Abstract
In the title compound, C11H15N3O2S, the dihedral angle between the mean planes of the benzene ring and hydrazinecarbothioamide group is 9.2 (1)°. An intramolecular O—H⋯N hydrogen bond is observed, serving to maintain an approximately planar conformation for the molecule. In the crystal, inversion dimers linked by C—H⋯O interactions occur. Further C—H⋯O contacts link dimers into (010) chains.
Related literature
For the synthesis and structure of thiosemicarbazones as ligands, see: Lobana et al. (2009 ▶, 2012 ▶). For palladium complexes with thiosemicarbazone ligands, see: Chellan et al. (2010 ▶). For related structures, see: Anderson et al. (2012 ▶, 2013 ▶). For bond lengths, see: Allen et al. (1987 ▶).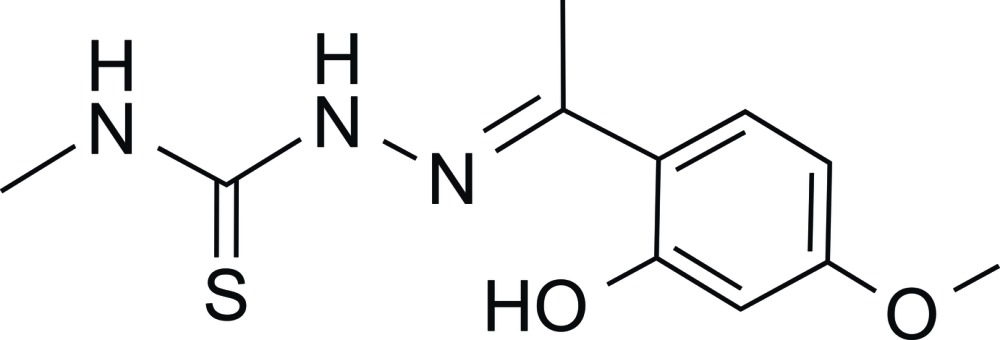
Experimental
Crystal data
C11H15N3O2S
M r = 253.32
Monoclinic,
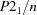
a = 10.9881 (8) Å
b = 9.1468 (6) Å
c = 12.5575 (9) Å
β = 109.400 (8)°
V = 1190.45 (15) Å3
Z = 4
Mo Kα radiation
μ = 0.27 mm−1
T = 173 K
0.42 × 0.38 × 0.14 mm
Data collection
Agilent Xcalibur (Eos, Gemini) diffractometer
Absorption correction: multi-scan (CrysAlis PRO and CrysAlis RED; Agilent, 2012 ▶) T min = 0.728, T max = 1.000
13894 measured reflections
4104 independent reflections
3320 reflections with I > 2σ(I)
R int = 0.043
Refinement
R[F 2 > 2σ(F 2)] = 0.076
wR(F 2) = 0.231
S = 1.16
4104 reflections
158 parameters
H-atom parameters constrained
Δρmax = 1.21 e Å−3
Δρmin = −0.46 e Å−3
Data collection: CrysAlis PRO (Agilent, 2012 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis RED (Agilent, 2012 ▶); program(s) used to solve structure: SUPERFLIP (Palatinus & Chapuis, 2007 ▶); program(s) used to refine structure: SHELXL2012 (Sheldrick, 2008 ▶); molecular graphics: OLEX2 (Dolomanov et al., 2009 ▶); software used to prepare material for publication: OLEX2.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813019831/hg5331sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813019831/hg5331Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813019831/hg5331Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯N1 | 0.82 | 1.85 | 2.566 (3) | 145 |
| C10—H10A⋯O2i | 0.96 | 2.59 | 3.301 (4) | 132 |
| C10—H10C⋯O1ii | 0.96 | 2.57 | 3.481 (4) | 158 |
Symmetry codes: (i) 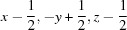 ; (ii)
; (ii) 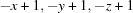 .
.
Acknowledgments
JPJ acknowledges the NSF–MRI program (grant No. CHE-1039027) for funds to purchase the X-ray diffractometer.
supplementary crystallographic information
Comment
Thiosemicarbazones are a versatile class of ligands that can adopt multiple modes of binding to a metal (Lobana, et al., 2009) and the synthesis and structure determination of these metal complexes is an active area of research. (Lobana, et al., 2012) Palladium complexes with thiosemicarbazone ligands have been shown to have a variety of biological activity including anti-fungal and anti-tumor activity. (Chellan, et al., 2010). We have previously reported the structure of two analogous novel thiosemicarbazones (Anderson, et al., 2012; Anderson, et al., 2013). Here, we report the synthesis and crystal structure of a novel thiosemicarbazone ligand, (I), C11H15N3O2S .
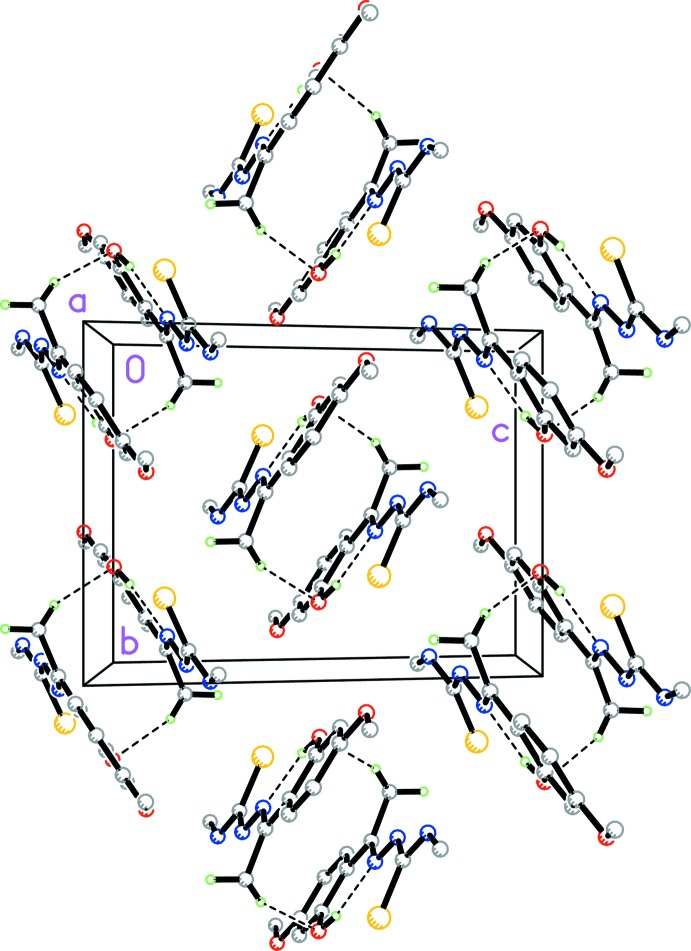
In (I), the dihedral angle between the mean planes of the benzene ring and hydrazinecarbothioamide group (N1/N2/C8/S1/N3) is 9.2 (1)° (Fig. 1). Bond lengths are in normal ranges (Allen et al., 1987). In the crystal, an intramolecular O—H···N hydrogen bond is observed serving to keep the molecule in a nearly planar conformation. Additional weak and C—H···O intermolecular interactions (Table 1) assist in linking the molecules into dimers along (010) and influence crystal packing (Fig. 2).
Experimental
A 50 mL round bottom flask was charged with 0.218 g (1.31 mmol) of 2'-hydroxy-4'-methoxyacetophenone, 0.138 g (1.31 mmol) of 4-methyl-3- thiosemicarbazide, dissolved in 20 mL of methanol. The resulting colorless solution was refluxed for 48 hours and then a drop of concentrated HCl was added and the solution was refluxed for an additional 48 hours. The resulting yellow solution was transferred to a 125 mL separatory funnel. Dichloromethane (10 mL) and water (10 mL) were added, and the organic layer was separated. The aqueous layer was extracted with an additional 10 mL of dichloromethane. The organic layers were combined, washed with brine (2 x 10 mL), dried with magnesium sulfate, and the solvent was removed in vacuo to give a yellow solid (Fig. 3). The solid was dissolved in hot acetonitrile, allowed to cool to room temperature and then stored at 273 K resulting in colorless crystals (58 mg, 18%) after 24 hours. M.p. 448-453 K.
Refinement
All of the H atoms were placed in their calculated positions and then refined using the riding model with atom—H lengths of 0.93Å (CH), 0.96Å (CH3), 0.86Å (NH) or 0.82Å (OH). Isotropic displacement parameters for these atoms were set to 1.2 (CH, NH) or 1.5 (CH3, OH) times Ueq of the parent atom. Idealised Me refined as rotating group: C9(H9A,H9B,H9C), C10(H10A,H10B,H10C), C11(H11A,H11B,H11C). Idealised tetrahedral OH refined as rotating group: O1(H1).
Figures
Fig. 1.
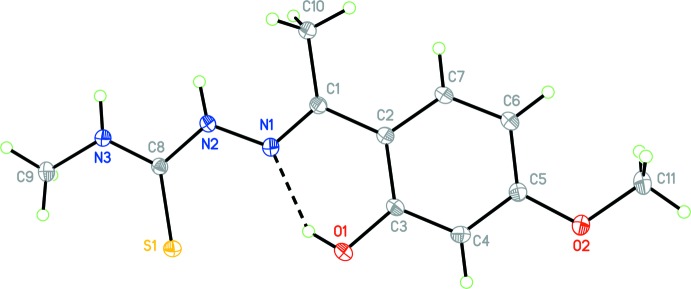
ORTEP drawing of (I) showing the atom labeling scheme and 30% probability displacement ellipsoids. Dashed lines indicate O1—H1···N1 intramolecular hydrogen bonds.
Fig. 2.
Molecular packing for (I) viewed along the b axis. Dashed lines indicate O—H···N intramolecular hydrogen bonds and weak C—H···O intermolecular interactions linking the molecules into dimers along (010).
Fig. 3.
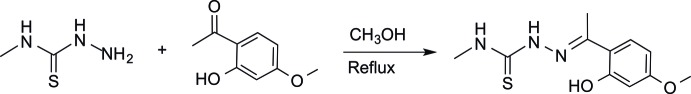
Synthesis of (I).
Crystal data
| C11H15N3O2S | F(000) = 536 |
| Mr = 253.32 | Dx = 1.413 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.7107 Å |
| a = 10.9881 (8) Å | Cell parameters from 4367 reflections |
| b = 9.1468 (6) Å | θ = 3.0–32.9° |
| c = 12.5575 (9) Å | µ = 0.27 mm−1 |
| β = 109.400 (8)° | T = 173 K |
| V = 1190.45 (15) Å3 | Irregular, colourless |
| Z = 4 | 0.42 × 0.38 × 0.14 mm |
Data collection
| Agilent Xcalibur (Eos, Gemini) diffractometer | 4104 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 3320 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.043 |
| Detector resolution: 16.0416 pixels mm-1 | θmax = 33.0°, θmin = 3.0° |
| ω scans | h = −15→16 |
| Absorption correction: multi-scan (CrysAlis PRO and CrysAlis RED; Agilent, 2012) | k = −12→13 |
| Tmin = 0.728, Tmax = 1.000 | l = −18→18 |
| 13894 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.076 | H-atom parameters constrained |
| wR(F2) = 0.231 | w = 1/[σ2(Fo2) + (0.0907P)2 + 2.429P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.16 | (Δ/σ)max < 0.001 |
| 4104 reflections | Δρmax = 1.21 e Å−3 |
| 158 parameters | Δρmin = −0.46 e Å−3 |
| 0 restraints |
Special details
| Experimental. 1H NMR [(CD3)2CO]: 11.7 (br s, 1H, NH) 9.52 (br s, 1H, OH) 7.73 (br s, 1H, NH) 7.5 (d, J = 8.6, 1H Ar) 6.47 (d, J = 8.6 1H Ar) 6.42 (d, 1H Ar) 3.80 (s, 3H, CH3) 3.13 (d, J = 4.7, 3H, CH3) 2.43 (s, 3H, CH3) |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.21259 (6) | 0.28834 (7) | 0.36426 (6) | 0.02286 (19) | |
| O1 | 0.55069 (19) | 0.2003 (2) | 0.50127 (19) | 0.0280 (4) | |
| H1 | 0.4936 | 0.2530 | 0.4608 | 0.042* | |
| O2 | 0.9881 (2) | 0.0943 (3) | 0.60376 (19) | 0.0309 (5) | |
| N1 | 0.4600 (2) | 0.4175 (3) | 0.36942 (19) | 0.0220 (4) | |
| N2 | 0.3540 (2) | 0.5019 (3) | 0.31878 (19) | 0.0220 (4) | |
| H2 | 0.3628 | 0.5869 | 0.2929 | 0.026* | |
| N3 | 0.1393 (2) | 0.5407 (3) | 0.2551 (2) | 0.0240 (4) | |
| H3 | 0.1595 | 0.6225 | 0.2317 | 0.029* | |
| C1 | 0.5729 (2) | 0.4652 (3) | 0.3762 (2) | 0.0198 (4) | |
| C2 | 0.6806 (2) | 0.3678 (3) | 0.4335 (2) | 0.0201 (5) | |
| C3 | 0.6655 (2) | 0.2410 (3) | 0.4935 (2) | 0.0207 (5) | |
| C4 | 0.7702 (3) | 0.1533 (3) | 0.5478 (2) | 0.0245 (5) | |
| H4 | 0.7585 | 0.0703 | 0.5862 | 0.029* | |
| C5 | 0.8923 (3) | 0.1875 (3) | 0.5459 (2) | 0.0232 (5) | |
| C6 | 0.9109 (3) | 0.3099 (3) | 0.4871 (2) | 0.0260 (5) | |
| H6 | 0.9923 | 0.3327 | 0.4844 | 0.031* | |
| C7 | 0.8055 (3) | 0.3967 (3) | 0.4329 (2) | 0.0250 (5) | |
| H7 | 0.8182 | 0.4786 | 0.3939 | 0.030* | |
| C8 | 0.2350 (2) | 0.4514 (3) | 0.3095 (2) | 0.0192 (4) | |
| C9 | 0.0037 (3) | 0.5110 (4) | 0.2322 (3) | 0.0299 (6) | |
| H9A | −0.0070 | 0.4535 | 0.2924 | 0.045* | |
| H9B | −0.0298 | 0.4582 | 0.1624 | 0.045* | |
| H9C | −0.0422 | 0.6016 | 0.2264 | 0.045* | |
| C10 | 0.5946 (3) | 0.6111 (3) | 0.3322 (3) | 0.0255 (5) | |
| H10A | 0.5542 | 0.6133 | 0.2517 | 0.038* | |
| H10B | 0.6856 | 0.6278 | 0.3508 | 0.038* | |
| H10C | 0.5580 | 0.6860 | 0.3658 | 0.038* | |
| C11 | 1.1161 (3) | 0.1303 (4) | 0.6095 (3) | 0.0349 (7) | |
| H11A | 1.1420 | 0.2195 | 0.6510 | 0.052* | |
| H11B | 1.1191 | 0.1425 | 0.5344 | 0.052* | |
| H11C | 1.1736 | 0.0529 | 0.6467 | 0.052* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0232 (3) | 0.0190 (3) | 0.0279 (3) | −0.0010 (2) | 0.0104 (2) | 0.0013 (2) |
| O1 | 0.0214 (9) | 0.0276 (10) | 0.0383 (11) | −0.0008 (8) | 0.0143 (8) | 0.0071 (8) |
| O2 | 0.0216 (9) | 0.0354 (11) | 0.0358 (11) | 0.0056 (8) | 0.0094 (8) | 0.0128 (9) |
| N1 | 0.0192 (10) | 0.0224 (10) | 0.0245 (10) | 0.0012 (8) | 0.0072 (8) | 0.0009 (8) |
| N2 | 0.0172 (9) | 0.0207 (10) | 0.0287 (11) | −0.0001 (8) | 0.0083 (8) | 0.0027 (8) |
| N3 | 0.0197 (10) | 0.0212 (10) | 0.0320 (11) | 0.0019 (8) | 0.0099 (8) | 0.0049 (8) |
| C1 | 0.0193 (11) | 0.0200 (11) | 0.0199 (10) | −0.0012 (8) | 0.0063 (8) | −0.0006 (8) |
| C2 | 0.0193 (11) | 0.0198 (10) | 0.0212 (10) | −0.0014 (9) | 0.0068 (8) | −0.0001 (8) |
| C3 | 0.0203 (11) | 0.0217 (11) | 0.0221 (11) | −0.0022 (9) | 0.0098 (9) | −0.0007 (9) |
| C4 | 0.0262 (12) | 0.0228 (12) | 0.0266 (12) | −0.0002 (10) | 0.0115 (10) | 0.0047 (9) |
| C5 | 0.0209 (11) | 0.0251 (12) | 0.0239 (11) | 0.0009 (9) | 0.0076 (9) | 0.0021 (9) |
| C6 | 0.0197 (11) | 0.0277 (13) | 0.0314 (13) | −0.0024 (10) | 0.0096 (10) | 0.0042 (10) |
| C7 | 0.0205 (11) | 0.0247 (12) | 0.0308 (13) | −0.0024 (9) | 0.0101 (10) | 0.0044 (10) |
| C8 | 0.0200 (11) | 0.0191 (10) | 0.0200 (10) | −0.0002 (8) | 0.0086 (8) | −0.0020 (8) |
| C9 | 0.0203 (12) | 0.0316 (14) | 0.0383 (15) | 0.0048 (10) | 0.0103 (11) | 0.0067 (12) |
| C10 | 0.0200 (11) | 0.0207 (11) | 0.0348 (14) | −0.0013 (9) | 0.0080 (10) | 0.0026 (10) |
| C11 | 0.0204 (12) | 0.0417 (17) | 0.0424 (17) | 0.0044 (12) | 0.0104 (11) | 0.0084 (14) |
Geometric parameters (Å, º)
| S1—C8 | 1.695 (3) | C3—C4 | 1.383 (4) |
| O1—H1 | 0.8200 | C4—H4 | 0.9300 |
| O1—C3 | 1.350 (3) | C4—C5 | 1.386 (4) |
| O2—C5 | 1.362 (3) | C5—C6 | 1.393 (4) |
| O2—C11 | 1.423 (4) | C6—H6 | 0.9300 |
| N1—N2 | 1.366 (3) | C6—C7 | 1.383 (4) |
| N1—C1 | 1.290 (3) | C7—H7 | 0.9300 |
| N2—H2 | 0.8600 | C9—H9A | 0.9600 |
| N2—C8 | 1.354 (3) | C9—H9B | 0.9600 |
| N3—H3 | 0.8600 | C9—H9C | 0.9600 |
| N3—C8 | 1.328 (3) | C10—H10A | 0.9600 |
| N3—C9 | 1.446 (4) | C10—H10B | 0.9600 |
| C1—C2 | 1.465 (4) | C10—H10C | 0.9600 |
| C1—C10 | 1.494 (4) | C11—H11A | 0.9600 |
| C2—C3 | 1.422 (4) | C11—H11B | 0.9600 |
| C2—C7 | 1.400 (4) | C11—H11C | 0.9600 |
| C3—O1—H1 | 109.5 | C7—C6—H6 | 120.8 |
| C5—O2—C11 | 117.3 (2) | C2—C7—H7 | 118.3 |
| C1—N1—N2 | 119.4 (2) | C6—C7—C2 | 123.4 (2) |
| N1—N2—H2 | 120.1 | C6—C7—H7 | 118.3 |
| C8—N2—N1 | 119.8 (2) | N2—C8—S1 | 122.13 (19) |
| C8—N2—H2 | 120.1 | N3—C8—S1 | 123.55 (19) |
| C8—N3—H3 | 117.4 | N3—C8—N2 | 114.3 (2) |
| C8—N3—C9 | 125.1 (2) | N3—C9—H9A | 109.5 |
| C9—N3—H3 | 117.4 | N3—C9—H9B | 109.5 |
| N1—C1—C2 | 115.4 (2) | N3—C9—H9C | 109.5 |
| N1—C1—C10 | 123.1 (2) | H9A—C9—H9B | 109.5 |
| C2—C1—C10 | 121.5 (2) | H9A—C9—H9C | 109.5 |
| C3—C2—C1 | 122.5 (2) | H9B—C9—H9C | 109.5 |
| C7—C2—C1 | 121.1 (2) | C1—C10—H10A | 109.5 |
| C7—C2—C3 | 116.4 (2) | C1—C10—H10B | 109.5 |
| O1—C3—C2 | 122.7 (2) | C1—C10—H10C | 109.5 |
| O1—C3—C4 | 116.6 (2) | H10A—C10—H10B | 109.5 |
| C4—C3—C2 | 120.7 (2) | H10A—C10—H10C | 109.5 |
| C3—C4—H4 | 119.6 | H10B—C10—H10C | 109.5 |
| C3—C4—C5 | 120.8 (2) | O2—C11—H11A | 109.5 |
| C5—C4—H4 | 119.6 | O2—C11—H11B | 109.5 |
| O2—C5—C4 | 115.5 (2) | O2—C11—H11C | 109.5 |
| O2—C5—C6 | 124.2 (2) | H11A—C11—H11B | 109.5 |
| C4—C5—C6 | 120.3 (2) | H11A—C11—H11C | 109.5 |
| C5—C6—H6 | 120.8 | H11B—C11—H11C | 109.5 |
| C7—C6—C5 | 118.5 (2) | ||
| O1—C3—C4—C5 | −179.1 (3) | C3—C2—C7—C6 | −0.4 (4) |
| O2—C5—C6—C7 | −179.2 (3) | C3—C4—C5—O2 | 179.1 (2) |
| N1—N2—C8—S1 | 2.8 (3) | C3—C4—C5—C6 | −1.2 (4) |
| N1—N2—C8—N3 | −177.6 (2) | C4—C5—C6—C7 | 1.0 (4) |
| N1—C1—C2—C3 | −9.8 (4) | C5—C6—C7—C2 | −0.2 (4) |
| N1—C1—C2—C7 | 171.0 (2) | C7—C2—C3—O1 | 179.8 (2) |
| N2—N1—C1—C2 | 179.1 (2) | C7—C2—C3—C4 | 0.3 (4) |
| N2—N1—C1—C10 | 0.7 (4) | C9—N3—C8—S1 | −1.7 (4) |
| C1—N1—N2—C8 | 178.8 (2) | C9—N3—C8—N2 | 178.7 (3) |
| C1—C2—C3—O1 | 0.6 (4) | C10—C1—C2—C3 | 168.6 (2) |
| C1—C2—C3—C4 | −179.0 (2) | C10—C1—C2—C7 | −10.6 (4) |
| C1—C2—C7—C6 | 178.9 (3) | C11—O2—C5—C4 | −176.4 (3) |
| C2—C3—C4—C5 | 0.5 (4) | C11—O2—C5—C6 | 3.8 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N1 | 0.82 | 1.85 | 2.566 (3) | 145 |
| C10—H10A···O2i | 0.96 | 2.59 | 3.301 (4) | 132 |
| C10—H10C···O1ii | 0.96 | 2.57 | 3.481 (4) | 158 |
Symmetry codes: (i) x−1/2, −y+1/2, z−1/2; (ii) −x+1, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG5331).
References
- Agilent (2012). CrysAlis PRO and CrysAlis RED Agilent Technologies, Yarnton, Oxfordshire, England.
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Anderson, B. J., Keeler, A. M., O’Rourke, K. A., Krauss, S. T. & Jasinski, J. P. (2013). Acta Cryst. E69, o11. [DOI] [PMC free article] [PubMed]
- Anderson, B. J., Kennedy, C. J. & Jasinski, J. P. (2012). Acta Cryst. E68, o2982. [DOI] [PMC free article] [PubMed]
- Chellan, P., Shunmoogam-Gounden, N., Hendricks, D. T., Gut, J., Rosenthal, P. J., Lategan, C., Smith, P. J., Chibale, K. & Smith, G. S. (2010). Eur. J. Inorg. Chem. pp. 3520–3528.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Lobana, T. S., Kumari, P., Bawa, G., Hundal, G., Butcher, R. J., Fernandez, F. J., Jasinski, J. P. & Golen, J. A. (2012). Z. Anorg. Allg. Chem. 638, 804–810.
- Lobana, T. S., Sharma, R., Bawa, G. & Khanna, S. (2009). Coord. Chem. Rev. 253, 977–1055.
- Palatinus, L. & Chapuis, G. (2007). J. Appl. Cryst. 40, 786–790.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813019831/hg5331sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813019831/hg5331Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813019831/hg5331Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report