Abstract
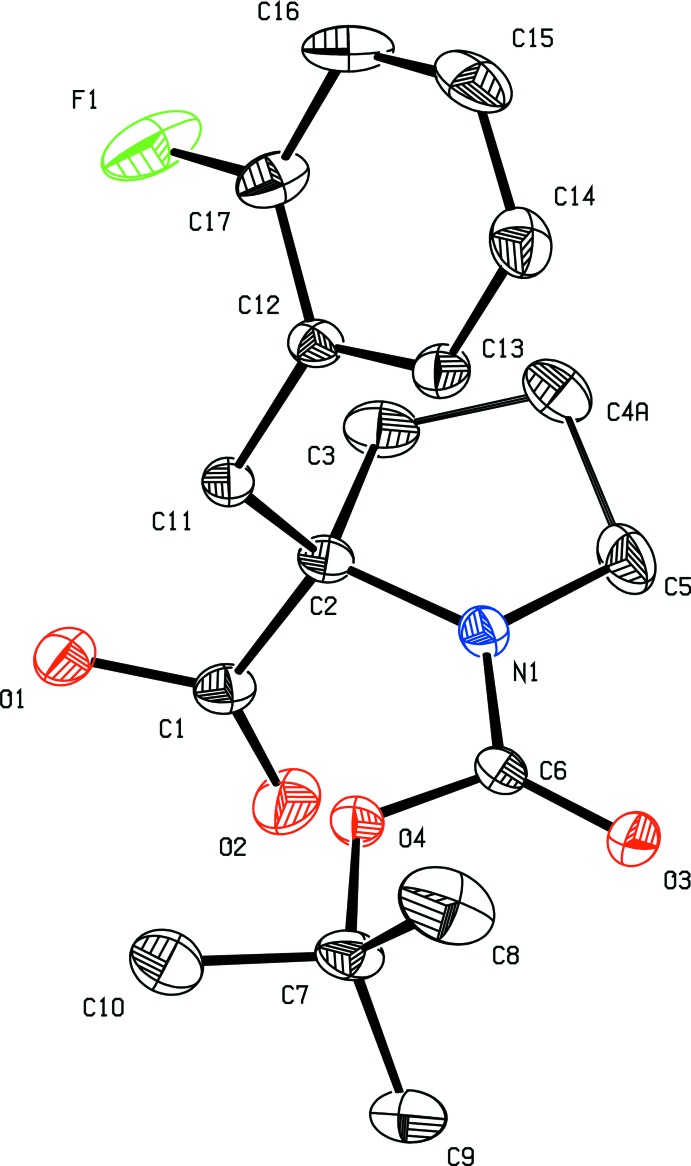
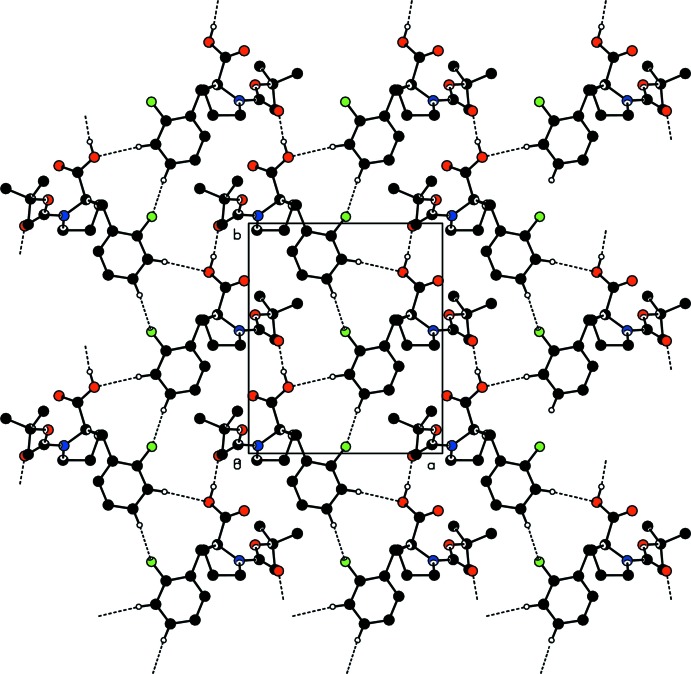
In the title compound, C17H22FNO4, the pyrrolidine ring adopts an envelope conformation with the disordered components of the methylene C atom, with site occupancies of 0.896 (7) and 0.104 (7), being the flap on either side of the mean plane involving the other atoms of the ring. The carboxylic acid group forms dihedral angles of 72.06 (11) and 45.44 (5)° with the N-tert-butoxycarbonyl group and the 2-fluorobenzyl group, respectively. In the crystal, two-dimensional layers of molecules parallel to (001) are built through an R 4 4(23) motif generated via O—H⋯O, C—H⋯O and C—H⋯F interactions, and an R 2 2(11) motif generated by C—H⋯O and C—H⋯F interactions.
Related literature
For general background, see: Taylor et al. (1998 ▶); Jeng et al. (2002 ▶); Anderson et al. (2004 ▶); Ryder et al. (2000 ▶). For biological activity of the title compound, see: Tamazyan et al. (2004 ▶). For graph-set notation of hydrogen bonding, see: Bernstein et al. (1995 ▶). For puckering parameters, see: Cremer & Pople (1975 ▶).
Experimental
Crystal data
C17H22FNO4
M r = 323.36
Orthorhombic,
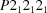
a = 10.4777 (1) Å
b = 12.4283 (2) Å
c = 13.1550 (2) Å
V = 1713.04 (4) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 100 K
0.54 × 0.34 × 0.24 mm
Data collection
Bruker Kappa APEXII diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2008 ▶) T min = 0.962, T max = 0.977
14001 measured reflections
3426 independent reflections
2712 reflections with I > 2σ(I)
R int = 0.031
Standard reflections: 0
Refinement
R[F 2 > 2σ(F 2)] = 0.050
wR(F 2) = 0.114
S = 1.06
3426 reflections
225 parameters
8 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.37 e Å−3
Δρmin = −0.32 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLUTON (Spek, 2009 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536813019788/gk2586sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813019788/gk2586Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C15—H15A⋯F1i | 0.95 | 2.59 | 3.378 (3) | 141 |
| C16—H16A⋯O1i | 0.95 | 2.60 | 3.541 (3) | 173 |
| O1—H1⋯O3ii | 0.89 (3) | 1.73 (3) | 2.611 (2) | 173 (3) |
Symmetry codes: (i) 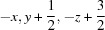 ; (ii)
; (ii) 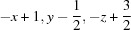 .
.
Acknowledgments
The authors thank Dr Mutharasu Devarajan, Associate Professor, and the staff of the X-ray Crystallography Unit, School of Physics, Universiti Sains Malaysia, for their help in the data collection.
supplementary crystallographic information
Comment
Modified amino acids are known to enhance the chemical, physical and biological properties of proteins (Anderson et al., 2004). Also, due to their structural diversity and functional versatility, they are widely used as chiral building blocks and molecular scaffolds in pharmaceutics (Taylor et al., 1998; Ryder et al., 2000; Jeng et al., 2002). N-Butoxycarbonyl-(S)-α-benzyl proline, a closely related analogue of the title compound N-tert-butoxycarbonyl-α-(2-fluorobenzyl)-L-proline, is a potential non-nucleoside reverse transcriptase inhibitor in anti-human-immunodeficiency virus type-1 (Tamazyan et al., 2004).
The present paper describes the crystal structure of the title compound (Fig. 1), which crystallizes in the orthorhombic space group P212121. It is a modified amino acid with the N-terminus protected by a tert-butyloxycarbonyl (Boc) group and the Cα (C2) H atom replaced by a 2-fluorobenzyl group. In the pyrrolidine ring, the C4 atom of the ring displays positional disorder with site-occupation factors of 0.896 (7) and 0.104 (7). The pyrrolidine ring (N1/C2/C3/C4A/C5) adopts the envelope conformation with the C4A atom deviating from the plane defined the remaining ring atoms by 0.5677 (4) Å. Puckering parameters calculated for this ring are of Q = 0.369 (3) Å, φ = 285.7 (3)° (Cremer & Pople, 1975). The dihedral angles between the mean plane of the carboxylic acid group, N-Boc and 2-methyl-2-fluorobenzene are 72.06 (11)° and 45.44 (5)°, respectively.
The molecules are linked by a combination of O—H···O, C—H···O and C—H···F hydrogen bonds. The carboxylic O1 acts as a donor to the carbonyl O3 at (-x + 1, y - 1/2, -z + 3/2) forming chains parallel to the b axis through C(7) motifs (Bernstein et al., 1995). These 21 screw-generated parallel chains are interconnected through C15—H15A···F1 (-x, y + 1/2, -z + 3/2) and C16—H16A···O1 (-x, y + 1/2, -z + 3/2) hydrogen bonds leading to a layer parallel to the ab plane. The characteristic building units of this layer are an R44(23) ring generated by all the hydrogen-bonds and an R22(11) generated exclusively by C—H···O and C—H···F hydrogen bonds (Fig. 2).
Experimental
A mixture of 2-(2-fluorobenzyl)-L-proline (1.0 mmol) and tetramethylammonium hydroxide pentahydrate (1.2 mmol) in acetonitrile (10 ml) was stirred for 30 min. After 30 min, Boc2O (2.0 mmol) was added and stirred continuously for 2 d. The acetonitrile was removed in vacuo and residue was partitioned between ether (20 ml) and water (10 ml). The aqueous layer was washed with ether (10 ml) and acidified with 10% aqueous citric acid to pH 3–4. The aqueous layer was extracted with ethyl acetate (3 × 10 ml) and combined organic extracts were washed with brine solution (1 × 10 ml), dried over Na2SO4, and concentrated to yield N-tert-butoxycarbonyl-α-(2-fluorobenzyl)-L-proline (m.p. 430-433 K) as a white solid. Crystals were grown from ethanolic solution by slow evaporation at room temperature.
Refinement
All H atoms, except hydroxy H1 atom, were placed at geometrically calculated positions (0.99 Å for methylene C—H, 0.98 Å for methyl C—H and 0.95 Å for aromatic C—H) and refined using a riding model. The Uiso values of all H atoms were constrained to 1.2Ueq (1.5 times for hydroxyl and methyl H atoms) of the respective atom to which the H atom bonds. The hydroxy H1 atom was freely refined. In the pyrrolidine ring, the C4 atom exhibits disorder (resolved into C4A and C4B) and the same was modelled using SIMU and SADI restraints leading to site-occupancies of 0.896 (7) and 0.104 (7). In the absence of significant anomalous scattering effects 1492 Friedel pairs were merged. The enantiomer has been assigned by reference to an unchanging chiral centre in the synthetic procedure.
Figures
Fig. 1.
Molecular structure of the title compound showing the major component of the disorder. Displacement ellipsoids for non-H atoms are drawn at the 50% probability level.
Fig. 2.
Part of the crystal structure of the title compound, showing R22(11) and R44(23) motifs through a combination of O—H···O, C—H···O and C—H···F hydrogen bonds and the formation of a two dimensional layer parallel to the ab plane. For the sake of clarity, H atoms not involved in hydrogen bonding have been omitted.
Crystal data
| C17H22FNO4 | F(000) = 688 |
| Mr = 323.36 | Dx = 1.254 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 3426 reflections |
| a = 10.4777 (1) Å | θ = 2.3–30.0° |
| b = 12.4283 (2) Å | µ = 0.10 mm−1 |
| c = 13.1550 (2) Å | T = 100 K |
| V = 1713.04 (4) Å3 | Block, colourless |
| Z = 4 | 0.54 × 0.34 × 0.24 mm |
Data collection
| Bruker Kappa APEXII diffractometer | 3426 independent reflections |
| Radiation source: fine-focus sealed tube | 2712 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.031 |
| φ and ω scans | θmax = 32.6°, θmin = 2.3° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2008) | h = −13→15 |
| Tmin = 0.962, Tmax = 0.977 | k = −18→18 |
| 14001 measured reflections | l = −18→18 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.114 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.043P)2 + 0.5231P] where P = (Fo2 + 2Fc2)/3 |
| 3426 reflections | (Δ/σ)max < 0.001 |
| 225 parameters | Δρmax = 0.37 e Å−3 |
| 8 restraints | Δρmin = −0.32 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| F1 | −0.00102 (13) | 0.97131 (13) | 0.77647 (14) | 0.0493 (4) | |
| O1 | 0.29559 (14) | 0.71232 (12) | 0.76893 (13) | 0.0272 (3) | |
| H1 | 0.320 (3) | 0.645 (3) | 0.756 (3) | 0.055 (9)* | |
| O2 | 0.47731 (16) | 0.74954 (12) | 0.68734 (14) | 0.0346 (4) | |
| O3 | 0.65381 (14) | 1.01134 (12) | 0.77232 (14) | 0.0296 (4) | |
| O4 | 0.53366 (13) | 0.89798 (11) | 0.86819 (11) | 0.0227 (3) | |
| N1 | 0.45214 (15) | 0.96664 (13) | 0.72598 (13) | 0.0191 (3) | |
| C1 | 0.38068 (19) | 0.77841 (16) | 0.72823 (16) | 0.0205 (4) | |
| C2 | 0.33880 (18) | 0.89677 (15) | 0.73257 (15) | 0.0177 (4) | |
| C3 | 0.2696 (2) | 0.92068 (18) | 0.63034 (17) | 0.0272 (5) | |
| H3A | 0.2918 | 0.8660 | 0.5786 | 0.033* | 0.896 (7) |
| H3B | 0.1759 | 0.9210 | 0.6398 | 0.033* | 0.896 (7) |
| H3C | 0.2326 | 0.8539 | 0.6017 | 0.033* | 0.104 (7) |
| H3D | 0.2001 | 0.9735 | 0.6407 | 0.033* | 0.104 (7) |
| C4A | 0.3162 (2) | 1.0299 (2) | 0.59876 (18) | 0.0271 (7) | 0.896 (7) |
| H4A | 0.2680 | 1.0875 | 0.6337 | 0.032* | 0.896 (7) |
| H4B | 0.3084 | 1.0398 | 0.5244 | 0.032* | 0.896 (7) |
| C4B | 0.3694 (15) | 0.9656 (18) | 0.5602 (4) | 0.058 (7) | 0.104 (7) |
| H4C | 0.3306 | 1.0131 | 0.5083 | 0.070* | 0.104 (7) |
| H4D | 0.4169 | 0.9072 | 0.5257 | 0.070* | 0.104 (7) |
| C5 | 0.4573 (2) | 1.02965 (19) | 0.63191 (17) | 0.0299 (5) | |
| H5A | 0.5122 | 0.9947 | 0.5804 | 0.036* | 0.896 (7) |
| H5B | 0.4887 | 1.1036 | 0.6447 | 0.036* | 0.896 (7) |
| H5C | 0.5453 | 1.0334 | 0.6049 | 0.036* | 0.104 (7) |
| H5D | 0.4248 | 1.1036 | 0.6427 | 0.036* | 0.104 (7) |
| C6 | 0.55438 (19) | 0.96175 (15) | 0.78805 (16) | 0.0212 (4) | |
| C7 | 0.6244 (2) | 0.89395 (17) | 0.95460 (17) | 0.0247 (4) | |
| C8 | 0.6362 (3) | 1.0048 (2) | 1.0019 (2) | 0.0452 (7) | |
| H8A | 0.6801 | 1.0531 | 0.9545 | 0.068* | |
| H8B | 0.6852 | 0.9998 | 1.0651 | 0.068* | |
| H8C | 0.5509 | 1.0332 | 1.0166 | 0.068* | |
| C9 | 0.7514 (2) | 0.8487 (2) | 0.9205 (2) | 0.0371 (6) | |
| H9A | 0.7944 | 0.9009 | 0.8764 | 0.056* | |
| H9B | 0.7373 | 0.7817 | 0.8828 | 0.056* | |
| H9C | 0.8048 | 0.8341 | 0.9801 | 0.056* | |
| C10 | 0.5574 (3) | 0.8169 (2) | 1.02650 (18) | 0.0337 (5) | |
| H10A | 0.5444 | 0.7476 | 0.9924 | 0.050* | |
| H10B | 0.4745 | 0.8470 | 1.0462 | 0.050* | |
| H10C | 0.6099 | 0.8063 | 1.0873 | 0.050* | |
| C11 | 0.25451 (19) | 0.92051 (15) | 0.82578 (15) | 0.0186 (4) | |
| H11A | 0.1825 | 0.8690 | 0.8263 | 0.022* | |
| H7B | 0.3054 | 0.9077 | 0.8880 | 0.022* | |
| C12 | 0.20116 (18) | 1.03351 (16) | 0.82987 (14) | 0.0182 (4) | |
| C13 | 0.2744 (2) | 1.12161 (15) | 0.85860 (16) | 0.0212 (4) | |
| H13 | 0.3613 | 1.1109 | 0.8764 | 0.025* | |
| C14 | 0.2240 (2) | 1.22470 (17) | 0.86194 (18) | 0.0292 (5) | |
| H14A | 0.2765 | 1.2837 | 0.8809 | 0.035* | |
| C15 | 0.0967 (2) | 1.24157 (19) | 0.83757 (19) | 0.0342 (6) | |
| H15A | 0.0619 | 1.3121 | 0.8403 | 0.041* | |
| C16 | 0.0208 (2) | 1.1563 (2) | 0.80940 (19) | 0.0357 (6) | |
| H16A | −0.0665 | 1.1670 | 0.7928 | 0.043* | |
| C17 | 0.0743 (2) | 1.05449 (18) | 0.80576 (18) | 0.0272 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F1 | 0.0207 (6) | 0.0479 (9) | 0.0792 (12) | 0.0004 (6) | −0.0048 (8) | −0.0278 (9) |
| O1 | 0.0206 (7) | 0.0158 (6) | 0.0451 (9) | 0.0020 (5) | 0.0062 (7) | −0.0057 (7) |
| O2 | 0.0263 (8) | 0.0254 (7) | 0.0522 (10) | 0.0054 (6) | 0.0151 (8) | −0.0038 (8) |
| O3 | 0.0184 (7) | 0.0204 (7) | 0.0501 (10) | −0.0006 (5) | 0.0024 (7) | 0.0084 (7) |
| O4 | 0.0185 (7) | 0.0237 (7) | 0.0259 (7) | −0.0023 (6) | −0.0066 (6) | 0.0078 (6) |
| N1 | 0.0188 (8) | 0.0191 (7) | 0.0193 (7) | 0.0021 (6) | 0.0023 (7) | 0.0041 (7) |
| C1 | 0.0184 (9) | 0.0200 (8) | 0.0232 (9) | 0.0035 (7) | −0.0021 (8) | −0.0037 (8) |
| C2 | 0.0177 (9) | 0.0168 (8) | 0.0185 (8) | 0.0039 (7) | 0.0000 (7) | −0.0016 (7) |
| C3 | 0.0272 (11) | 0.0329 (11) | 0.0214 (10) | 0.0072 (9) | −0.0055 (9) | −0.0034 (9) |
| C4A | 0.0359 (14) | 0.0264 (12) | 0.0189 (11) | 0.0043 (10) | −0.0072 (10) | 0.0021 (10) |
| C4B | 0.094 (16) | 0.047 (13) | 0.034 (12) | 0.037 (13) | 0.001 (12) | 0.011 (11) |
| C5 | 0.0379 (13) | 0.0284 (11) | 0.0233 (10) | 0.0024 (10) | 0.0057 (10) | 0.0093 (9) |
| C6 | 0.0192 (9) | 0.0134 (8) | 0.0310 (11) | 0.0031 (7) | 0.0015 (8) | 0.0030 (8) |
| C7 | 0.0222 (10) | 0.0232 (9) | 0.0288 (11) | 0.0031 (8) | −0.0111 (9) | −0.0023 (9) |
| C8 | 0.0569 (17) | 0.0303 (12) | 0.0485 (16) | 0.0053 (12) | −0.0199 (14) | −0.0135 (12) |
| C9 | 0.0251 (12) | 0.0356 (12) | 0.0506 (16) | 0.0086 (10) | −0.0058 (11) | 0.0071 (12) |
| C10 | 0.0347 (13) | 0.0404 (13) | 0.0258 (11) | 0.0022 (11) | −0.0086 (10) | 0.0069 (10) |
| C11 | 0.0175 (9) | 0.0159 (8) | 0.0226 (10) | 0.0030 (7) | 0.0039 (7) | 0.0000 (8) |
| C12 | 0.0175 (9) | 0.0189 (8) | 0.0180 (9) | 0.0044 (7) | 0.0027 (7) | −0.0007 (8) |
| C13 | 0.0195 (9) | 0.0205 (9) | 0.0236 (10) | 0.0018 (7) | 0.0026 (8) | −0.0019 (8) |
| C14 | 0.0350 (12) | 0.0199 (9) | 0.0329 (12) | 0.0015 (9) | 0.0066 (10) | −0.0026 (9) |
| C15 | 0.0430 (14) | 0.0242 (10) | 0.0354 (12) | 0.0173 (10) | 0.0045 (11) | 0.0014 (10) |
| C16 | 0.0260 (12) | 0.0402 (13) | 0.0407 (13) | 0.0178 (10) | −0.0068 (10) | −0.0068 (11) |
| C17 | 0.0184 (10) | 0.0304 (11) | 0.0326 (12) | 0.0034 (8) | −0.0004 (9) | −0.0091 (10) |
Geometric parameters (Å, º)
| F1—C17 | 1.356 (3) | C5—H5C | 0.9900 |
| O1—C1 | 1.325 (2) | C5—H5D | 0.9900 |
| O1—H1 | 0.89 (3) | C7—C9 | 1.513 (3) |
| O2—C1 | 1.201 (2) | C7—C8 | 1.517 (3) |
| O3—C6 | 1.228 (2) | C7—C10 | 1.518 (3) |
| O4—C6 | 1.337 (2) | C8—H8A | 0.9800 |
| O4—C7 | 1.483 (2) | C8—H8B | 0.9800 |
| N1—C6 | 1.348 (3) | C8—H8C | 0.9800 |
| N1—C5 | 1.465 (3) | C9—H9A | 0.9800 |
| N1—C2 | 1.474 (3) | C9—H9B | 0.9800 |
| C1—C2 | 1.536 (3) | C9—H9C | 0.9800 |
| C2—C11 | 1.540 (3) | C10—H10A | 0.9800 |
| C2—C3 | 1.557 (3) | C10—H10B | 0.9800 |
| C3—C4A | 1.501 (3) | C10—H10C | 0.9800 |
| C3—C4B | 1.502 (4) | C11—C12 | 1.513 (3) |
| C3—H3A | 0.9900 | C11—H11A | 0.9900 |
| C3—H3B | 0.9900 | C11—H7B | 0.9900 |
| C3—H3C | 0.9900 | C12—C17 | 1.391 (3) |
| C3—H3D | 0.9900 | C12—C13 | 1.389 (3) |
| C4A—C5 | 1.541 (4) | C13—C14 | 1.386 (3) |
| C4A—H4A | 0.9900 | C13—H13 | 0.9500 |
| C4A—H4B | 0.9900 | C14—C15 | 1.388 (4) |
| C4B—C5 | 1.540 (4) | C14—H14A | 0.9500 |
| C4B—H4C | 0.9900 | C15—C16 | 1.376 (4) |
| C4B—H4D | 0.9900 | C15—H15A | 0.9500 |
| C5—H5A | 0.9900 | C16—C17 | 1.385 (3) |
| C5—H5B | 0.9900 | C16—H16A | 0.9500 |
| C1—O1—H1 | 108 (2) | N1—C5—H5D | 111.2 |
| C6—O4—C7 | 121.37 (16) | C4B—C5—H5D | 111.2 |
| C6—N1—C5 | 120.42 (17) | C4A—C5—H5D | 73.1 |
| C6—N1—C2 | 125.31 (16) | H5A—C5—H5D | 134.9 |
| C5—N1—C2 | 113.21 (17) | H5C—C5—H5D | 109.1 |
| O2—C1—O1 | 124.25 (19) | O3—C6—O4 | 124.63 (19) |
| O2—C1—C2 | 122.92 (19) | O3—C6—N1 | 123.33 (19) |
| O1—C1—C2 | 112.72 (16) | O4—C6—N1 | 112.04 (17) |
| N1—C2—C1 | 109.38 (15) | O4—C7—C9 | 110.42 (18) |
| N1—C2—C11 | 113.33 (16) | O4—C7—C8 | 109.64 (18) |
| C1—C2—C11 | 112.15 (16) | C9—C7—C8 | 112.8 (2) |
| N1—C2—C3 | 102.24 (16) | O4—C7—C10 | 101.69 (17) |
| C1—C2—C3 | 106.49 (16) | C9—C7—C10 | 110.92 (19) |
| C11—C2—C3 | 112.60 (15) | C8—C7—C10 | 110.8 (2) |
| C4A—C3—C2 | 105.08 (18) | C7—C8—H8A | 109.5 |
| C4B—C3—C2 | 106.1 (6) | C7—C8—H8B | 109.5 |
| C4A—C3—H3A | 110.7 | H8A—C8—H8B | 109.5 |
| C4B—C3—H3A | 70.7 | C7—C8—H8C | 109.5 |
| C2—C3—H3A | 110.7 | H8A—C8—H8C | 109.5 |
| C4A—C3—H3B | 110.7 | H8B—C8—H8C | 109.5 |
| C4B—C3—H3B | 140.0 | C7—C9—H9A | 109.5 |
| C2—C3—H3B | 110.7 | C7—C9—H9B | 109.5 |
| H3A—C3—H3B | 108.8 | H9A—C9—H9B | 109.5 |
| C4A—C3—H3C | 141.3 | C7—C9—H9C | 109.5 |
| C4B—C3—H3C | 110.5 | H9A—C9—H9C | 109.5 |
| C2—C3—H3C | 110.5 | H9B—C9—H9C | 109.5 |
| H3B—C3—H3C | 70.3 | C7—C10—H10A | 109.5 |
| C4A—C3—H3D | 71.2 | C7—C10—H10B | 109.5 |
| C4B—C3—H3D | 110.5 | H10A—C10—H10B | 109.5 |
| C2—C3—H3D | 110.5 | C7—C10—H10C | 109.5 |
| H3A—C3—H3D | 136.3 | H10A—C10—H10C | 109.5 |
| H3C—C3—H3D | 108.7 | H10B—C10—H10C | 109.5 |
| C3—C4A—C5 | 103.42 (19) | C12—C11—C2 | 114.72 (16) |
| C3—C4A—H4A | 111.1 | C12—C11—H11A | 108.6 |
| C5—C4A—H4A | 111.1 | C2—C11—H11A | 108.6 |
| C3—C4A—H4B | 111.1 | C12—C11—H7B | 108.6 |
| C5—C4A—H4B | 111.1 | C2—C11—H7B | 108.6 |
| H4A—C4A—H4B | 109.0 | H11A—C11—H7B | 107.6 |
| C3—C4B—C5 | 103.4 (2) | C17—C12—C13 | 116.22 (19) |
| C3—C4B—H4C | 111.1 | C17—C12—C11 | 121.26 (19) |
| C5—C4B—H4C | 111.1 | C13—C12—C11 | 122.51 (17) |
| C3—C4B—H4D | 111.1 | C14—C13—C12 | 121.8 (2) |
| C5—C4B—H4D | 111.1 | C14—C13—H13 | 119.1 |
| H4C—C4B—H4D | 109.0 | C12—C13—H13 | 119.1 |
| N1—C5—C4B | 102.7 (6) | C15—C14—C13 | 119.9 (2) |
| N1—C5—C4A | 101.83 (18) | C15—C14—H14A | 120.1 |
| N1—C5—H5A | 111.4 | C13—C14—H14A | 120.1 |
| C4B—C5—H5A | 72.6 | C16—C15—C14 | 120.1 (2) |
| C4A—C5—H5A | 111.4 | C16—C15—H15A | 120.0 |
| N1—C5—H5B | 111.4 | C14—C15—H15A | 120.0 |
| C4B—C5—H5B | 141.5 | C15—C16—C17 | 118.6 (2) |
| C4A—C5—H5B | 111.4 | C15—C16—H16A | 120.7 |
| H5A—C5—H5B | 109.3 | C17—C16—H16A | 120.7 |
| N1—C5—H5C | 111.2 | F1—C17—C16 | 118.07 (19) |
| C4B—C5—H5C | 111.2 | F1—C17—C12 | 118.55 (19) |
| C4A—C5—H5C | 142.4 | C16—C17—C12 | 123.4 (2) |
| H5B—C5—H5C | 73.0 | ||
| C6—N1—C2—C1 | −55.2 (2) | C3—C4A—C5—N1 | −35.4 (2) |
| C5—N1—C2—C1 | 113.02 (19) | C3—C4A—C5—C4B | 60.5 (8) |
| C6—N1—C2—C11 | 70.8 (2) | C7—O4—C6—O3 | 9.2 (3) |
| C5—N1—C2—C11 | −121.03 (18) | C7—O4—C6—N1 | −170.18 (16) |
| C6—N1—C2—C3 | −167.78 (18) | C5—N1—C6—O3 | 2.7 (3) |
| C5—N1—C2—C3 | 0.4 (2) | C2—N1—C6—O3 | 170.11 (19) |
| O2—C1—C2—N1 | −26.3 (3) | C5—N1—C6—O4 | −177.96 (18) |
| O1—C1—C2—N1 | 157.34 (17) | C2—N1—C6—O4 | −10.5 (3) |
| O2—C1—C2—C11 | −152.9 (2) | C6—O4—C7—C9 | −65.0 (2) |
| O1—C1—C2—C11 | 30.7 (2) | C6—O4—C7—C8 | 59.9 (3) |
| O2—C1—C2—C3 | 83.5 (2) | C6—O4—C7—C10 | 177.21 (18) |
| O1—C1—C2—C3 | −92.9 (2) | N1—C2—C11—C12 | 60.3 (2) |
| N1—C2—C3—C4A | −23.3 (2) | C1—C2—C11—C12 | −175.22 (17) |
| C1—C2—C3—C4A | −138.06 (18) | C3—C2—C11—C12 | −55.1 (2) |
| C11—C2—C3—C4A | 98.6 (2) | C2—C11—C12—C17 | 103.8 (2) |
| N1—C2—C3—C4B | 21.3 (9) | C2—C11—C12—C13 | −76.9 (2) |
| C1—C2—C3—C4B | −93.5 (9) | C17—C12—C13—C14 | −0.7 (3) |
| C11—C2—C3—C4B | 143.2 (9) | C11—C12—C13—C14 | 180.0 (2) |
| C4B—C3—C4A—C5 | −60.8 (8) | C12—C13—C14—C15 | 0.9 (3) |
| C2—C3—C4A—C5 | 36.8 (2) | C13—C14—C15—C16 | −0.5 (4) |
| C4A—C3—C4B—C5 | 60.9 (8) | C14—C15—C16—C17 | −0.2 (4) |
| C2—C3—C4B—C5 | −34.1 (16) | C15—C16—C17—F1 | −179.1 (2) |
| C6—N1—C5—C4B | 147.8 (9) | C15—C16—C17—C12 | 0.5 (4) |
| C2—N1—C5—C4B | −21.1 (9) | C13—C12—C17—F1 | 179.50 (19) |
| C6—N1—C5—C4A | −169.45 (18) | C11—C12—C17—F1 | −1.2 (3) |
| C2—N1—C5—C4A | 21.7 (2) | C13—C12—C17—C16 | 0.0 (3) |
| C3—C4B—C5—N1 | 33.3 (16) | C11—C12—C17—C16 | 179.3 (2) |
| C3—C4B—C5—C4A | −60.4 (8) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C15—H15A···F1i | 0.95 | 2.59 | 3.378 (3) | 141 |
| C16—H16A···O1i | 0.95 | 2.60 | 3.541 (3) | 173 |
| O1—H1···O3ii | 0.89 (3) | 1.73 (3) | 2.611 (2) | 173 (3) |
Symmetry codes: (i) −x, y+1/2, −z+3/2; (ii) −x+1, y−1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: GK2586).
References
- Anderson, J. C., Wu, N., Santoro, S. W., Lakshman, V., King, D. S. & Schultz, P. G. (2004). Proc. Natl Acad. Sci. USA, 101, 7566–7571. [DOI] [PMC free article] [PubMed]
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruker (2009). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Jeng, A. Y., Savage, P., Beil, M. E., Bruseo, C. W., Hoyer, D., Fink, C. A. & Trapani, A. J. (2002). Clin. Sci. 103, 98–101. [DOI] [PubMed]
- Ryder, T. R., Hu, L. Y., Rafferty, M. F., Lotarski, S. M., Rock, D. M., Stoehr, S. J. & Szoke, B. G. (2000). Drug Des. Discov. 16, 317–322. [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Tamazyan, R., Karapetyan, H., Martirisyan, A., Martirosyan, V., Harutyunyan, G. & Gasparyan, S. (2004). Acta Cryst. C60, o390–o392. [DOI] [PubMed]
- Taylor, P. P., Pantaleone, D. P., Senkpeil, R. F. & Fotheringham, I. G. (1998). Trends Biotechnol. 16, 412–418. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536813019788/gk2586sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813019788/gk2586Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report