.068* | |
| H18C | 0.2077 | 0.9486 | 0.3081 | 0.068* | |
| C19 | 0.1235 (4) | 0.6996 (3) | 0.4357 (2) | 0.0453 (7) | |
| H19A | 0.1213 | 0.6108 | 0.4554 | 0.068* | |
| H19B | 0.1938 | 0.7076 | 0.3822 | 0.068* | |
| H19C | −0.0028 | 0.7184 | 0.4140 | 0.068* | |
| C20 | 0.2548 (4) | 0.6297 (3) | 0.6466 (2) | 0.0546 (8) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0602 (5) | 0.0394 (4) | 0.0360 (4) | 0.0018 (3) | 0.0067 (3) | −0.0035 (3) |
| S2 | 0.0552 (5) | 0.0359 (4) | 0.0397 (4) | 0.0042 (3) | 0.0079 (3) | 0.0063 (3) |
| S3 | 0.0538 (5) | 0.0395 (4) | 0.0488 (5) | 0.0045 (3) | 0.0102 (4) | −0.0032 (3) |
| S4 | 0.0534 (5) | 0.0508 (5) | 0.0343 (4) | 0.0019 (4) | 0.0001 (3) | 0.0040 (3) |
| N1 | 0.100 (2) | 0.0396 (16) | 0.075 (2) | −0.0008 (16) | 0.0267 (18) | −0.0085 (15) |
| N2 | 0.096 (2) | 0.0385 (16) | 0.074 (2) | 0.0009 (15) | 0.0168 (18) | −0.0122 (15) |
| N3 | 0.126 (3) | 0.067 (2) | 0.096 (3) | 0.034 (2) | 0.044 (2) | 0.036 (2) |
| N4 | 0.094 (2) | 0.059 (2) | 0.083 (2) | −0.0117 (17) | −0.0187 (18) | 0.0285 (18) |
| C1 | 0.0446 (16) | 0.0374 (16) | 0.0437 (17) | 0.0029 (13) | 0.0064 (13) | −0.0007 (13) |
| C2 | 0.0362 (15) | 0.0378 (15) | 0.0390 (16) | 0.0041 (12) | 0.0023 (12) | 0.0039 (13) |
| C3 | 0.0340 (14) | 0.0327 (14) | 0.0331 (15) | 0.0031 (11) | 0.0038 (11) | 0.0025 (12) |
| C4 | 0.0351 (15) | 0.0411 (16) | 0.0375 (16) | 0.0034 (12) | 0.0030 (12) | 0.0016 (13) |
| C5 | 0.0426 (16) | 0.0355 (15) | 0.0387 (16) | 0.0024 (12) | 0.0038 (13) | −0.0002 (13) |
| C6 | 0.0396 (15) | 0.0341 (15) | 0.0364 (16) | 0.0044 (12) | 0.0046 (12) | −0.0001 (12) |
| C7 | 0.063 (2) | 0.0372 (17) | 0.055 (2) | 0.0003 (15) | 0.0145 (16) | −0.0051 (15) |
| C8 | 0.0604 (19) | 0.0353 (16) | 0.0450 (18) | 0.0048 (14) | 0.0022 (14) | 0.0143 (14) |
| C9 | 0.0579 (18) | 0.0425 (16) | 0.0303 (15) | 0.0027 (14) | 0.0015 (13) | −0.0020 (13) |
| C10 | 0.0543 (18) | 0.0351 (16) | 0.0538 (19) | 0.0085 (14) | 0.0114 (15) | 0.0007 (15) |
| C11 | 0.0480 (17) | 0.0458 (18) | 0.0489 (18) | 0.0131 (14) | 0.0175 (14) | 0.0102 (15) |
| C12 | 0.0351 (15) | 0.0507 (18) | 0.0424 (17) | 0.0122 (13) | 0.0152 (13) | 0.0097 (14) |
| C13 | 0.0290 (13) | 0.0421 (16) | 0.0314 (15) | 0.0047 (12) | 0.0072 (11) | 0.0005 (12) |
| C14 | 0.0345 (14) | 0.0447 (17) | 0.0368 (16) | 0.0044 (12) | 0.0060 (12) | 0.0004 (13) |
| C15 | 0.0454 (16) | 0.0437 (17) | 0.0422 (17) | 0.0028 (13) | 0.0050 (13) | 0.0048 (14) |
| C16 | 0.0367 (15) | 0.0398 (15) | 0.0370 (16) | 0.0055 (12) | 0.0077 (12) | −0.0003 (12) |
| C17 | 0.070 (2) | 0.050 (2) | 0.067 (2) | 0.0178 (17) | 0.0297 (19) | 0.0122 (18) |
| C18 | 0.0427 (16) | 0.063 (2) | 0.0307 (15) | 0.0129 (14) | 0.0007 (13) | 0.0102 (14) |
| C19 | 0.0459 (17) | 0.0402 (16) | 0.0440 (17) | −0.0025 (13) | 0.0009 (13) | −0.0084 (13) |
| C20 | 0.057 (2) | 0.050 (2) | 0.052 (2) | −0.0026 (16) | −0.0051 (15) | 0.0129 (16) |
Geometric parameters (Å, º)
| S1—C6 | 1.714 (3) | C8—H8A | 0.9600 |
| S1—C1 | 1.750 (3) | C8—H8B | 0.9600 |
| S2—C6 | 1.716 (3) | C8—H8C | 0.9600 |
| S2—C5 | 1.745 (3) | C9—H9A | 0.9600 |
| S3—C16 | 1.716 (3) | C9—H9B | 0.9600 |
| S3—C11 | 1.741 (3) | C9—H9C | 0.9600 |
| S4—C16 | 1.703 (3) | C11—C12 | 1.360 (4) |
| S4—C15 | 1.742 (3) | C11—C17 | 1.423 (4) |
| N1—C7 | 1.136 (4) | C12—C13 | 1.439 (3) |
| N2—C10 | 1.130 (4) | C12—C18 | 1.501 (4) |
| N3—C17 | 1.140 (4) | C13—C16 | 1.377 (3) |
| N4—C20 | 1.137 (4) | C13—C14 | 1.427 (4) |
| C1—C2 | 1.351 (4) | C14—C15 | 1.365 (4) |
| C1—C7 | 1.421 (4) | C14—C19 | 1.495 (4) |
| C2—C3 | 1.428 (3) | C15—C20 | 1.420 (4) |
| C2—C8 | 1.502 (4) | C18—H18A | 0.9600 |
| C3—C6 | 1.384 (3) | C18—H18B | 0.9600 |
| C3—C4 | 1.425 (4) | C18—H18C | 0.9600 |
| C4—C5 | 1.356 (4) | C19—H19A | 0.9600 |
| C4—C9 | 1.495 (4) | C19—H19B | 0.9600 |
| C5—C10 | 1.428 (4) | C19—H19C | 0.9600 |
| C6—S1—C1 | 89.13 (13) | H9B—C9—H9C | 109.5 |
| C6—S2—C5 | 88.80 (13) | N2—C10—C5 | 175.0 (4) |
| C16—S3—C11 | 88.88 (14) | C12—C11—C17 | 125.3 (3) |
| C16—S4—C15 | 88.73 (13) | C12—C11—S3 | 115.1 (2) |
| C2—C1—C7 | 126.0 (3) | C17—C11—S3 | 119.6 (2) |
| C2—C1—S1 | 114.5 (2) | C11—C12—C13 | 110.0 (3) |
| C7—C1—S1 | 119.5 (2) | C11—C12—C18 | 125.1 (3) |
| C1—C2—C3 | 110.7 (2) | C13—C12—C18 | 124.9 (3) |
| C1—C2—C8 | 124.6 (3) | C16—C13—C14 | 111.9 (2) |
| C3—C2—C8 | 124.6 (2) | C16—C13—C12 | 112.0 (2) |
| C6—C3—C4 | 111.9 (2) | C14—C13—C12 | 136.0 (3) |
| C6—C3—C2 | 112.3 (2) | C15—C14—C13 | 110.0 (2) |
| C4—C3—C2 | 135.9 (2) | C15—C14—C19 | 123.4 (3) |
| C5—C4—C3 | 110.8 (2) | C13—C14—C19 | 126.5 (3) |
| C5—C4—C9 | 124.2 (3) | C14—C15—C20 | 127.2 (3) |
| C3—C4—C9 | 125.0 (2) | C14—C15—S4 | 114.8 (2) |
| C4—C5—C10 | 123.0 (3) | C20—C15—S4 | 118.0 (2) |
| C4—C5—S2 | 114.7 (2) | C13—C16—S4 | 114.5 (2) |
| C10—C5—S2 | 122.2 (2) | C13—C16—S3 | 114.0 (2) |
| C3—C6—S1 | 113.3 (2) | S4—C16—S3 | 131.48 (17) |
| C3—C6—S2 | 113.8 (2) | N3—C17—C11 | 179.2 (4) |
| S1—C6—S2 | 132.84 (17) | C12—C18—H18A | 109.5 |
| N1—C7—C1 | 178.6 (4) | C12—C18—H18B | 109.5 |
| C2—C8—H8A | 109.5 | H18A—C18—H18B | 109.5 |
| C2—C8—H8B | 109.5 | C12—C18—H18C | 109.5 |
| H8A—C8—H8B | 109.5 | H18A—C18—H18C | 109.5 |
| C2—C8—H8C | 109.5 | H18B—C18—H18C | 109.5 |
| H8A—C8—H8C | 109.5 | C14—C19—H19A | 109.5 |
| H8B—C8—H8C | 109.5 | C14—C19—H19B | 109.5 |
| C4—C9—H9A | 109.5 | H19A—C19—H19B | 109.5 |
| C4—C9—H9B | 109.5 | C14—C19—H19C | 109.5 |
| H9A—C9—H9B | 109.5 | H19A—C19—H19C | 109.5 |
| C4—C9—H9C | 109.5 | H19B—C19—H19C | 109.5 |
| H9A—C9—H9C | 109.5 | N4—C20—C15 | 177.4 (4) |
| C6—S1—C1—C2 | 0.4 (2) | C16—S3—C11—C12 | 0.6 (2) |
| C6—S1—C1—C7 | −178.9 (2) | C16—S3—C11—C17 | 180.0 (2) |
| C7—C1—C2—C3 | 179.1 (3) | C17—C11—C12—C13 | −179.8 (3) |
| S1—C1—C2—C3 | −0.1 (3) | S3—C11—C12—C13 | −0.4 (3) |
| C7—C1—C2—C8 | 0.5 (5) | C17—C11—C12—C18 | 0.3 (5) |
| S1—C1—C2—C8 | −178.6 (2) | S3—C11—C12—C18 | 179.7 (2) |
| C1—C2—C3—C6 | −0.3 (3) | C11—C12—C13—C16 | 0.0 (3) |
| C8—C2—C3—C6 | 178.3 (2) | C18—C12—C13—C16 | 179.8 (2) |
| C1—C2—C3—C4 | −179.5 (3) | C11—C12—C13—C14 | 178.1 (3) |
| C8—C2—C3—C4 | −1.0 (5) | C18—C12—C13—C14 | −2.0 (5) |
| C6—C3—C4—C5 | −0.6 (3) | C16—C13—C14—C15 | 0.6 (3) |
| C2—C3—C4—C5 | 178.7 (3) | C12—C13—C14—C15 | −177.5 (3) |
| C6—C3—C4—C9 | 179.9 (2) | C16—C13—C14—C19 | −178.2 (2) |
| C2—C3—C4—C9 | −0.9 (5) | C12—C13—C14—C19 | 3.7 (5) |
| C3—C4—C5—C10 | −176.6 (3) | C13—C14—C15—C20 | 178.6 (3) |
| C9—C4—C5—C10 | 3.0 (4) | C19—C14—C15—C20 | −2.5 (5) |
| C3—C4—C5—S2 | 0.9 (3) | C13—C14—C15—S4 | −0.4 (3) |
| C9—C4—C5—S2 | −179.5 (2) | C19—C14—C15—S4 | 178.5 (2) |
| C6—S2—C5—C4 | −0.8 (2) | C16—S4—C15—C14 | 0.1 (2) |
| C6—S2—C5—C10 | 176.8 (3) | C16—S4—C15—C20 | −179.0 (2) |
| C4—C3—C6—S1 | 180.00 (18) | C14—C13—C16—S4 | −0.6 (3) |
| C2—C3—C6—S1 | 0.5 (3) | C12—C13—C16—S4 | 177.97 (17) |
| C4—C3—C6—S2 | 0.0 (3) | C14—C13—C16—S3 | −178.12 (18) |
| C2—C3—C6—S2 | −179.51 (18) | C12—C13—C16—S3 | 0.5 (3) |
| C1—S1—C6—C3 | −0.5 (2) | C15—S4—C16—C13 | 0.3 (2) |
| C1—S1—C6—S2 | 179.6 (2) | C15—S4—C16—S3 | 177.3 (2) |
| C5—S2—C6—C3 | 0.5 (2) | C11—S3—C16—C13 | −0.6 (2) |
| C5—S2—C6—S1 | −179.6 (2) | C11—S3—C16—S4 | −177.5 (2) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: RZ5077).
References
- Bruker (2000). SADABS, SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Cornel, A. & Kirsch, G. (2001). J. Heterocycl. Chem. 38, 1167–1171.
- Gunasekaran, B., Sureshbabu, R., Mohanakrishnan, A. K., Chakkaravarthi, G. & Manivannan, V. (2009). Acta Cryst. E65, o2455. [DOI] [PMC free article] [PubMed]
- Mabkhot, Y. N., Barakat, A., Al-Majid, A. & Choudhary, M. I. (2013). Int. J. Mol. Sci. 14, 5712–5722. [DOI] [PMC free article] [PubMed]
- Mashraqui, S. H., Asharf, M., Hariharasubrahmanian, H., Kellogg, R. K. & Meetsma, A. (2004). J. Mol. Struct. 689, 107–113.
- Mashraqui, S. H., Hariharasubrahmanian, H. & Kumar, S. (1999). Synthesis, pp. 2030–2033.
- Mishra, R., Jha, K. K., Kumar, S. & Tomer, S. (2011). Pharma Chem. 3, 38–54.
- Nardelli, M. (1995). J. Appl. Cryst. 28, 659.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813017960/rz5077sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813017960/rz5077Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813017960/rz5077Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report