Abstract
Two systematic series of increasingly hydrophilic derivatives of duocarmycin SA are described that feature the incorporation of ethylene glycol units (n = 1–5) into the methoxy substituents of the trimethoxyindole subunit. These derivatives exhibit progressively increasing water solubility, along with progressive decreases in cell growth inhibitory activity and DNA alkylation efficiency with the incremental ethylene glycol unit incorporations. A linear relationship between cLogP and –logIC50 for cell growth inhibition and –logAE (AE = cell free DNA alkylation efficiency) is observed where cLogP values span the productive range of 2.5–0.49 and –logIC50 values span the range of 11.2–6.4, representing IC50 values covering a 105 range (0.008 to 370 nM). The results quantify a fundamental role the compound hydrophobic character plays in the expression of the biological activity of members in this class, driving the intrinsically reversible DNA alkylation reaction, and define the stunning magnitude of its effect.
Introduction
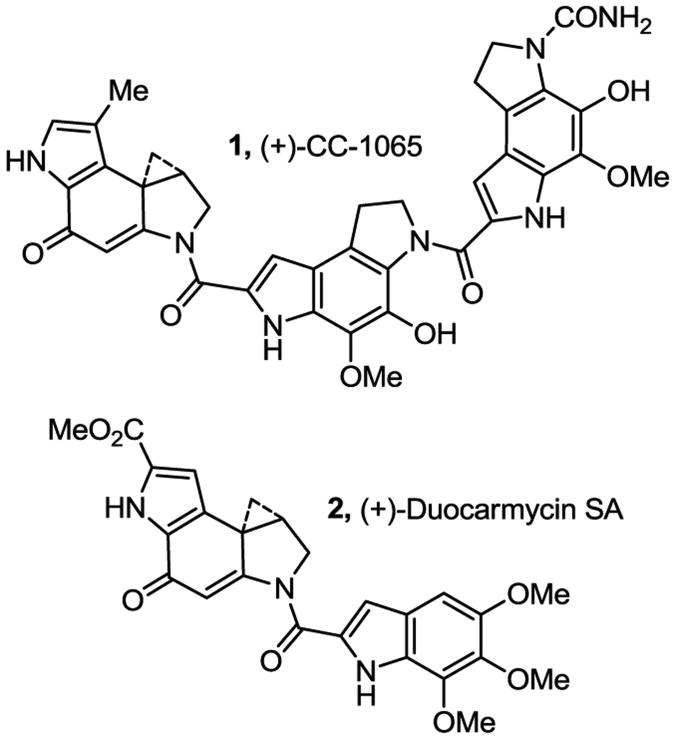
CC-1065 (1) and duocarmycin SA (2) represent the parent members of a class of antitumor compounds that derive their biological activity from their ability to selectively alkylate duplex DNA (Figure 1).1–3 The study of the natural products, their synthetic unnatural enantiomers,4 and key analogues has defined many of the fundamental structural features that control their DNA alkylation selectivity, efficiency, rate, and catalysis,5 providing a detailed understanding of the relationships between structure, reactivity, and biological activity.3–5 Despite the extensive efforts conducted over the more than 30 years since the report of the initial member of this of natural products, herein we report studies that define an additional previously unappreciated and fundamental property integrated into the structure of this class of compounds that contributes to their DNA alkylation properties and biological activity and the stunning magnitude of its impact.
Figure 1.
Representative natural products in class.
The alkylation subunits of the natural products contain a vinylogous amide, which confers stability to what would otherwise be a reactive cyclopropane.6 Disruption of this key vinylogous amide occurs through a DNA minor groove binding-induced conformational change, which brings the cyclopropane into conjugation with the cyclohexadienone ring system and activates it for nucleophilic attack. Thus, the compounds are typically unreactive, but they are selectively activated for adenine N3 alkylation upon target DNA binding.7,8 Pertinent to the work detailed herein, this reactivity is still attenuated,9 allowing selective capture by appropriately positioned adenines within the preferred AT-rich non-covalent binding sites such that it is the non-covalent binding selectivity of the compounds that controls the alkylation site selectivity2,5.
An additional unique feature of this class of natural products is the observation that the seco phenol synthetic precursors possess indistinguishable biological properties (DNA alkylation, in vitro cytotoxic activity, in vivo antitumor activity) in comparison to the cyclopropane derivatives themselves. Such seco phenol derivatives, like those disclosed herein, undergo facile in situ Ar-3′ spirocyclization with the displacement of an appropriate leaving group to afford the cyclopropane found in the natural products.
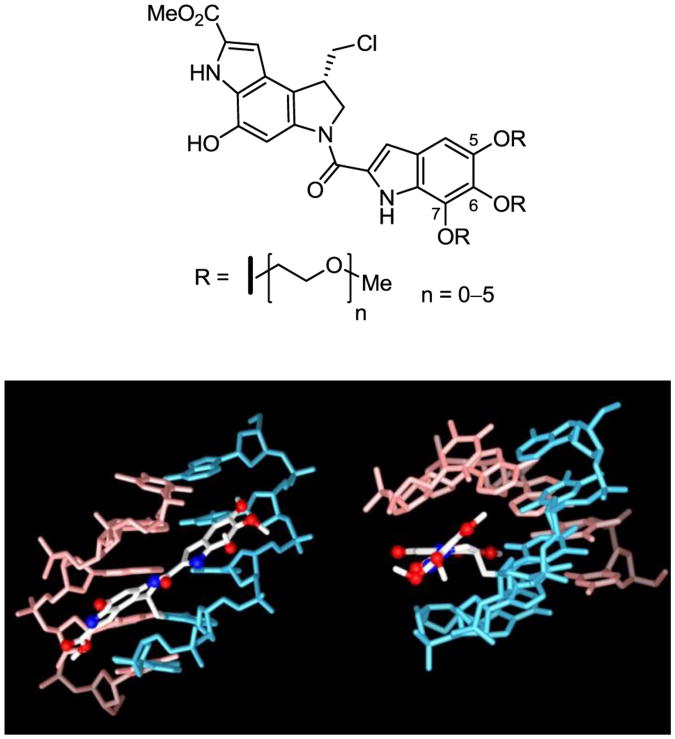
In recent efforts,10,11 we became interested in the preparation of a series of more water soluble derivatives of duocarmycin SA derived through the systematic incorporation of polyethylene glycol units into the trimethoxyindole DNA binding subunit (Figure 2). The C6 and C7 sites are located on the face of the trimethoxyindole that extends away from DNA minor groove and the two methoxy substituents located at these sites can be removed without significantly impacting the biological properties of duocarmycin SA.5 The C5 methoxy substituent lies at a peripheral site at the end of the drug-bound DNA complex.12 This site constitutes one that is not only capable of accommodating substituents that enhance activity,13 but it represents a site widely used to introduce large substituents including long linkers for antibody-drug conjugation.11 As a result, all three positions (C5–C7) represent ideal sites for introduction of structural modifications. Herein, we describe the synthesis and biological properties of a series of duocarmycin SA derivatives modified at these sites, replacing all three methoxy substituents or just the central C6 methoxy group with a systematic series of polyethylene glycol (PEG) substituents (Figure 2). Their examination revealed an additional and previously unappreciated property of this class of compounds that contributes in a remarkably substantial and fundamental way to their DNA alkylation capabilities and biological properties that likely has implications for other DNA minor groove binding compounds.14
Figure 2.
Top: PEG modified duocarmycin SA analogs (n = 1–5) and seco-duocarmycin SA (n = 0). Bottom: Structure of duocarmycin SA bound to DNA (ref 12a) highlighting the disposition of the C5–C7 methoxy groups.
Results and Discussion
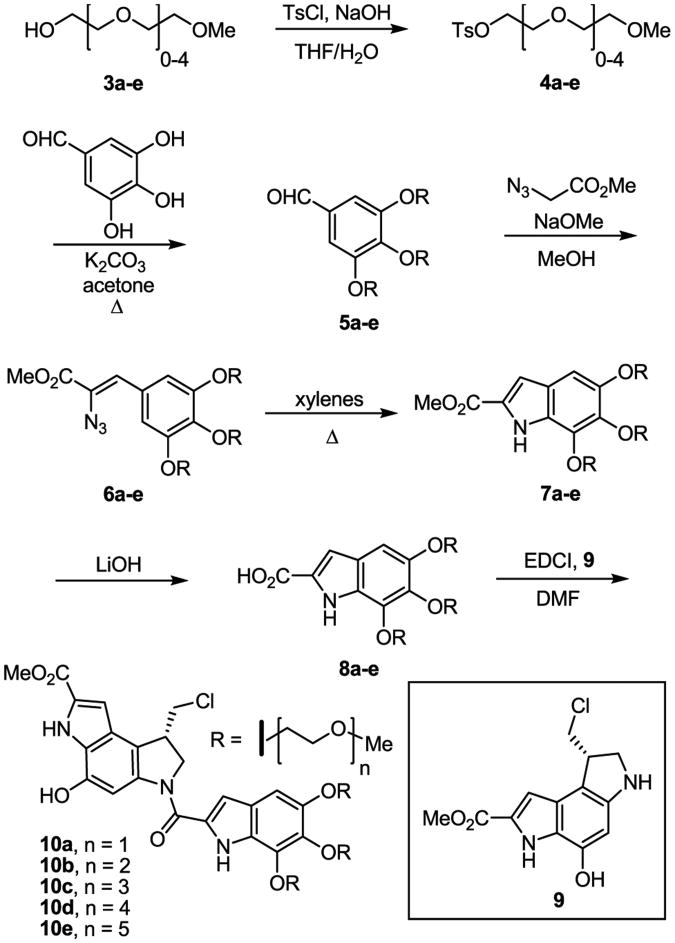
Synthesis of 10a–e
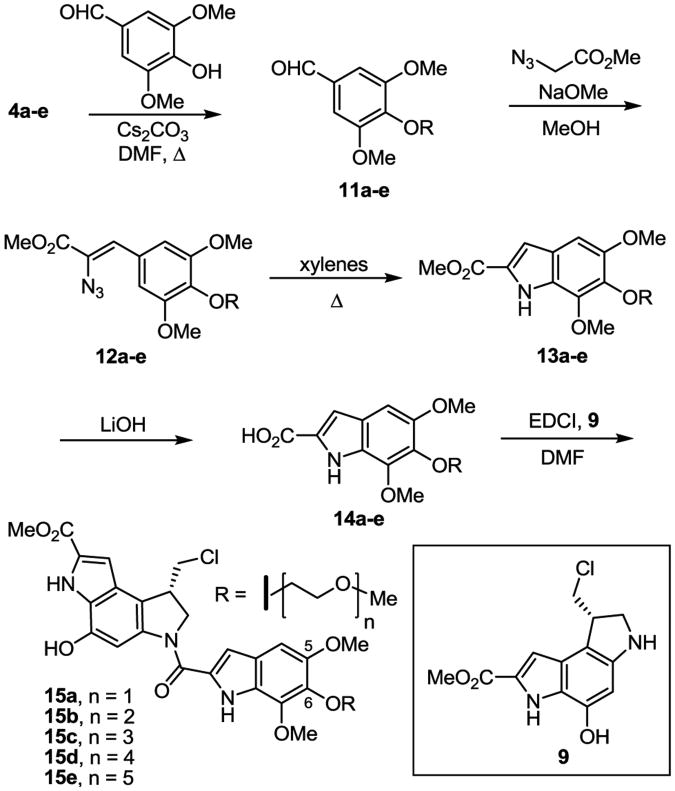
The preparation of the derivatives began with the tosylation of the commercially available alcohols 3a–e according to a published procedure15 to provide 4a–e in near quantitative yields (Scheme 1). Without optimization, alkylation of 3,4,5-trihydroxybenzaldehyde with 4a–e afforded the desired tri-alkylated products 5a–e as the precursors to the initial series. Treatment of the aldehydes 5a–e with methyl 2-azidoacetate and sodium methoxide provided the styryl azides 6a–e in high yield. Warming a solution of 6a–e in xylenes initiated a Hemetsberger indole cyclization to afford 7a–e.16 The methyl esters of 7a–e were hydrolyzed to provide the carboxylic acids 8a–e, which were subsequently coupled with the optically active duocarmycin SA alkylation subunit 917 to provide the seco-duocarmycin SA derivatives 10a–e.
Scheme 1.
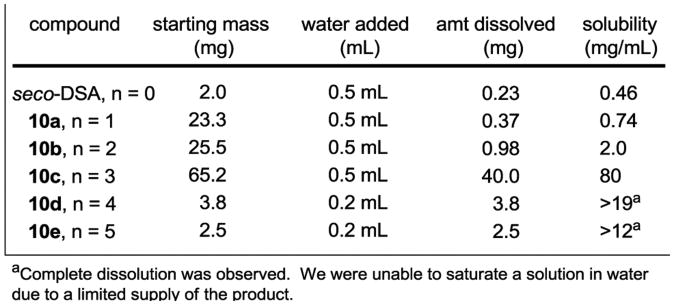
Water Solubility of 10a–e
The water solubility of the PEG modified duocarmycin SA analogs was determined by treating samples of 10a–e with known volumes of distilled water and stirring for 40 h to ensure saturated dissolution. The solutions were then filtered, concentrated, and the resulting samples were dried and weighed to provide an accurate measurement of the dissolved sample (Figure 3). While the parent molecule seco-duocarmycin SA exhibited low water solubility (0.46 mg/mL), the modified derivatives 10a–e showed incrementally and much improved solubility. As anticipated, as the number of ethylene glycol units incorporated into the molecules increased, the water solubility also increased. Although we were not able to saturate an aqueous solution of 10d and 10e due to a limited supply of the products themselves, the solubility of these compounds in water was determined to be greater than 19 mg/mL and 12 mg/mL respectively, and are assuredly much higher due to the excellent solubility of 10c (80 mg/mL).
Figure 3.
Water solubility of seco-duocarmycin SA and 10a–e.
Biological Activity of 10a–e
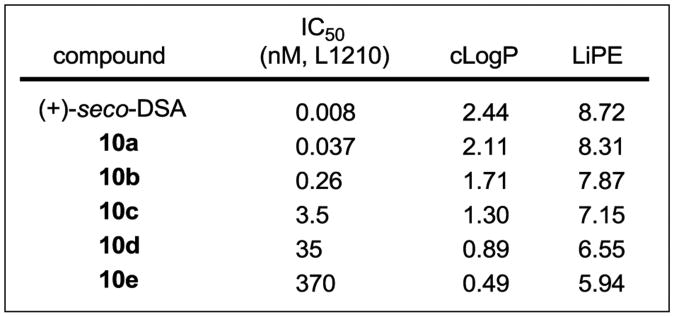
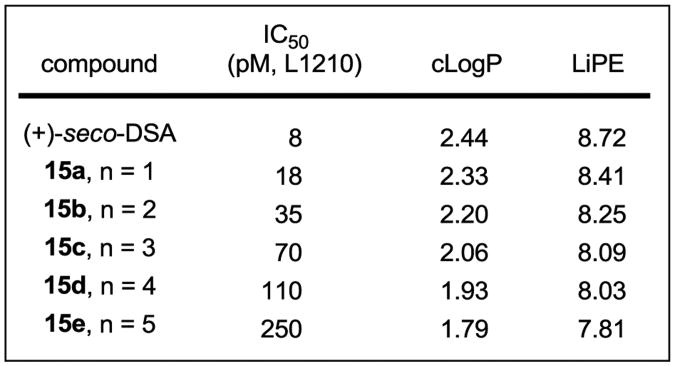
Compounds 10a–e were tested for cell growth inhibition in a cytotoxic assay traditionally used to compare members of this class, enlisting mouse lymphocytic leukemia cells (L1210, Figure 4). Incremental changes in activity were observed for compounds 10a–e, representing an approximately 10-fold change in potency for each of the three-fold incorporations of an ethylene glycol unit into the DNA binding subunit. Thus, compound 10a exhibited the most potent activity of the new series with an IC50 of 37 pM, being approximately 5-fold less active than duocarmycin SA, whereas compounds 10d and 10e were the least potent (IC50 = 35 and 370 nM, respectively). Through the series, a remarkably large 50,000-fold range in activity was observed.
Figure 4.
Cell growth inhibition activity (L1210), cLogP, and LiPE for seco-duocarmycin SA and 10a–e.
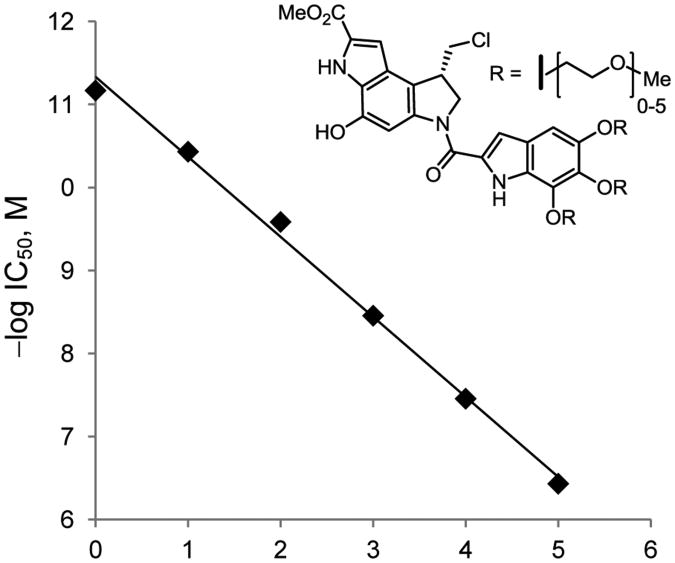
A plot of the incremental additions of the ethylene glycol units (n = 0–5) versus the –log IC50 (M) provides a well-defined linear relationship (Figure 5). Moreover, the changes are of a remarkable magnitude, representing 10-fold changes in activity for each three-fold ethylene glycol unit introduction and a stunning 105-fold change in IC50 values over the six compounds examined (n = 0–5).
Figure 5.
Plot of n (0–5) versus –log IC50, slope = –0.96, r2 = 0.996.
DNA Alkylation Properties of 10a–e
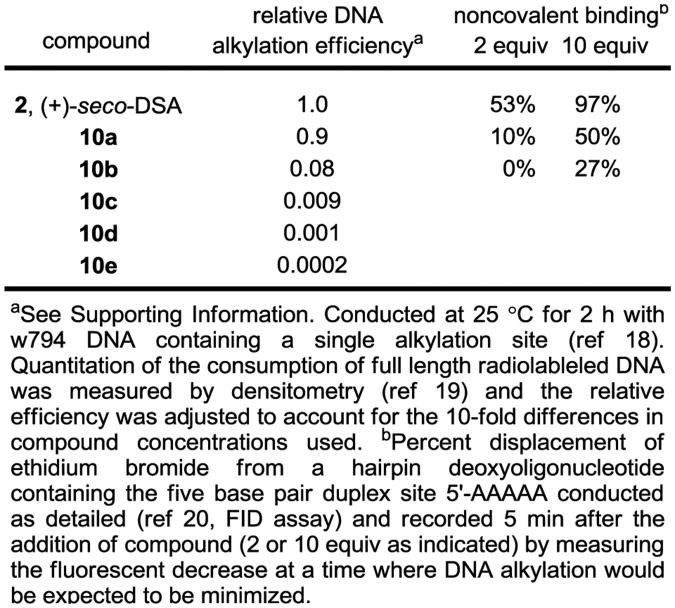
The reduced cell growth inhibitory activity of the compounds against L1210 may be due to either a diminished interaction with their biological target (a reduced DNA alkylation rate and efficiency) or the result of some other alteration of their properties. In order to define the factors contributing to the reduced activity, the cell free DNA alkylation properties of 10a–e were examined using procedures previously described in detail.18,19 Consistent with expectations, the DNA alkylation selectivity observed with duocarmycin SA was unaltered by the structural modifications found in 10a–e. However, 10a–e exhibited incrementally and substantially diminished rates and efficiencies of DNA alkylation (2 > 10a > 10b > 10c > 10d > 10e) directly correlating with the diminished cell growth inhibition potency (Supporting Information Figures S1 and S2). Moreover, the differences in the efficiency of cell free DNA alkylation observed (ca. 10-fold between each derivative) are of a magnitude to suggest that they alone account for the progressive 10-fold loss in biological activity in each iteration in the series (Figure 6).
Figure 6.
Relative DNA alkylation efficiency and binding affinity.
cLogP Correlations
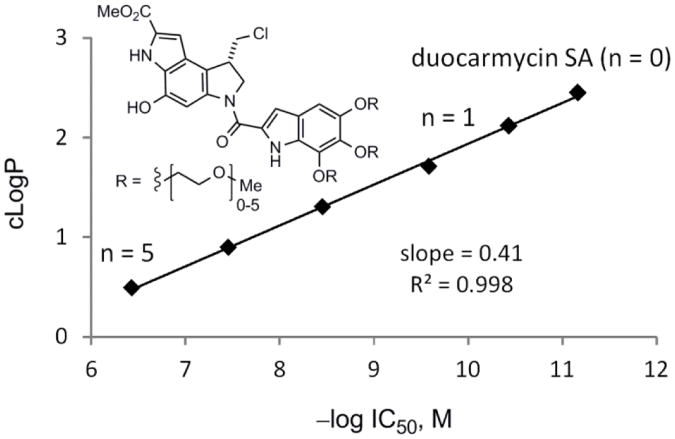
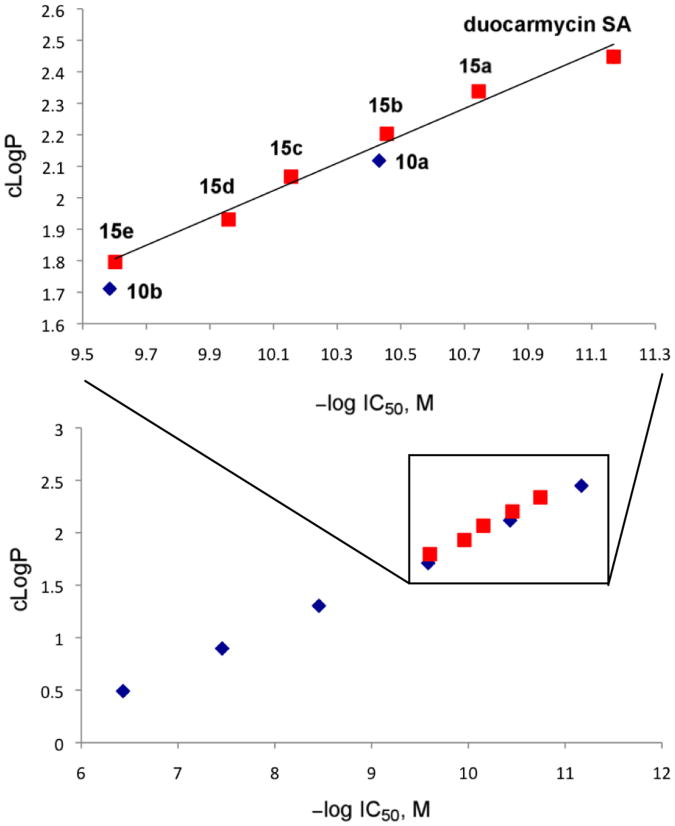
Since it is unlikely that the systematic substituent changes sterically preclude DNA minor groove binding or that they do so in such a defined incremental manner, the results suggest that it may be the incremental increases in water solubility itself, or more appropriately compared as decreases in cLogP, that decrease the efficiency of DNA alkylation. Presumably, this reduction is a consequence of progressively diminished non-covalent DNA binding affinity of the derivatives, where they increasingly partition into the aqueous buffer versus hydrophobic DNA minor groove. Although it is difficult to accurately experimentally assess the non-covalent DNA binding properties of compounds that covalently alkylate DNA, especially those whose range of affinities may be as large as those of 10a–e, the examination of 2, 10a, and 10b in a fluorescent intercalator displacement (FID) assay20 provided results consistent with each displaying large incremental differences in affinity for a typical five base-pair AT-rich alkylation site (Figure 6). It has often been observed that the more insoluble DNA binding compounds in a series are the most effective, although we are not aware of a study capable of systematically probing such a relationship. With the assumption that the PEG extensions of C5–C7 methoxy substituents in duocarmycin SA do not sterically impede DNA binding incrementally, the compounds 10a–e represent an ideal series on which to establish such a relationship. Beautifully, a direct linear relationship between cLogP21 and –log IC50 is observed for the compounds in the series (Figure 7). Notably, the cLogP values span the productive and perfectly acceptable range of 2.5–0.49 for seco-duocarmycin SA to 10e with incremental changes of approximately 0.4 and the corresponding –log IC50 values span the range of 11.2– 6.4, representing IC50 values for the series covering a 105 range (0.008 to 370 nM).
Figure 7.
Plot of cLogP versus –log IC50, slope = 0.41, r2 = 0.998.
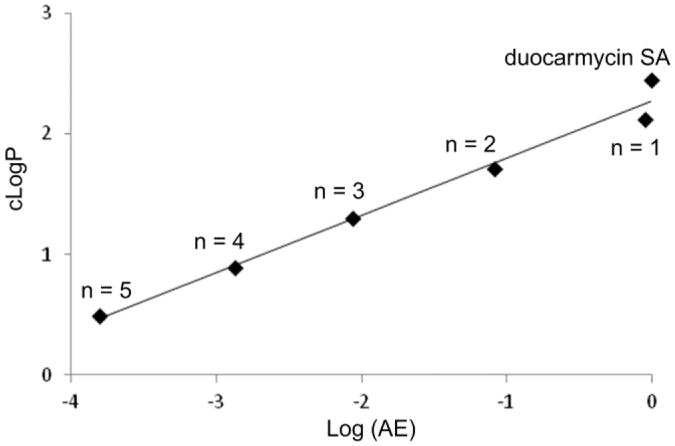
Another common measure used to assess the quality of compounds, to establish the extent to which the hydrophobic properties of a compound is productively used, and to link a compound's potency and lipophilicity is the lipophilic efficiency (LiPE = –log IC50 – cLogP), sometimes referred to as ligand–lipophilicity efficiency (LLE).22 By this measure, there is a clear indication of a well-defined, incrementally improved efficiency as the lipophilic character of the compounds increase, quantifying the increasingly productive use of the hydrophobic character of the molecules and its unmistakable magnitude (see Figure 4). Thus, the LiPE smoothly increases across the series (10e to 2) where the –log IC50 is increasing more than the increases in cLogP. As reflected in this LiPE measure and a feature that is especially notable in this series, the increase in cLogP in going from 10e to 2 is not derived from added hydrophobic structure content, but rather from systematic removal of hydrophilic components. In each stepwise removal of the hydrophilic components, the efficiency with which the hydrophobic interactions are utilized in target engagement is increased. Moreover, this occurs within the perfectly acceptable cLogP range of 0.49–2.5. Although there are additional parameters that also change across this series of compounds including incremental changes in molecular weight, number of hydrogen-bond acceptors, and polar surface area, they all also contribute to the overall composite property that make up the cLogP.22 Since the correlation and its magnitude are also observed for the target DNA alkylation efficiency conducted under cell free conditions, the relationship does not appear to reflect a significant contribution from the relative cell permeability of the compounds.23 Rather, the relationship can be accounted for and may be a direct reflection of the compound's interaction with the biological target itself. In fact, a plot of cLogP versus Log (AE), where AE = the relative cell free DNA alkylation efficiency, provides an analogous linear relationship that displays the same slope using either the data derived from densitometry measurements on the unreacted full length DNA (Figure 8, data in Figure 6) or from the resulting cleavage band (Supporting Information Figure S4). Just as significantly, a plot of –log IC50 versus Log (AE) displays a linear relationship with a slope of 0.85 (r2 = 0.99, Supporting Information Figure S5), not only establishing this direct correlation between functional activity and cell free target activity, but quantifying it as one that alone can account for nearly all of the effects (slope of 0.85 vs 1) observed in the cell based functional assay. This is the case because the target DNA alkylation effects are exceptionally large and outweigh any other effects of the structural changes including the effects on permeability.23
Figure 8.
Plot of cLogP versus Log (AE), (AE = relative DNA alkylation efficiency, Fig. 6), slope = 0.48, r2 = 0.98.
Synthesis of 15a–e
As highlighted in Figure 2, the duocarmycin SA trimethoxyindole substituents at the C6 and C7 positions extend away from DNA, indicating that modifications at these sites cannot sterically impede target binding, whereas those at C5 lie at a peripheral site at the end of the drug-bound DNA complex. Unlike the C6 and C7 sites, substituents or even additional DNA binding subunits attached at the C5 site impact target binding and have been used to enhance, not diminish, DNA binding and biological potency. As a result, the C5 PEG substitution in 10a–e would not be expected to sterically impede the behavior of the initial series nor would they be expected to do so in an incremental manner. However and because the preceding results were so stunning, we decided to establish whether the modifications at C5 in the 10a–e series were contributing to or responsible for the observed effects. The series most easily accessible and ideal for the study is 15a–e, where the systematic PEG modification is carrie out at a single site (C6) central to the location of the three methoxy groups. Not only is this the least intrusive of the three sites (C5–C7), but it is the easiest of the three sites to selectively modify. Thus, alkylation of the symmetrical 3,5-dimethoxy-4-hydroxybenzaldehyde with 4a–e afforded the desired alkylated products 11a–e as the precursors to this second series (Scheme 2). Treatment of aldehydes 11a–e with methyl 2-azidoacetate and sodium methoxide provided the cinnamate azides 12a–e, which were subsequently warmed in xylenes to initiate the Hemetsberger indole cyclization to afford 13a–e. The methyl esters were hydrolyzed to provide the carboxylic acids 14a–e, which were subsequently coupled with the duocarmycin SA alkylation subunit 917 to provide the seco-duocarmycin SA derivatives 15a–e.
Scheme 2.
Biological Activity of 15a–e
The series of compounds 15a–e was tested for cell growth inhibition, enlisting mouse lymphocytic leukemia cells (L1210, Figure 9). Like the observations made with 10a–e, incremental changes in activity were observed for compounds 15a–e, albeit representing smaller losses in potency for the sequential incorporations of an ethylene glycol unit since each stepwise structural change is much smaller. Compound 15a exhibited the most potent activity of the new series with an IC50 of 18 pM, being approximately 2-fold less active than duocarmycin SA, whereas compound 15e was the least potent (IC50 = 250 pM), experiencing a 30-fold loss in activity.
Figure 9.
Cell growth inhibition activity (L1210), cLogP, and LiPE for 15a–e.
cLogP Correlation Including 15a–e
The incorporation of the data for 15a–e into the plot of cLogP versus –log IC50 revealed that the series conformed to its expectations, indicating the generality of the observations, confirming that the C5 modifications to the original 10a–e series were not responsible for the observed trends, and representing a target non-obtrusive series that experiences smaller incremental losses in activity with each single ethylene glycol subunit introduction. These observations were further generalized by examination of 15a–e and 2 in a second tumor cell line, HCT116 (human colorectal cancer), where the series was found to exhibit an analogous direct linear relationship between cLogP and –log IC50 (Supporting Information Table S1 and Figure S3, r2 = 0.98).
Finally and despite this iterative loss in activity in both series, it is worth noting that the compounds examined are still extraordinarily potent. Up to six ethylene glycol units can be incorporated and still provide derivatives with IC50 values of approximately 250 pM or less and the incorporation of nine ethylene glycol units can still provide derivatives with single digit nM IC50 values. Thus, such modifications can be used to alter properties including solubility while still maintaining predictable and remarkable biological potencies.
Hydrophobic Binding-Driven-Bonding
As the properties of this class of molecules were first being defined, we often referred to the DNA alkylation reaction by this class of molecules as “hydrophobic binding-driven-bonding”,2h,24 reflecting a recognition of the dominance of the hydrophobic interactions in stabilizing their non-covalent complex with DNA that in turn drives and stabilizes the potentially reversible alkylation reaction that is not observed outside the structure of duplex DNA. Although the source of catalysis5 for the DNA alkylation reaction was not recognized at the time and the reversibility of the reaction was only demonstrated many years later,25 the results herein define and quantitate the fundamental role that the hydrophobic character of the molecules plays in both driving the intrinsically reversible DNA alkylation reaction and in the expression of their biological properties and establish the extraordinary magnitude of its effect.
Preceding studies have (1) shown that the DNA binding subunits control of the non-covalent minor groove binding selectivity and defined its impact on the resulting DNA alkylation selectivity,2,5,26 (2) characterized the importance of the vinylogous amide stabilization of the alkylation subunit and delineated the role that the DNA binding subunits play in its DNA binding-induced disruption and catalysis of the alkylation reaction,5-7 (3) identified alkylation subunit structural features that can markedly influence its intrinsic reactivity and impose stereoelectronic control on its reaction regioselectivity,9,27 (4) discovered the subtle impact of strategically placed substituents,28 and (5) delineated a fundamental relationship between the alkylation subunit intrinsic reactivity and biological potency.29 Herein, we define yet another fundamental property that Nature has integrated into their structures that profoundly influences their cell free DNA alkylation efficiency and functional activity.
Conclusion
A series of PEG-derivatized duocarmycin SA analogs were examined that demonstrated progressively increasing and ultimately excellent water solubility, and progressively diminished cell growth inhibition and DNA alkylation properties. The changes across the initial series examined (10a–e) are of a remarkable magnitude, representing 10-fold reductions in biological activity for each three-fold ethylene glycol unit introduction and providing an extraordinary 105-fold change in IC50 values over the series. A direct linear relationship between cLogP and –log IC50 (cell growth inhibition) and –Log AE (AE = cell free DNA alkylation efficiency) is observed for the compounds in the series where the cLogP values span the productive and effective range of 2.5–0.49 with incremental changes of approximately 0.4, and the –log IC50 values span the range of 11.2–6.4, representing IC50 values for the series covering a 105 range (0.008 to 370 nM). A second series (15a–e), entailing smaller stepwise PEG incorporations at a single site incapable of sterically impeding an interaction with the biological target, exhibit identical qualitative and quantitative trends, establish the generality of the observations, and rule out substituent effects unique to substitution at the C5 site. The results of the combined series define and quantitate a fundamental role that the hydrophobic character of the molecules play in the expression of the biological activity of members in this class, driving the intrinsically reversible DNA alkylation reaction (hydrophobic binding-driven-bonding), and define the stunning magnitude of its effect. In addition to providing a predicable relationship on which to design duocarmycin analogs, the implications of the observations likely extend to other classes of minor groove DNA binding compounds, and the approach may represent a general design protocol by which to assess the importance of hydrophobic properties in driving other small molecule/target interactions. Finally, it is remarkable to recognize that, more than 30 years after the discovery of the initial member of this class of natural products and despite the extensive efforts to date, there are still unrecognized fundamental structural features integrated in the natural product structures that are integral to the expression of their biological properties.
Experimental Section
General
Reagents and solvents were purchased reagent-grade and used without further purification. THF was freshly distilled from sodium benzophenone ketyl. All reactions were performed in oven-dried glassware under an Ar atmosphere. Evaporation and concentration in vacuo was performed at 20 °C. TLC was conducted using precoated SiO2 60 F254 glass plates from EMD with visualization by UV light (254 or 366 nm). Optical rotations were determined on a Rudolf Research Analytical Autopol III Automatic Polarimeter (λ = 589 nm, 25 °C). NMR (1H or 13C) were recorded on Bruker DRX-500 and DRX-600 NMP spectrophotometers at 298K. Residual solvent peaks were used as an internal reference. Coupling constants (J) (H,H) are given in Hz. Coupling patterns are designated as singlet (s), doublet (d), triplet (t), quadruplet (q), multiplet (m), or broad singlet (br). IR spectra were recorded on a Thermo Scientific Nicolet 380 FT-IR spectrophotometer and measured neat. High resolution mass spectral data were acquired on an Agilent Technologies high resolution LC/MSD-TOF, and the detected masses are given as m/z with m representing the molecular ion. The purity of each tested compound (>95%) was determined on an Agilent 1100 LC/MS instrument using a ZORBAX SBC18 column (3.5 mm, 4.6 mm × 50 mm, with a flow rate of 0.75 mL/min and detection at 220 and 254 nm) with a 10-98% acetonitrile/water/0.1% formic acid gradient.
Compounds 4a-e.13
A solution of NaOH (686 mg, 17.2 mmol) and alcohol (3a, 12.0 mmol) in 1:1 THF:H2O (8 mL) at 0 °C was treated with a solution of TsCl (2.13 g, 11.2 mmol) in 4 mL of THF. The solution was stirred at 0 °C for 8 h, after which the reaction mixture was poured into ice-water. The water mixture was extracted with ethyl acetate (×2), and the combined organic extracts were washed with saturated aqueous NaCl, dried (Na2SO4), and concentrated to provide 4a (1.94 g, 70%): 1H NMR (acetone-d6, 600 MHz) δ 7.81 (d, J = 7.8 Hz, 2H), 7.49 (d, J = 7.8 Hz, 2H), 4.15 (t, J = 7.8 Hz, 2H), 3.55 (t, J = 7.8 Hz, 2H), 3.23 (s, 3H), 2.46 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 146.8, 135.3, 131.8, 129.7, 71.5, 71.4, 59.7, 22.5; IR (film) vmax 2892, 1597 cm−1; ESI-TOF HRMS m/z 231.0690 (M+H+, C10H14O4S requires 231.0686). For 4b: (2.72 g, 83%); 1H NMR (acetone-d6, 600 MHz) δ 7.81 (d, J = 8.4 Hz, 2H), 7.48 (d, J = 7.8 Hz, 2H), 4.16 (t, J = 3.6 Hz, 2H), 3.65 (t, J = 3.6 Hz, 2H), 3.51 (t, J = 4.2 Hz, 2H), 3.41 (t, J = 3.9 Hz, 2H), 3.26 (s, 3H), 2.46 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 146.7, 135.3, 131.8, 129.7, 73.5, 72.0, 71.6, 70.2, 59.8, 22.5; IR (film) vmax 2879, 1598 cm−1; ESI-TOF HRMS m/z 275.0957 (M+H+, C12H18O5S requires 275.0948). For 4c: (3.34 g, 87%); 1H NMR (acetone-d6, 600 MHz) δ 7.82 (d, J = 8.4 Hz, 2H), 7.48 (d, J = 7.8 Hz, 2H), 4.16 (t, J = 3.6 Hz, 2H), 3.66 (t, J = 3.6 Hz, 2H), 3.53-3.50 (m, 6H), 3.45 (t, J = 3.6 Hz, 2H), 3.28 (s, 3H), 2.46 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 146.7, 135.3, 131.8, 129.7, 73.6, 72.2, 72.1, 72.0, 71.6, 70.2, 59.8, 22.5; IR (film) vmax 2872, 1598 cm– 1; ESI-TOF HRMS m/z 319.1217 (M+H+, C14H22O6S requires 319.1210). For 4d: (4.30 g, 99%); 1H NMR (acetone-d6, 600 MHz) δ 7.82 (d, J = 7.8 Hz, 2H), 7.49 (d, J = 7.8 Hz, 2H), 4.16 (t, J = 3.6 Hz, 2H), 3.66 (t, J = 3.6 Hz, 2H), 3.56–3.50 (m, 10H), 3.46 (t, J = 3.0 Hz, 2H), 3.28 (s, 3H), 2.46 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 146.7, 135.3, 131.8, 129.7, 73.6, 72.21, 72.17, 72.1, 72.0, 71.7, 70.2, 59.7, 22.5; IR (film) vmax 2874, 1597 cm−1; ESI-TOF HRMS m/z 363.1466 (M+H+, C16H26O7S requires 363.1472). For 4e: (4.81 g, 98%); 1H NMR (acetone-d6, 600 MHz) δ 7.82 (d, J = 7.8 Hz, 2H), 7.49 (d, J = 7.8 Hz, 2H), 4.16 (t, J = 4.2 Hz, 2H), 3.67 (t, J = 3.6 Hz, 2H), 3.58-3.50 (m, 14H), 3.46 (t, J = 3.6 Hz, 2H), 3.28 (s, 3H), 2.46 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 146.7, 135.3, 131.8, 129.7, 73.6, 72.21, 72.16, 72.1, 72.0, 71.7, 70.2, 59.8, 22.5; IR (film) vmax 2873, 1597 cm−1; ESI-TOF HRMS m/z 407.1732 (M+H+, C18H30O8S requires 407.1730).
Compounds 5a-e
A solution of 4a (1.94 g, 8.42 mmol) and 3,4,5-trihydroxybenzaldehyde (433 mg, 2.81 mmol) in acetone (14 mL) was treated with K2CO3 (1.94 g, 14.0 mmol) and warmed at 65 °C for 40 h. The reaction mixture was diluted with ethyl acetate, washed with H2O and saturated aqueous NaCl, and dried (Na2SO4). The solution was concentrated under reduced pressure and the resulting residue was purified by flash chromatography (SiO2, 50% EtOAc/hexanes) to afford 5a as a colorless oil (205 mg, 22%): 1H NMR (acetone-d6, 600 MHz) δ 9.87 (s, 1H), 7.25 (s, 2H), 4.26–4.20 (m, 6H), 3.76 (t, J = 4.4 Hz, 4H), 3.68 (t, J = 4.8 Hz, 2H), 3.38 (s, 6H), 3.35 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 192.6, 155.0, 145.6, 133.8, 110.1, 74.1, 73.5, 72.6, 70.6, 59.9, 59.7; IR (film) vmax 2929, 2882, 2820, 1690, 1582 cm−1; ESI-TOF HRMS m/z 329.1595 (M+H+, C16H24O7 requires 329.1595). For 5b: (344 mg, 23%); 1H NMR (acetone-d6, 600 MHz) δ 9.87 (s, 1H), 7.26 (s, 2H), 4.27-4.23 (m, 6H), 3.86 (t, J = 4.8 Hz, 4H), 3.78 (t, J = 4.8 Hz, 2H), 3.68 (t, J = 4.8 Hz, 4H), 3.65 (t, J = 5.2 Hz, 2H), 3.51 (t, J = 5.2 Hz, 4H), 3.48 (t, J = 4.8 Hz, 2H), 3.30 (s, 6H), 3.29 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 192.6, 155.0, 145.8, 133.8, 110.3, 74.3, 73.7, 72.3, 72.2, 72.0, 71.2, 70.8, 59.8; IR (film) vmax 2875, 1690, 1582 cm−1; ESI-TOF HRMS m/z 461.2379 (M+H+, C22H36O10 requires 461.2381). For 5c: (402 mg, 19%); 1H NMR (acetone-d6, 600 MHz) δ 9.87 (s, 1H), 7.26 (s, 2H), 4.27–4.18 (m, 6H), 3.88–3.83 (m, 4H), 3.80–3.76 (m, 2H), 3.69–3.64 (m, 6H), 3.62–3.54 (m, 12H), 3.48–3.44 (m, 6H), 3.28 (s, 9H); 13C NMR (acetone-d6, 150 MHz) δ 192.6, 155.0, 144.9, 133.8, 110.3, 74.3, 74.2, 73.6, 72.4, 72.32, 72.30, 72.28, 72.21, 72.20, 72.07, 72.05, 71.4, 71.3, 70.8, 70.7, 59.8; IR (film) vmax 2872, 1689, 1580 cm−1; ESI-TOF HRMS m/z 593.3165 (M+H+, C28H48O13 requires 593.3168). For 5d: (144 mg, 49%); 1H NMR (acetone-d6, 600 MHz) δ 9.87 (s, 1H), 7.27 (s, 2H), 4.28–4.19 (m, 6H), 3.89–3.83 (m, 4H), 3.81–3.76 (m, 2H), 3.69–3.64 (m, 6H), 3.63– 3.54 (m, 24H), 3.48–3.44 (m, 6H), 3.28 (s, 9H); 13C NMR (acetone-d6, 150 MHz) δ 192.7, 155.0, 144.9, 133.8, 110.3, 74.5, 74.3, 74.2, 73.6, 72.4, 72.3, 72.24, 72.21, 72.1, 72.03, 71.97, 71.34, 71.26, 70.8, 70.7, 59.8; IR (film) vmax 2870, 1689, 1581 cm−1; ESI-TOF HRMS m/z 725.3951 (M+H+, C34H60O16 requires 725.3954). For 5e: (149 mg, 39%); 1H NMR (acetone-d6, 600 MHz) δ 9.88 (s, 1H), 7.27 (s, 2H), 4.28–4.19 (m, 6H), 3.89–3.83 (m, 4H), 3.81–3.77 (m, 2H), 3.70–3.65 (m, 6H), 3.64–3.55 (m, 36H), 3.47–3.44 (m, 6H), 3.28 (s, 9H); 13C NMR (acetone-d6, 150 MHz) δ 192.7, 155.0, 145.7, 133.8, 110.3, 74.3, 74.2, 73.6, 73.5, 72.4, 72.3, 72.2, 72.0, 71.9, 71.33, 71.26, 70.8, 70.7, 59.8; IR (film) vmax 2871, 1688, 1579 cm−1; ESI-TOF HRMS m/z 857.4734 (M+H+, C40H72O19 requires 857.4740).
Compounds 6a-e
A solution of 5a (105 mg, 0.320 mmol) and methyl 2-azidoacetate (0.312 mL, 3.20 mmol) in MeOH (6.0 mL) at 0 °C was treated with NaOMe (138 mg, 2.56 mmol). The solution was stirred for 4 h at 0 °C, after which the reaction mixture was diluted with ethyl acetate, washed with H2O and saturated aqueous NaCl, and dried (Na2SO4). The solution was concentrated under reduced pressure and purified by flash chromatography (SiO2, 100% EtOAc) to afford 6a as a colorless oil (84 mg, 62%): 1H NMR (acetone-d6, 600 MHz) δ 7.28 (s, 2H), 6.88 (s, 1H), 4.20–4.14 (m, 6H), 3.88 (s, 3H), 3.74 (t, J = 4.4 Hz, 4H), 3.66 (t, J = 4.4 Hz, 2H), 3.38 (s, 6H), 3.35 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 165.5, 154.3, 141.8, 130.3, 127.1, 126.2, 112.1, 74.0, 73.5, 72.7, 70.6, 60.0, 59.7, 54.2; IR (film) vmax 2927, 2881, 1712, 1618, 1573 cm−1; ESI-TOF HRMS m/z 426.1861 (M+H+, C19H27N3O8 requires 426.1871). For 6b: (265 mg, 64%); 1H NMR (acetone-d6, 600 MHz) δ 7.28 (s, 2H), 6.88 (s, 1H), 4.20–4.16 (m, 6H), 3.88 (s, 3H), 3.83 (t, J = 4.4 Hz, 4H), 3.76 (t, J = 4.4 Hz, 2H), 3.69–3.64 (m, 6H), 3.52–3.47 (m, 6H), 3.30 (s, 9H); 13C NMR (acetone-d6, 150 MHz) δ 165.5, 154.3, 141.8, 130.2, 127.1, 126.1, 112.2, 74.2, 73.7, 72.2, 72.0, 71.3, 70.8, 59.8, 54.2; IR (film) vmax 2874, 1712, 1618, 1573 cm−1; ESI-TOF HRMS m/z 558.2648 (M+H+, C25H39N3O11 requires 558.2657). For 6c: (140 mg, 30%); 1H NMR (acetone-d6, 600 MHz) δ 7.29 (s, 2H), 6.89 (s, 1H), 4.22–4.18 (m, 6H), 3.89 (s, 3H), 3.86–3.83 (m, 4H), 3.79–3.76 (m, 2H), 3.70–3.64 (m, 6H), 3.61–3.56 (m, 12H), 3.49–3.45 (m, 6H), 3.28 (s, 9H); 13C NMR (acetone-d6, 150 MHz) δ 165.5, 154.3, 141.8, 130.2, 127.1, 126.1, 112.2, 74.2, 73.6, 72.4, 72.30, 72.28, 72.26, 72.2, 72.07, 72.06, 71.3, 70.8, 59.8, 54.2; IR (film) vmax 2873, 1712, 1619, 1575 cm−1; ESI-TOF HRMS m/z 690.3461 (M+H+, C31H51N3O14 requires 690.3444). For 6d: (26.5 mg, 38%); 1H NMR (acetone-d6, 600 MHz) δ 7.30 (s, 2H), 6.90 (s, 1H), 4.22–4.18 (m, 6H), 3.89 (s, 3H), 3.88–3.83 (m, 4H), 3.79–3.76 (m, 2H), 3.71–3.65 (m, 6H), 3.63–3.54 (m, 24H), 3.48–3.45 (m, 6H), 3.28 (s, 9H); 13C NMR (acetone-d6, 150 MHz) δ 165.5, 154.3, 141.6, 130.3, 127.1, 126.2, 112.1, 74.2, 73.6, 72.4, 72.3, 72.24, 72.20, 72.17, 72.15, 72.11, 72.03, 72.02, 71.3, 70.7, 59.8, 54.2; IR (film) vmax 2871, 1713, 1577 cm−1; ESI-TOF HRMS m/z 822.4229 (M+H+, C37H63N3O17 requires 822.4230). For 6e: (22.0 mg, 26%); 1H NMR (acetone-d6, 600 MHz) δ 7.30 (s, 2H), 6.90 (s, 1H), 4.23–4.19 (m, 6H), 3.89 (s, 3H), 3.88–3.84 (m, 4H), 3.79–3.76 (m, 2H), 3.71–3.65 (m, 6H), 3.63–3.52 (m, 36H), 3.48–3.45 (m, 6H), 3.28 (s, 9H); 13C NMR (acetone-d6, 150 MHz) δ 165.6, 154.3, 141.7, 130.2, 127.1, 126.2, 112.2, 74.2, 73.6, 72.4, 72.33, 72.28, 72.25, 72.23, 72.21, 72.0, 71.4, 70.8, 59.8, 54.2; IR (film) vmax 2870, 1714, 1579 cm−1; ESI-TOF HRMS m/z 976.4808 (M+Na+, C43H75N3O20 requires 976.4836).
Compounds 7a-e
A solution of 6a (84 mg, 0.197 mmol) in xylenes (10 mL) was warmed at 140 °C for 6 h, then loaded directly onto a silica gel column. The xylenes was eluted with hexanes, and then the residue was purified (100% EtOAc) to afford 7a (73.6 mg, 94%): 1H NMR (acetone-d6, 600 MHz) δ 10.66 (s, 1H), 7.06 (s, 1H), 6.96 (s, 1H), 4.32 (m, 2H), 4.19 (m, 2H), 4.14 (m, 2H), 3.88 (s, 3H), 3.75 (m, 4H), 3.70 (m, 2H), 3.50 (s, 3H), 3.39 (s, 3H), 3.38 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 163.4, 151.0, 142.5, 140.2, 129.5, 129.0, 125.1, 110.1, 102.0, 75.1, 74.7, 73.7, 73.5, 72.8, 70.6, 59.9, 59.7, 52.9; IR (film) vmax 2927, 2880, 1707, 1536 cm−1; ESI-TOF HRMS m/z 398.1809 (M+H+, C19H27NO8 requires 398.1809). For 7b: (250 mg, 99%); 1H NMR (acetone-d6, 600 MHz) δ 10.42 (s, 1H), 7.06 (s, 1H), 6.97 (s, 1H), 4.36 (m, 2H), 4.22 (m, 2H), 4.15 (m, 2H), 3.87 (s, 3H), 3.85 (m, 2H), 3.82–3.78 (m, 4H), 3.74 (m, 2H), 3.68–3.63 (m, 6H), 3.51 (m, 4H), 3.32 (s, 3H), 3.30 (s, 6H); 13C NMR (acetone-d6, 150 MHz) δ 163.4, 151.1, 142.4, 140.1, 129.6, 129.0, 125.1, 110.3, 101.8, 75.0, 74.8, 73.72, 73.70, 73.4, 72.2, 72.1, 72.03, 72.01, 71.4, 70.7, 59.82, 59.78, 52.9; IR (film) vmax 2877, 1711, 1536 cm−1; ESI-TOF HRMS m/z 530.2588 (M+H+, C25H39NO11 requires 530.2596). For 7c: (129 mg, 96%); 1H NMR (acetone-d6, 600 MHz) δ 10.48 (s, 1H), 7.06 (s, 1H), 6.98 (s, 1H), 4.37 (m, 2H), 4.23 (m, 2H), 4.16 (m, 2H), 3.87 (s, 3H), 3.86 (m, 2H), 3.81 (m, 2H), 3.74 (m, 4H), 3.72-3.66 (m, 4H), 3.62-3.56 (m, 12H), 3.46 (m, 6H), 3.28 (s, 9H); 13C NMR (acetone-d6, 150 MHz) δ 163.4, 151.1, 142.4, 140.1, 129.6, 129.0, 125.1, 110.2, 101.9, 75.0, 74.8, 73.7, 73.6, 72.43, 72.40, 72.33, 72.31, 72.29, 72.2, 72.1, 72.04, 72.00, 71.4, 70.8, 59.8, 59.7, 52.9; IR (film) vmax 2875, 1710, 1536 cm−1; ESI-TOF HRMS m/z 662.3360 (M+H+, C31H51NO14 requires 662.3382). For 7d: (22.6 mg, 88%); 1H NMR (acetone-d6, 600 MHz) δ 10.53 (s, 1H), 7.07 (s, 1H), 6.98 (s, 1H), 4.37 (m, 2H), 4.24 (m, 2H), 4.17 (m, 2H), 3.88 (s, 3H), 3.86 (m, 2H), 3.82 (m, 2H), 3.75 (m, 4H), 3.72–3.66 (m, 4H), 3.62–3.54 (m, 24H), 3.46 (m, 6H), 3.27 (s, 9H); 13C NMR (acetone-d6, 150 MHz) δ 163.4, 151.1, 142.4, 140.1, 129.6, 129.0, 125.1, 110.2, 101.8, 75.1, 74.8, 73.62, 73.59, 72.41, 72.38, 72.32, 72.29, 72.24, 72.20, 72.17, 72.15, 72.1, 72.03, 72.00, 71.4, 70.7, 59.8, 59.7, 52.9; IR (film) vmax 2872, 1710, 1582 cm−1; ESI-TOF HRMS m/z 794.4159 (M+H+, C37H63NO17 requires 794.4169). For 7e: (20.0 mg, 93%); 1H NMR (acetone-d6, 600 MHz) δ 10.54 (s, 1H), 7.07 (s, 1H), 6.98 (s, 1H), 4.37 (m, 2H), 4.24 (m, 2H), 4.17 (m, 2H), 3.88 (s, 3H), 3.86 (m, 2H), 3.81 (m, 2H), 3.75 (m, 4H), 3.72–3.66 (m, 4H), 3.62–3.52 (m, 36H), 3.46 (m, 6H), 3.28 (s, 9H); 13C NMR (acetone-d6, 150 MHz) δ 163.4, 151.1, 142.4, 140.1, 129.6, 129.0, 125.1, 110.2, 101.8, 75.1, 74.8, 73.6, 72.41, 72.39, 72.32, 72.30, 72.27, 72.24, 72.21, 72.16, 72.03, 72.00, 71.4, 70.7, 59.8, 59.7, 52.9; IR (film) vmax 2872, 1711, 1584 cm−1; ESI-TOF HRMS m/z 926.4936 (M+H+, C43H75NO20 requires 926.4955).
Compounds 8a-e
A solution of 7a (78 mg, 0.197 mmol) in 3:2:1 THF:MeOH:H2O (4.0 mL) was treated with LiOH (23.6 mg, 0.985 mmol) and was stirred for 16 h at room temperature. The reaction mixture was diluted with ethyl acetate, washed with 0.1 M HCl, H2O, and saturated aqueous NaCl, and dried (Na2SO4). The solution was concentrated under reduced pressure to afford 8a as a white solid (66.4 mg, 88%): 1H NMR (acetone-d6, 500 MHz) δ 10.66 (s, 1H), 7.06 (d, J = 2.0 Hz, 1H), 6.98 (s, 1H), 4.33 (t, J = 4.5 Hz, 2H), 4.22–4.18 (m, 4H), 4.15 (t, J = 4.5 Hz, 2H), 3.77–3.73 (m, 6H), 3.71 (t, J = 5.0 Hz, 2H), 3.49 (s, 3H), 3.40 (s, 3H), 3.38 (s, 3H); 13C NMR (acetone-d6, 125 MHz) δ 163.8, 151.0, 142.4, 140.1, 129.6, 129.5, 125.3, 110.1, 102.1, 75.1, 74.7, 73.7, 73.5, 72.8, 72.7, 70.6, 59.9, 59.7; IR (film) vmax 3407, 2927, 1676, 1537 cm−1; ESI-TOF HRMS m/z 384.1650 (M+H+, C18H25NO8 requires 384.1653). For 8b: (219 mg, 89%); 1H NMR (acetone-d6, 500 MHz) δ 10.54 (s, 1H), 7.08 (d, J = 2.0 Hz, 1H), 6.97 (s, 1H), 4.37 (t, J = 4.5 Hz, 2H), 4.23 (t, J = 4.5 Hz, 2H), 4.16 (t, J = 4.5 Hz, 2H), 3.87–3.83 (m, 2H), 3.81 (t, J = 5.0 Hz, 2H), 3.74–3.67 (m, 4H), 3.63–3.56 (m, 6H), 3.48–3.44 (m, 4H), 3.282 (s, 3H), 3.280 (s, 3H), 3.26 (s, 3H); 13C NMR (acetone-d6, 125 MHz) δ 163.9, 150.9, 142.2, 140.1, 129.7, 129.5, 125.2, 110.3, 101.9, 74.9, 74.8, 73.62, 73.55, 72.4, 72.3, 72.24, 72.19, 72.04, 72.00, 71.9, 71.4, 70.8, 59.8, 59.7; IR (film) vmax 3420, 2875, 1701, 1538 cm−1; ESI-TOF HRMS m/z 516.2440 (M+H+, C24H37NO11 requires 516.2439). For 8c: (120 mg, 92%); 1H NMR (acetone-d6, 600 MHz) δ 10.60 (s, 1H), 7.08 (d, J = 1.8 Hz, 1H), 6.97 (s, 1H), 4.37 (t, J = 4.2 Hz, 2H), 4.23 (t, J = 4.2 Hz, 2H), 4.16 (t, J = 4.2 Hz, 2H), 3.87–3.83 (m, 2H), 3.81 (t, J = 4.8 Hz, 2H), 3.76–3.66 (m, 10H), 3.63–3.55 (m, 12H), 3.48–3.44 (m, 4H), 3.29 (s, 3H), 3.28 (s, 3H), 3.26 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 163.9, 150.9, 142.1, 140.0, 129.7, 129.5, 125.2, 110.2, 101.8, 74.9, 74.8, 73.6, 73.5, 72.4, 72.33, 72.25, 72.24, 72.21, 72.16, 72.14, 72.02, 71.99, 71.97, 71.9, 71.4, 71.3, 71.2, 70.8, 59.8, 59.7; IR (film) vmax 3390, 2874, 1701, 1539 cm−1; ESI-TOF HRMS m/z 648.3232 (M+H+, C30H49NO14 requires 648.3226). For 8d: (21 mg, 97%); 1H NMR (acetone-d6, 600 MHz) δ 10.47 (s, 1H), 7.06 (d, J = 1.8 Hz, 1H), 6.97 (s, 1H), 4.38 (t, J = 4.2 Hz, 2H), 4.23 (t, J = 4.2 Hz, 2H), 4.16 (t, J = 4.2 Hz, 2H), 3.87–3.81 (m, 4H), 3.74–3.68 (m, 4H), 3.63–3.54 (m, 24H), 3.48–3.44 (m, 4H), 3.28 (s, 6H), 3.27 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 165.0, 150.9, 142.1, 140.1, 129.4, 129.3, 125.3, 110.0, 101.9, 75.0, 74.8, 73.6, 73.5, 72.40, 72.38, 72.33, 72.30, 72.29, 72.20, 72.15, 72.12, 72.09, 72.03, 71.93, 70.8, 59.8; IR (film) vmax 3408, 2874, 1692, 1539 cm−1; ESI-TOF HRMS m/z 780.3996 (M+H+, C36H61NO17 requires 780.4012). For 8e: (19.7 mg, 98%); 1H NMR (acetone-d6, 600 MHz) δ 10.50 (s, 1H), 7.06 (d, J = 1.8 Hz, 1H), 6.96 (s, 1H), 4.38 (t, J = 4.2 Hz, 2H), 4.25 (t, J = 4.2 Hz, 2H), 4.16 (t, J = 4.2 Hz, 2H), 3.87–3.83 (m, 4H), 3.74– 3.68 (m, 4H), 3.63–3.55 (m, 36H), 3.47–3.44 (m, 4H), 3.2 (s, 9H); 13C NMR (acetone-d6, 150 MHz) δ 164.8, 150.8, 142.3, 140.1, 129.31, 129.3, 125.3, 110.3, 101.9, 75.0, 74.4, 73.6, 73.5, 72.41, 72.39, 72.3, 72.24, 72.21, 72.14, 72.13, 72.09, 72.07, 72.03, 71.98, 71.9, 71.5, 71.4, 71.34, 71.28, 70.8, 59.8; IR (film) vmax 3400, 2874, 1582 cm−1; ESI-TOF HRMS m/z 912.4777 (M+H+, C42H73NO20 requires 780.4012).
Compounds 10a-e
A solution of 8a (33.2 mg, 0.0866 mmol), 9 (38.5 mg, 0.0866 mmol), and EDCI (49.8 mg, 0.260 mmol) in DMF (2.0 mL) was stirred at room temperature for 16 h. The reaction mixture was diluted with ethyl acetate, washed with aqueous 1 M HCl, saturated aqueous NaHCO3, H2O, and saturated aqueous NaCl, and dried (Na2SO4). The solvent was removed under reduced pressure and the residue was purified by flash chromatography (SiO2) to provide 10a as a yellow solid (22.8 mg, 41%): 1H NMR (acetone-d6, 500 MHz) δ 10.69 (s, 1H), 10.53 (s, 1H), 8.92 (s, 1H), 8.04 (s, 1H), 7.27 (d, J = 2.0 Hz, 1H), 7.06 (d, J = 2.0 Hz, 1H), 7.01 (s, 1H), 4.78 (t, J = 9.5 Hz, 1H), 4.60 (dd, J = 11, 4.0 Hz, 1H), 4.36 (t, J = 4.5 Hz, 2H), 4.21–4.15 (m, 6H), 3.91 (t, J = 8.0 Hz, 1H), 3.90 (s, 3H), 3.78–3.71 (m, 6H), 3.54 (s, 3H), 3.41 (s, 3H), 3.39 (s, 3H); 13C NMR (acetone-d6, 125 MHz) δ 163.3, 161.2, 150.8, 145.5, 142.1, 140.9, 140.0, 133.0, 130.4, 128.1, 127.8, 126.3, 125.9, 115.3, 107.8, 107.6, 103.1, 102.3, 75.2, 74.7, 73.7, 73.6, 72.9, 70.7, 60.1, 60.0, 59.8, 56.7, 53.1, 49.0, 44.8; IR (film) vmax 3228, 2923, 2882, 1711, 1587 cm−1; ESI-TOF HRMS m/z 646.2153 (M+H+, C31H36ClN3O10 requires 646.2162). For 10b: (26.1 mg, 43%); 1H NMR (acetone-d6, 500 MHz) δ 10.70 (s, 1H), 10.28 (s, 1H), 8.93 (s, 1H), 8.02 (s, 1H), 7.27 (d, J = 2.0 Hz, 1H), 7.05 (d, J = 2.0 Hz, 1H), 7.01 (s, 1H), 4.77 (t, J = 9.5 Hz, 1H), 4.59 (dd, J = 11, 4.0 Hz, 1H), 4.39 (t, J = 4.5 Hz, 2H), 4.24–4.14 (m, 6H), 3.90 (s, 3H), 3.86 (t, J = 5.0 Hz, 2H), 3.81 (q, J = 5.5 Hz, 4H), 3.75 (t, J = 5.5 Hz, 2H), 3.72–3.64 (m, 6H), 3.54–3.51 (m, 4H), 3.31 (s, 3H), 3.30 (s, 3H), 3.28 (s, 3H); 13C NMR (acetone-d6, 125 MHz) δ 163.3, 161.3, 150.9, 145.5, 142.0, 140.8, 139.9, 133.0, 130.4, 128.1, 127.8, 126.3, 125.7, 115.3, 107.8, 107.6, 103.1, 102.1, 75.1, 74.8, 73.7, 73.5, 72.23, 72.17, 72.13, 72.07, 71.5, 70.8, 59.8, 56.8, 53.1, 49.0, 44.8; IR (film) vmax 3239, 2922, 2883, 1714, 1612 cm−1; ESI-TOF HRMS m/z 800.2770 (M+Na+, C37H48ClN3O13 requires 800.2768). For 10c: (64.6 mg, 45%); 1H NMR (acetone-d6, 600 MHz) δ 10.77 (s, 1H), 10.37 (s, 1H), 9.05 (s, 1H), 8.04 (s, 1H), 7.26 (d, J = 1.8 Hz, 1H), 7.04 (d, J = 2.4 Hz, 1H), 7.00 (s, 1H), 4.76 (t, J = 10.2 Hz, 1H), 4.57 (dd, J = 10.2, 3.6 Hz, 1H), 4.39 (t, J = 4.2 Hz, 2H), 4.24–4.13 (m, 6H), 3.89 (s, 3H), 3.87–3.79 (m, 7H), 3.76 (m, 2H), 3.69 (t, J = 4.8 Hz, 4H), 3.65–3.57 (m, 14H), 3.48 (t, J = 4.8 Hz, 4H), 3.30 (s, 3H), 3.29 (s, 3H), 3.22 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 163.3, 161.3, 150.8, 145.5, 141.9, 140.8, 139.8, 132.9, 130.3, 128.1, 127.8, 126.2, 125.7, 115.3, 107.8, 107.7, 103.1, 102.0, 75.1, 74.8, 73.64, 73.63, 73.5, 72.4, 72.29, 72.26, 72.2, 72.07, 72.06, 72.04, 71.96, 71.4, 70.8, 59.8, 56.7, 53.1, 48.9, 44.7; IR (film) vmax 3256, 2924, 2878, 1713, 1613 cm−1; ESI-TOF HRMS m/z 910.3701 (M+H+, C43H60ClN3O16 requires 910.3735). For 10d: (4.5 mg, 14%); 1H NMR (acetone-d6, 600 MHz) δ 11.00 (s, 1H), 10.37 (s, 1H), 9.16 (br s, 1H), 8.00 (s, 1H), 7.27 (s, 1H), 7.07 (s, 1H), 7.04 (s, 1H), 4.79 (t, J = 10.2 Hz, 1H), 4.60 (dd, J = 10.2, 3.6 Hz, 1H), 4.40 (t, J = 4.2 Hz, 2H), 4.26–4.16 (m, 6H), 3.90 (s, 3H), 3.87–3.79 (m, 7H), 3.76–3.70 (m, 6H), 3.65–3.55 (m, 26H), 3.47 (m, 4H), 3.29 (s, 3H), 3.27 (s, 3H), 3.22 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 163.4, 161.2, 150.7, 145.6, 141.8, 140.8, 139.9, 133.1, 130.3, 128.1, 127.9, 126.2, 125.8, 115.1, 107.7, 107.6, 103.1, 102.1, 75.2, 74.9, 73.6, 73.5, 72.7, 72.6, 72.4, 72.3, 72.22, 72.19, 72.04, 71.97, 71.9, 71.8, 71.42, 71.37, 71.2, 70.8, 59.8, 56.7, 53.1, 49.0, 44.8; IR (film) vmax 3260, 2921, 2874, 1716, 1617 cm−1; ESI-TOF HRMS m/z 1042.4492 (M+H+, C49H72ClN3O19 requires 1042.4521). For 10e: (3.0 mg, 12%); 1H NMR (acetone-d6, 600 MHz) δ 11.06 (s, 1H), 10.37 (s, 1H), 9.32 (br s, 1H), 8.00 (s, 1H), 7.27 (s, 1H), 7.08 (s, 1H), 7.04 (s, 1H), 4.79 (t, J = 9.6 Hz, 1H), 4.60 (dd, J = 10.2, 3.6 Hz, 1H), 4.40 (t, J = 4.2 Hz, 2H), 4.27–4.16 (m, 6H), 3.90 (s, 3H), 3.86–3.79 (m, 7H), 3.71–3.54 (m, 44H), 3.45 (m, 4H), 3.28 (s, 3H), 3.27 (s, 3H), 3.25 (s, 3H); 13C NMR (acetone-d6, 150 MHz) δ 163.4, 161.3, 150.7, 145.7, 141.8, 140.8, 139.9, 133.0, 130.3, 128.1, 127.9, 126.2, 125.8, 115.1, 107.7, 107.6, 103.1, 102.1, 75.1, 74.9, 73.6, 73.5, 72.36, 72.35, 72.34, 72.30, 72.28, 72.26, 72.25, 72.21, 72.19, 72.18, 72.16, 72.15, 72.10, 72.06, 72.02, 72.00, 71.9, 71.4, 70.8, 59.8, 56.7, 53.0, 49.0, 44.8; IR (film) vmax 3245, 2869, 1712, 1617 cm−1; ESI-TOF HRMS m/z 1174.5289 (M+H+, C55H84ClN3O22 requires 1174.5307).
Compounds 11a-e
A solution of 4a (277 mg, 1.20 mmol) and 4-hydroxy-3,5-dimethoxybenzaldehyde (182 mg, 1.00 mmol) in DMF (3.3 mL) was treated with Cs2CO3 (326 mg, 1.00 mmol) and the reaction mixture was warmed at 160 °C for 20 min under microwave irradiation. After cooling, the reaction mixture was poured into water, extracted with ethyl acetate, washed with saturated aqueous NaCl and dried over Na2SO4. The solution was concentrated under reduced pressure and the resulting residue was purified by flash chromatography (SiO2) to afford 11a as a colorless oil (187 mg, 78%): 1H NMR (CDCl3, 400 MHz) δ 9.85 (s, 1H), 7.11 (s, 2H), 4.21 (t, J = 4.8 Hz, 2H), 3.90 (s, 6H), 3.71 (t, J = 4.8 Hz, 2H), 3.41 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 190.6, 153.3, 142.0, 131.4, 106.1, 71.7, 71.3, 58.4, 55.7; IR (film) vmax 2934, 1691, 1588, 1462, 1329, 1120, 1035 cm−1; ESI-TOF HRMS m/z 241.1080 (M+H+, C12H16O5 requires 241.1076). For 11b: (220 mg, 73%); 1H NMR (CDCl3, 400 MHz) δ 9.85 (s, 1H), 7.11 (s, 2H), 4.25 (t, J = 4.8 Hz, 2H), 3.90 (s, 6H), 3.81 (t, J = 4.8 Hz, 2H), 3.69 (t, J = 4.8 Hz, 2H), 3.54 (t, J = 4.8 Hz, 2H), 3.35 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 190.7, 153.3, 142.2, 131.4, 106.2, 71.9, 71.5, 70.1, 70.0, 58.6, 55.8; IR (film) vmax 2941, 1691, 1587, 1462, 1423, 1328, 1126, 1038 cm−1; ESI-TOF HRMS m/z 285.1335 (M+H+, C14H20O6 requires 285.1333). For 11c: (230 mg, 83%); 1H NMR (CDCl3, 400 MHz) δ 9.80 (s, 1H), 7.04 (s, 2H), 4.17 (t, J = 4.8 Hz, 2H), 3.90 (s, 6H), 3.81 (t, J = 4.8 Hz, 2H), 3.73 (t, J = 4.8 Hz, 2H), 3.64 (t, J = 4.8 Hz, 2H), 3.54–3.60 (m, 4H), 3.46 (t, J = 4.8 Hz, 2H), 3.29 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 190.9, 153.5, 142.4, 131.5, 106.4, 72.1, 71.6, 70.4, 70.3, 70.2, 70.1, 58.7, 56.0; IR (film) vmax 2872, 1688, 1496, 1459, 1422, 1325, 1118, 1033 cm−1; ESI-TOF HRMS m/z 329.1584 (M+H+, C16H24O7 requires 329.1595). For 11d: (580 mg, 78%); 1H NMR (CDCl3, 400 MHz) δ 9.78 (s, 1H), 7.04 (s, 2H), 4.16 (t, J = 4.8 Hz, 2H), 3.83 (s, 6H), 3.73 (t, J = 4.8 Hz, 2H), 3.62 (t, J = 4.8 Hz, 2H), 3.52-3.60 (m, 8H), 3.44 (t, J = 4.8 Hz, 2H), 3.27 (s, 3H); 13C NMR (CDCl3, 100 MHz) 5 190.9, 153.5, 142.4, 131.5, 106.4, 72.1, 71.6, 70.4, 70.3, 70.2, 70.1, 58.8, 56.0; IR (film) vmax 2870, 1689, 1584, 1460, 1422, 1325, 1229, 1119, 1033 cm−1; ESI-TOF HRMS m/z 373.1865 (M+H+, C18H28O8 requires 373.1857). For 11e: (280 mg, 72%); 1H NMR (CDCl3, 400 MHz) δ 9.84 (s, 1H), 7.09 (s, 2H), 4.22 (t, J = 4.8 Hz, 2H), 3.89 (s, 6H), 3.79 (t, J = 4.8 Hz, 2H), 3.68 (t, J = 4.8 Hz, 2H), 3.58–3.66 (m, 12H), 3.52 (t, J = 4.8 Hz, 2H), 3.34 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 191.0, 153.7, 142.6, 131.7, 106.6, 72.3, 71.8, 70.6, 70.5, 70.4, 70.3, 58.9, 56.2; IR (film) vmax 2869, 1689, 1585, 1496, 1460, 1326, 1119, 1034 cm−1; ESI-TOF HRMS m/z 417.2123 (M+H+, C20H32O9 requires 417.2119).
Compounds 13a-e
A solution of 11a (132 mg, 0.55 mmol) and methyl 2-azidoacetate (632 mg, 5.50 mmol) in MeOH (2.75 mL) at 0 °C was treated with NaOMe (4.4 M in MeOH, 1.0 ml, 4.4 mmol). The solution was stirred for 3 h at 0 °C, after which the reaction mixture was diluted with ethyl acetate, washed with water and saturated aqueous NaCl, and dried over Na2SO4. The solution was concentrated under reduced pressure to afford crude 12a. The crude material 12a was dissolved in xylenes (10 mL), warmed at 140 °C for 12 h, then loaded directly onto a silica gel column. The xylenes was eluted with hexane, and then the residue was purified by flash chromatography to afford 13a as a colorless oil (93 mg, 55%): 1H NMR (CDCl3, 400 MHz) δ 9.09 (s, 1H), 7.11 (s, 1H), 6.82 (s, 1H), 4.20 (t, J = 4.8 Hz, 2H), 4.09 (s, 3H), 3.92 (s, 3H), 3.88 (s, 3H), 3.75 (t, J = 4.8 Hz, 2H), 3.46 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 162.1, 150.2, 139.3, 139.1, 126.7, 126.5, 123.1, 108.7, 97.5, 72.6, 71.7, 61.2, 58.9, 56.1, 51.8; IR (film) vmax 3302, 2932, 1701, 1570, 1493, 1300, 1220, 1198, 1110, 1058 cm−1; ESI-TOF HRMS m/z 310.1295 (M+H+, C15H19NO6 requires 310.1291). For 13b: (120 mg, 51%); 1H NMR (CDCl3, 400 MHz) δ 8.98 (s, 1H), 7.09 (s, 1H), 6.81 (s, 1H), 4.21 (t, J = 4.8 Hz, 2H), 4.08 (s, 3H), 3.91 (s, 3H), 3.86 (s, 3H), 3.83 (t, J = 4.8 Hz, 2H), 3.72 (t, J = 4.8 Hz, 2H), 3.58 (t, J = 4.8 Hz, 2H), 3.38 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 162.1, 150.2, 139.2, 139.1, 126.7, 126.5, 123.0, 108.7, 97.5, 72.7, 71.9, 70.4, 70.3, 61.3, 59.0, 56.1, 51.8; IR (film) vmax 3305, 2934, 1702, 1537, 1496, 1302, 1223, 1196, 1108, 1055 cm−1; ESI-TOF HRMS m/z 354.1549 (M+H+, C17H23NO7 requires 354.1547). For 13c: (89 mg, 58%); 1H NMR (CDCl3, 400 MHz) δ 9.01 (s, 1H), 7.08 (s, 1H), 6.78 (s, 1H), 4.19 (t, J = 4.8 Hz, 2H), 4.06 (s, 3H), 3.90 (s, 3H), 3.84 (s, 3H), 3.81 (t, J = 4.8 Hz, 2H), 3.72 (t, J = 4.8 Hz, 2H), 3.68 (t, J = 4.8 Hz, 2H), 3.64 (t, J = 4.8 Hz, 2H), 3.52 (t, J = 4.8 Hz, 2H), 3.35 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 162.0, 150.2, 139.3, 139.0, 126.7, 126.5, 123.0, 108.7, 97.6, 72.7, 71.8, 70.6, 70.5, 70.4, 70.3, 61.2, 58.9, 56.1, 51.8; IR (film) vmax 3307, 2873, 1707, 1536, 1436, 1302, 1222, 1092, 1053 cm−1; ESI-TOF HRMS m/z 398.1802 (M+H+, C19H27NO8 requires 398.1809). For 13d: (319 mg, 61%); 1H NMR (CDCl3, 400 MHz) δ 9.02 (s, 1H), 7.09 (s, 1H), 6.80 (s, 1H), 4.19 (t, J = 4.8 Hz, 2H), 4.07 (s, 3H), 3.91 (s, 3H), 3.86 (s, 3H), 3.82 (t, J = 4.8 Hz, 2H), 3.71 (t, J = 4.8 Hz, 2H), 3.60–3.68 (m, 8H), 3.56 (t, J = 4.8 Hz, 2H), 3.36 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 162.0, 150.2, 139.3, 139.1, 126.7, 126.4, 123.1, 108.8, 97.6, 72.8, 71.9, 70.6, 70.5, 70.4, 70.3, 61.3, 59.0, 56.1, 51.9; IR (film) vmax 3304, 2884, 1710, 1538, 1436, 1254, 1222, 1113, 1057 cm−1; ESI-TOF HRMS m/z 442.2077 (M+H+, C21H31NO9 requires 442.2071). For 13e: (130 mg, 59%); 1H NMR (CDCl3, 400 MHz) δ 8.94 (s, 1H), 7.09 (s, 1H), 6.80 (s, 1H), 4.20 (t, J = 4.8 Hz, 2H), 4.09 (s, 3H), 3.92 (s, 3H), 3.87 (s, 3H), 3.82 (t, J = 4.8 Hz, 2H), 3.72 (t, J = 4.8 Hz, 2H), 3.68 (t, J = 4.8 Hz, 2H), 3.60–3.66 (m, 10H), 3.57 (t, J = 4.8 Hz, 2H), 3.37 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 162.0, 150.2, 139.2, 139.0, 126.6, 126.4, 123.0, 108.7, 97.5, 72.7, 71.8, 70.5, 70.4, 70.3, 70.2, 61.2, 58.9, 56.1, 51.8; IR (film) vmax 3321, 2873, 1710, 1538, 1462, 1303, 1228, 1196, 1113, 1057 cm−1; ESI-TOF HRMS m/z 486.2330 (M+H+, C23H35NO10 requires 486.2334).
Compounds 14a-e
A solution of 13a (100 mg, 0.32 mmol) in 3:2:1 THF:MeOH:H2O (6.4 mL) was treated with LiOH (39 mg, 1.62 mmol). The reaction mixture was stirred for 16 h at room temperature. The reaction mixture was diluted with ethyl acetate, washed with aqueous 0.1 M HCl, water, saturated aqueous NaCl, and dried over Na2SO4. The solution was concentrated under reduced pressure to afford 14a as a white solid (80 mg, 83%): 1H NMR (CDCl3, 400 MHz) δ 9.06 (s, 1H), 7.24 (s, 1H), 6.83 (s, 1H), 4.20 (t, J = 4.8 Hz, 2H), 4.11 (s, 3H), 3.88 (s, 3H), 3.76 (t, J = 4.8 Hz, 2H), 3.47 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 166.2, 150.4, 139.8, 139.1, 127.2, 125.9, 123.2, 110.7, 97.7, 72.7, 71.8, 61.4, 58.9, 56.2; IR (film) vmax 3286, 2938, 1679, 1548, 1302, 1260, 1115, 1057 cm−1; ESI-TOF HRMS m/z 296.1141 (M+H+, C14H17NO6 requires 296.1129). For 14b: (105 mg, 87%); 1H NMR (CDCl3, 400 MHz) δ 9.19 (s, 1H), 7.22 (s, 1H), 6.81 (s, 1H), 4.22 (t, J = 4.8 Hz, 2H), 4.10 (s, 3H), 3.86 (s, 3H), 3.84 (t, J = 4.8 Hz, 2H), 3.74 (t, J = 4.8 Hz, 2H), 3.60 (t, J = 4.8 Hz, 2H), 3.40 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 165.9, 150.3, 139.6, 139.1, 127.1, 126.2, 123.2, 110.4, 97.6, 72.8, 71.9, 70.4, 61.3, 59.0, 56.1; IR (film) vmax 3270, 2940, 1686, 1540, 1302, 1225, 1113, 1057 cm−1; ESI-TOF HRMS m/z 340.1405 (M+H+, C16H21NO7 requires 340.1391). For 14c: (45 mg, 89%); 1H NMR (CDCl3, 400 MHz) δ 9.28 (s, 1H), 7.19 (s, 1H), 6.79 (s, 1H), 4.20 (t, J = 4.8 Hz, 2H), 4.08 (s, 3H), 3.85 (s, 3H), 3.83 (t, J = 4.8 Hz, 2H), 3.74 (t, J = 4.8 Hz, 2H), 3.65–3.70 (m, 4H), 3.55 (t, J = 4.8 Hz, 2H), 3.37 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 165.6, 150.2, 139.6, 139.0, 127.0, 126.4, 123.2, 110.2, 97.6, 72.8, 70.6, 70.5, 70.4, 70.3, 70.2, 61.3, 58.9, 56.1; IR (film) vmax 3274, 2932, 1692, 1498, 1301, 1220, 1112, 1056 cm−1; ESI-TOF HRMS m/z 384.1650 (M+H+, C18H25NO8 requires 384.1653). For 14d: (110 mg, 88%); 1H NMR (CDCl3, 400 MHz) δ 9.28 (s, 1H), 7.19 (s, 1H), 6.79 (s, 1H), 4.20 (t, J = 4.8 Hz, 2H), 4.08 (s, 3H), 3.86 (s, 3H), 3.84 (t, J = 4.8 Hz, 2H), 3.72 (t, J = 4.8 Hz, 2H), 3.62–3.70 (m, 8H), 3.55 (t, J = 4.8 Hz, 2H), 3.37 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 165.5, 150.2, 139.5, 139.0, 127.0, 126.2, 123.1, 110.2, 97.6, 72.7, 71.8, 70.5, 70.4, 70.3, 70.2, 61.3, 58.9, 56.1; IR (film) vmax 3273, 2934, 1699, 1498, 1462, 1301, 1220, 1114, 1057 cm−1; ESI-TOF HRMS m/z 428.1910 (M+H+, C20H29NO9 requires 428.1915). For 14e: (80 mg, 91%); 1H NMR (CDCl3, 400 MHz) δ 9.14 (s, 1H), 7.21 (s, 1H), 6.98 (s, 1H), 6.81 (s, 1H), 4.22 (t, J = 4.8 Hz, 2H), 4.07 (s, 3H), 3.87 (s, 3H), 3.83 (t, J = 4.8 Hz, 2H), 3.73 (t, J = 4.8 Hz, 2H), 3.63–3.70 (m, 12H), 3.55 (t, J = 4.8 Hz, 2H), 3.38 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ 165.3, 150.2, 139.5, 139.0, 127.0, 126.4, 123.1, 110.1, 97.6, 72.7, 71.8, 70.5, 70.4, 70.3, 61.3, 58.9, 56.1; IR (film) νmax 3273, 2875, 1700, 1540, 1461, 1301, 1219, 1113, 1057 cm−1; ESI-TOF HRMS m/z 472.2180 (M+H+, C22H33NO10 requires 472.2177).
Compounds 15a-e
A solution of 14a (21 mg, 0.070 mmol), 9 (15.5 mg, 0.035 mmol), and EDCI (20 mg, 0.105 mmol) in DMF (0.6 mL) was stirred at room temperature for 16 h. The reaction mixture was diluted with ethyl acetate, washed with aqueous 1 M HCl, saturated aqueous NaHCO3, water and saturated aqueous NaCl, and dried over Na2SO4. The solvent was removed under reduced pressure and the residue was purified by flash chromatography (SiO2) to provide 15a as a yellow solid (13.1 mg, 62%): 1H NMR (CDCl3, 400 MHz) δ 10.59 (s, 1H), 9.43 (s, 1H), 8.02 (s, 1H), 7.02 (s, 1H), 6.94 (s, 1H), 6.83 (s, 1H), 4.64 (t, J = 10.0 Hz, 1H), 4.61 (dd, J = 10.0, 4.0 Hz, 1H), 4.21 (t, J = 4.8 Hz, 2H), 4.07 (s, 3H), 3.92–3.96 (m, 1H), 3.91 (s, 3H), 3.87 (s, 3H), 3.80– 3.85 (m, 1H), 3.75 (t, J = 4.8 Hz, 2H), 3.52 (t, J = 10.4 Hz, 1H), 3.45 (s, 3H); 13C NMR (CDCl3, 150 MHz) δ 161.7, 159.9, 152.5, 150.2, 145.6, 142.0, 139.2, 132.5, 129.8, 125.6, 125.5, 123.6, 123.3, 113.9, 106.3, 105.9, 97.6, 87.9, 72.7, 71.8, 61.3, 58.9, 56.2, 55.0, 52.6, 46.5, 42.8; IR (film) νmax 3212, 2937, 1733, 1702, 1616, 1459, 1312, 1220, 1149, 1107 cm−1; ESI-TOF HRMS m/z 558.1650 (M+H+, C27H28ClN3O8 requires 558.1638), [α]20D +8.0 (c 0.5, CHCl3). For 15b: (14.6 mg, 57%); 1H NMR (CDCl3, 400 MHz) δ 10.59 (s, 1H), 9.43 (s, 1H), 8.02 (s, 1H), 7.02 (s, 1H), 6.93 (s, 1H), 6.82 (s, 1H), 4.63 (t, J = 10.0 Hz, 1H), 4.60 (dd, J = 10.0, 4.0 Hz, 1H), 4.23 (t, J = 4.8 Hz, 2H), 4.09 (s, 3H), 3.90–3.96 (m, 1H), 3.93 (s, 3H), 3.88 (s, 3H), 3.85 (t, J = 4.8 Hz, 2H), 3.79–3.83 (m, 1H), 3.74 (t, J = 4.8 Hz, 2H), 3.60 (t, J = 4.8 Hz, 2H), 3.51 (t, J = 10.0 Hz, 1H), 3.40 (s, 3H); 13C NMR (CDCl3, 150 MHz) δ 161.8, 159.9, 152.5, 150.2, 145.7, 142.1, 139.2, 132.6, 129.8, 125.6, 125.5, 123.6, 123.3, 113.9, 106.2, 105.9, 97.4, 87.9, 72.7, 72.0, 70.5, 70.4, 61.3, 59.1, 56.2, 55.0, 52.6, 46.5, 42.9; IR (film) νmax 3214, 2937, 1730, 1702, 1617, 1460, 1311, 1220, 1150, 1107 cm−1; ESI-TOF HRMS m/z 602.1899 (M+H+, C29H32ClN3O9 requires 602.1900); [α]20D +5.8 (c 0.6, CHCl3). For 15c: (10.1 mg, 47%); 1H NMR (CDCl3, 600 MHz) δ 10.59 (s, 1H), 9.40 (s, 1H), 8.03 (s, 1H), 7.03 (s, 1H), 6.94 (s, 1H), 6.85 (s, 1H), 4.63 (t, J = 10.2 Hz, 1H), 4.60 (dd, J = 10.2, 3.6 Hz, 1H), 4.23 (t, J = 4.8 Hz, 2H), 4.09 (s, 3H), 3.90–3.96 (m, 1H), 3.93 (s, 3H), 3.88 (s, 3H), 3.85 (t, J = 4.8 Hz, 2H), 3.80–3.84 (m, 1H), 3.75 (t, J = 4.8 Hz, 2H), 3.70 (t, J = 4.8 Hz, 2H), 3.67 (t, J = 4.8 Hz, 2H), 3.56 (t, J = 4.8 Hz, 2H), 3.52 (t, J = 10.2 Hz, 1H), 3.39 (s, 3H); 13C NMR (CDCl3, 150 MHz) δ 161.7, 159.9, 152.6, 150.2, 145.7, 142.2, 139.2, 132.6, 129.9, 125.7, 125.5, 123.7, 123.4, 113.9, 106.2, 105.9, 97.5, 87.9, 72.9, 71.9, 70.7, 70.6, 70.5, 70.4, 61.3, 59.0, 56.2, 55.0, 52.6, 46.5, 42.9; IR (film) νmax 3213, 2925, 1732, 1700, 1617, 1460, 1313, 1257, 1150, 1107 cm−1; ESI-TOF HRMS m/z 646.2144 (M+H+, C31H36ClN3O10 requires 646.2162); [α]20D +5.6 (c 0.3, CHCl3). For 15d: (11.2 mg, 44%); 1H NMR (CDCl3, 600 MHz) δ 10.60 (s, 1H), 9.41 (s, 1H), 8.03 (s, 1H), 7.03 (s, 1H), 6.94 (s, 1H), 6.85 (s, 1H), 4.63 (t, J = 10.2 Hz, 1H), 4.60 (dd, J = 10.2, 3.6 Hz, 1H), 4.22 (t, J = 4.8 Hz, 2H), 4.09 (s, 3H), 3.90–3.96 (m, 1H), 3.94 (s, 3H), 3.89 (s, 3H), 3.85 (t, J = 4.8 Hz, 2H), 3.80–3.84 (m, 1H), 3.74 (t, J = 4.8 Hz, 2H), 3.62–3.72 (m, 8H), 3.55 (t, J = 4.8 Hz, 2H), 3.51 (t, J = 10.2 Hz, 1H), 3.38 (s, 3H); 13C NMR (CDCl3, 150 MHz) δ 161.7, 159.9, 152.5, 150.1, 145.6, 142.0, 139.1, 132.5, 129.8, 125.6, 125.4, 123.6, 123.3, 113.9, 106.2, 105.9, 97.4, 87.9, 72.8, 71.8, 70.6, 70.5, 70.4, 70.3, 61.3, 59.0, 56.2, 55.0, 52.6, 46.5, 42.8; IR (film) νmax 3193, 2875, 1732, 1700, 1617, 1459, 1310, 1218, 1147, 1108 cm−1; ESI-TOF HRMS m/z 690.2423 (M+H+, C33H40ClN3O11 requires 690.2424); [α]20D +15.4 (c 0.4, CHCl3). For 15e: (12.4 mg, 57%); 1H NMR (CDCl3, 600 MHz) δ 10.60 (s, 1H), 9.41 (s, 1H), 8.03 (s, 1H), 7.03 (s, 1H), 6.94 (s, 1H), 6.85 (s, 1H), 4.63 (t, J = 10.2 Hz, 1H), 4.60 (dd, J = 10.2, 3.6 Hz, 1H), 4.22 (t, J = 4.8 Hz, 2H), 4.09 (s, 3H), 3.90-3.96 (m, 1H), 3.94 (s, 3H), 3.89 (s, 3H), 3.85 (t, J = 4.8 Hz, 2H), 3.80–3.84 (m, 1H), 3.74 (t, J = 4.8 Hz, 2H), 3.62-3.72 (m, 8H), 3.55 (t, J = 4.8 Hz, 2H), 3.51 (t, J = 10.2 Hz, 1H), 3.38 (s, 3H); 13C NMR (CDCl3, 150 MHz) δ 161.7, 159.9, 152.5, 150.2, 145.7, 142.1, 139.2, 132.6, 129.9, 125.7, 125.5, 123.6, 123.3, 113.9, 106.2, 105.9, 97.6, 87.9, 72.8, 71.9, 70.6, 70.5, 70.4, 70.3, 61.3, 59.0, 56.2, 55.0, 52.5, 46.5, 42.9; IR (film) νmax 3230, 2890, 1731, 1701, 1617, 1459, 1312, 1258, 1219, 1148, 1108 cm−1; ESI-TOF HRMS m/z 734.2653 (M+H+, C35H44ClN3O12 requires 734.2686); [α]20D +12.6 (c 0.6, CHCl3).
Water Solubility Measurements
Compounds 10a–e and the natural enantiomer of seco-duocarmycin SA were treated with distilled water and stirred at 23 °C for 40 h. The solutions were taken up in a 1 mL syringe and filtered through a micro filter into a tared vial. The water was removed under a stream of nitrogen and the resulting residues were azeotroped with toluene (×3). This material was then dried under vacuum for 72 h to ensure complete dryness before weighing the sample present.
Cell Growth Inhibition Assay
Compounds were tested for their cell growth inhibition of L1210 (ATCC CCL-219) mouse lymphocytic leukemia cells or HCT116 (ATCC #CCL-247, human colorectal carcinoma) cells. A population of cells (>1 × 106 cells/mL as determined with a hemocytometer) was diluted with an appropriate amount of Dulbecco-modified Eagle Medium (DMEM, Gibco) containing 10% fetal bovine serum (FBS, Gibco) to a final concentration of 30,000 cells/mL. To each well of a 96-well plate (Corning Costar), 100 μL of the cell-media solution was added with a multichannel pipette. The cultures were incubated at 37 °C in an atmosphere of 5% CO2 and 95% humidified air for 24 h. The test compounds were added to the plate as follows: test substances were diluted in DMSO to a concentration of 1 mM and 10-fold serial dilutions were performed on a separate 96-well plate. Fresh culture medium (100 μL) was added to each well of cells to constitute 200 μL of medium per well followed by 2 μL of each test agent. Compounds were tested in duplicate (≥2–6 times) at six concentrations between 0–100 nM or 0–1000 nM. Following addition, cultures were incubated for an additional 72 h.
A phosphatase assay was used to establish the IC50 values as follows: the media in each cell was removed and 100 μL of phosphatase solution (100 mg phosphatase substrate in 30 mL 0.1 M NaOAc, pH 5.5, 0.1% Triton X-100 buffer) was added to each well. The plates were incubated at 37 °C for either 5 min (L1210) or 15 min (HCT116). After the given incubation time, 50 μL of 0.1 N NaOH was added to each well and the absorption at 405 nm was determined using a 96 well plate reader. As the absorption is directly proportional to the number of living cells, the IC50 values were calculated and the reported values represent of the average of ≥4 determinations (SD ±10%).
DNA Alkylation Studies
See reference 18a for full details of the DNA alkylation procedure. DNA alkylation was conducted at 25 °C for 2 h with 5′-32P-end-labeled w794 DNA containing a single alkylation site. Quantitation of the consumption of full length radiolableled DNA was measured by densitometry of the amount of remaining non-alkylated DNA using NIH ImageJ software25 and the relative efficiency of DNA alkylation was adjusted to account for the 10-fold differences in compound concentrations used (Supporting Information Figure S2).
Supplementary Material
Figure 10.
Plot of cLogP versus –log IC50 including 15a–e (red), slope = 0.43, r2 = 0.98.
Acknowledgments
Dedicated to Robert M. Williams on the occasion of his 60th birthday. We gratefully acknowledge the financial support of the National Institute of Health (CA041986, CA042056) and Bristol-Myers Squibb. J.P.L., A.L.W. and K.K.D. are Skaggs Fellows, J.P.L. was a BMS fellowship recipient (2011-2012), and A.L.W. was a NSF predoctoral fellowship recipient (2009–2012). We thank Michael Cameron of Scripps, Florida for conducting the PAMPA permeability assays.
Abbreviations Used
- DMF
N,N-dimethylformamide
- DSA
duocarmycin SA
- EDCI
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- LiPE
lipophilic efficiency
- PEG
polyethylene glycol
- THF
tetrahydrofuran
- TsCl
p-toluenesulfonyl chloride
Footnotes
Notes: The authors declare no competing financial interest.
Supporting Information: DNA alkylation gels of 10a–10e in w794 DNA and cell growth inhibition studies with HCT116 are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Martin DG, Biles C, Gerpheide SA, Hanka LJ, Krueger WC, McGovren JP, Mizsak SA, Neil GL, Stewart JC, Visser J. CC-1065 (NSC 298223), A Potent New Antitumor Agent. Improved Production and Isolation, Characterization and Antitumor Activity. J Antibiot. 1981;34:1119–1125. doi: 10.7164/antibiotics.34.1119. [DOI] [PubMed] [Google Scholar]; (b) Ichimura M, Ogawa T, Takahashi K, Kobayashi E, Kawamoto I, Yasuzawa T, Takahashi I, Nakano H, Duocarmycin SA. A New Antitumor Antibiotic From Streptomyces sp. J Antibiot. 1990;43:1037–1038. doi: 10.7164/antibiotics.43.1037. [DOI] [PubMed] [Google Scholar]; (c) Takahashi I, Takahashi K, Ichimura M, Morimoto M, Asano K, Kawamoto I, Tomita F, Nakano HDuocarmycin. A New Antitumor Antibiotic From Streptomyces. J Antibiot. 1988;41:1915–1917. doi: 10.7164/antibiotics.41.1915. [DOI] [PubMed] [Google Scholar]; (d) Igarashi Y, Futamata K, Fujita T, Sekine A, Senda H, Naoki H, Furumai TYatakemycin. A Novel Antifungal Antibiotic Produced by Streptomyces sp. TP-A0356. J Antibiot. 2003;56:107–113. doi: 10.7164/antibiotics.56.107. [DOI] [PubMed] [Google Scholar]
- 2.For duocarmycin SA, see: Boger DL, Johnson DS, Yun W. (+)- and ent-(−)-Duocarmycin SA and (+)- and ent-(−)-N-Boc-DSA DNA Alkylation Properties. Alkylation Site Models That Accommodate the Offset AT-Rich Adenine N3 Alkylation Selectivity of the Enantiomeric Agents. J Am Chem Soc. 1994;116:1635–1656. doi: 10.1016/s0968-0896(00)82007-6.For yatakemycin, see: Parrish JP, Kastrinsky DB, Wolkenberg SE, Igarashi Y, Boger DL. DNA Alkylation Properties of Yatakemycin. J Am Chem Soc. 2003;125:10971–10976. doi: 10.1021/ja035984h.Trzupek JD, Gottesfeld JM, Boger DL. Alkylation of Duplex DNA in Nucleosome Core Particles by Duocarmycin SA and Yatakemycin. Nat Chem Biol. 2006;2:79–82. doi: 10.1038/nchembio761.Tichenor MS, MacMillan KS, Trzupek JD, Rayl TJ, Hwang I, Boger DL. Systematic Exploration of the Structural Features of Yatakemycin Impacting DNA Alkylation and Biological Activity. J Am Chem Soc. 2007;129:10858–10869. doi: 10.1021/ja072777z.For CC-1065, see: Hurley LH, Lee CS, McGovren JP, Warpehoski MA, Mitchell MA, Kelly RC, Aristoff PA. Molecular Basis for Sequence-Specific DNA Alkylation by CC-1065. Biochemistry. 1988;27:3886–3892. doi: 10.1021/bi00410a054.Hurley LH, Warpehoski MA, Lee CS, McGovren JP, Scahill TA, Kelly RC, Mitchell MA, Wicnienski NA, Gebhard I, Johnson PD, Bradford VS. Sequence Specificity of DNA Alkylation by the Unnatural Enantiomer of CC-1065 and Its Synthetic Analogues. J Am Chem Soc. 1990;112:4633–4649.Boger DL, Johnson DS, Yun W, Tarby CM. Molecular Basis for Sequence Selective DNA Alkylation by (+)- and ent-(−)-CC-1065 and Related Agents: Alkylation Site Models that Accommodate the Offset AT-Rich Adenine N3 Alkylation Selectivity. Bioorg Med Chem. 1994;2:115–135. doi: 10.1016/s0968-0896(00)82007-6.Boger DL, Coleman RS, Invergo BJ, Sakya SM, Ishizaki T, Munk SA, Zarrinmayeh H, Kitos PA, Thompson SC. Synthesis and Evaluation of Aborted and Extended CC-1065 Functional Analogs: (+)- and (−)-CPI-PDE-I1, (+)- and (−)-CPI-CDPI1, and (+)- and (−)-CPI-CDPI3. Preparation of Key Partial Structures and Definition of an Additional Functional Role of the CC-1065 Central and Right-Hand Subunits. J Am Chem Soc. 1990;112:4623–4632.For duocarmycin A, see: Boger DL, Ishizaki T, Zarrinmayeh H, Munk SA, Kitos PA, Suntornwat O. Duocarmycin–Pyrindamycin DNA Alkylation Properties and Identification, Synthesis, and Evaluation of Agents Incorporating the Pharmacophore of the Duocarmycin-Pyrindamycin Alkylation Subunit. Identification of the CC-1065 Duocarmycin Common Pharmacophore. J Am Chem Soc. 1990;112:8961–8971.Boger DL, Ishizaki T, Zarrinmayeh H. Isolation and Characterization of the Duocarmycin– Adenine DNA Adduct. J Am Chem Soc. 1991;113:6645–6649.Boger DL, Yun W, Terashima S, Fukuda Y, Nakatani K, Kitos PA, Jin Q. DNA Alkylation Properties of the Duocarmycins: (+)-Duocarmycin A, epi-(+)-Duocarmycin A, ent-(−)-Duocarmycin A and epi,ent-(−)-Duocarmycin A. Bioorg Med Chem Lett. 1992;2:759–765.
- 3.Reviews: Boger DL, Johnson DS. CC-1065 and the Duocarmycins: Understanding Their Biological Function Through Mechanistic Studies. Angew Chem Int Ed Engl. 1996;35:1438–1474.Boger DL. The Duocarmycins: Synthetic and Mechanistic Studies. Acc Chem Res. 1995;28:20–29.Boger DL, Johnson DS. CC-1065 and the Duocarmycins: Unraveling the Keys to a New Class of Naturally Derived DNA Alkylating Agents. Proc Natl Acad Sci USA. 1995;92:3642–3649. doi: 10.1073/pnas.92.9.3642.Tichenor MS, Boger DL. Yatakemycin: Total Synthesis, DNA Alkylation, and Biological Properties. Natural Prod Rep. 2008;25:220–226. doi: 10.1039/b705665f.MacMillan KS, Boger DL. Fundamental Relationships Between Structure, Reactivity, and Biological Activity for the Duocarmycins and CC-1065. J Med Chem. 2009;52:5771. doi: 10.1021/jm9006214.Searcey M. Duocarmycins: Nature's Prodrugs? Curr Pharm Des. 2002;8:1375–1389. doi: 10.2174/1381612023394539.
- 4.Boger DL, Coleman RS. Total Synthesis of (+)-CC-1065 and ent-(−)-CC-1065. J Am Chem Soc. 1988;110:1321–1323.Boger DL, Coleman RS. Total Synthesis of CC-1065, and the Precise, Functional Agent CPI-CDPI2. J Am Chem Soc. 1988;110:4796–4807.Boger DL, Machiya K. Total Synthesis of (+)-Duocarmycin SA. J Am Chem Soc. 1992;114:10056–10058.Boger DL, Machiya K, Hertzog DL, Kitos PA, Holmes D. Total Synthesis and Preliminary Evaluation of (+)- and ent-(−)-Duocarmycin SA. J Am Chem Soc. 1993;115:9025–9036.Boger DL, Yun W. CBI-TMI: Synthesis and Evaluation of a Key Analog of the Duocarmycins. Validation of a Direct Relationship between Chemical Solvolytic Stability and Cytotoxic Potency and Confirmation of the Structural Features Responsible for the Distinguishing Behavior of Enantiomeric Pairs of Agents. J Am Chem Soc. 1994;116:7996–8006.Boger DL, McKie JA, Nishi T, Ogiku T. Enantioselective Total Synthesis of (+)-Duocarmycin A, epi-(+)-Duocarmycin A, and Their Unnatural Enantiomers. J Am Chem Soc. 1996;118:2301–2302.Boger DL, McKie JA, Nishi T, Ogiku T. Total Synthesis of (+)-Duocarmycin A and epi-(+)-Duocarmycin A and Their Unnatural Enantiomers: Assessment of Chemical and Biological Properties. J Am Chem Soc. 1997;119:311–325.Tichenor MS, Trzupek JD, Kastrinsky DB, Shiga F, Hwang I, Boger DL. Asymmetric Total Synthesis of (+)- and ent-(−)-Yatakemycin and Duocarmycin SA. Evaluation of Yatakemycin Key Partial Structures and Its Unnatural Enantiomer. J Am Chem Soc. 2006;128:15683–15696. doi: 10.1021/ja064228j.MacMillan KS, Nguyen T, Hwang I, Boger DL. Total Synthesis and Evaluation of iso-Duocarmycin SA and iso-Yatakemycin. J Am Chem Soc. 2009;131:1187–1194. doi: 10.1021/ja808108q.Review: Boger DL, Boyce CE, Garbaccio RM, Goldberg JA. CC-1065 and the Duocarmycins: Synthetic Studies. Chem Rev. 1997;97:787–828. doi: 10.1021/cr960095g.
- 5.(a) Boger DL, Hertzog DL, Bollinger B, Johnson DS, Cai H, Goldberg J, Turnbull P. Duocarmycin SA Shortened, Simplified, and Extended Agents: A Systematic Examination of the Role of the DNA Binding Subunit. J Am Chem Soc. 1997;119:4977–4986. [Google Scholar]; (b) Boger DL, Bollinger B, Hertzog DL, Johnson DS, Cai H, Mesini P, Garbaccio RM, Jin Q, Kitos PA. Reversed and Sandwiched Analogs of Duocarmycin SA: Establishment of the Origin of the Sequence-Selective Alkylation of DNA and New Insights into the Source of Catalysis. J Am Chem Soc. 1997;119:4987–4998. [Google Scholar]
- 6.(a) Boger DL, Garbaccio RM. Catalysis of the CC-1065 and Duocarmycin DNA Alkylation Reaction: DNA Binding Induced Conformational Change in the Agent Results in Activation. Bioorg Med Chem. 1997;5:263–276. doi: 10.1016/s0968-0896(96)00238-6. [DOI] [PubMed] [Google Scholar]; (b) Boger DL, Turnbull P. Synthesis and Evaluation of a Carbocyclic Analogue of the CC-1065 and Duocarmycin Alkylation Subunits: Role of the Vinylogous Amide and Implications on DNA Alkylation Catalysis. J Org Chem. 1998;63:8004–8011. [Google Scholar]
- 7.Boger DL, Garbaccio RM. Shape-Dependent Catalysis: Insights into the Source of Catalysis for the CC-1065 and Duocarmycin DNA Alkylation Reaction. Acc Chem Res. 1999;32:1043–1052. [Google Scholar]
- 8.Wolkenberg SE, Boger DL. Mechanisms of in situ Activation for DNA Targeting Antitumor Agents. Chem Rev. 2002;102:2477–2495. doi: 10.1021/cr010046q. [DOI] [PubMed] [Google Scholar]
- 9.Boger DL, Turnbull P. Synthesis and Evaluation of CC-1065 and Duocarmycin Analogs Incorporating the 1,2,3,4,11,11a-Hexahydrocyclopropa[c]naphtho[2,1-b]azepin-6-one (CNA) Alkylation Subunit: Structural Features that Govern Reactivity and Reaction Regioselectivity. J Org Chem. 1997;62:5849–5863. [Google Scholar]
- 10.For representative examples, see: Aristoff PA. CC-1065 Analogs: Sequence Specific DNA-Alkylating Antitumor Agents. Adv Med Chem. 1993;2:67–110.Kobayashi E, Okamoto A, Asada M, Okabe M, Nagamura S, Asai A, Saito H, Gomi K, Hirata T. Characteristics of Antitumor Activity of KW-2189, A Novel Water-Soluble Derivative of Duocarmycin, Against Murine and Human Tumors. Cancer Res. 1994;54:2404–2410.Amishiro N, Nagamura S, Kobayashi E, Gomi K, Saito H. New Water-Soluble Duocarmycin Derivatives: Synthesis and Antitumor Activity of A-Ring Pyrrole Compounds Bearing β-Heteroarylacryloyl Groups. J Med Chem. 1999;42:669–676. doi: 10.1021/jm980559y.Nagamura S, Asai A, Kanda Y, Kobayashi E, Gomi K, Saito H. Synthesis and Antitumor Activity of Duocarmycin Derivatives: Modification of Segment A of Duocarmycin B2. Chem Pharm Bull. 1996;44:1723–1730. doi: 10.1248/cpb.44.1723.Nagamura S, Kanda Y, Kobayashi E, Gomi K, Saito H. Synthesis and Antitumor Activity of Duocarmycin Derivatives. Chem Pharm Bull. 1995;43:1530–1535. doi: 10.1248/cpb.43.1530.Boger DL, Boyce CW, Garbaccio RM, Searcey M, Jin Q. CBI Prodrug Analogs of CC-1065 and the Duocarmycins. Synthesis. 1999:1505–1509.Tietze LF, Schuster HJ, Schmuck K, Schuberth I, Alves F. Duocarmycin-based Prodrugs for Cancer Prodrug Monotherapy. Bioorg Med Chem. 2008;16:6312–6318. doi: 10.1016/j.bmc.2008.05.009.Tietze LF, Lieb M, Herzig T, Haunert F, Schuberth I. Strategy for Tumor-Selective Chemotherapy by Enzymatic Liberation of seco-Duocarmycin SA Derivatives from Nontoxic Prodrugs. Bioorg Med Chem. 2001;9:1929–1939. doi: 10.1016/s0968-0896(01)00098-0.Li LS, Sinha SC. Studies Toward Duocarmycin Prodrugs for the Antibody Prodrug Therapy Approach. Tetrahedron Lett. 2009;50:2932–2935. doi: 10.1016/j.tetlet.2009.03.205.Asai A, Nagamura S, Kobayashi E, Gomi K, Saito H. Synthesis and Antitumor Activity of Water-Soluble Duocarmycin B1 Prodrugs. Bioorg Med Chem Lett. 1999;9:2995–2998. doi: 10.1016/s0960-894x(99)00518-1.Jin W, Trzupek JD, Rayl TJ, Broward MA, Vielhauer GA, Weir SJ, Hwang I, Boger DL. A Unique Class of Duocarmycin and CC-1065 Analogues Subject to Reductive Activation. J Am Chem Soc. 2007;129:15391–15397. doi: 10.1021/ja075398e.Lajiness JP, Robertson WM, Dunwiddie I, Broward MA, Vielhauer GA, Weir SJ, Boger DL. Design, Synthesis, and Evaluation of Duocarmycin O-Amino Phenol Prodrugs Subject to Tunable Reductive Activation. J Med Chem. 2010;53:7731–7738. doi: 10.1021/jm1010397.Wolfe AL, Duncan KK, Parelkar N, Weir SJ, Vielhauer GA, Boger DL. A Novel, Unusually Efficacious Duocarmycin Carbamate Prodrug that Releases No Residual Byproduct. J Med Chem. 2012;55:5878–5886. doi: 10.1021/jm300330b.Pors K, Loadman PM, Shnyder SD, Sutherland M, Sheldrake HM, Guino M, Kiakos K, Hartley JA, Searcey M, Patterson LH. Modification of the Duocarmycin Pharmacophore Enables CYP1A1 Targeting for Biological Activity. Chem Commun. 2011;47:12062–12064. doi: 10.1039/c1cc15638a.
- 11.For representative examples, see: Lillo AM, Sun C, Gao C, Ditzel H, Parrish JP, Gauss CM, Moss J, Felding-Habermann B, Wirsching P, Boger DL, Janda KD. A Human Single-Chain Antibody Specific for Integrin α3β1 Capable of Cell Internalization and Delivery of Antitumor Agents. Chem Biol. 2004;11:897–906. doi: 10.1016/j.chembiol.2004.04.018.Goldmacher VS, Kovtun YV. Antibody–Drug Conjugates: Using Monoclonal Antibodies for Delivery of Cytotoxic Payloads to Cancer Cells. Therapeutic Delivery. 2011;2:397–416. doi: 10.4155/tde.10.98.Ducry L, Stump B. Antibody–Drug Conjugates: Linking Cytotoxic Payloads to Monoclonal Antibodies. Bioconjugate Chem. 2010;21:5–13. doi: 10.1021/bc9002019.Zhao RH, Erickson HK, Leece BA, Reid EE, Goldmacher VS, Lambert JM, Chari RVJ. Antibody– Drug Conjugates: Linking Cytotoxic Payloads to Monoclonal Antibodies. J Med Chem. 2012;55:766–782. doi: 10.1021/jm201284m.
- 12.(a) Eis PS, Smith JA, Rydzewski JM, Case DA, Boger DL, Chazin WJ. High Resolution Solution Structure of a DNA Duplex Alkylated by the Antitumor Agent Duocarmycin SA. J Mol Biol. 1997;272:237–252. doi: 10.1006/jmbi.1997.1223. [DOI] [PubMed] [Google Scholar]; (b) Schnell JR, Ketchem RR, Boger DL, Chazin WJ. Binding-Induced Activation of DNA Alkylation by Duocarmycin SA: Insights from the Structure of an Indole Derivative–DNA Adduct. J Am Chem Soc. 1999;121:5645–5652. [Google Scholar]; (c) Smith JA, Bifulco G, Case DA, Boger DL, Gomez-Paloma L, Chazin WJ. The Structural Basis for in situ Activation of DNA Alkylation by Duocarmycin SA. J Mol Biol. 2000;300:1195–1204. doi: 10.1006/jmbi.2000.3887. [DOI] [PubMed] [Google Scholar]; (d) Bassarello C, Cimino P, Bifulco G, Boger DL, Smith JA, Chazin WJ, Paloma LG. NMR Structure of the (+)-CPI-indole/d(GACTAATTGAC)-d(GTCAATTAGTC) Covalent Complex. ChemBioChem. 2003;4:1188–1193. doi: 10.1002/cbic.200300642. [DOI] [PubMed] [Google Scholar]
- 13.Robertson WM, Kastrinsky DB, Hwang I, Boger DL. Synthesis and Evaluation of a Series of C5′-Substituted Duocarmycin SA Analogs. Bioorg Med Chem Lett. 2010;20:2722–2725. doi: 10.1016/j.bmcl.2010.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tse WC, Boger DL. Sequence-Selective DNA Recognition: Natural Products and Nature's Lessons. Chem Biol. 2004;11:1607–1617. doi: 10.1016/j.chembiol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Ouchi M, Inoue Y, Liu Y, Nagamune S, Nakamura S, Wada K, Hakaushi T. Convenient and Efficient Tosylation of Oligoethylene Glycols and the Related Alcohols in Tetrahydrofuran-Water in the Presence of Sodium Hydroxide. Bull Chem Soc Jpn. 1990;63:1260–1262. [Google Scholar]
- 16.(a) Boger DL, Ishizaki T, Zarrinmayeh H, Kitos PA, Suntornwat O. Synthesis and Preliminary Evaluation of Agents Incorporating the Pharmacophore of the Duocarmycin/Pyrindamycin Alkylation Subunit: Identification of the CC-1065/Duocarmycin Common Pharmacophore. J Org Chem. 1990;55:4499–4502. [Google Scholar]; (b) Boger DL, Coleman RS. Diels–Alder Reactions of Heterocyclic Azadienes: Total Synthesis of PDE I, PDE II, and PDE I Dimer Methyl Ester. J Am Chem Soc. 1987;109:2717–2727. [Google Scholar]; (c) Boger DL, Coleman RS, Invergo BJ. Studies on the Total Synthesis of CC-1065: Preparation of a Synthetic Simplified 3-Carbamoyl-1,2-dihydro-3H-pyrrolo[3,2-e]indole Dimer/Trimer/Tetramer (CDPI Dimer/Trimer/Tetramer) and Development of Methodology for PDE-I Dimer Methyl Ester Formation. J Org Chem. 1987;52:1521–1530. [Google Scholar]
- 17.Tichenor MS, Kastrinsky DB, Boger DL. Total Synthesis, Structure Revision, and Absolute Configuration of (+)-Yatakemycin. J Am Chem Soc. 2004;126:8396–8398. doi: 10.1021/ja0472735. [DOI] [PubMed] [Google Scholar]
- 18.(a) Boger DL, Munk SA, Zarrinmayeh H, Ishizaki T, Haught J, Bina M. An Alternative and Convenient Strategy for Generation of Substantial Quantities of Singly 5′-P32-End-Labeled Double-Stranded DNA for Binding Studies. Development of a Protocol for Examination of Functional Features of (+)-CC-1065 and the Duocarmycins That Contribute to Their Sequence-Selective DNA Alkylation Properties. Tetrahedron. 1991;47:2661–2682. [PubMed] [Google Scholar]; (b) Boger DL, Munk SA. DNA Alkylation Properties of Enhanced Functional Analogs of CC-1065 Incorporating the 1,2,9,9a-Tetrahydrocyclopropa[1,2-c]benz[1,2-e]indol-4-one (CBI) Alkylation Subunit. J Am Chem Soc. 1992;114:5487–5496. [Google Scholar]
- 19.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of Image Analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(a) Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. A Simple, High Resolution Method for Establishing DNA Binding Affinity and Sequence Selectivity. J Am Chem Soc. 2001;123:5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]; (b) Tse WC, Boger DL. A Fluorescent Intercalator Displacement (FID) Assay for Establishing DNA Binding Selectivity and Affinity. Acc Chem Res. 2004;37:61–69. doi: 10.1021/ar030113y. [DOI] [PubMed] [Google Scholar]; (c) Boger DL, Fink BE, Hedrick MP. Total Synthesis of Distamycin A and 2640 Analogues: Solution-Phase Combinatorial Approach to the Discovery of New, Bioactive DNA Binding Agents and Development of a Rapid, High-Throughput Screen for Determining Relative DNA Binding Affinity or DNA Binding Sequence Selectivity. J Am Chem Soc. 2000;122:6382–6394. [Google Scholar]; (d) Boger DL, Tse WC. Thiazole Orange as the Fluorescent Intercalator in a High Resolution FID Assay for Determining DNA Binding Affinity and Sequence Selectivity of Small Molecules. Bioorg Med Chem. 2001;9:2511–2518. doi: 10.1016/s0968-0896(01)00243-7. [DOI] [PubMed] [Google Scholar]; (e) Yeung BKS, Tse WC, Boger DL. Determination of Binding Affinities of Triplex Forming Oligonucleotides Using a Fluorescent Intercalator Displacement (FID) Assay. Bioorg Med Chem Lett. 2003;13:3801–3804. doi: 10.1016/j.bmcl.2003.07.005. [DOI] [PubMed] [Google Scholar]; (f) Ham YW, Tse WC, Boger DL. High Resolution Assessment of Protein-DNA Binding Affinity and Selectivity Utilizing a Fluorescent Intercalator Displacement (FID) Assay. Bioorg Med Chem Lett. 2003;13:3805–3807. doi: 10.1016/j.bmcl.2003.06.002. [DOI] [PubMed] [Google Scholar]; (g) Tse WC, Ishii T, Boger DL. Comprehensive High Resolution Analysis of Hairpin Polyamides Utilizing a Fluorescent Intercalator Displacement (FID) Assay. Bioorg Med Chem. 2003;11:4479–4486. doi: 10.1016/s0968-0896(03)00455-3. [DOI] [PubMed] [Google Scholar]; (h) Woods CR, Ishii T, Wu B, Bair KW, Boger DL. Hairpin versus Extended DNA Binding of a Substituted β-Alanine Linked Polyamide. J Am Chem Soc. 2002;124:2148–2152. doi: 10.1021/ja0122039. [DOI] [PubMed] [Google Scholar]; (i) Woods CR, Ishii T, Boger DL. Synthesis and DNA Binding Properties of Iminodiacetic Acid (IDA) Linked Polyamides: Characterization of Cooperative Extended 2:1 Side-by-Side Parallel Binding. J Am Chem Soc. 2002;124:10676–10682. doi: 10.1021/ja026588m. [DOI] [PubMed] [Google Scholar]
- 21.ChemDraw 12.0, specific algorithms for calculating LogP from fragment-based methods were developed by the Medicinal Chemistry Project and BioByte.
- 22.(a) Lessen PD, Springthorpe B. The Influence of Drug-like Concepts on Decision-Making in Medicinal Chemistry. Nat Rev Drug Discov. 2007;6:881–890. doi: 10.1038/nrd2445. [DOI] [PubMed] [Google Scholar]; (b) Edwards MP, Price DA. Role of Physicochemical Properties and Ligand Lipophilicity Efficiency in Addressing Drug Safety Risks. Annu Rep Med Chem. 2010;45:381–391. [Google Scholar]; (c) Mitchell RC. Hydrogen Bonding. 32. An Analysis of Water-Octanol and Water-Alkane Partitioning and the ΔLog P Parameter of Seiler. J Pharm Sci. 1994;83:1085–1100. doi: 10.1002/jps.2600830806. [DOI] [PubMed] [Google Scholar]; (d) A value for cLog P of approximately 2 or lower is often consistent with reasonable in vivo clearance, solubility and protein binding. High LiPE compounds should outperform low LiPE compounds as these will be compromised by reduced potency or increased lipophilicity. A recent analysis of animal safety studies revealed an increased risk of adverse outcome for compounds with cLogP >3. While achievable LiPE values are target and series specific, compounds with LiPE above 5 are usually considered highly optimized.
- 23.Measured permeability was established in the PAMPA assay (parallel artificial membrane permeability assay), where a 1 μM compound solution was prepared in 300 μL of phosphate buffered saline (pH = 7.4) and the ability of the compound to cross a lipid layer (plates from BD biosciences, 5 h, room temperature) into a second chamber containing buffered saline was quantified. The results indicate that the structural changes have a much less significant, but inhibitory impact on permeability: Papp = 4.0, 3.6, 4.2, 10, and 2.1 × 10−6 cm/s for 15a–15e, respectively, versus 9.5 × 10−6 cm/s for 2, representing 0- to 4.5-fold reductions in permeability within or throughout the series. Similarly, the permeability of 10e was established to be 0.11 × 10−6 cm/s, representing an 85-fold reduction relative to 2 and across the entire series (10a–10e) where the inhibitory cell growth IC50 values and relative DNA alkylation efficiencies progressively span the much larger 104–105 range (2 vs 10e).Kansy M, Senner F, Gubernator K. Physicochemical High Throughput Screening: Parallel Artificial Membrane Permeability Assay in the Description of Passive Absorption Processes. J Med Chem. 1998;41:1007–1010. doi: 10.1021/jm970530e.
- 24.Boger DL, Coleman RS, Invergo BJ, Zarrinmayeh H, Kitos PA, Thompson SC, Leong T, McLaughlin LW. A Demonstration of the Intrinsic Importance of Stabilizing Hydrophobic Binding and Non-Covalent van der Waals Contacts Dominant in the Non-Covalent CC-1065/B-DNA Binding. Chem Biol Interactions. 1990;73:29–52. doi: 10.1016/0009-2797(90)90107-x. [DOI] [PubMed] [Google Scholar]
- 25.Boger DL, Yun W. Reversibility of the Duocarmycin A and SA DNA Alkylation Reaction. J Am Chem Soc. 1993;115:9872–9873. [Google Scholar]
- 26.(a) Boger DL, Zarrinmayeh H, Munk SA, Kitos PA, Suntornwat O. Demonstration of a Pronounced Effect of Noncovalent Binding Selectivity on the (+)-CC-1065 DNA Alkylation and Identification of the Pharmacophore of the Alkylation Subunit. Proc Natl Acad Sci USA. 1991;88:1431–1435. doi: 10.1073/pnas.88.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Boger DL, Munk SA, Zarrinmayeh H. (+)-CC-1065 DNA Alkylation: Key Studies Demonstrating a Noncovalent Binding Selectivity Contribution to the Alkylation Selectivity. J Am Chem Soc. 1991;113:3980–3983. [Google Scholar]; (c) Boger DL, Johnson DS. Second Definitive Test of Proposed Models for the Origin of the CC-1065 and Duocarmycin DNA Alkylation Selectivity. J Am Chem Soc. 1995;117:1443–1444. [Google Scholar]; (d) Boger DL, Zhou J, Cai H. Demonstration and Definition of the Noncovalent Binding Selectivity of Agents Related to CC-1065 by an Affinity Cleavage Agent: Noncovalent Binding Coincidental with Alkylation. Bioorg Med Chem. 1996;4:859–868. doi: 10.1016/0968-0896(96)00073-9. [DOI] [PubMed] [Google Scholar]; (e) Boger DL, Wysocki RJ, Ishizaki T. Synthesis of N-Phenylsulfonyl-CI, N-Boc-CI, CI-CDPI1, and CI-CDPI2: CC-1065 Functional Analogs Incorporating the Parent 1,2,7,7a-Tetrahydrocyclopropa[1,2-c]indol-4-one (CI) Left-Hand Subunit. J Am Chem Soc. 1990;112:5230–5240. [Google Scholar]
- 27.Boger DL, Mesini P. Design, Synthesis, and Evaluation of CC-1065 and Duocarmycin Analogs Incorporating the 2,3,10.10a-Tetrahydro-1H-cyclopropa[d]benzo[f]quinol-5-one (CBQ) Alkylation Subunit: Identification and Structural Origin of Subtle Stereoelectronic Features that Govern Reactivity and Regioselectivity. J Am Chem Soc. 1994;116:11335–11348. [Google Scholar]
- 28.(a) Boger DL, Santillan AJr, Searcey M, Brunette SR, Wolkenberg SE, Hedrick MP, Jin Q. Synthesis and Evaluation of 1,2,8,8a-Tetrahydrocyclopropa[c]pyrrolo[3,2-e]indol-4(5H)-one, the Parent Alkylation Subunit of CC-1065 and the Duocarmycins: Impact of the Alkylation Subunit Substituents and Its Implications for DNA Alkylation Catalysis. J Org Chem. 2000;65:4101–4111. doi: 10.1021/jo000297j. [DOI] [PubMed] [Google Scholar]; (b) Boger DL, Stauffer F, Hedrick MP. Substituent Effects Within the DNA Binding Subunit of CBI Analogues of the Duocarmycins and CC-1065. Bioorg Med Chem Lett. 2001;11:2021–2024. doi: 10.1016/s0960-894x(01)00372-9. [DOI] [PubMed] [Google Scholar]
- 29.(a) Parrish JP, Hughes TV, Hwang I, Boger DL. Establishing the Parabolic Relationship Between Reactivity and Activity for Derivatives and Analogues of the Duocarmycin and CC-1065 Alkylation Subunits. J Am Chem Soc. 2004;126:80–81. doi: 10.1021/ja038162t. [DOI] [PubMed] [Google Scholar]; (b) Boger DL, Ishizaki T. Resolution of a CBI Precursor and Incorporation into the Synthesis of (+)- and (−)-CBI, (+)- and (−)-CBI-CDPI1, (+)- and (−)-CBI-CDPI2: Enhanced Functional Analogs of (+)-CC-1065: A Critical Appraisal of the Proposed Relationship Between Electrophile Reactivity, DNA Binding Properties, and Cytotoxic Potency. Tetrahedron Lett. 1990;31:793–796. [Google Scholar]; (c) Boger DL, Ishizaki T, Sakya SM, Munk SA, Kitos PA, Jin Q, Besterman JM. Synthesis and Preliminary Evaluation of (+)-CBI-indole2: An Enhanced Functional Analog of (+)-CC-1065. Bioorg Med Chem Lett. 1991;1:115–120. [Google Scholar]; (d) Boger DL, Munk SA, Ishizaki T. The (+)-CC-1065 DNA Alkylation: Observation of an Unexpected Relationship Between Cyclopropane Electrophile Reactivity and the Efficiency of DNA Alkylation. J Am Chem Soc. 1991;113 [Google Scholar]; (e) Boger DL, Yun W. Role of the CC-1065 and Duocarmycin N2 Substituent: Validation of a Direct Relationship Between Solvolysis Chemical Stability and In Vitro Biological Potency. J Am Chem Soc. 1994;116:5523–5524. [Google Scholar]; (f) Tichenor MS, MacMillan KS, Stover JS, Wolkenberg SE, Pavani MG, Zanella L, Zaid AN, Spalluto G, Rayl TJ, Hwang I, Baraldi PG, Boger DL. Rational Design, Synthesis, and Evaluation of Key Analogues of CC-1065 and the Duocarmycins. J Am Chem Soc. 2007;129:14092–14099. doi: 10.1021/ja073989z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.