Summary
Staphylococcus aureus (S. aureus) is one of the most common organisms associated with infections among burn patients and has shown a frequent and rapid development of antibiotic resistance. The presence of genes encoding aminoglycoside modifying enzymes (AME) and tetracycline resistance were detected by PCR and multiplex-PCR. Among the 151 S. aureus isolates recovered from the burn patients, 96 (63.6%) were detected to have mecA gene. The rate of tetracycline resistance genes associated with mecA was 61% (92/151). Forty nine isolates (32.4%) contained tetM, 26 (17.2%) possessed only tetK and 21 (13.9%) contained both tetM and tetK. The presence of the aac(6’)-Ie-aph(2’’)-I gene was determined in 18 isolates, aph(3’)-IIIa in 8 isolates, both the aac(6’)-Ie-aph(2’’)-I, aph(3’)-IIIa and the ant(4’)-Ia genes in 69 isolates, both aac(6’)-Ie-aph(2’’)-I and ant(4’)-Ia in 6 isolates, and both the aph(3’)-IIIa and the ant(4’)-Ia genes in 8 isolates. Most of the strains which harboured the mecA gene also contained the tet and AME genes.
Keywords: Staphylococcus aureus, burn, antibiotic resistance
Abstract
Staphylococcus aureus (S. aureus) est l’un des organismes les plus communs associés à des infections chez les patients souffrant de brûlures et a montré une évolution fréquente et plus rapide de la résistance aux antibiotiques. La présence de gènes codant pour des enzymes de modification de l’aminoglycoside et de la résistance tétracycline a été détectée par PCR et PCR multiplexe. Parmi les 151 isolats de S. aureus récupérés des patients brûlés, 96 (63,6%) contenaient le gène mecA. Le taux de gènes de résistance à la tétracycline associés à mecA était de 61% (92/151). Quarante-neuf isolats (32,4%) contenaient tetM, 26 (17,2%) ne possédaient que tetK et 21 (13,9%) contenaient tetM et tetK. La présence du gène aac(6 ‘)-Ie-aph (‘’ 2)-I a été identifiée dans 18 isolats, et le gène aph(3’)-IIIa dans 8 isolats. Les gènes aac(6 ‘)-Ie-aph(2’’)-I, aph(3')-IIIa et ant(4’)-Ia ont été identifiés dans 69 isolats, aaC (6 ‘)-Ie-aph (2’’)-I et ant(4’)-Ia dans 6 isolats, et l'aph (3')-IIIa et ant(4’)-Ia dans 8 isolats. La plupart des souches qui hébergeaient le gène mecA contenait également des gènes tet et AME.
Introduction
Infection is one of the most serious problems in patients hospitalized with thermal injury, and still remains a leading cause of death among these patients.1 Infections caused by antibiotic resistant bacteria should be considered as a potential risk and their resistance pattern should be identified as soon as possible. Staphylococcus aureus is known to be one of the major causes of infections acquired in hospitals and communities worldwide, and is one of the most common organisms associated with infections among burn patients.2,3 Since the introduction of antibiotics in medicine, S. aureus has shown a frequent and rapid development and spread of antibiotic resistance, and has developed resistance to all types of antibiotics.4
Tetracyclines are broad-spectrum antibiotics used in the treatment and prevention of bacterial infections.5 Most tetracycline resistant bacteria have acquired tetracycline resistance genes (tet). Two main mechanisms of resistance to tetracycline have been described in S. aureus: active efflux, resulting from the acquisition of the plasmid- located tetK and tetL genes and ribosomal protection by elongation factor-like proteins that are encoded by chromosomal or transposonal tetM or tetO determinants.6,7
Aminoglycosides are broad-spectrum antibiotics that are used in combination with other antibiotics such as ß- lactams for treatment of S. aureus infections.8 Inactivation of aminoglycoside antibiotics by aminoglycoside modifying enzymes (AME), such as aminoglycoside phosphotransferase (APH), acetyl-transferases (AAC), and nucleotidyl- transferase (ANT) enzymes, is the most common mechanism of aminoglycoside resistance. 8,9 The most common AME encoding genes among S. aureus are aac(6’)-Ie-aph(2’’), aph(3’)-IIIa, and ant(4’)-Ia, which can be harbored on plasmid or chromosome and are often harbored on transposable elements.10
The results of an earlier investigation at the burn unit in Tehran have shown that the incidence of S. aureus is high and the majority of the isolates were resistant to tetracycline and aminoglycoside antibiotics phenotypically.3 This study was conducted to investigate the incidence of tetracycline and aminoglycoside resistance determinants among a collection of S. aureus strains isolated from burn patients.
Materials and methods
Bacterial isolates
A total of 151 non duplicate (i.e., only one isolate per patient) and non-consecutive S. aureus isolates were collected, during September 2010 and June 2012, from clinical samples of burn patients, admitted to the burn ward at Shahid Motahari Hospital, Tehran, Iran. Isolates were identified to the species level by Gram staining, production of coagulase, catalase, DNase and oxidation, and fermentation of mannitol.
Antimicrobial susceptibility testing
Tests for susceptibility to oxacillin (1 μg), gentamicin (10 μg), amikacin (30 μg), kanamycin (30 μg), netilmicin (30 μg), tetracycline (30 μg), doxycycline (30 μg), and minocycline (30 μg) were performed by the disc diffusion method as recommended by the Clinical Laboratory Standards Institute (CLSI).11 S. aureus ATCC 25923 was used as the control strain.
Amplification of mecA, AMEs and tet genes
Total DNA template was extracted from S. aureus isolates by the boiling method previously outlined.12 Under the standard polymerase chain reaction (PCR) conditions, a series of primers (Table I) were used, according to the methods described below, for the detection of mecA, tet and AME genes. Recognition of genes encoding the AMEs enzymes (aac(6’)-Ie-aph(2’’)-I, aph(2’’)-Ib, aph(2’’)-Ic, aph(2’’)-Id, aph(3’)-IIIa, and ant(4’)-Ia) and methicillin resistance (mecA) was performed as described previously.9,10 Amplification of tet genes (tetK, tetL, tetM and tetO) was performed using CinnaGen PCR master kit (Cinna- Gen Co.). PCR was performed in a final volume of 25 ml containing 0.5 µM of each primer. The PCR protocol consisted of an initial denaturation step at 94°C for 5 min; 30 amplification cycles at 94°C for 1 min, at 57°C for 1 min (for tetO, at 51°C for tetM, tetL, and tetK), and at 72°C for 1 min, followed by a final extension step at 72°C for 5 min. Amplified products were analyzed by electrophoresis on 1% agarose gel containing 0.5 μg/ml ethidium bromide and photographed under UV illumination.
Table I. The oligonucleotide primers used for PCR amplified antimicrobial resistance.
Statistical analysis
Differences in the prevalence of genes among S. aureus isolates were calculated using the chi-square test for each gene. A P value of ≤ 0.05 was considered as statistically significant
Results
All the isolates were investigated for the presence of genes encoding tetracycline, aminoglycoside and methicillin resistance. The results are summarized in Tables II and III.
Table II. Distribution of tetracycline resistance genes among S. aureus isolates.
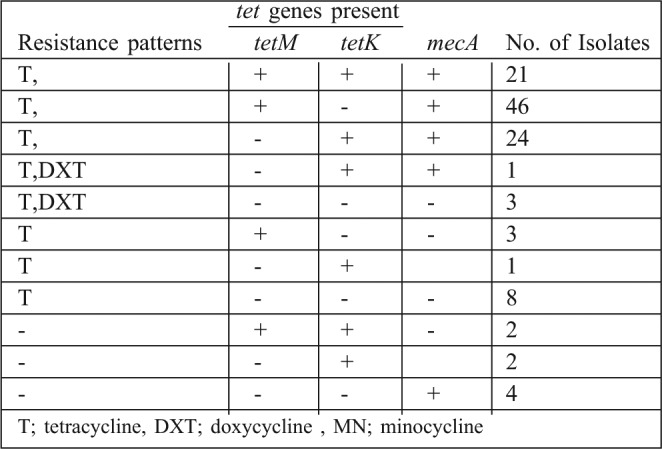
Table III. Distribution of aminoglycoside resistance genes among the S. aureus isolates.
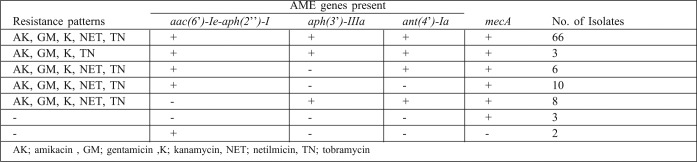
The mecA gene encoding methicillin resistance was determined in 63.6% (96/151) of the S. aureus isolates included in this study.
The rate of tetracycline resistance genes associated with mecA was 61% (92/151). All the tetracycline resistant isolates, excluding 11 isolates, gave positive results for tet genes. Forty nine isolates (32.4%) contained tetM, 26 (17.2%) possessed tetK and 21(13.9%) included both tetM and tetK genes. Four isolates were sensitive to tetracycline but positive for the tet genes (two isolates contained both tetM and tetK and two isolates contained only tetK). None of the isolates were positive for tetL and tetO in the PCR assay.
The presence of the aac(6’)-Ie-aph(2’’)-I gene was determined in 18 isolates, the aph(3’)-IIIa in 8 isolates, both the aac(6’)-Ie-aph(2’’)-I, aph(3’)-IIIa and the ant(4’)-Ia genes in 69 isolates, both aac(6’)-Ie-aph(2’’)-I and the ant(4’)-Ia in 6 isolates, and both the aph(3’)-IIIa and the ant(4’)-Ia genes in 8 isolates. The gene encoding AME was encountered in at least 95 isolates whilst it was absent in 56 of the isolates. Amplification of the aph(2’’)-Ib, aph(2’’)-Ic and aph(2’’)-Id genes was negative for all the isolates.
Coexistence of the tet and AME genes were encountered in 55 of the isolates.
Discussion
Tetracyclines still remain the first-line treatment for a number of infections in many parts of the world, including Iran.3,16,17 According to the literature review, there is a limited amount of data regarding the prevalence rate of tet genes among S. aureus strains isolated from burn patients in Iran. In this study, the coexistence rate of mecA and tet genes was 61%, which is slightly higher than similar reports from Europe (57.1%) and lower than those from Japan (100%).16-19
In the current study, the most common tetracycline resistance mechanism is mediated by tetM and tetK genes, respectively. The resistance mechanism mediated by these genes is predominant among tetracycline resistant MRSA from Turkey, Malaysia, and most European countries, but not so in North America where there is a high prevalence rate of the tetK gene among MRSA isolates.5,16,18-21 The tet genes are contained within conjugative transposons that can be transferred horizontally and expressed in Gram-positive and Gram-negative bacteria.22
Coexistence of tetM and tetK genes among the S. aureus isolates was detected in this study, as well as in the study of Trzcinski et al.14 Our results are also consistent with those reporting the tetL and the tetO genes to be rarely detected in S. aureus isolates.18-21
The aac(6’)-Ie-aph(2’’) gene encodes the AAC(6’)-APH(2’’) enzyme, a bifunctional enzyme with kinase activity, that inactivates a broad range of aminoglycosides and confers concomitant resistance to gentamicin and the majority of aminoglycosides commonly used in medical practice.8,9,23 As also reported in other studies, the aac(6’)-Ie-aph(2’’) gene was the most common AME gene among the S. aureus isolates and was found either alone or with other AME or tet genes.9,23
Remarkably, most of the strains which harboured the mecA gene also contained the tet and AME genes. These genes might be located on the same or associated genetic element. In MRSA, methicillin resistance is mediated by mecA, which is carried on Staphylococcal Cassette Chromosome mec (SCCmec). SCCmec type III is the dominant SCCmec type in Iran; this type includes not only the mecA gene, but also the genes encoding resistance to several non-beta-lactam antibiotic classes, such as tetracyclines and aminoglycosides.24
Conclusion
This study has shown that the S. aureus isolates recovered from burn patients contain a variety of tetracycline and amino glycoside resistance genes, and tetM and aac(6’)-Ie-aph(2’’) genes were the most common tet and AME genes among the S. aureus isolates, respectively.
Acknowledgments
This research has been supported by Tehran University of Medical Sciences & Health Services grant 20041/30-4-91.
References
- 1.Ribeiro NF, Heath CH, Kierath J, et al. Burn wounds infected by contaminated water: Case reports, review of the literature and recommendations for treatment. Burns. 2010;36:9–22. doi: 10.1016/j.burns.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Chen X, Yang HH, Huangfu YC, et al. Molecular epidemiologic analysis of Staphylococcus aureus isolated from four burn centers. Burns. 2012;38:738–42. doi: 10.1016/j.burns.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 3.Shahsavan S, Emaneini M, Noorazar Khoshgnab B, et al. A high prevalence of mupirocin and macrolide resistance determinant among Staphylococcus aureus strains isolated from burnt patients. Burns. 2012;38:378–82. doi: 10.1016/j.burns.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Boucher HW, Corey GR. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2008;46:344–9. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- 5.Ardic N, Ozyurt M, Sareyyupoglu B, et al. Investigation of erythromycin and tetracycline resistance genes in methicillin-resistant staphylococci. Int J Antimicrob Agents. 2005;26:213–8. doi: 10.1016/j.ijantimicag.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Esposito S, Leone S, Petta E, et al. Treatment options for skin and soft tissue infections caused by meticillin-resistant Staphylococcus aureus: oral vs.parenteral; home vs. hospital. Int J Antimicrob Agents. 2009;34:30–5. doi: 10.1016/S0924-8579(09)70547-3. [DOI] [PubMed] [Google Scholar]
- 7.McCallum N, Berger-Bächi B, Senn MM, et al. Regulation of antibiotic resistance in Staphylococcus aureus. Int J Med Microbiol. 2010;300:118–29. doi: 10.1016/j.ijmm.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resistance Updates. 2010;13:151–71. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fatholahzadeh B, Emaneini M, Feizabadi MM, et al. Characterisation of genes encoding aminoglycoside-modifying enzymes among meticillin-resistant Staphylococcus aureus isolated from two hospitals in Tehran, Iran. Int J Antimicrob Agents. 2009;33:264–5. doi: 10.1016/j.ijantimicag.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 10.Emaneini M, Taherikalani M, Eslampour MA, et al. Phenotypic and genotypic evaluation of aminoglycoside resistance in clinical isolates of staphylococci in Tehran, Iran. Microb Drug Resist. 2009;15:129–32. doi: 10.1089/mdr.2009.0869. [DOI] [PubMed] [Google Scholar]
- 11.Performance Standards for Antimicrobial Susceptibility Testing; Twenty-First Informational Supplement. Vol. 31. Clinical and Laboratory Standards Institute.; 2011. pp. 100–21. [Google Scholar]
- 12.Fatholahzadeh B, Hashemi FB, Emaneini M, et al. Detection of Vancomycin Resistant Enterococci (VRE) isolated from urinary tract infections (UTI) in Tehran, Iran. Daru J Pharm Sci. 2006;14:141–5. [Google Scholar]
- 13.Strommenger B, Kettlitz C, Werner G, et al. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J Clin Microbiol. 2003;41:4089–94. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trzcinski K, Cooper BS, Hryniewicz W, et al. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2000;45:763–70. doi: 10.1093/jac/45.6.763. [DOI] [PubMed] [Google Scholar]
- 15.Malhotra-Kumar S, Lammens C, Piessens J, et al. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob Agents Chemother. 2005;49:4798–800. doi: 10.1128/AAC.49.11.4798-4800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones CH, Tuckman M, Howe AY, et al. Diagnostic PCR analysis of the occurrence of methicillin and tetracycline resistance genes among Staphylococcus aureus isolates from phase 3 clinical trials of tigecycline for complicated skin and skin structure infections. Antimicrob Agents Chemother. 2006;50:505–10. doi: 10.1128/AAC.50.2.505-510.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts MC. Tetracycline therapy: Update. Clin Infect Dis. 2003;36:462–7. doi: 10.1086/367622. [DOI] [PubMed] [Google Scholar]
- 18.Schmitz FJ, Krey A, Sadurski R, et al. European SENTRY Participants: Resistance to tetracycline and distribution of tetracycline resistance genes in European Staphylococcus aureus isolates. J Antimicrob Chemother. 2001;47:239–40. doi: 10.1093/jac/47.2.239. [DOI] [PubMed] [Google Scholar]
- 19.Sekiguchi J, Fujino T, Saruta K, et al. Prevalence of erythromycin-, tetracycline-, and aminoglycoside- resistance genes in methicillinresistant Staphylococcus aureus in hospitals in Tokyo and Kumamoto. Jpn J Infect Dis. 2004;57:74–7. [PubMed] [Google Scholar]
- 20.Lozano C, Porres-Osante N, Crettaz J, et al. Changes in genetic lineages, resistance, and virulence in clinical methicillin-resistant Staphylococcus aureus in a Spanish hospital. J Infect Chemother. 2013;19:233–42. doi: 10.1007/s10156-012-0486-4. [DOI] [PubMed] [Google Scholar]
- 21.Lim KT, Hanifah YA, Yusof M, et al. ermA, ermC , tetM and tetK are essential for erythromycin and tetracycline resistance among methicillin-resistant Staphylococcus aureus strains isolated from a tertiary hospital in Malaysia. Indian J Med Microbiol. 2012;30:203–7. doi: 10.4103/0255-0857.96693. [DOI] [PubMed] [Google Scholar]
- 22.Chopra I, Roberts M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–60. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carneiro LA, Queiroz ML, Merquior VL, et al. Antimicrobial-resistance and enterotoxin-encoding genes among staphylococci isolated from expressed human breast milk. J Med Microbiol. 2004;53:761–8. doi: 10.1099/jmm.0.05453-0. [DOI] [PubMed] [Google Scholar]
- 24.Fatholahzadeh B, Emaneini M, Gilbert G, et al. Staphylococcal cassette chromosome mec (SCCmec) analysis and antimicrobial susceptibility patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolates in Tehran, Iran. Microb Drug Resist. 2008;14:217–20. doi: 10.1089/mdr.2008.0822. [DOI] [PubMed] [Google Scholar]



