Abstract
The Cystic Fibrosis Transmembrane Conductance Regulator protein (CFTR) is a member of the ABC transporter superfamily. CFTR is distinguished from all other members of this superfamily by its status as an ion channel as well as the presence of its unique regulatory (R) domain. We investigated the origin and subsequent evolution of the R domain along the CFTR evolutionary lineage. The R domain protein coding sequence originated via the loss of a splice donor site at the 3′ end of exon 14, leading to the subsequent read-through and capture of formerly intronic sequence as novel coding sequence. Inclusion of the remaining part of the R domain coding sequence in the CFTR transcript involved a lineage-specific gain of exonic sequence with no homology to protein coding sequences outside of CFTR and loss of two exons conserved among ABC family members. These events occurred at the base of the Gnathostome evolutionary lineage ~550–650 million years ago. The apparent origination of the R domain de novo from previously non-coding sequence is consistent with its lack of sequence similarity to other domains as well as its intrinsically disordered structure, which has important implications for its function. In particular, this lack of structure may provide for a dynamic and inducible regulatory activity based on transient physical interactions with more structured domains of the protein. Since its acquisition along the CFTR evolutionary lineage, the R domain has evolved more rapidly than any other CFTR domain; however, there is no evidence for positive (adaptive) selection in the evolution of the domain. The R domain does show a distinct pattern of relative evolutionary rates compared to other CFTR domains, which sheds additional light on the connection between its function and evolution. The regulatory function of the R domain is dependent upon a fairly small number of sites that are subject to phosphorylation, and these sites were fixed very early in R domain evolution and have remained largely invariant since that time. In contrast, the rest of the R domain has been free to drift in sequence space leading to a more star-like phylogeny than seen for the other CFTR domains. The case of the R domain suggests that domain acquisition via the de novo creation of coding sequence, and the novel functional utility that such an event would seemingly entail, can be one route by which neo-functionalization is favored to occur.
Keywords: Cystic fibrosis, R domain, Molecular evolution, Coding sequence, Neo-functionalization
1. Introduction
The human Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) is a transmembrane protein that forms a channel for the transport of chloride ions in epithelial cells (Gadsby et al., 2006). Mutations to the CFTR encoding gene disable this ion channel function and lead to Cystic Fibrosis, which is among the most common lethal genetic diseases affecting Caucasians in the United States and Europe (Rommens et al., 1989). The CFTR transmembrane ion channel pore is made up of two separate domains (TMD1 & TMD2), each of which contains six membrane spanning helices (Supplementary Fig. 1). On the cytoplasmic side of the plasma membrane, the CFTR structure is characterized by two globular nucleotide binding domains (NBD1 & NBD2), which interact with each other and with an unstructured regulatory region known as the R domain.
Despite the fact that it functions as an ion channel, CFTR +is a member of the ABC superfamily of membrane transporters (Dassa and Bouige, 2001; Dean and Annilo, 2005). The human genome encodes seven distinct families of ABC transporters (ABCA–ABCG), and CFTR is most closely related with the ABCC family (Supplementary Fig. 2A) (Jordan et al., 2008). This similarity can be seen both at the level of sequence identity and domain architecture (Supplementary Fig. 2B). In fact, CFTR is also referred to by the alternate gene symbol ABCC7 (http://www.ncbi.nlm.nih.gov/gene/1080), consistent with its identity as a modified ABC transporter and a member of the ABCC family. Nevertheless, CFTR is distinguished from all other members of the ABCC family, and all other ABC transporters for that matter, by the presence of the R domain (Supplementary Fig. 2B).
Given that the R domain is a defining characteristic of CFTR, one which distinguishes it from all related ABC transporters, it has been the target of a number of functional studies aimed at understanding its contribution to the CFTR-specific ion channel function. CFTR is an ATP-dependent chloride channel with activity that is jointly regulated by the R domain and the NBDs (Gadsby and Nairn, 1994). Full channel activity requires protein kinase A dependent phosphorylation at multiple sites in the R domain (Chang et al., 1993;Cheng et al., 1991;Rich et al., 1993) along with the binding and hydrolysis of ATPs by the NBDs (Gadsby and Nairn, 1999). This combinatorial activation of the channel is achieved via highly dynamic physical interactions within and between the R domain and the NBDs. In particular, the R domain is thought to stimulate channel opening via direct phosphorylation-dependent dissociation from the NBDs, which in turn facilitates NBD dimerization and subsequent ATP binding and hydrolysis at the dimer interface (Baker et al., 2007). Interestingly, the R domain may also play an inhibitory role in CFTR channel activity, both in the unphosphorylated state, and also when specific serine residues are phosphorylated (Baldursson et al., 2001; Vais et al., 2004; Wilkinson et al., 1997; Xie et al., 2002).
The CFTR-specific R domain is further distinguished from the TMDs and NBDs shared with ABC transporters in that it does not appear to adopt any stable structural conformation (Dulhanty and Riordan, 1994; Ostedgaard et al., 2000). The intrinsically disordered state of the R domain may facilitate its dynamic physical association with the NBDs by allowing for multiple binding events, depending on which residues are phosphorylated, and by facilitating the reversibility needed for such serially inducible binding events (Baker et al., 2007;Wright and Dyson, 1999).
Despite the functional knowledge that has been accumulated for the CFTR R domain, very little is known about its origin and subsequent evolution. There is little or no demonstrable sequence homology between the R domain and any other known domains, which makes it difficult to determine how and from where the domain may have been acquired by CFTR. Furthermore, the kinds of selective forces that have shaped the R domain evolution since its acquisition by CFTR remain largely unexplored. In this study, we sought to explore where the R domain came from and how it has evolved since that time. Specifically, we sought to understand: 1) the timing of and molecular mechanisms that underlie the acquisition of the R domain along the CFTR lineage, and 2) the role of natural selection in the subsequent evolution of the R domain as it relates to the known function of the domain.
2. Results and discussion
2.1. Part I: origin of the R domain
We first attempted to evaluate the origin of the CFTR R domain via a series of comparative sequence analyses with closely related ABC transporter sequences.
2.2. Comparative analysis of CFTR gene sequence and structure
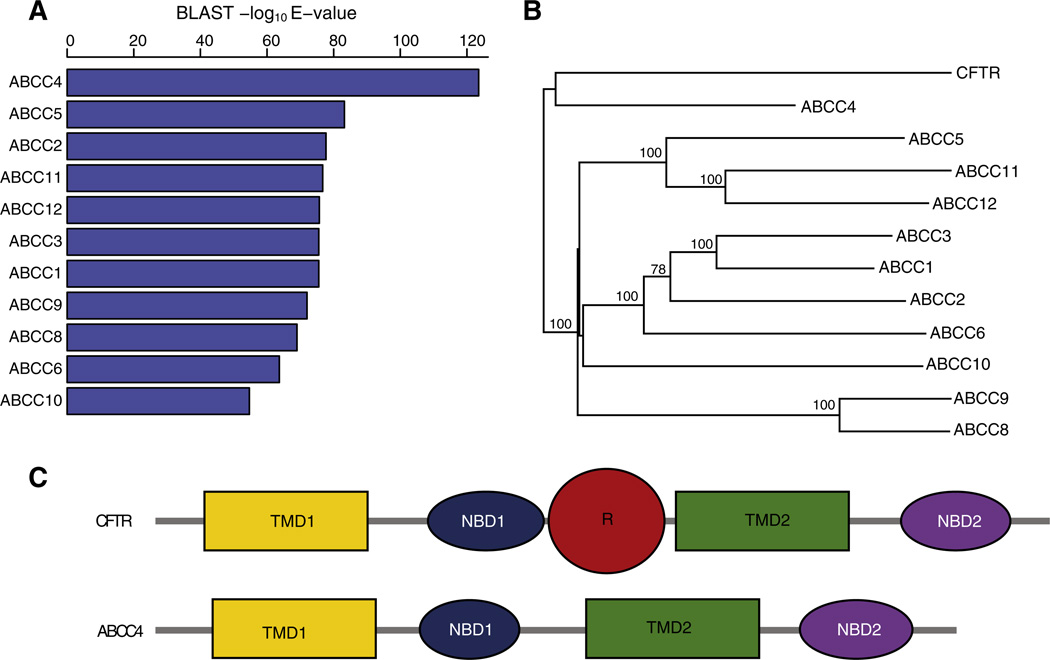
Sequence similarity comparison and phylogenetic analysis show that CFTR is most closely related to the ABCC4 member of the ABCC family (Figs. 1A & B). CFTR and ABCC4 share a common ancestor that is distinct from the remaining family members and their close similarity can also be seen at the level of domain architecture (Fig. 1C, Supplementary Fig. 2).
Fig. 1.
ABCC4 is the closest relative of CFTR. (A) Statistical significance of BLASTP hits of the human CFTR protein sequence against human ABCC subfamily members. (B) Protein sequence based phylogeny showing relationships between human CFTR and ABCC subfamily members. (C) Domain architecture of human CFTR and ABCC4 proteins.
The protein sequence and structural similarity between CFTR and ABCC4 also extends to the level of gene sequence architecture in terms of the exon–intron structures of their genes (Supplementary Fig. 3). In order to evaluate the similarities of gene exon–intron structures between the genes that encode CFTR and the other ABCC family members, we devised a simple quantitative metric that accounts for the percentage of exons that show complete overlap between gene pairs. For example, 22 of the 27 CFTR exons (81%) show complete overlap with the corresponding (orthologous) exons of ABCC4, and conversely 27 of the 31 ABCC4 exons (87%) show complete overlap with their CFTR counterparts (Supplementary Fig. 4A). The average exon conservation between CFTR and ABCC4 (84%) is substantially higher than seen for the gene that encodes the next closest protein sequence in the family ABCC5 (23%; Supplementary Fig. 4B). Furthermore, the average exon conservation between CFTR and ABCC4 (84%) is far higher than seen between CFTR and all other members of the ABCC family (13–27%; Supplementary Fig. 4C).
The high similarity seen for the CFTR and ABCC4 gene exon–intron structures, along with the marked differences between the gene structures of CFTR and the other ABCC family members, is consistent with the observation that CFTR and ABCC4 share a recent common ancestor to the exclusion of all other family members (Fig. 1B). When considered together with their respective domain architectures (Fig. 1C), these results indicate that the R domain emerged late in the evolution of the ABCC family, after the divergence of the CFTR and ABCC4 lineages, via a change in the exon–intron structure of the CFTR gene. These results also suggest that careful comparison of the CFTR and ABCC4 gene sequences, contrasted against the background of the other ABCC family member gene sequences, could provide valuable clues as to how and when the R domain emerged.
2.3. A lineage-specific extension of exon 14 gave rise to the R domain
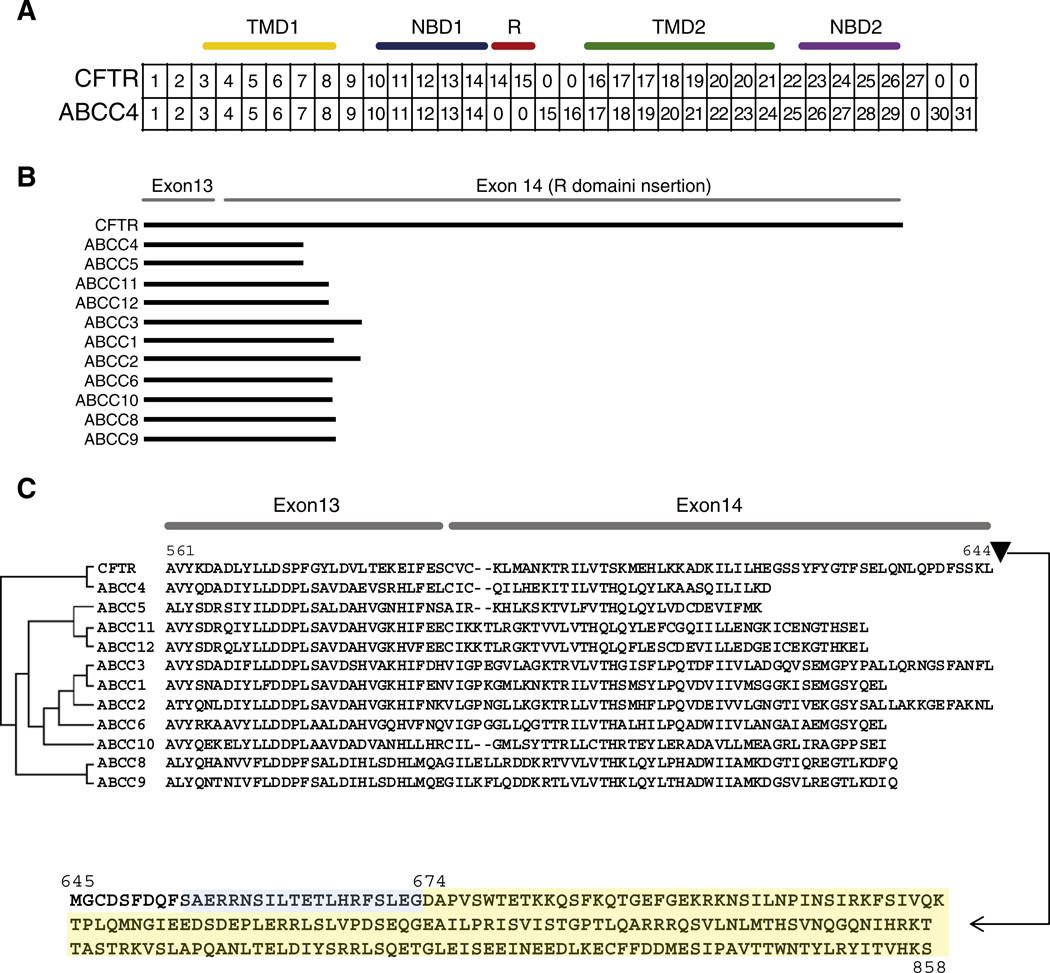
The CFTR and ABCC4 exon–intron structures are highly conserved through the first part of the protein coding sequences for the TMD1 and NBD1 domains; exons 1–13 are completely correspondent between the two genes in this region (Fig. 2A). The R domain is encoded by an apparent extension of CFTR exon 14 and an additional CFTR exon 15. At this point in the CFTR-ABCC4 alignment, the exon–intron correspondence drops off precipitously (Fig. 2B). The 5′ region of exon 14 that encodes part of the NBD1 domain is conserved among both genes, but the R domain encoding portions of exon 14 and the entire exon 15 are unique to CFTR. When all other human members of the ABCC family are compared in a similar way, the extension of CFTR exon 14 leading to the addition of the R domain is even more apparent (Fig. 2B), and in fact this extension occurred in a highly conserved region of these genes/proteins (Fig. 2C). Taken together, these results suggest that a single mutational event may have been responsible for the emergence of the R domain along the CFTR lineage.
Fig. 2.
The R domain is encoded by a lineage-specific expansion of exons 14 and 15 in CFTR relative to ABCC4. (A) Correspondence between CFTR and ABCC4 exons (based on the alignment shown in Supplementary Fig. 3). Corresponding exonic regions are placed in the same column, and exons (or exonic regions) that do not have corresponding sequences are marked with 0. Locations of CFTR domains are indicated above. (B) Visual scheme of BLAST results showing local sequence similarity between CFTR and ABCC subfamily members in exon 13 and the 5′ end of exon 14 along with the R domain insertion in CFTR exon 14. (C) Amino acid sequence alignment between CFTR and ABCC subfamily members corresponding to the protein region encoded by exon 13 and the 5′ end of exon 14. The location of the CFTR-specific R domain extension and its sequence are shown.
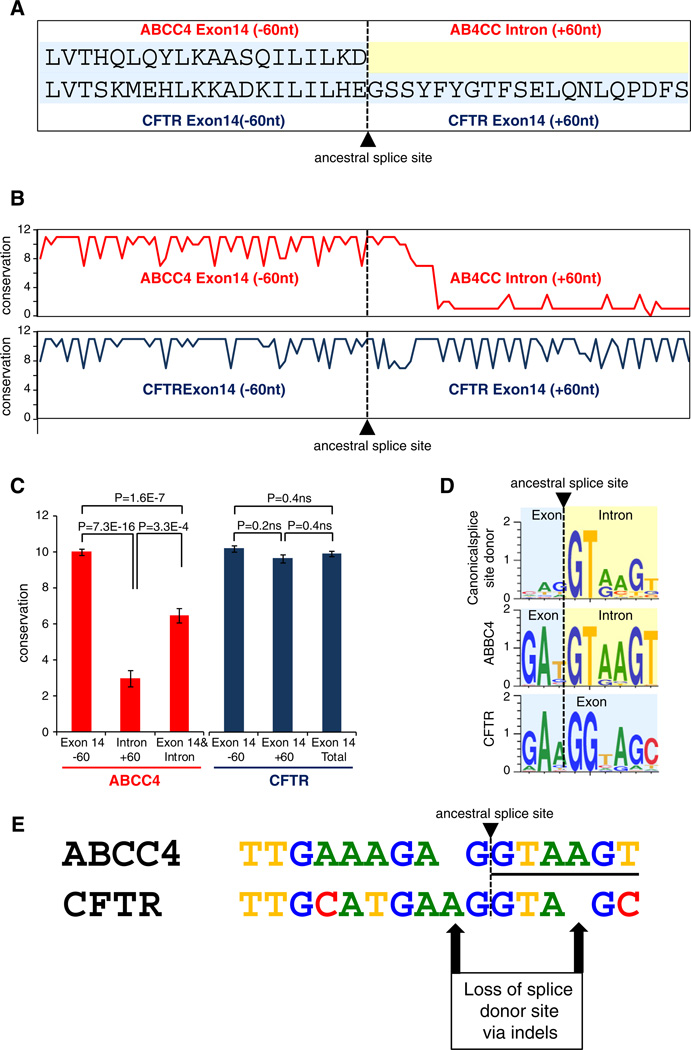
To better understand how the R domain may have emerged along the CFTR lineage after its divergence from the common ancestor with ABCC4, we evaluated gene sequence alignment of the exon 14 extension region among orthologous CFTR sequences and orthologous ABCC4 sequences from a diverse set of vertebrates. The specific region analyzed consisted of 60 bp upstream and 60 bp downstream of the position that marks the end of exon 14 in ABBC4 and the beginning of the R domain extension in CFTR exon 14 (Fig. 3A). The level of sequence conservation among ABCC4 orthologs drops off precipitously at this point and in to the intron, whereas the corresponding sequence is conserved among CFTR orthologs at this point (Figs. 3B and C). This indicates that formerly intronic sequence in CFTR has been conserved by virtue of functional constraint, consistent with its status as newly acquired protein coding sequence exon, whereas the corresponding intronic region in ABCC4 remained free from constraint.
Fig. 3.
Loss of a splice donor site in CFTR exon14led to the capture of formerly intronic sequence as R domain coding sequence. (A) Amino acid sequence alignment for the region centered on the ancestral ABCC4 splice donor site and the corresponding exon 14 extension in CFTR. (B & C) ABCC4 (red) and CFTR (blue) nucleotide sequence conservation levels for the same region. (D) Sequence motifs representing the site-specific sequence variation for canonical human splice site donor sequences, the ABCC4 exon 14 splice site donor sequence, and the corresponding region in CFTR. (E) Nucleotide sequence alignment for the region corresponding to the ABCC4 ancestral splice donor site and the CFTR exon 14 extension.
Position-specific sequence conservation at the ABCC4 exon 14 3′ exon–intron junction site reveals that ABCC4 vertebrate sequences encode a fairly canonical splice donor site AG|GTAA (Fig. 3D). However, at the same point on the CFTR alignment the sequences have been shifted one position downstream AA|GGTA. This change appears to have been based on a pair of insertion/deletion events in the sequence just adjacent to the splice donor site, which resulted in a loss of the site and subsequent read-through and capture of formerly intronic sequence as additional exon 14 sequence in the CFTR lineage (Fig. 3E). It is not possible to determine which of these events occurred first, or if they occurred in very close proximity in time, but this slight and discrete pair of changes had profound consequences with respect to the functional distinction of the CFTR ion channel from its close ABC transporter relatives.
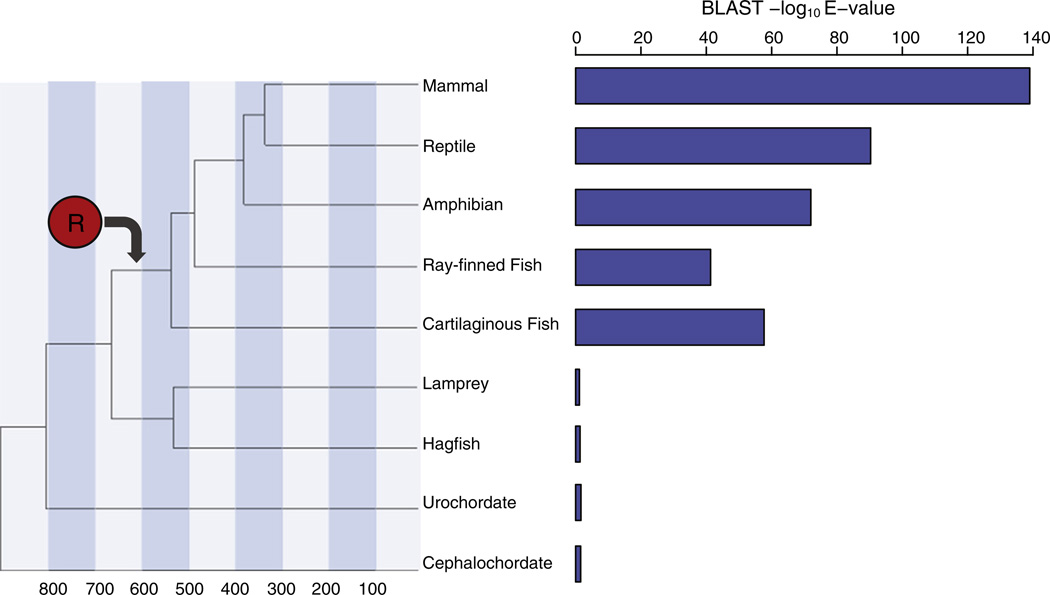
In order to determine approximately when the R domain emerged along the CFTR lineage, we evaluated the phyletic distribution of R domain homologous sequences among chordates. Mammals, reptiles, amphibians and fish can all be seen to possess R domain sequences, whereas basal vertebrates and chordates do not encode sequences with homology to the R domain (Fig. 4). Thus, the CFTR R domain appears to have emerged just prior to the diversification of the vertebrate superclass Gnathostomata, which comprises all jawed vertebrates including fish, between ~650 and 550 mya.
Fig. 4.
The R domain emerged prior to the diversification of the vertebrate super class Gnathostomata between ~650 and 550 mya. Phylogeny of the major vertebrate groups along with approximate divergence times in millions of years. Statistical significance of the best BLAST hit for each group using the human CFTR R domain sequence as a query. Approximate emergence time of the R domain (red circle) on the phylogeny as indicated by the BLAST results is shown.
While the majority of the R domain is encoded by the extended exon 14, the carboxy terminus of the domain is encoded by CFTR exon 15. This exonic region is also found exclusively in CFTR and missing from related ABC genes (Supplementary Figs. 4 & 5). There are also no homologous regions for exon 15 or its encoded amino acid sequence outside of its CFTR orthologs, further consistent with its evolutionary novelty. Exon 15 also shows no evidence of having originated from a mobile genetic element or repetitive sequence of any kind. Still, the incorporation of the novel CFTR exon 15 into the full-length CFTR transcript is supported by the presence of a canonical splice donor site at the 3′ end of exon 14 along with paired splice acceptor and splice donor sites at the 5′ and 3′ ends of exon 15 (Supplementary Fig. 6). And despite the fact that exon 15 is evolutionarily younger than the remaining CFTR exons, its splice sites show similar levels of conservation to those from the rest of the gene, indicating equally strong selective constraint for the incorporation of this lineage-specific exon into the CFTR transcript (Supplementary Fig. 6).
Although CFTR exon 15 is lineage-specific, it is found in a genomic region that is smaller than the corresponding regions in related ABC genes. In fact, ABC genes encode additional exons 15 & 16 in this region that are missing from CFTR. These data point to a lineage-specific loss of protein coding genomic sequence in the CFTR gene. In other words, CFTR added an additional protein-coding domain despite an overall loss of genomic sequence in the region, i.e. the origin of the R domain cannot be attributed to a CFTR lineage-specific genome sequence insertion as may have been expected a priori.
2.4. Part II: evolution of the R domain
Having explored the timing and the mutational events that led to the origin of the CFTR R domain, we next attempted to understand the nature of the evolutionary forces that have acted on the domain since its emergence.
2.5. Selective constraint on the R domain
Multiple sequence alignments of both CFTR amino acid sequences and protein coding nucleotide sequences were evaluated in order to assess the levels of selective constraint on the R domain compared to the other CFTR domains. The R domain can be seen to show the highest levels of amino acid and nucleotide sequence diversities along with the highest ratio of non-synonymous-to-synonymous substitution rates (dN/dS) indicative of relatively low levels of selective constraint for this domain relative to all other CFTR domains (Figs. 5A, B). This observation, along with the fact that the R domain evolved from 3 intronic sequence into exonic protein coding sequence, raises the possibility that the domain has experienced positive selection to accommodate its novel function. Nevertheless, several lines of evidence seem to argue against a role for positive selection in the evolution of the R domain. First, when averaged across the entire R domain and for all lineages considered, pairwise levels of dN are significantly lower than levels of dS, consistent with purifying selection (Fig. 5C). Second, when R domain dN versus dS levels are considered for individual branches on the CFTR phylogeny, all branches show dN ≪ dS (Fig. 5D). Third, when dN versus dS levels are considered for individual codons within the R domain, all codons show dN ≪ dS (Fig. 5E). In other words, despite the relatively low levels of selective constraint across the R domain, we were unable to find any statistically significant evidence for positive selection based on branch-specific or site-specific analyses of dN and dS.
Fig. 5.
The R domain sequence is more variable than other CFTR domains but does not show evidence of positive selection. Average (±standard error) amino acid (A) and nucleotide (B) diversity levels for CFTR domains. (C) Average (±standard error) dN/dS ratios for CFTR domains. (D) CFTR vertebrate phylogeny showing branch specific values of dN/dS for the R domain sequences. Branch-specific dN/dS values are color coded according to the legend shown. (E) dN/dS ratios for all codons in the R domain, from CFTR residue 654 to 858, are shown (blue line) in comparison to the neutral expectation dN/dS = 1 (red dashed line).
It is worth noting that absence of evidence for positive selection on the R domain can not necessarily be taken as evidence of absence of such selection at some point in the history of the domain. Indeed, it is difficult to imagine that intronic sequence could be acquired and assimilated as functionally constrained protein coding exonic sequence as happened in the case of the R domain without at least some nonsynonymous mutations being swept to fixation by positive selection. However, it may simply be the case that these particular changes happened too early, or too periodically, in the evolution of the domain to be detected by the extant sequences available for analysis. In addition, if only a few positions of the domain are critical for its functional utility, then these sites may have pre-existed in the sequences of the founder population, and as a consequence they would simply show evidence of strong selective constraint subsequent to the emergence of the domain on the CFTR lineage. Finally, these two scenarios are not mutually exclusive and some aspect of both may have been at play in the evolution of the domain.
Previously, it has been shown that genes that are expressed in a more tissue-specific manner, as well as exons that are alternatively expressed ( Chen et al., 2006; Ramensky et al., 2008), show lower levels of selective constraint (i.e. higher dN/dS) than more constitutively expressed genes/ exons. While there is no evidence of alternative splicing for the R domain, it may be the case that R domain exons are expressed in a more tissue-specific manner than the remaining CFTR domains. This could also explain their relatively low levels of selective constraint. We evaluated this possibility by comparing the levels of tissue/cell type-specificity for the CFTR R domain encoding exons compared to the remaining exons. R domain encoding exons do not show different levels of cell type-specificity than the remaining exons indicating that differences in expression profiles between exons do not explain the observed differences in the levels of selective constraint (Supplementary Fig. 7).
2.6. Anomalous evolutionary patterns of the R domain
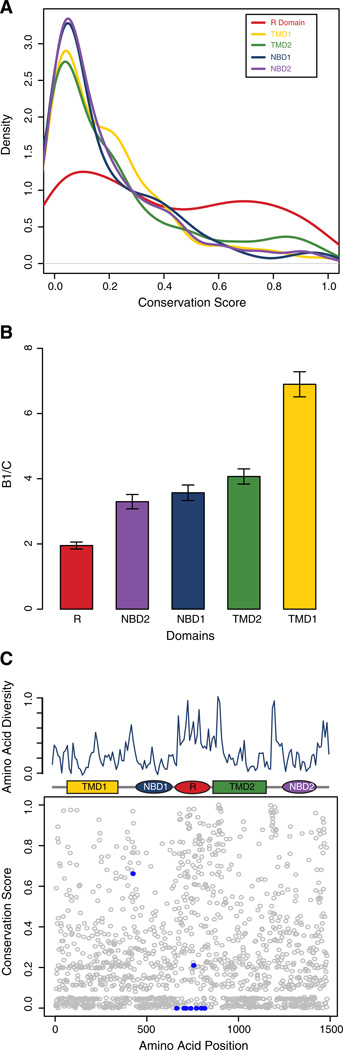
Despite the lack of evidence for the action of positive selection on the CFTR R domain, sequences for this domain show distinctly anomalous patterns of evolution compared to other CFTR domains. First of all, the relative rates of site-specific sequence conservation across the R domain differ markedly from the other four domains. Conservation levels are fairly evenly distributed across R domain sites as can be seen from the relatively flat density distribution of conservation scores in Fig. 6A. In contrast, the other domains show a peak corresponding to highly conserved sites (low scores) and the distributions then fall off steeply to less conserved sites. These differences indicate that the R domain experiences a very different mode of selective constraint across individual sites with relatively few sites being highly conserved compared to the other domains.
Fig. 6.
The R domain shows anomalous evolutionary patterns of sequence evolution. (A) Density distribution of site-specific conservation scores for CFTR domains (color coded as shown in the legend). (B) Ratios of the between group branch length divided by the average within group branch length (B1/C) for CFTR domain-specific phylogenies (see Supplementary Fig. 8). (C) Site-specific amino acid diversity along the length of the CFTR protein. The conservation levels of each individual amino acid position are shown (circles below), along with a sliding window of amino acid diversity (line above), in comparison to the CFTR domain architecture. CFTR residues that are subject to phosphorylation are shown in blue.
Independent phylogenetic analyses of the five CFTR domains also show that the R domain has very distinct patterns of evolution with respect to branch-specific rates of change. The phylogeny of the CFTR sequences analyzed here shows two distinct groups of sequences, with fish on one side and terrestrial vertebrates on the other, separated by a long internal branch (Supplementary Fig. 8). Branches leading to sequences within the groups are relatively short especially for the mammalian CFTR sequences. This same pattern can be seen for the TMD and NBD domains, whereas the R domain phylogeny is far more star-like without a long internal branch and with relatively long external branches distributed throughout the tree (Supplementary Fig. 8). When this pattern for the domain specific phylogenies is quantified by taking the ratio of the length of this internal branch (B1) over the average length of all other branches (C), the anomalous pattern of R domain evolution becomes even more apparent (Fig. 6B). This unique pattern for R domain evolution further underscores the possibility that a distinct set of functional constraints and selective forces have been at play in its evolution.
The R domain is known to be unstructured (Dulhanty and Riordan, 1994;Ostedgaard et al., 2000), which is consistent with its emergence from formerly non-coding intronic sequence and its anomalous patterns of evolution compared to the other structured domains. Nevertheless, its overall levels of sequence divergence clearly indicate that the R domain is subject to selective constraint based on some functional utility. R domain regulatory function is predicated upon the phosphorylation of specific serine residues, which facilitates dissociation with NBD domains and their subsequent dimerization and activation of the channel (Baker et al., 2007). Given the demonstrated functional importance of such sites, we expected them to be highly conserved compared to other sites in the domain. Indeed, R domain-based sites that have been experimentally demonstrated to be phosphorylated are highly conserved, and in fact all but one are totally invariant (Fig. 6C). The high levels of conservation for these sites stand in stark contrast to the overall levels of evolution for the domain.
2.7. Model for the evolution of the R domain
Considered together, the anomalous patterns of R domain evolution and its relative levels of selective constraint allow us to pose a model for its initial emergence, its acquisition of functional utility and its subsequent evolution. First, the R domain can be seen to have emerged from previously non-coding intronic sequence. It has long been held that it is extremely rare to evolve protein coding sequences from non-coding sequences de novo in this way, and that it is far more common that new protein sequences evolve from duplication of existing protein coding sequences and/or from recombination of existing protein coding domains (Jacob, 1977; Ohno, 1970). However, recent studies suggest that de novo evolution of protein coding sequences may be more prevalent and important than previously imagined (Carvunis et al., 2012; Tautz and Domazet-Loso, 2011), and this may be particularly true for the human evolutionary lineage (Wu et al., 2011). In any case, this particular aspect of the R domain origin had important implications for its function and evolution. The fact that the R domain originated from intronic sequence made it extremely unlikely that it would be able to adopt a highly ordered structural confirmation, and indeed the domain is known to comprise a largely unstructured random coil (Dulhanty and Riordan, 1994; Ostedgaard et al., 2000). Thus, the function of the R domain is not dependent on its structure per se, as with the other domains in CFTR, but rather on the regulatory potential encoded by a handful of key residues, i.e. those serines that are phosphorylated and enable R domain interaction with the NBDs to activate the channel (Gadsby and Nairn, 1999). Indeed, these particular sites are largely invariant among CFTR sequences, suggesting that they either existed at the time of the emergence of the domain or were swept to fixation shortly thereafter. Since that time the phosphorylated residues have been highly conserved across CFTR evolution. In this way, the evolution of the R domain was likely to consist of a short period of intense and profound change, leading to its phosphorylation-based regulatory capacity, followed by high conservation of the handful of phosphorylated residues and a simultaneous slow and steady drift for the rest of its sequence. This model is consistent with the more star-like phylogeny seen for the R domain compared to the other CFTR domains as well as its relatively flat distribution of site-specific conservation levels.
The rapid and profound evolutionary change represented by the acquisition of the R domain allowed CFTR, formerly an alternating-access transporter, to become locked into a given conformational state for longer periods of time thus fundamentally altering its activity and allowing it to explore novel functional space while the ancestral function was maintained by the existing repertoire of ABC transporters. Thus, CFTR may be considered to represent a case of neo-functionalization whereby gene duplication allows for a new paralog to take on a completely different function (Force et al., 1999). The combination of its emergence from non-coding sequence and its neo-functionalization make the CFTR R domain a particularly fascinating case of molecular evolution.
3. Materials and methods
3.1. Sequence and structure comparison within and between CFTR and other ABC transporter family members
The CFTR protein structure was obtained from the published CFTR homology model (Serohijos et al., 2008). Protein sequence similarity comparisons between CFTR and related ABC transporters were done using the BLASTP program (Altschul et al., 1997). Human protein sequences of CFTR and members of the ABCC transporter subfamily were obtained from the NCBI RefSeq database (Pruitt et al., 2009) (Supplementary Table 1). Protein sequences were aligned using ClustalW (Thompson et al., 1994), and Neighbor-Joining trees (Saitou and Nei, 1987) were constructed using the program MEGA (Tamura et al., 2011). Protein domain architectures for the sequences were characterized with the SMART (Schultz et al., 1998) tool.
CFTR and ABCC gene models (i.e. exon–intron structures) were taken from the NCBI Gene database (Maglott et al., 2011) (Supplementary Table 1). Each of the CFTR exons were compared to that of the representative member of the ABCC family using local pair wise alignment with Blast2Seq (Altschul et al., 1990) and EMBOSS Needle optimal global Alignment (Rice et al., 2000), and the set of individual exon alignments were considered together to characterize pairwise similarities in exon–intron structures. For each member in an alignment pair the number of exons that show complete overlap with the corresponding (i.e. orthologous) exons of the other member is normalized by its total number of exons to compute a percent exon conservation score. The average exon conservation score is taken as the average of these two percentages for both members in the pair. For the purposes of analyzing both CFTR and ABCC4 orthologous sequences from exons 13 to 15, i.e. at the point of the R domain extension in CFTR, separate CFTR and ABCC4 alignments of orthologous vertebrate nucleotide sequences were taken from the ‘17-Way Cons’ track of the Mar. 2006 (NCBI36/hg18) human genome reference sequence at the UCSC genome browser (Fujita et al., 2011). Nucleotide sequence identity for these regions in the CFTR and ABCC4 alignments was computed based on the Kimura 2-parameter model implemented in MEGA. Sequence motif analysis of the alignments for this region spanning the R domain extension, or the ABCC4 exon–intron junction, were performed using the Weblogo tool (Crooks et al., 2004).
For the purposes of identifying the timing of the origin of the CFTR R domain, PSI-BLAST was used to search all chordate sequences in the Genbank non-redundant database (Sayers et al., 2012). The specific time estimate reported is based on the deuterostome divergence time estimates reported in (Blair and Hedges, 2005).
3.2. Evolutionary forces on CFTR domains
All available vertebrate CFTR NCBI RefSeq mRNAs, both protein coding nucleotide sequences and their corresponding amino acid (protein) sequences, were analyzed in order to characterize the relative selective forces acting on the five CFTR domains (Supplementary Table 1). The ClustalW algorithm implemented in the program MEGA was used to align CFTR protein sequences and the corresponding protein coding nucleotide sequences were then aligned in-frame based on the protein sequence alignment. Human CFTR domain boundaries were determined using the SMART program and Neighbor-Joining phylogenies for each of the five domains were computed with the program MEGA. A Neighbor-Joining phylogeny based on the CFTR protein coding nucleotide sequence alignment was also computed using the program MEGA.
CFTR amino acid conservation levels were characterized using the Consurf webserver (Ashkenazy et al., 2010;Berezin et al., 2004), and Consurf scores were normalized to the interval [0, 1 ] with 0 being the most conserved and 1 being the least conserved. The locations and identities of experimentally characterized CFTR phosphorylation sites in the R domain were taken from (Baker et al., 2007). Overall CFTR nucleotide diversity levels along with dN and dS values were computed using MEGA. Codon-specific dN/dS values and branch-specific dN/dS values were computed using the GABranch analysis tool implemented on the Data Monkey web server (Delport et al., 2010; Pond and Frost, 2005a,2005b; Pond et al., 2005).
The relationship between CFTR exon-specific expression levels and dN/dS was evaluated using expression data taken from exon tiling arrays. Expression levels for CFTR exons across 67 tissues/cell-types, generated by the University of Washing ENCODE group using the Affymetrix Human Exon 1.0 GeneChip, were taken from the UCSC Genome Browser ENCODE UW Affy All-Exon Arrays track. For each CFTR exon, tissue/cell-type-specific expression levels were computed as: where N is the number of tissues/cell-types and Xi is expression level in tissue i (Yanai et al., 2005).
Supplementary Material
Abbreviations
- ABC
transporters
- ATP
binding cassette transporters
- ABCC
ATP-binding cassette, sub-family C
- ABCC4
ATP-binding cassette, sub-family C, member 4
- ABCC7
ATP-binding cassette, sub-family C, member 7
- CFTR
Cystic Fibrosis Transmembrane Conductance Regulator protein
- NBD
Nucleotide binding domain
- R domain
Regulatory domain
- TMD
Transmembrane Domain.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gene.2013.02.050.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JM, et al. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat. Struct. Mol. Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldursson O, Ostedgaard LS, Rokhlina T, Cotten JF, Welsh MJ. Cystic fibrosis transmembrane conductance regulator Cl−channels with R domain deletions and translocations show phosphorylation-dependent and -independent activity. J. Biol. Chem. 2001;276:1904–1910. doi: 10.1074/jbc.M006934200. [DOI] [PubMed] [Google Scholar]
- Berezin C, et al. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol. Biol. Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Carvunis AR, et al. Proto-genes and de novo gene birth. Nature. 2012;487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang XB, et al. Protein kinase A (PKA) still activates CFTR chloride channel after mutagenesis of all 10 PKA consensus phosphorylation sites. J. Biol. Chem. 1993;268:11304–11311. [PubMed] [Google Scholar]
- Chen FC, Wang SS, Chen CJ, Li WH, Chuang TJ. Alternatively and constitutively spliced exons are subject to different evolutionary forces. Mol. Biol. Evol. 2006;23:675–682. doi: 10.1093/molbev/msj081. [DOI] [PubMed] [Google Scholar]
- Cheng SH, Rich DP, Marshall J, Gregory RJ, Welsh MJ, Smith AE. Phosphorylation of the R domain by cAMP-dependent protein kinase regulates the CFTR chloride channel. Cell. 1991;66:1027–1036. doi: 10.1016/0092-8674(91)90446-6. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassa E, Bouige P. The ABC of ABCS: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 2001;152:211–229. doi: 10.1016/s0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulhanty AM, Riordan JR. Phosphorylation by cAMP-dependent protein kinase causes a conformational change in the R domain of the cystic fibrosis transmembrane conductance regulator. Biochemistry. 1994;33:4072–4079. doi: 10.1021/bi00179a036. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadsby DC, Nairn AC. Regulation of CFTR channel gating. Trends Biochem. Sci. 1994;19:513–518. doi: 10.1016/0968-0004(94)90141-4. [DOI] [PubMed] [Google Scholar]
- Gadsby DC, Nairn AC. Control of CFTR channel gating by phosphorylation and nucleotide hydrolysis. Physiol. Rev. 1999;79:S77–S107. doi: 10.1152/physrev.1999.79.1.S77. [DOI] [PubMed] [Google Scholar]
- Gadsby DC, Vergani P, Csanady L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Kota KC, Cui G, Thompson CH, McCarty NA. Evolutionary and functional divergence between the cystic fibrosis transmembrane conductance regulator and related ATP-binding cassette transporters. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18865–18870. doi: 10.1073/pnas.0806306105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- Ostedgaard LS, Baldursson O, Vermeer DW, Welsh MJ, Robertson AD. A functional R domain from cystic fibrosis transmembrane conductance regulator is predominantly unstructured in solution. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5657–5662. doi: 10.1073/pnas.100588797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005a;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD. A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol. Biol. Evol. 2005b;22:478–485. doi: 10.1093/molbev/msi031. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI reference sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky VE, Nurtdinov RN, Neverov AD, Mironov AA, Gelfand MS. Positive selection in alternatively spliced exons of human genes. Am. J. Hum. Genet. 2008;83:94–98. doi: 10.1016/j.ajhg.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rich DP, et al. Regulation of the cystic fibrosis transmembrane conductance regulator Cl− channel by negative charge in the R domain. J. Biol. Chem. 1993;268:20259–20267. [PubMed] [Google Scholar]
- Rommens JM, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serohijos AW, et al. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3256–3261. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Loso T. The evolutionary origin of orphan genes. Nat. Rev. Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vais H, Zhang R, Reenstra WW. Dibasic phosphorylation sites in the R domain of CFTR have stimulatory and inhibitory effects on channel activation. Am. J. Physiol. Cell Physiol. 2004;287:C737–C745. doi: 10.1152/ajpcell.00504.2003. [DOI] [PubMed] [Google Scholar]
- Wilkinson DJ, et al. CFTR activation: additive effects of stimulatory and inhibitory phosphorylation sites in the R domain. Am. J. Physiol. 1997;273:L127–L133. doi: 10.1152/ajplung.1997.273.1.L127. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure–function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Wu DD, Irwin DM, Zhang YP. De novo origin of human protein-coding genes. PLoS Genet. 2011;7:e1002379. doi: 10.1371/journal.pgen.1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Adams LM, Zhao J, Gerken TA, Davis PB, Ma J. A short segment of the R domain of cystic fibrosis transmembrane conductance regulator contains channel stimulatory and inhibitory activities that are separable by sequence modification. J. Biol. Chem. 2002;277:23019–23027. doi: 10.1074/jbc.M201661200. [DOI] [PubMed] [Google Scholar]
- Yanai I, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.