Abstract
The title compound, C14H20N4O4·0.5H2O [systematic name: (2S)-5-{[amino(iminiumyl)methyl]amino}-2-{[(1Z)-4-methoxy-2-oxidobenzylidene]azaniumyl}pentanoate hemihydrate], has been synthesized by the reaction of l-arginine and 4-methoxysalicylaldehyde and crystallizes with two independent substituted l-arginine molecules and one water molecule of solvation in the asymmetric unit. Each molecule exists as a zwitterion and adopts a Z configuration about the central C=N. The molecular conformation is stabilized by strong intramolecular N—H⋯O hydrogen bonds that generate S(6) and S(10) ring motifs. Intermolecular N—H⋯O and O—H⋯O hydrogen bonds involving also the water molecule and weak intermolecular C—H⋯Owater interactions link the molecules into an infinite one-dimensional ribbon structure extending along the b axis. The known (2S) absolute configuration for l-arginine was invoked. Weak intermolecular C—H⋯π interactions are also present.
Related literature
For the synthesis of similar compounds, see: Srinivasan et al. (1986 ▶); Moutet & Ourari (1997 ▶). For general background on Schiff bases, see: von Konig et al. (1982 ▶); Lewis et al. (2009 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶). For related structures, see: Oueslati et al. (2007 ▶).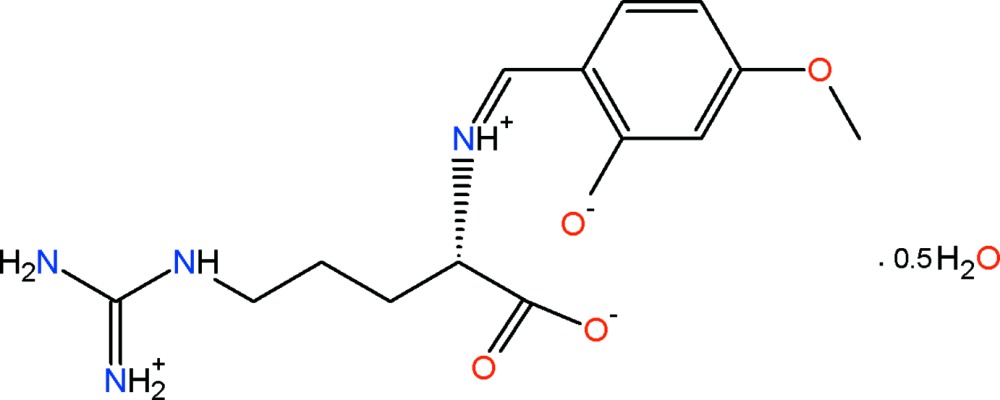
Experimental
Crystal data
C14H20N4O4·0.5H2O
M r = 317.35
Monoclinic,
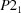
a = 10.1828 (11) Å
b = 10.3414 (11) Å
c = 15.5542 (16) Å
β = 102.688 (2)°
V = 1597.9 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 293 K
0.30 × 0.30 × 0.25 mm
Data collection
Bruker Kappa APEXII CCD-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2004 ▶) T min = 0.971, T max = 0.976
18648 measured reflections
7483 independent reflections
5859 reflections with I > 2σ(I)
R int = 0.036
Refinement
R[F 2 > 2σ(F 2)] = 0.072
wR(F 2) = 0.167
S = 1.10
7483 reflections
452 parameters
14 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.25 e Å−3
Δρmin = −0.21 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: APEX2 and SAINT (Bruker, 2004 ▶); data reduction: SAINT and XPREP (Bruker, 2004 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶) and Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813019727/zs2267sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813019727/zs2267Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O2 | 0.89 (1) | 1.94 (3) | 2.638 (4) | 134 (3) |
| N4—H4A⋯O4 | 0.90 (1) | 2.06 (1) | 2.935 (4) | 165 (3) |
| N5—H5⋯O6 | 0.90 (1) | 1.90 (3) | 2.600 (4) | 134 (3) |
| N8—H8A⋯O7 | 0.90 (1) | 2.03 (1) | 2.914 (4) | 166 (3) |
| N2—H2⋯O2i | 0.86 | 1.92 | 2.758 (4) | 166 |
| N3—H3A⋯O3i | 0.89 (1) | 2.58 (3) | 3.333 (4) | 142 (3) |
| N3—H3B⋯O4ii | 0.89 (1) | 1.93 (1) | 2.817 (4) | 175 (4) |
| N4—H4B⋯O3ii | 0.89 (1) | 2.03 (1) | 2.912 (4) | 171 (3) |
| N6—H6⋯O6iii | 0.86 | 1.89 | 2.705 (4) | 158 |
| N7—H7A⋯O8iii | 0.89 (1) | 2.50 (3) | 3.202 (4) | 135 (3) |
| N7—H7B⋯O7iv | 0.90 (1) | 1.91 (1) | 2.800 (4) | 173 (3) |
| N8—H8B⋯O8iv | 0.89 (1) | 2.05 (2) | 2.911 (4) | 161 (3) |
| O1W—H2W⋯O3v | 0.94 (1) | 1.98 (2) | 2.881 (5) | 159 (6) |
| C15—H15C⋯O1W i | 0.96 | 2.56 | 3.451 (7) | 155 |
| C22—H22⋯O1W | 0.93 | 2.53 | 3.359 (6) | 149 |
| C1—H1C⋯Cg1vi | 0.96 | 2.96 | 3.669 (4) | 132 |
| C15—H15C⋯Cg2vii | 0.96 | 2.98 | 3.762 (5) | 139 |
Symmetry codes: (i)  ; (ii)
; (ii) 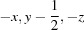 ; (iii)
; (iii) 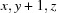 ; (iv)
; (iv) 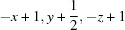 ; (v)
; (v) 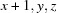 ; (vi)
; (vi) 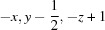 ; (vii)
; (vii) 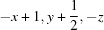 .
.
Acknowledgments
MS thanks the UGC Networking Centre, School of Chemistry, University of Hyderabad, India, for the award of a Visiting Research Fellowship to use the facilities at the School. The authors also thank for access to the X-ray diffraction equipment.
supplementary crystallographic information
Comment
The Schiff base ligands derived from salicylaldehyde derivatives have been found to be excellent chelating agents for most applications in coordination chemistry such as in catalysis (Srinivasan et al., 1986) and electrocatalysis (Moutet & Ourari, 1997). Schiff bases of the general type p-R'-C6H4—CH═N—C6H4—R"-p are well known reagents that find their practical application in various areas, e.g. photography (von Konig et al., 1982) and medicinal and pharmaceutical chemistry (Lewis et al., 2009). Here, we report the synthesis of the title compound, C14H20N4O4. 0.5H2O [systematic name: (2S)-5- {[amino(iminio)methyl]amino}-2-{[(1Z)-(4-methoxy-2-oxidophenyl) methylene]ammonio}pentanoate hemihydrate] and the structure is reported herein.
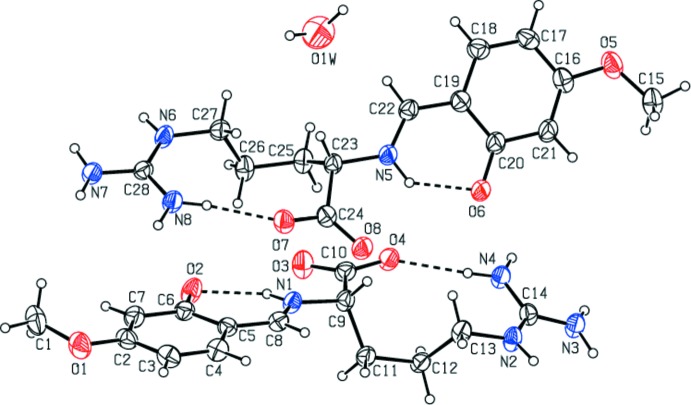
This compound crystallizes with two independent substituted L-arginine molecules (A and B), together with one water molecule of solvation in the asymmetric unit (Fig. 1). Each molecule exists as a zwitterion and adopts a Z configuration about the central iminium C═N functional group which is coplanar with the adjacent benzene ring. The known (2S) absolute configuration for L-arginine was invoked for the trivially named chiral centres at C9 and C23. The C—N bond distances of the NH2 groups(N3—C14, N4—C14, N7—C28 and N8—C28) are 1.332 (6), 1.319 (6), 1.328 (6) and 1.322 (5) Å, respectively, which is short for a C—N single bond, but still not quite as contracted as one would expect for a fully established C═N. These bond length features are consistent with an imino resonance form as is commonly found for C—N single bonds involving sp2 hybridized C and N atoms (Oueslati et al., 2007). The bond distances C6—O2, C10—O3, C10—O4, C20—O6, C24—O7 and C24—O8 [1.284 (5), 1.248 (6), 1.239 (5), 1.285 (5), 1.253 (5) and 1.237 (5) Å, respectively], clearly indicate the presence of C═O double bonds, including those also generated through resonance. The H atoms attached to the phenolic groups (O2 and O6) are transferred to the basic centres N1 and N5 respectively, generating the iminium groups. Also, the carboxylic H-atoms on O4 and O7 have been transferred to N4 and N8, respectively, to generate the common amino acid zwitterions.
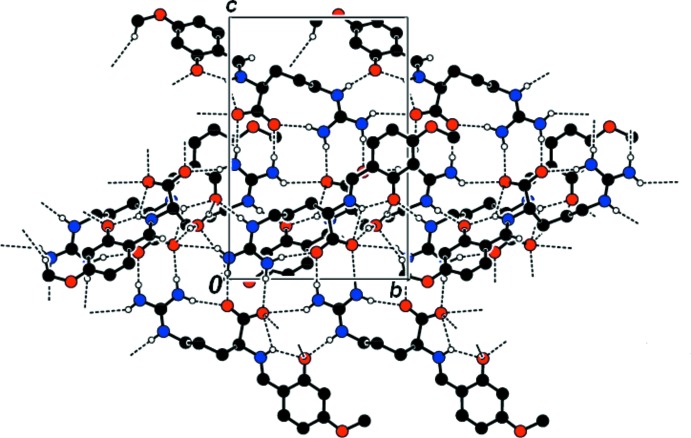
In both molecules A and B, all nitrogen H-atoms are involved in hydrogen bonding (Table 1). In each, intramolecular N—H···O hydrogen bonds lead to the formation of a six- and a ten-membered ring motif [S(6) and S(10), respectively (Bernstein et al., 1995)] (Fig.1). Intermolecular N—H···O and O—H···O hydrogen bonds involving also the water molecule and weak intermolecular C—H···Owater interactions link the molecules into an infinite one-dimensional ribbon structure extending along the b axis (Fig. 2). Present also are weak intermolecular C—H..π interactions.
Experimental
L-Arginine and 4-methoxy salicylaldehyde (E-Merck- analar grades) were mixed in 1:1 stoichiometric proportions and dissolved in a triply distilled water–ethanol mixture using a mechanical stirrer for about four hours. The raw reaction product was removed by filtration, then re-dissolved in a water–ethanol solvent mixture and kept aside to allow crystal growth at ambient temperature. Bright yellowish crystals formed in 3 days and on removal were recrystallized several times to obtain the crystal specimen used in the X-ray analysis.
Refinement
The H atoms were positioned geometrically, with methyl C—H distances of 0.96 Å (methylene), 0.93 Å (aromatic) and the N2—H and N6—H distances of 0.86 Å, and were refined as riding on their parent atoms, with Uiso(H) = 1.2–1.5 Ueq of the parent atom. The remaining N—H atoms and water molecule H atoms were located from a difference Fourier map and refined with distance restraints [N—H = 0.90 (2) and O—H = 0.91 (2) Å] with Uiso(H) = 1.2Ueq(N) and Uiso(H) = 1.5Ueq(O). The known (2S) absolute configuration for L-arginine was invoked at the trivially numbered chiral centres of the A and B molecules (C9 and C23, respectively) (Flack parameter: 0.01 (14) for 3448 Friedel pairs).
Figures
Fig. 1.
Molecular configuration and atom numbering scheme for the two independent substituted L-arginine molecules and the water molecule of solvation in the asymmetric unit of the title compound. Displacement ellipsoids are drawn at the 30% probability level and intramolecular hydrogen bonds are shown as dashed lines.
Fig. 2.
Packing diagram of the title compound viewed down the a axis. Dashed lines indicate intra and intermolecular N—H···O and O—H···O hydrogen bonds and weak C—H···O intermolecular interactions.
Crystal data
| C14H20N4O4·0.5H2O | F(000) = 676 |
| Mr = 317.35 | Dx = 1.319 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2yb | Cell parameters from 3158 reflections |
| a = 10.1828 (11) Å | θ = 2.5–31.2° |
| b = 10.3414 (11) Å | µ = 0.10 mm−1 |
| c = 15.5542 (16) Å | T = 293 K |
| β = 102.688 (2)° | Block, yellow |
| V = 1597.9 (3) Å3 | 0.30 × 0.30 × 0.25 mm |
| Z = 4 |
Data collection
| Bruker Kappa APEXII CCD-detector diffractometer | 7483 independent reflections |
| Radiation source: fine-focus sealed tube | 5859 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.036 |
| ω and φ scans | θmax = 28.4°, θmin = 2.1° |
| Absorption correction: multi-scan (SADABS; Bruker, 2004) | h = −13→13 |
| Tmin = 0.971, Tmax = 0.976 | k = −13→13 |
| 18648 measured reflections | l = −20→20 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.072 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.167 | w = 1/[σ2(Fo2) + (0.0716P)2 + 0.2262P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.10 | (Δ/σ)max < 0.001 |
| 7483 reflections | Δρmax = 0.25 e Å−3 |
| 452 parameters | Δρmin = −0.21 e Å−3 |
| 14 restraints | Absolute structure: Flack (1983), 3448 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.1 (14) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.1564 (4) | 1.2584 (4) | 0.5422 (3) | 0.0774 (13) | |
| H1A | 0.1669 | 1.3231 | 0.5874 | 0.116* | |
| H1B | 0.0660 | 1.2607 | 0.5074 | 0.116* | |
| H1C | 0.2185 | 1.2750 | 0.5051 | 0.116* | |
| C2 | 0.1744 (3) | 1.0292 (4) | 0.5285 (2) | 0.0536 (8) | |
| C3 | 0.2238 (4) | 0.9149 (4) | 0.5704 (2) | 0.0566 (9) | |
| H3 | 0.2572 | 0.9129 | 0.6310 | 0.068* | |
| C4 | 0.2224 (3) | 0.8064 (4) | 0.5214 (2) | 0.0515 (8) | |
| H4 | 0.2571 | 0.7303 | 0.5492 | 0.062* | |
| C5 | 0.1703 (3) | 0.8047 (3) | 0.4296 (2) | 0.0444 (7) | |
| C6 | 0.1152 (4) | 0.9209 (3) | 0.3871 (2) | 0.0487 (8) | |
| C7 | 0.1196 (4) | 1.0328 (4) | 0.4389 (2) | 0.0559 (9) | |
| H7 | 0.0853 | 1.1101 | 0.4127 | 0.067* | |
| C8 | 0.1721 (3) | 0.6905 (3) | 0.3831 (2) | 0.0471 (7) | |
| H8 | 0.2109 | 0.6185 | 0.4146 | 0.057* | |
| C9 | 0.1152 (3) | 0.5543 (3) | 0.2513 (2) | 0.0462 (7) | |
| H9 | 0.2055 | 0.5167 | 0.2598 | 0.055* | |
| C10 | 0.0613 (4) | 0.5805 (3) | 0.1526 (2) | 0.0499 (8) | |
| C11 | 0.0248 (4) | 0.4611 (3) | 0.2890 (2) | 0.0521 (8) | |
| H11A | 0.0645 | 0.4484 | 0.3510 | 0.063* | |
| H11B | −0.0618 | 0.5025 | 0.2849 | 0.063* | |
| C12 | 0.0006 (4) | 0.3291 (3) | 0.2457 (2) | 0.0496 (8) | |
| H12A | −0.0534 | 0.2779 | 0.2773 | 0.059* | |
| H12B | −0.0510 | 0.3401 | 0.1859 | 0.059* | |
| C13 | 0.1279 (4) | 0.2552 (3) | 0.2433 (2) | 0.0524 (8) | |
| H13A | 0.1827 | 0.3057 | 0.2120 | 0.063* | |
| H13B | 0.1792 | 0.2420 | 0.3030 | 0.063* | |
| C14 | 0.0664 (3) | 0.1157 (3) | 0.1148 (2) | 0.0474 (7) | |
| C15 | 0.6713 (5) | −0.0130 (4) | 0.0132 (3) | 0.0757 (12) | |
| H15A | 0.6971 | −0.0757 | −0.0255 | 0.114* | |
| H15B | 0.5757 | −0.0169 | 0.0082 | 0.114* | |
| H15C | 0.7165 | −0.0313 | 0.0728 | 0.114* | |
| C16 | 0.6801 (4) | 0.2145 (4) | 0.0364 (2) | 0.0574 (9) | |
| C17 | 0.7369 (4) | 0.3301 (4) | 0.0165 (2) | 0.0575 (9) | |
| H17 | 0.7866 | 0.3324 | −0.0270 | 0.069* | |
| C18 | 0.7196 (4) | 0.4378 (4) | 0.0602 (2) | 0.0551 (8) | |
| H18 | 0.7587 | 0.5146 | 0.0472 | 0.066* | |
| C19 | 0.6423 (3) | 0.4371 (3) | 0.1264 (2) | 0.0494 (8) | |
| C20 | 0.5796 (4) | 0.3217 (3) | 0.1448 (2) | 0.0573 (9) | |
| C21 | 0.6024 (4) | 0.2094 (4) | 0.0976 (2) | 0.0631 (10) | |
| H21 | 0.5639 | 0.1313 | 0.1086 | 0.076* | |
| C22 | 0.6243 (3) | 0.5520 (3) | 0.1693 (2) | 0.0480 (7) | |
| H22 | 0.6677 | 0.6254 | 0.1550 | 0.058* | |
| C23 | 0.5306 (4) | 0.6853 (3) | 0.2695 (2) | 0.0463 (7) | |
| H23 | 0.6141 | 0.7357 | 0.2800 | 0.056* | |
| C24 | 0.4951 (3) | 0.6572 (3) | 0.3579 (2) | 0.0470 (8) | |
| C25 | 0.4207 (4) | 0.7613 (3) | 0.2061 (2) | 0.0532 (8) | |
| H25A | 0.4490 | 0.7738 | 0.1510 | 0.064* | |
| H25B | 0.3392 | 0.7097 | 0.1935 | 0.064* | |
| C26 | 0.3882 (4) | 0.8927 (3) | 0.2403 (2) | 0.0550 (9) | |
| H26A | 0.3201 | 0.9348 | 0.1956 | 0.066* | |
| H26B | 0.3509 | 0.8803 | 0.2920 | 0.066* | |
| C27 | 0.5096 (4) | 0.9796 (3) | 0.2639 (2) | 0.0582 (9) | |
| H27A | 0.5780 | 0.9366 | 0.3078 | 0.070* | |
| H27B | 0.5460 | 0.9925 | 0.2119 | 0.070* | |
| C28 | 0.4773 (3) | 1.1249 (3) | 0.3797 (2) | 0.0456 (7) | |
| N1 | 0.1241 (3) | 0.6759 (3) | 0.29926 (18) | 0.0476 (6) | |
| N2 | 0.0979 (3) | 0.1312 (3) | 0.20033 (17) | 0.0545 (7) | |
| H2 | 0.1009 | 0.0637 | 0.2330 | 0.065* | |
| N3 | 0.0323 (4) | −0.0009 (3) | 0.0810 (2) | 0.0585 (8) | |
| N4 | 0.0709 (3) | 0.2127 (3) | 0.06010 (18) | 0.0525 (7) | |
| N5 | 0.5525 (3) | 0.5643 (3) | 0.22704 (17) | 0.0482 (6) | |
| N6 | 0.4817 (4) | 1.1051 (3) | 0.29785 (19) | 0.0597 (8) | |
| H6 | 0.4674 | 1.1695 | 0.2620 | 0.072* | |
| N7 | 0.4545 (3) | 1.2428 (3) | 0.40673 (19) | 0.0524 (7) | |
| N8 | 0.4954 (4) | 1.0294 (3) | 0.43757 (19) | 0.0575 (8) | |
| O1 | 0.1831 (3) | 1.1333 (3) | 0.58193 (18) | 0.0720 (8) | |
| O2 | 0.0628 (3) | 0.9232 (2) | 0.30397 (14) | 0.0621 (7) | |
| O3 | 0.0088 (3) | 0.6877 (3) | 0.13073 (16) | 0.0692 (8) | |
| O4 | 0.0710 (3) | 0.4893 (2) | 0.10247 (15) | 0.0584 (6) | |
| O5 | 0.7073 (3) | 0.1124 (3) | −0.01041 (17) | 0.0733 (8) | |
| O6 | 0.5054 (3) | 0.3186 (3) | 0.20185 (19) | 0.0779 (9) | |
| O7 | 0.5093 (3) | 0.7516 (3) | 0.40963 (15) | 0.0587 (6) | |
| O8 | 0.4554 (3) | 0.5477 (2) | 0.37124 (16) | 0.0586 (6) | |
| O1W | 0.8113 (4) | 0.8224 (5) | 0.2043 (3) | 0.1180 (13) | |
| H1W | 0.834 (6) | 0.892 (5) | 0.244 (4) | 0.177* | |
| H2W | 0.889 (4) | 0.783 (6) | 0.194 (5) | 0.177* | |
| H4A | 0.081 (3) | 0.2932 (15) | 0.082 (2) | 0.044 (9)* | |
| H7A | 0.459 (4) | 1.306 (3) | 0.3681 (19) | 0.058 (11)* | |
| H4B | 0.056 (4) | 0.205 (4) | 0.0015 (7) | 0.060 (11)* | |
| H7B | 0.461 (3) | 1.251 (3) | 0.4649 (8) | 0.047 (9)* | |
| H8A | 0.492 (4) | 0.9466 (15) | 0.420 (2) | 0.055 (10)* | |
| H8B | 0.492 (4) | 1.045 (4) | 0.4935 (10) | 0.056 (10)* | |
| H3A | 0.021 (4) | −0.064 (3) | 0.118 (2) | 0.065 (12)* | |
| H3B | 0.003 (3) | −0.009 (4) | 0.0228 (8) | 0.054 (10)* | |
| H5 | 0.513 (3) | 0.493 (2) | 0.241 (2) | 0.053 (10)* | |
| H1 | 0.083 (3) | 0.744 (2) | 0.271 (2) | 0.049 (9)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.063 (2) | 0.064 (3) | 0.102 (3) | 0.002 (2) | 0.011 (2) | −0.035 (3) |
| C2 | 0.0496 (19) | 0.061 (2) | 0.0531 (19) | −0.0094 (17) | 0.0164 (15) | −0.0168 (17) |
| C3 | 0.055 (2) | 0.069 (2) | 0.0425 (17) | −0.0028 (18) | 0.0048 (15) | −0.0087 (18) |
| C4 | 0.0464 (18) | 0.055 (2) | 0.0472 (18) | −0.0038 (15) | −0.0021 (14) | −0.0025 (16) |
| C5 | 0.0464 (17) | 0.0452 (17) | 0.0428 (16) | −0.0023 (14) | 0.0126 (14) | −0.0029 (14) |
| C6 | 0.0536 (19) | 0.0472 (19) | 0.0475 (18) | −0.0096 (15) | 0.0158 (15) | −0.0057 (15) |
| C7 | 0.065 (2) | 0.0477 (19) | 0.056 (2) | −0.0052 (17) | 0.0158 (17) | −0.0047 (16) |
| C8 | 0.0481 (18) | 0.0470 (17) | 0.0459 (17) | −0.0023 (15) | 0.0097 (14) | 0.0040 (15) |
| C9 | 0.0590 (19) | 0.0395 (17) | 0.0390 (16) | −0.0023 (15) | 0.0085 (14) | −0.0018 (13) |
| C10 | 0.062 (2) | 0.048 (2) | 0.0410 (17) | −0.0091 (16) | 0.0149 (15) | −0.0017 (15) |
| C11 | 0.075 (2) | 0.0420 (18) | 0.0416 (17) | −0.0030 (16) | 0.0188 (16) | 0.0003 (14) |
| C12 | 0.067 (2) | 0.0421 (17) | 0.0416 (16) | −0.0115 (16) | 0.0158 (15) | −0.0018 (14) |
| C13 | 0.072 (2) | 0.0427 (17) | 0.0383 (16) | −0.0027 (17) | 0.0039 (15) | −0.0006 (14) |
| C14 | 0.0542 (19) | 0.0400 (17) | 0.0479 (18) | 0.0086 (15) | 0.0110 (15) | −0.0007 (15) |
| C15 | 0.092 (3) | 0.056 (2) | 0.077 (3) | 0.007 (2) | 0.016 (2) | −0.017 (2) |
| C16 | 0.074 (2) | 0.057 (2) | 0.0395 (17) | 0.0196 (18) | 0.0091 (16) | 0.0011 (15) |
| C17 | 0.062 (2) | 0.070 (2) | 0.0436 (18) | 0.0046 (19) | 0.0186 (16) | −0.0002 (17) |
| C18 | 0.058 (2) | 0.058 (2) | 0.0509 (19) | −0.0003 (17) | 0.0150 (16) | 0.0013 (17) |
| C19 | 0.0538 (19) | 0.0484 (19) | 0.0457 (17) | 0.0080 (15) | 0.0103 (14) | 0.0053 (15) |
| C20 | 0.085 (3) | 0.0418 (18) | 0.0497 (19) | 0.0124 (18) | 0.0257 (18) | 0.0095 (16) |
| C21 | 0.098 (3) | 0.0403 (18) | 0.057 (2) | 0.0063 (19) | 0.030 (2) | 0.0084 (16) |
| C22 | 0.0561 (19) | 0.0436 (17) | 0.0435 (16) | 0.0008 (15) | 0.0090 (14) | 0.0040 (14) |
| C23 | 0.062 (2) | 0.0387 (16) | 0.0384 (16) | 0.0033 (15) | 0.0125 (14) | 0.0014 (13) |
| C24 | 0.0542 (19) | 0.0458 (19) | 0.0395 (17) | 0.0143 (15) | 0.0068 (14) | 0.0091 (14) |
| C25 | 0.071 (2) | 0.0432 (17) | 0.0397 (17) | 0.0071 (17) | 0.0006 (15) | 0.0000 (14) |
| C26 | 0.077 (2) | 0.0447 (18) | 0.0380 (17) | 0.0116 (17) | 0.0017 (16) | 0.0011 (14) |
| C27 | 0.091 (3) | 0.0456 (18) | 0.0443 (18) | 0.0001 (19) | 0.0285 (18) | 0.0043 (15) |
| C28 | 0.0564 (19) | 0.0407 (17) | 0.0414 (17) | −0.0062 (15) | 0.0144 (14) | 0.0009 (14) |
| N1 | 0.0623 (18) | 0.0359 (14) | 0.0433 (15) | 0.0001 (13) | 0.0085 (13) | 0.0033 (12) |
| N2 | 0.086 (2) | 0.0386 (14) | 0.0359 (14) | 0.0072 (14) | 0.0062 (14) | 0.0056 (12) |
| N3 | 0.090 (2) | 0.0406 (16) | 0.0440 (17) | −0.0011 (15) | 0.0131 (16) | −0.0001 (14) |
| N4 | 0.082 (2) | 0.0396 (15) | 0.0363 (14) | −0.0004 (14) | 0.0131 (14) | 0.0004 (12) |
| N5 | 0.0685 (18) | 0.0377 (14) | 0.0424 (14) | 0.0028 (13) | 0.0205 (13) | 0.0037 (11) |
| N6 | 0.104 (2) | 0.0340 (14) | 0.0457 (16) | 0.0022 (15) | 0.0258 (16) | 0.0065 (12) |
| N7 | 0.0753 (19) | 0.0424 (16) | 0.0406 (15) | −0.0044 (14) | 0.0153 (14) | −0.0013 (13) |
| N8 | 0.093 (2) | 0.0435 (17) | 0.0388 (15) | −0.0007 (16) | 0.0215 (15) | 0.0025 (13) |
| O1 | 0.083 (2) | 0.0680 (18) | 0.0646 (16) | −0.0093 (15) | 0.0160 (14) | −0.0276 (14) |
| O2 | 0.0966 (19) | 0.0432 (13) | 0.0405 (13) | −0.0050 (13) | 0.0020 (12) | 0.0025 (11) |
| O3 | 0.108 (2) | 0.0544 (16) | 0.0412 (13) | 0.0161 (15) | 0.0071 (13) | 0.0039 (12) |
| O4 | 0.0903 (19) | 0.0466 (13) | 0.0413 (12) | −0.0059 (13) | 0.0210 (12) | −0.0030 (11) |
| O5 | 0.107 (2) | 0.0646 (17) | 0.0525 (15) | 0.0202 (16) | 0.0257 (15) | −0.0042 (13) |
| O6 | 0.133 (3) | 0.0394 (13) | 0.0811 (19) | 0.0060 (16) | 0.0668 (19) | 0.0087 (14) |
| O7 | 0.0839 (18) | 0.0523 (14) | 0.0415 (12) | 0.0089 (13) | 0.0170 (12) | 0.0003 (11) |
| O8 | 0.0798 (17) | 0.0455 (14) | 0.0527 (14) | 0.0036 (13) | 0.0189 (12) | 0.0084 (11) |
| O1W | 0.097 (3) | 0.116 (3) | 0.136 (4) | 0.000 (2) | 0.015 (2) | 0.010 (3) |
Geometric parameters (Å, º)
| C1—O1 | 1.434 (5) | C17—C18 | 1.336 (5) |
| C1—H1A | 0.9600 | C17—H17 | 0.9300 |
| C1—H1B | 0.9600 | C18—C19 | 1.428 (5) |
| C1—H1C | 0.9600 | C18—H18 | 0.9300 |
| C2—O1 | 1.351 (4) | C19—C22 | 1.394 (5) |
| C2—C7 | 1.383 (5) | C19—C20 | 1.412 (5) |
| C2—C3 | 1.390 (5) | C20—O6 | 1.286 (4) |
| C3—C4 | 1.355 (5) | C20—C21 | 1.420 (5) |
| C3—H3 | 0.9300 | C21—H21 | 0.9300 |
| C4—C5 | 1.409 (4) | C22—N5 | 1.282 (4) |
| C4—H4 | 0.9300 | C22—H22 | 0.9300 |
| C5—C8 | 1.388 (5) | C23—N5 | 1.454 (4) |
| C5—C6 | 1.426 (5) | C23—C24 | 1.525 (4) |
| C6—O2 | 1.286 (4) | C23—C25 | 1.535 (5) |
| C6—C7 | 1.405 (5) | C23—H23 | 0.9800 |
| C7—H7 | 0.9300 | C24—O8 | 1.235 (4) |
| C8—N1 | 1.297 (4) | C24—O7 | 1.253 (4) |
| C8—H8 | 0.9300 | C25—C26 | 1.523 (5) |
| C9—N1 | 1.455 (4) | C25—H25A | 0.9700 |
| C9—C11 | 1.535 (5) | C25—H25B | 0.9700 |
| C9—C10 | 1.537 (4) | C26—C27 | 1.507 (6) |
| C9—H9 | 0.9800 | C26—H26A | 0.9700 |
| C10—O4 | 1.241 (4) | C26—H26B | 0.9700 |
| C10—O3 | 1.245 (4) | C27—N6 | 1.452 (4) |
| C11—C12 | 1.518 (5) | C27—H27A | 0.9700 |
| C11—H11A | 0.9700 | C27—H27B | 0.9700 |
| C11—H11B | 0.9700 | C28—N6 | 1.300 (4) |
| C12—C13 | 1.512 (5) | C28—N8 | 1.321 (4) |
| C12—H12A | 0.9700 | C28—N7 | 1.326 (4) |
| C12—H12B | 0.9700 | N1—H1 | 0.891 (10) |
| C13—N2 | 1.448 (4) | N2—H2 | 0.8600 |
| C13—H13A | 0.9700 | N3—H3A | 0.894 (10) |
| C13—H13B | 0.9700 | N3—H3B | 0.892 (10) |
| C14—N2 | 1.309 (4) | N4—H4A | 0.898 (10) |
| C14—N4 | 1.323 (4) | N4—H4B | 0.894 (10) |
| C14—N3 | 1.330 (4) | N5—H5 | 0.896 (10) |
| C15—O5 | 1.418 (5) | N6—H6 | 0.8600 |
| C15—H15A | 0.9600 | N7—H7A | 0.893 (10) |
| C15—H15B | 0.9600 | N7—H7B | 0.896 (10) |
| C15—H15C | 0.9600 | N8—H8A | 0.898 (10) |
| C16—O5 | 1.346 (4) | N8—H8B | 0.893 (10) |
| C16—C21 | 1.366 (5) | O1W—H1W | 0.940 (10) |
| C16—C17 | 1.393 (6) | O1W—H2W | 0.937 (10) |
| O1—C1—H1A | 109.5 | C19—C18—H18 | 119.4 |
| O1—C1—H1B | 109.5 | C22—C19—C20 | 120.7 (3) |
| H1A—C1—H1B | 109.5 | C22—C19—C18 | 119.5 (3) |
| O1—C1—H1C | 109.5 | C20—C19—C18 | 119.7 (3) |
| H1A—C1—H1C | 109.5 | O6—C20—C19 | 121.4 (3) |
| H1B—C1—H1C | 109.5 | O6—C20—C21 | 121.5 (3) |
| O1—C2—C7 | 123.8 (4) | C19—C20—C21 | 117.1 (3) |
| O1—C2—C3 | 114.9 (3) | C16—C21—C20 | 120.9 (4) |
| C7—C2—C3 | 121.3 (3) | C16—C21—H21 | 119.5 |
| C4—C3—C2 | 118.8 (3) | C20—C21—H21 | 119.5 |
| C4—C3—H3 | 120.6 | N5—C22—C19 | 125.1 (3) |
| C2—C3—H3 | 120.6 | N5—C22—H22 | 117.5 |
| C3—C4—C5 | 122.4 (3) | C19—C22—H22 | 117.5 |
| C3—C4—H4 | 118.8 | N5—C23—C24 | 109.6 (3) |
| C5—C4—H4 | 118.8 | N5—C23—C25 | 108.2 (3) |
| C8—C5—C4 | 119.6 (3) | C24—C23—C25 | 113.1 (3) |
| C8—C5—C6 | 121.6 (3) | N5—C23—H23 | 108.6 |
| C4—C5—C6 | 118.8 (3) | C24—C23—H23 | 108.6 |
| O2—C6—C7 | 121.0 (3) | C25—C23—H23 | 108.6 |
| O2—C6—C5 | 121.1 (3) | O8—C24—O7 | 127.0 (3) |
| C7—C6—C5 | 117.9 (3) | O8—C24—C23 | 118.6 (3) |
| C2—C7—C6 | 120.8 (4) | O7—C24—C23 | 114.5 (3) |
| C2—C7—H7 | 119.6 | C26—C25—C23 | 114.6 (3) |
| C6—C7—H7 | 119.6 | C26—C25—H25A | 108.6 |
| N1—C8—C5 | 125.3 (3) | C23—C25—H25A | 108.6 |
| N1—C8—H8 | 117.3 | C26—C25—H25B | 108.6 |
| C5—C8—H8 | 117.3 | C23—C25—H25B | 108.6 |
| N1—C9—C11 | 108.8 (3) | H25A—C25—H25B | 107.6 |
| N1—C9—C10 | 109.1 (3) | C27—C26—C25 | 112.8 (3) |
| C11—C9—C10 | 112.3 (3) | C27—C26—H26A | 109.0 |
| N1—C9—H9 | 108.8 | C25—C26—H26A | 109.0 |
| C11—C9—H9 | 108.8 | C27—C26—H26B | 109.0 |
| C10—C9—H9 | 108.8 | C25—C26—H26B | 109.0 |
| O4—C10—O3 | 126.4 (3) | H26A—C26—H26B | 107.8 |
| O4—C10—C9 | 115.6 (3) | N6—C27—C26 | 114.0 (3) |
| O3—C10—C9 | 117.9 (3) | N6—C27—H27A | 108.8 |
| C12—C11—C9 | 116.7 (3) | C26—C27—H27A | 108.8 |
| C12—C11—H11A | 108.1 | N6—C27—H27B | 108.8 |
| C9—C11—H11A | 108.1 | C26—C27—H27B | 108.8 |
| C12—C11—H11B | 108.1 | H27A—C27—H27B | 107.7 |
| C9—C11—H11B | 108.1 | N6—C28—N8 | 121.3 (3) |
| H11A—C11—H11B | 107.3 | N6—C28—N7 | 120.1 (3) |
| C13—C12—C11 | 114.1 (3) | N8—C28—N7 | 118.6 (3) |
| C13—C12—H12A | 108.7 | C8—N1—C9 | 125.7 (3) |
| C11—C12—H12A | 108.7 | C8—N1—H1 | 116 (2) |
| C13—C12—H12B | 108.7 | C9—N1—H1 | 118 (2) |
| C11—C12—H12B | 108.7 | C14—N2—C13 | 123.9 (3) |
| H12A—C12—H12B | 107.6 | C14—N2—H2 | 118.0 |
| N2—C13—C12 | 111.3 (3) | C13—N2—H2 | 118.0 |
| N2—C13—H13A | 109.4 | C14—N3—H3A | 118 (3) |
| C12—C13—H13A | 109.4 | C14—N3—H3B | 119 (2) |
| N2—C13—H13B | 109.4 | H3A—N3—H3B | 122 (4) |
| C12—C13—H13B | 109.4 | C14—N4—H4A | 118 (2) |
| H13A—C13—H13B | 108.0 | C14—N4—H4B | 125 (3) |
| N2—C14—N4 | 121.7 (3) | H4A—N4—H4B | 117 (3) |
| N2—C14—N3 | 119.8 (3) | C22—N5—C23 | 124.8 (3) |
| N4—C14—N3 | 118.4 (3) | C22—N5—H5 | 117 (2) |
| O5—C15—H15A | 109.5 | C23—N5—H5 | 118 (2) |
| O5—C15—H15B | 109.5 | C28—N6—C27 | 123.2 (3) |
| H15A—C15—H15B | 109.5 | C28—N6—H6 | 118.4 |
| O5—C15—H15C | 109.5 | C27—N6—H6 | 118.4 |
| H15A—C15—H15C | 109.5 | C28—N7—H7A | 115 (2) |
| H15B—C15—H15C | 109.5 | C28—N7—H7B | 115 (2) |
| O5—C16—C21 | 124.5 (4) | H7A—N7—H7B | 127 (3) |
| O5—C16—C17 | 114.1 (3) | C28—N8—H8A | 121 (2) |
| C21—C16—C17 | 121.4 (3) | C28—N8—H8B | 120 (2) |
| C18—C17—C16 | 119.6 (3) | H8A—N8—H8B | 118 (3) |
| C18—C17—H17 | 120.2 | C2—O1—C1 | 118.2 (3) |
| C16—C17—H17 | 120.2 | C16—O5—C15 | 118.8 (3) |
| C17—C18—C19 | 121.2 (3) | H1W—O1W—H2W | 110.4 (17) |
| C17—C18—H18 | 119.4 | ||
| O1—C2—C3—C4 | −178.2 (3) | C18—C19—C20—C21 | 2.6 (5) |
| C7—C2—C3—C4 | 2.5 (5) | O5—C16—C21—C20 | 179.4 (4) |
| C2—C3—C4—C5 | −1.2 (5) | C17—C16—C21—C20 | −1.6 (6) |
| C3—C4—C5—C8 | 179.9 (3) | O6—C20—C21—C16 | 179.2 (4) |
| C3—C4—C5—C6 | −1.0 (5) | C19—C20—C21—C16 | −1.0 (6) |
| C8—C5—C6—O2 | 1.2 (5) | C20—C19—C22—N5 | 0.0 (5) |
| C4—C5—C6—O2 | −177.9 (3) | C18—C19—C22—N5 | 176.9 (3) |
| C8—C5—C6—C7 | −178.9 (3) | N5—C23—C24—O8 | −18.7 (4) |
| C4—C5—C6—C7 | 2.0 (4) | C25—C23—C24—O8 | 102.2 (4) |
| O1—C2—C7—C6 | 179.3 (3) | N5—C23—C24—O7 | 161.9 (3) |
| C3—C2—C7—C6 | −1.5 (5) | C25—C23—C24—O7 | −77.3 (4) |
| O2—C6—C7—C2 | 179.1 (3) | N5—C23—C25—C26 | −178.7 (3) |
| C5—C6—C7—C2 | −0.8 (5) | C24—C23—C25—C26 | 59.7 (4) |
| C4—C5—C8—N1 | 178.1 (3) | C23—C25—C26—C27 | 57.0 (4) |
| C6—C5—C8—N1 | −1.1 (5) | C25—C26—C27—N6 | −179.2 (3) |
| N1—C9—C10—O4 | 167.8 (3) | C5—C8—N1—C9 | −174.4 (3) |
| C11—C9—C10—O4 | −71.5 (4) | C11—C9—N1—C8 | 60.9 (4) |
| N1—C9—C10—O3 | −14.6 (4) | C10—C9—N1—C8 | −176.2 (3) |
| C11—C9—C10—O3 | 106.2 (4) | N4—C14—N2—C13 | 6.0 (5) |
| N1—C9—C11—C12 | 178.7 (3) | N3—C14—N2—C13 | −175.9 (3) |
| C10—C9—C11—C12 | 57.7 (4) | C12—C13—N2—C14 | 79.9 (4) |
| C9—C11—C12—C13 | 55.3 (4) | C19—C22—N5—C23 | −177.9 (3) |
| C11—C12—C13—N2 | −179.3 (3) | C24—C23—N5—C22 | −155.8 (3) |
| O5—C16—C17—C18 | −178.4 (3) | C25—C23—N5—C22 | 80.4 (4) |
| C21—C16—C17—C18 | 2.5 (6) | N8—C28—N6—C27 | −1.6 (6) |
| C16—C17—C18—C19 | −0.8 (6) | N7—C28—N6—C27 | 178.4 (3) |
| C17—C18—C19—C22 | −178.7 (3) | C26—C27—N6—C28 | 83.0 (5) |
| C17—C18—C19—C20 | −1.8 (5) | C7—C2—O1—C1 | −11.4 (5) |
| C22—C19—C20—O6 | −0.7 (5) | C3—C2—O1—C1 | 169.3 (3) |
| C18—C19—C20—O6 | −177.6 (4) | C21—C16—O5—C15 | −9.9 (6) |
| C22—C19—C20—C21 | 179.5 (3) | C17—C16—O5—C15 | 171.0 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O2 | 0.89 (1) | 1.94 (3) | 2.638 (4) | 134 (3) |
| N4—H4A···O4 | 0.90 (1) | 2.06 (1) | 2.935 (4) | 165 (3) |
| N5—H5···O6 | 0.90 (1) | 1.90 (3) | 2.600 (4) | 134 (3) |
| N8—H8A···O7 | 0.90 (1) | 2.03 (1) | 2.914 (4) | 166 (3) |
| N2—H2···O2i | 0.86 | 1.92 | 2.758 (4) | 166 |
| N3—H3A···O3i | 0.89 (1) | 2.58 (3) | 3.333 (4) | 142 (3) |
| N3—H3B···O4ii | 0.89 (1) | 1.93 (1) | 2.817 (4) | 175 (4) |
| N4—H4B···O3ii | 0.89 (1) | 2.03 (1) | 2.912 (4) | 171 (3) |
| N6—H6···O6iii | 0.86 | 1.89 | 2.705 (4) | 158 |
| N7—H7A···O8iii | 0.89 (1) | 2.50 (3) | 3.202 (4) | 135 (3) |
| N7—H7B···O7iv | 0.90 (1) | 1.91 (1) | 2.800 (4) | 173 (3) |
| N8—H8B···O8iv | 0.89 (1) | 2.05 (2) | 2.911 (4) | 161 (3) |
| O1W—H1W···O2v | 0.94 (1) | 2.33 (6) | 2.882 (5) | 117 (4) |
| O1W—H2W···O3v | 0.94 (1) | 1.98 (2) | 2.881 (5) | 159 (6) |
| C15—H15C···O1Wi | 0.96 | 2.56 | 3.451 (7) | 155 |
| C22—H22···O1W | 0.93 | 2.53 | 3.359 (6) | 149 |
| C1—H1C···Cg1vi | 0.96 | 2.96 | 3.669 (4) | 132 |
| C15—H15C···Cg2vii | 0.96 | 2.98 | 3.762 (5) | 139 |
Symmetry codes: (i) x, y−1, z; (ii) −x, y−1/2, −z; (iii) x, y+1, z; (iv) −x+1, y+1/2, −z+1; (v) x+1, y, z; (vi) −x, y−1/2, −z+1; (vii) −x+1, y+1/2, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: ZS2267).
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruker (2004). APEX2, SAINT, XPREP and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Konig, A. von, Moll, F. & Rosenhahn, L. (1982). US Patent 4358531.
- Lewis, J., Matteucci, M., Chen, T. & Jiao, H. (2009). WO Patent 2009140553 A2.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Moutet, J. C. & Ourari, A. (1997). Electrochim. Acta, 42, 2525–2531.
- Oueslati, A., Kefi, R., Ben Nasr, C. & Lefebvre, F. (2007). J. Mol. Struct. 871, 49–58.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Srinivasan, K., Michaud, P. & Kochi, J. K. (1986). J. Am. Chem. Soc. 108, 2309–2320. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813019727/zs2267sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813019727/zs2267Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report