SUMMARY
Gene duplication results in two identical paralogs that diverge through mutation, leading to loss or gain of interactions with other biomolecules. Here, we comprehensively characterize such network rewiring for C. elegans transcription factors (TFs) within and across four newly delineated molecular networks. Remarkably, we find that even highly similar TFs often have different interaction degree and partners. In addition, we find that most TF families have a member that is highly connected in multiple networks. Further, different TF families have opposing correlations between network connectivity and phylogenetic age, suggesting that they are subject to different evolutionary pressures. Finally, TFs that have similar partners in one network generally do not in another, indicating a lack of pressure to retain cross-network similarity. Our multiparameter analyses provide an unprecedented glimpse into the evolutionary dynamics that shaped TF networks.
INTRODUCTION
Gene duplication is a major driving force in evolution (Ohno, 1970). After duplication, two identical paralogs emerge that each mutate over evolutionary time. Paralog divergence may be illustrated in terms of sequence and function (Figure 1A), where recently duplicated genes share perfect identity in sequence and, over time, both change in sequence leading to their functional divergence. When one paralog acquires a new function (neo-functionalization), this can lead to functional innovation, and as a result, new organismal complexity. When both paralogs share at least some aspects of the function of their ancestor (subfunctionalization), this can maintain redundancy, which can be beneficial for the organism, for instance to buffer genetic or environmental perturbations (Burga et al., 2011; MacNeil and Walhout, 2011).
Figure 1. A C. elegans PDI Network of TF-Encoding Gene Promoters.
(A) Conceptual diagram of study. Each dot represents a TF paralog pair. The red dot at the top-right corner indicates a pair immediately after duplication when the sequences and function of the two TFs are, by definition, identical. As evolutionary time progresses (indicated by the arrows), both TFs acquire mutations that result in changes in protein sequence similarity, that in turn confer changes in function, and therefore, functional similarity.
(B) We experimentally delineated three physical TF networks involving protein-DNA and protein-protein interactions (PDI and PPI), as well as computationally inferred a co-expression network using available transcriptome data.
(C) A gene-centered C. elegans PDI network, delineated by eY1H assays. Major C. elegans TF families are grouped into brown supernodes. ATH: AT Hook; BED: BEAF/DREF-like; bZIP: basic leucine zipper; DM: Doublesex/Mab-3; ETS: Erythroblast Transformation Specific; FKH: Forkhead; HMG: High Mobility Group; MH1: MAD Homology-1; PD: Paired Domain. PDIs are indicated by arrows between the nodes, with the arrowhead pointing to the family of DNA baits. The thickness of the arrows reflects the number of PDIs detected between the TF families. PDIs between paralogs are shown in red, others in green.
(D) eY1H PDIs overlap significantly with interaction detected by ChIP in vivo. The Venn diagram on the left illustrates the number of interactions that overlap. The graph on the right illustrates that this overlap is significant. The eY1H network was randomized 20,000 times, preserving the network topology, and the overlap in each randomized network was tabulated and plotted. The dotted line indicates the average overlap of 35.3 interactions in the randomized networks. The red arrow indicates the 95 interactions that overlap in the real network. The same result was obtained when the ChIP network was randomized (data not shown).
(E) eY1H PDIs overlap significantly with the occurrence of TF binding sites. The Venn diagram on the left illustrates the number of interactions that overlap. The graph on the right illustrates that this overlap is significant. The eY1H network was randomized 20,000 times, preserving the network topology, and the overlap in each randomized network was tabulated and plotted. The dotted line indicates the average overlap of 45.9 interactions in the randomized networks. The red arrow indicates the 103 interactions that overlap in the real network. The same result was obtained when the binding site-TF annotations were randomized (data not shown).
(F) Cumulative density plot of co-expression values for various groups of TF pairs, including those involved in PDIs. Each point on each line represents the cumulative proportion of pairs that have co-expression scores lower than the value on the x-axis. Interaction pairs are significantly more co-expressed than screened pairs (Spearman Rank: p=2.5×10−22).
See also Figure S1, and Tables S1, S2, S3 and S4.
Genes and proteins work together in the context of intricate molecular interaction networks, and it is the rewiring of these networks during evolution that drives functional change. For example, new interactions can be indicative of neo-functionalization while loss of ancestral interactions can result in sub-functionalization. While different types of networks are rapidly being mapped for numerous model organisms, little is known about the scale of network rewiring and how this relates to sequence divergence. Studies of molecular interaction divergence have so far focused on a few genes (Ow et al., 2008), a single interaction type (Hollenhorst et al., 2007), and/or small gene families (Grove et al., 2009; Tan et al., 2008). In order to gain systems-level insights into network rewiring, it is important to take a “multiparameter” approach and comprehensively characterize and compare networks that consist of different interaction types.
Transcription factors (TFs) are among the most duplicated genes, and comprise 5–10% of eukaryotic proteomes (Levine and Tjian, 2003). They function by engaging in different types of interactions, including protein-DNA interactions (PDIs) with genomic regulatory elements and protein-protein interactions (PPIs) with other TFs and transcriptional cofactors (CFs). Thus TFs are premier candidates to study network evolution. TFs are grouped into families based on their type of DNA binding domain(s). Major TF families include nuclear hormone receptors (NHRs), homeodomains, C2H2 zinc finger proteins and basic helix-loop-helix proteins (bHLHs) (Reece-Hoyes et al., 2005; Vaquerizas et al., 2009).
The comprehensive study of TF network rewiring requires the systematic comparison of all PDIs and PPIs. Yeast one- and two-hybrid assays (Y1H and Y2H) provide a convenient biophysical tool for measuring direct PDIs and PPIs respectively (Deplancke et al., 2004; Walhout, 2006; Walhout et al., 2000). These assays are carried out in the milieu of the yeast nucleus and each protein is expressed under the control of the same promoter. Thus, while these methods are not without caveats (Walhout, 2011), they uniquely enable the direct comparison of interactions involving paralogous proteins measured under identical conditions. This is in contrast to in vivo methods such as chromatin immunoprecipitation (ChIP) that are confounded by native expression patterns that affect protein abundance. In addition, these methods do not necessarily solely measure direct physical interactions, and are affected greatly by antibody quality (Walhout, 2011). Thus, while in vivo methods are extremely useful, they are not desirable when one aims to directly compare the interactions a TF can engage in and to relate these interactions to protein sequence.
We recently developed enhanced Y1H (eY1H) assays for the highthroughput, pair-wise interrogation of PDIs involving C. elegans, Arabidopsis and human TFs (Gaudinier et al., 2011; Reece-Hoyes et al., 2011a; Reece-Hoyes et al., 2011b). These assays utilize a high-density colony array in which TFs are tested in quadruplicate for their capacity to interact with a promoter of interest, together with the robotic handling of assay plates. We have adapted this platform to create enhanced Y2H (eY2H) assays for the identification of PPIs with a protein of interest.
Here, we present four comprehensive networks that capture five interaction types for most C. elegans TFs (Figure 1B). We experimentally delineate a PDI network between TF proteins and promoters of TF genes, which provides two types of interactions: the TFs that bind each promoter (TF-in) and the promoters bound by each TF (TF-out). We also comprehensively measure PPIs among TFs (TF-TF PPI) and between CFs and TFs (CF-TF PPI). Finally, we computationally infer a TF-TF co-expression network based on 123 expression profiles from the SPELL database (Chikina et al., 2009). The integration of these networks enables the correlation between paralog sequence and interaction divergence at an unprecedented scale. The multiparameter analyses of these networks reveal extensive network rewiring and provide a unique glimpse into the evolutionary dynamics that have shaped TF networks. This work provides a rich resource for further functional analysis as well as structural analysis of the molecular determinants that dictate the gain or loss of particular interactions.
RESULTS
A Gene-Centered Core PDI Network for C. elegans TFs
The C. elegans genome encodes 937 predicted TFs, including 271 NHR, 217 C2H2 zinc finger, 101 homeodomain, and 41 bHLH proteins (Reece-Hoyes et al., 2005). Throughout this study, we refer to TFs belonging to the same family (i.e. possessing the same DNA binding domain) as “TF paralogs”. In total, we have generated 834 TF protein expression clones (89%), and 659 (70%) TF gene promoter clones (Figure S1). We used eY1H assays to systematically interrogate pair-wise PDIs between all available TF promoters and TF proteins. For further analyses, we focused on a high-confidence PDI network of 4,453 involving 489 TF promoters and 291 TFs (Figure 1C, Tables S1, S2).
Three analyses indicate that our PDI network is of high quality. First, we found a significant overlap between PDIs detected by eY1H assays and those found by ChIP in vivo (p<0.0001, Figure 1D) (Gerstein et al., 2010; Tabuchi et al., 2011). Second, we found a significant overlap between eY1H interactions and the occurrence of known and newly determined TF binding sites (p<0.0001, Figure 1E, Table S3). Third, we computationally inferred a co-expression network by using 123 expression-profiling experiments to generate a probabilistic score for each TF-TF pair that illustrates the similarity in changes in expression across multiple experiments (Table S4), which is different than spatiotemporal co-expression that reflects expression in the same tissue. Integration of the PDI network with the co-expression network revealed a significant increase in co-expression similarity between TFs and their target genes compared to screened TF-target pairs (Figure 1F). Together, these observations underscore the high quality of the PDI network.
A Comprehensive PPI Network Among C. elegans TFs
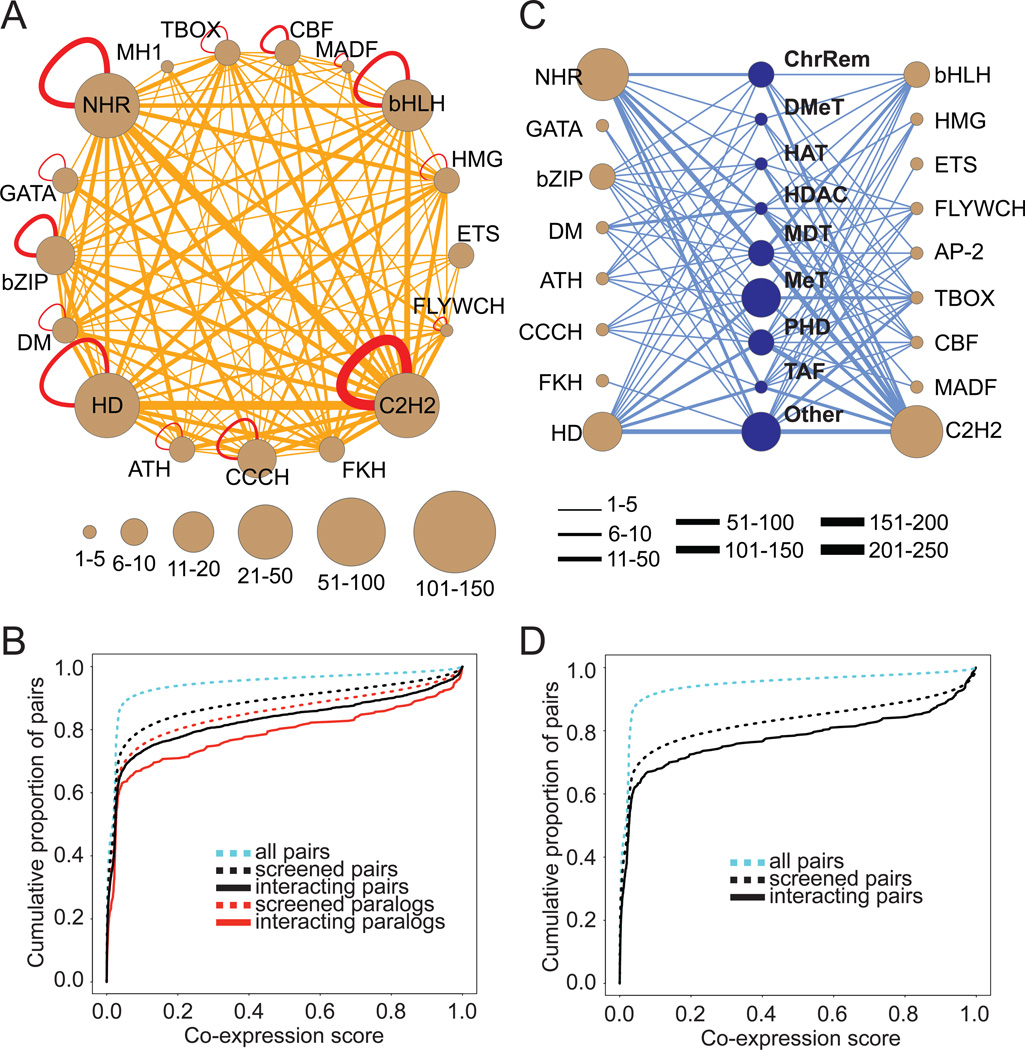
TFs physically interact with other TFs to combinatorially regulate gene expression (Ravasi et al., 2010; Walhout, 2006). We used eY2H assays to comprehensively identify PPIs among the 834 available TFs (Table S5), and detected 2,253 high-confidence interactions among 437 TFs (Figure 2A, Table S6). We retrieved 40 of 47 (85%) of previously reported PPIs that were measured in a pair-wise manner (i.e. each of these pairs was systematically examined before) (Grove et al., 2009; Vermeirssen et al., 2007b), and 50% of those measured by a pooling strategy (i.e. where proteins are tested in pools and it is not certain that each pair was actually examined) (Simonis et al., 2009). These recovery rates of published interactions are similar to that observed with eY1H (Reece-Hoyes et al., 2011b). As in the PDI network, interacting TFs in the PPI network are significantly more co-expressed (Figure 2B). Together, these observations provide support for the quality of the TF-TF PPI network.
Figure 2. Two C. elegans TF Protein-Protein Interaction Networks.
(A) A C. elegans TF-TF PPI network, delineated by eY2H assays. Major C. elegans TF families are grouped into brown supernodes. CBF: CCAAT-Binding Factor; MADF: Mothers Against Dpp Factor. PPIs are indicated by edges between the nodes, with the thickness of the edge reflecting the number of PPIs detected between the TF families. PPIs between paralogs are shown in red, others in orange.
(B) Cumulative density plot of co-expression values for various groups of TF pairs, including those involved in TF-TF PPIs. Interaction pairs are significantly more co-expressed than screened pairs (Spearman Rank: p=2.5×10−18).
(C) A C. elegans CF-TF PPI network, delineated by eY2H assays. Major C. elegans CF and TF families are grouped into blue and brown supernodes, respectively. ChrRem: Chromatin Remodeler; DMet: Histone Demethylase; HAT: Histone Acetyl Transferase; HDAC: Histone Deacetylase; MDT: Mediator; PHD: Plant Homeodomain; TAF: TBP-Associated Factor; AP-2: Activator Protein-2. PPIs are indicated by blue edges between the nodes, with the thickness of the edge reflecting the number of PPIs detected between the CF and TF families.
(D) Cumulative density plot of co-expression values for various groups of TF pairs, including those involved in CF-TF PPIs. Interaction pairs are significantly more co-expressed than screened pairs (Spearman Rank: p=4×10−5).
See also Tables S5, S6 and S7.
A Comprehensive PPI Network between C. elegans CFs and TFs
TFs interact with CFs to regulate the expression of their downstream target genes. To map such interactions, we first defined a compendium of 228 putative C. elegans CFs (Table S7) that includes the following: 1) proteins possessing a plant homeodomain, chromodomain or bromodomain, 2) TBP-associated factors, 3) components of Mediator complex, 4) histone modifiers (only acetyl-transferases, deacetylases, methyl-transferases, demethylases), and 5) proteins identified as CFs in the literature (noted as “other” in Table S7). In total, we screened 196 CFs (86%) in eY2H assays versus all 834 TFs and detected 436 PPIs involving 65 co-factors and 152 TFs (Figure 2C, Table S6). We retrieved 50% of Y2H interactions detected previously (Arda et al., 2010; Simonis et al., 2009). Again, interacting pairs were significantly more co-expressed than screened pairs (Figure 2D).
Rapid Rewiring of Interaction Degree
As a first step into the characterization of network rewiring, we asked if the number of interactions (interaction degree) is conserved between close paralogs. We plotted the degree of all TFs for the four physical interaction types, and as expected, most TFs engage in few interactions while a small number are highly connected (Figure 3A). Such degree asymmetry is common, and has been postulated to provide network robustness to random node perturbation (Albert et al., 2000). The degree distribution for each TF family in each network is similar to that of the global distribution (Figure 3B). By plotting the degree based on protein sequence similarity rather than ranked from most to least connected, we found that in each network even the closest paralogs have remarkably different interaction degrees. For example, NHR-49 and NHR-114 each bind many TFs, but most of their close paralogs associate with only one or few TFs (Figure 3C, D). In addition, while both NHR-49 and NHR-114 bind many TFs, only NHR-49 is an intra-family hub that binds many other NHRs. Altogether, these observations indicate that interaction degrees diverge rapidly after gene duplication.
Figure 3. Interaction Degree Rewiring.
(A) Plots of TF degree for the four interaction types (TF-in, TF-out, TF-TF and CF-TF) delineated by the three physical TF networks. The y-axis indicates the number of interactions. Each bar represents a single TF.
(B) Plots of TF degree for the four major C. elegans TF families for each of the interaction types assayed. The y-axis indicates the number of interactions. Each bar represents a single TF.
(C) TF-TF PPI degree of NHR-49 or NHR-114 and their ten closest paralogs. TFs are ordered by similarity to NHR-49 (left) or NHR-114 (right). The y-axis indicates the number of interactions. Each bar represents a single TF.
(D) Local TF-TF PPI network of NHR-49 (red node) and its interaction partners. Orange nodes indicate other NHRs; gray nodes indicate other types of TFs, with numbers referring to how many proteins if greater than one. Red edges indicate PPIs between NHRs; black edges indicate PPIs between a NHR and another type of TF.
(E) Four examples of TFs that are highly connected in three or more of the datasets, indicated by the red asterisks. The numbers indicate the number of interactions for each dataset in which these TFs participate. Arrows indicate directional PDIs. LSY-2 is a C2H2 zinc finger TF.
(F) Graph depicting how often in one million randomized datasets the given number of TF families (x-axis) has a member that is highly connected in three or more of the four interaction type degree distributions. The red arrow indicates the observed number of the real TF families: p<0.001.
Remarkably, for eight of the nine major TF families, at least one member was among the most highly connected nodes in at least three of the four interaction types (p<0.001; Figures 3E, F, see Extended Experimental Procedures). This suggests that the evolutionary pressures that result in the retention or acquisition of large numbers of interactions in one network also increase the connectivity in other networks. Such coupling of cross-network connectivity may further increase system robustness, because a randomly perturbed TF would be less likely to have many connections in any of the multiple networks.
Differential Rewiring in TF Families with Respect to Evolutionary Age
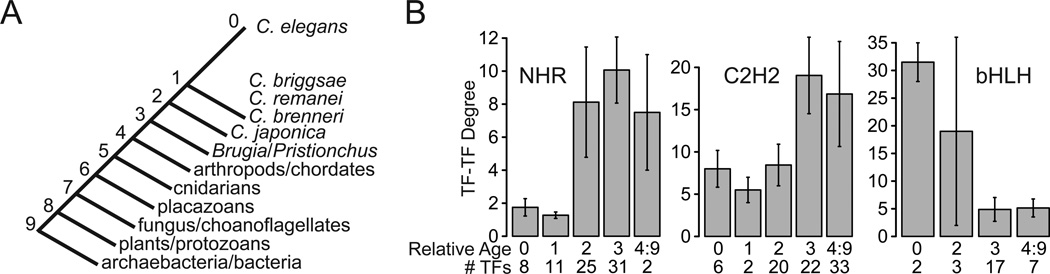
To explore the evolutionary dynamics that result in asymmetric degree distributions within each family, we estimated the duplication time of all C. elegans TFs using a phylogenetic approach (Table S8, Figure 4A, see Extended Experimental Procedures). Globally, we found that younger TFs (produced by more recent duplication events) tend to interact with fewer proteins in the TF-TF PPI network (Spearman correlation, r=0.173, p=0.005; Figure S2). Strikingly, however, we uncovered opposing trends for individual TF families within this same network (Figure 4B, Figure S2, Table S9). Older NHRs and C2H2 TFs have a higher degree (Figure 4B; Fisher’s exact test, p=0.046 and 0.008, respectively), which explains the global correlation for this PPI network, as these two families represent a large proportion of TFs. In contrast, younger bHLH proteins are more highly connected than older family members (Figure 4B, Fisher’s exact test, p=0.046), suggesting that younger bHLH proteins may have rapidly gained interactions. Consistent with this interpretation, members of the bHLH family, particularly those with highest degree, have a much larger fraction of unique interactions (i.e. those not shared by any other family members) than similarly highly connected NHRs (Table S6).
Figure 4. Differential Relationships Between Interaction Degree and Duplication Time.
(A) Stylized tree of life used to determine relative age of TFs. Numbers count tree bifurcations between C. elegans and the common ancestor of organisms listed on the branches.
(B) Comparison of TF-TF PPI network degree and relative age. The average degree of the TFs in each age group is displayed, with error bars indicating the standard error of the mean. Relative age values refer to branches on the tree in (A). The number of TFs in each age group is indicated.
See also Tables S8 and S9.
Noise Analysis of Interaction Profile Similarity Detection
After studying global interaction degrees, we compared the interaction profiles between pairs of paralogous TFs in more detail, by directly comparing individual interaction partners. Such an approach critically depends on low noise in the eY1H and eY2H data. In other words, if two independent screens of the same TF do not have highly overlapping interaction profiles, then interpreting overlap between paralogs will be extremely challenging. We previously demonstrated that eY1H assays exhibit low experimental noise levels: using multiple screens of two highly connected DNA baits, we observed that 90 percent of interactions seen in one eY1H screen are observed in another, and that any one screen captures about 90 percent of all interactions detection after multiple screens (Reece-Hoyes et al., 2011b). To further investigate assay reproducibility, we screened 77 TF promoters and 34 TF proteins twice in eY1H and eY2H assays, respectively. For both types of assay we observed ∼90% reproducibility (data not shown). This more broadly empirically determined rate of technical noise provided us with additional confidence to model the range of profile similarities that two proteins with truly identical interaction profiles would exhibit in the presence of experimental noise. Specifically, we removed 10% of the interactions in each network and replaced them with randomly generated new ones (preserving the degree distribution), effectively generating several replicates of the network. For all interaction types the median interaction profile similarity score for the same TF between replicate screens was 1.0, with at least 75% of these scores being greater than 0.85 (Figure 5A). Taken together, the low experimental noise enables the accurate comparison of interaction profiles between paralogs.
Figure 5. Sequence Similarity is a Poor Predictor of Interaction Profile Similarity.
(A) Boxplots indicating the interaction profile similarity scores generated when comparing original screens to simulated datasets with 10% experimental noise, based on the empirical observation that 90% of interactions found by eY1H and eY2H assays are reproduced in independent screens. The heavy horizontal black line indicates the median, the top and bottom of the box indicate the first and third quartiles, respectively, and the horizontal lines at the ends of the dotted lines below and above the box indicate the minimum and maximum values, respectively.
(B) Proportion of TF pairs with zero or non-zero interaction profile similarity scores.
(C) Scatter plots of full-length protein identity (%) versus interaction profile similarity score for TF pairs in which both members have at least one interaction. Paralog pairs are indicated by red dots, pairs of non-paralogs are in gray. Spearman rank correlations (r) are indicated for all non-paralog pairs (“r(np)”), all paralog pairs (“r(p)”), or close paralog pairs (“r(p+)”; >20% sequence identity).
(D) Boxplots indicating the range of interaction profile similarity scores for various sets of TF pairs, including paralog pairs binned by sequence identity. The heavy horizontal black line indicates the median, the top and bottom of the box indicate the first and third quartiles, respectively, and the horizontal lines at the ends of the dotted lines below and above the box indicate the minimum and maximum values, respectively.
(E) Graph depicting the normalized average interaction profile similarity score for paralogs binned by full-length protein identity. Scores were normalized relative to the average interaction profile similarity score for all non-paralog pairs.
Sequence Similarity is a Poor Predictor of Interaction Profile Similarity
We computed interaction profile similarity scores for all pairwise TF-TF combinations, involving TFs with at least one interaction. In each of the four physical interaction types, 50–70% of paralogs do not share any partners (i.e. interaction profile similarity score is zero; Figure 5B). Strikingly, this is similar to that observed for TFs from different families (non-paralogous TF pairs). Interaction profile similarity scores were then compared to protein sequence similarity, defined as the percentage of identical amino acids in either the full-length protein (Figure 5C) or only the DNA binding domain (Figure S3). Remarkably, there was little global correlation between sequence similarity and interaction profile similarity. We observed a slightly higher correlation for TF-out, TF-TF, and CF-TF interaction profiles when we restricted our analysis to close paralogs, defined as those pairs with 20% sequence identity or greater in the full-length protein. We observed much less of an increase for TF-in sequence-interaction correlation for close paralogs, likely because regulatory regions diverge much more rapidly than coding regions (Castillo-Davis et al., 2004). Importantly, for all four physical interaction types, interaction profile similarity scores for even the most similar paralogs do not approximate those for simulated replicate screens with empirically defined levels of experimental noise (compare Figures 5A, D). This demonstrates that the low correlation between sequence and interaction profile similarity is not due to experimental noise, but rather indicates dramatic and rapid interaction rewiring after gene duplication. Further, it reveals that protein sequence similarity is generally a poor predictor of interaction profile similarity.
Next, we asked whether there were any differences in network rewiring for the four interaction types. To do so, we averaged interaction profile similarity scores of paralogous TF pairs in six bins of protein identity and normalized these scores to the average score of all non-paralog pairs in each bin. Relating these normalized paralog interaction profile scores to sequence similarity revealed striking differences in how quickly interaction profile similarity decreased with increasing sequence divergence (Figure 5E). For instance, TF-out similarity is greater and persists much longer than the other interaction types: for paralogs with 25–50% sequence identity, the average interaction profile similarity score for TF-out interactions is ∼9-fold greater than the average similarity for non-paralog pairs, whereas it is ∼5-fold for TF-TF and CF-TF interactions, and no different from background in the TF-in network. Similar results were obtained when we used DNA binding domain identity as a measure of sequence similarity (data not shown). These results indicate that rewiring is different in the different interaction types with TF-out being the least sensitive and TF-in the most sensitive to sequence changes.
Cross-Network Analysis of Interaction Profile Similarities
Our comprehensive multiparameter study uniquely enabled us to broadly assess TF paralog divergence not only within, but also across different networks. Specifically, we asked whether paralog pairs with high similarity in one parameter (i.e. interaction types and co-expression) were also highly similar in another, or whether they evolved independently across the different networks. For each TF paralog pair we counted the networks in which it retained high similarity, defined as the top 10% of scores (Table S10). Notably, TF pairs with such high similarity scores were mostly non-paralogs (Figure 6A). The numbers of paralog pairs exhibiting high similarity in multiple networks were small. However, comparing the observed occurrence of cross-network high similarity to what would be expected when each network evolves independently revealed a slightly greater number of pairs with similarity in two or three parameters (Figure 6B). This suggested that there may be (small) levels of co-dependence between some parameters.
Figure 6. Lack of Evolutionary Pressure to Retain High Cross-Network Similarity.
(A) Proportion of paralog and non-paralog pairs that occur in the top 10% of scores for each interaction type and co-expression. In each network, only TFs with at least one interaction were considered.
(B) Proportions of paralog pairs with high similarity in zero, one or multiple parameters. Only pairs in which both members are functional in all assays were considered. The gray bars show the average results of 1,000 samplings in which 10% of the pairs were randomly selected from each parameter. The error bars show the standard deviation for each bin.
(C) Comparison of the number of paralog pairs observed in the top 10% for both listed parameters to the number expected when divergence is independent. See also Tables S10 and S11.
To identify the source of this putative co-evolution, we analyzed each pairwise combination of parameters independently by testing for association (using Fisher’s exact test) between high interaction similarity in one network and high similarity for the same paralog pair in another network (see Extended Experimental Procedures). This analysis revealed that most networks have diverged independently as we failed to reject the null hypothesis for many of them (Table S11). Thus, when a TF pair shares similar interactions in one network, it tends not to in another. However, there were some notable exceptions. For example, paralogs tend to exhibit high similarity in both the TF-TF and CF-TF networks as well as in both the TF-TF and TF-out networks more than expected based on a model of independent divergence (Figure 6C; Fisher’s exact p=0.0003 and p=0.01, respectively). The former may be a result of the fact that common domains mediate both types of protein-protein interactions, and the latter may reflect co-binding of target promoters. Surprisingly, we found anti-coupling between TF-TF and co-expression similarity as fewer than expected paralog pairs had high similarity in both of these parameters relative to independent divergence (Figure 6C; Fisher’s exact p=0.01). This suggests that paralogs that share interacting TFs tend to diverge more extensively in their expression patterns.
Examples of Network Rewiring
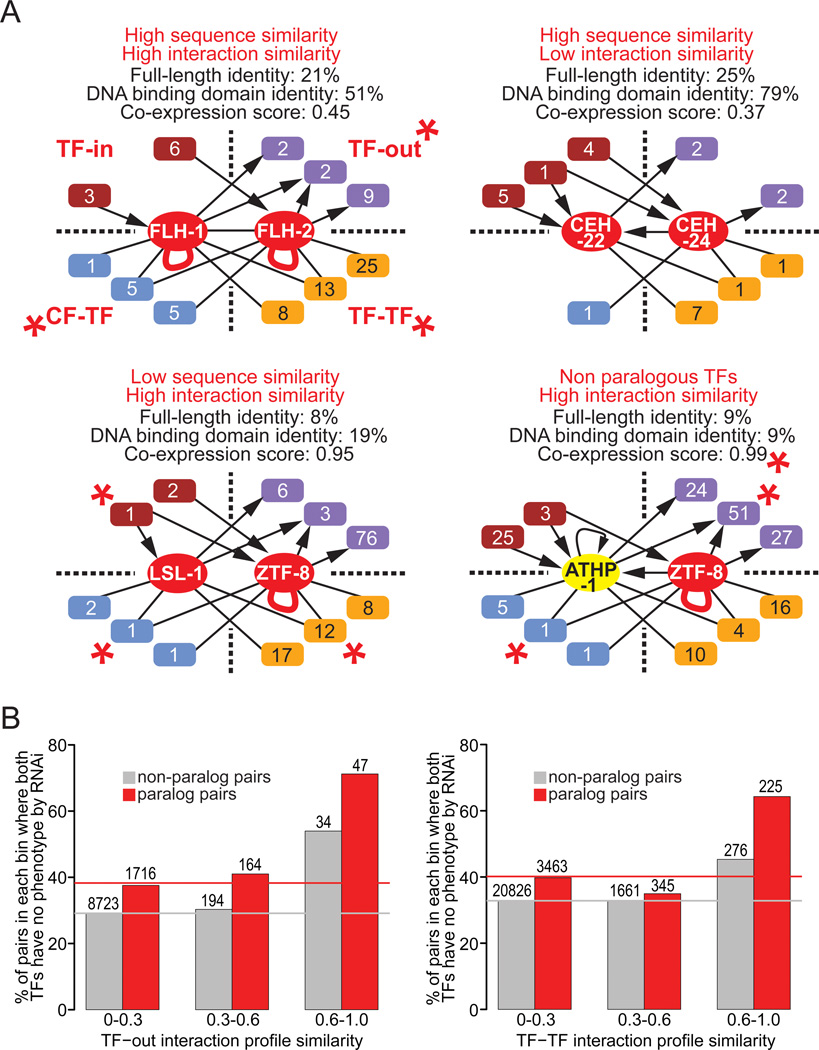
Several examples of TF pairs illustrate our overall findings (Figure 7A). A rare example of a pair with high sequence and overall network similarity is FLH-1 and FLH-2: TFs that are known to function redundantly in vivo (Ow et al., 2008). More common are paralog pairs that exhibit high sequence but low interaction profile similarity and vice versa. For instance, CEH-22 and CEH-24 share 25% full-length protein sequence identity (79% in the DNA binding domain), but share no promoter targets or CF partners and only a single interacting TF. An example of the converse is LSL-1 and ZTF-8, a paralog pair that share overlapping interactions in all networks but have low protein sequence similarity (8% full protein identity). These TFs may have retained functional similarity despite substantial sequence divergence, or alternatively, may have first diverged in sequence and function, and subsequently converged. Interactions shared by paralogs may contribute to redundant functions whereas unique interactions likely indicate sub- or neo-functionalization. Finally, the non-paralogous pair ATHP-1/ZTF-8 exhibits high co-expression and shares significant overlap in promoter and CF partners.
Figure 7. Examples of Multiparameter TF Pair Comparisons.
(A) Four examples of TF pairs with varying levels of protein sequence and parameter (interaction profile and co-expression) similarity. Parameters in which these pairs occurred in the top 10% are indicated by a red asterisk. FLH-1 and FLH-2 are paralogs that function partly redundantly in vivo (Ow et al., 2008), and are highly similar in their protein sequence and three functional parameters. CEH-22 and CEH-24 are paralogs with high protein sequence similarity but no highly similar parameters. LSL-1 and ZTF-8 are paralogs with low protein similarity but high similarity in three functional parameters. ATHP-1 and ZTF-8 are from different TF families but share high similarity in three parameters.
(B) Graphs depicting the proportion of TF pairs (paralogs in red, non-paralogs in gray) within three bins of interaction profile similarity scores (TF-out on left, TF-TF on right) for which both members have no phenotype by RNAi. The number above each column describes how many pairs represented.
TF Pairs with High Interaction Profile Similarity Are More Likely to Have No Phenotype
It is likely that redundant TFs exhibit high overlap in their interaction profiles. Indeed, FLH-1 and FLH-2 are highly similar in the different networks (Figure 7A). However, very few C. elegans TFs are known to function redundantly in vivo, making a global analysis infeasible. We hypothesized that the converse would also occur: that TFs with high interaction profile similarity are more likely to function redundantly in vivo. To address this we collated RNAi phenotype data for C. elegans TFs (www.wormbase.org), much of which was generated by genome-scale screens. In C. elegans, 80% of TFs are dispensable for viability when perturbed individually, which is slightly lower than that for all genes (89%). We asked whether TF pairs with high interaction profile similarity in any of the networks would not be annotated with any known phenotype. We first found that for paralog pairs both members more often have no phenotype detected than is the case for non-paralog pairs (compare gray and red horizontal lines in Figure 7B), which is indicative of redundancy between duplicated genes. Further, we found that a higher proportion of both paralog and non-paralog pairs with high interaction profile similarity scores exhibit no phenotype for TF-TF and TF-out parameters (Figure 7B), and that this trend is stronger for paralogs. Thus, our global analysis suggests that TFs with highly overlapping interaction profiles are more likely to be functionally redundant than TFs with dissimilar interaction profiles.
DISCUSSION
We present the first comprehensively mapped PDI and PPI networks for multiple large protein families in a metazoan organism. Our multiparameter study enabled unprecedented analysis of gene family evolution that produced numerous novel insights into the evolutionary dynamics that have shaped C. elegans TF networks. In addition, these networks provide a rich resource for future functional studies of individual TFs.
The networks we present are not complete. There are several reasons why we will have missed PDIs, including those involving TFs that bind DNA as obligate heterodimers or that need to be post-translationally modified (Walhout, 2011). In addition, it is possible that not all predicted TFs actually function as such. For instance, C2H2 zinc finger proteins may instead bind RNA, or other proteins (Tanaka Hall, 2005). However, and importantly, we only focus our analyses on TFs that do work in the assays and that have been comprehensively tested, enabling their direct comparison. Thus, our overall insights are not affected by technical false negatives.
A unique aspect of our study is the fact that we have mapped both PPI and PDI networks comprehensively by testing hundreds of thousands of pairwise combinations in a highly standardized manner, enabling direct comparisons across, as well as within, the different networks. This would not have been feasible with in vivo methods, largely because they are significantly affected by spatial TF expression as well as cellular TF concentrations. For instance, two highly similar TFs may exhibit very different interaction profiles because they are expressed at very different levels/tissues rather than because of their differences in protein sequence. Further, published datasets were not sufficient for this type of study. Available C. elegans Y1H and Y2H networks (Arda et al., 2010; Deplancke et al., 2006; Simonis et al., 2009; Vermeirssen et al., 2007a) are not suitable for paralog comparisons because they did not examine all interactions directly as these studies utilized library screens or pooling rather than direct pairwise testing. Further, data generated by the modENCODE consortium (Gerstein et al., 2010) are not useful for this type of study since they identify only PDIs for a minority of TFs and because, as noted before, in vivo interactions are affected not only by protein sequence, but also by expression levels. In the future, we envision that in vivo data need to be combined with biophysical data such as presented here to provide a complete picture of gene family divergence and the relative contribution of protein sequence and expression.
Intuitively, large changes in protein sequence are expected to confer large changes in function and small changes are expected to cause little functional change. However, we find that network rewiring occurs rapidly and is extensive for all TF families. These observations expand upon our previous findings for the bHLH TF family, where most family members are markedly different in multiple interaction types (Grove et al., 2009). This rapid divergence is broadly consistent with studies in yeast, which focused largely on duplicate gene pairs or smaller families (Wagner, 2005), as well as a recent large PPI study in Arabidopsis (2011).
In yeast, PPIs and genetic interactions are asymmetrically distributed between paralogs (VanderSluis et al., 2010; Wagner, 2002). Our results suggest a similar asymmetry for molecular interactions of C. elegans TFs: in all networks, most families are characterized by one, or a few highly connected hubs, while most family members have relatively few interactions. We also find that most TF families have at least one member with a large number of interactions in at least three of the four networks. The evolutionary forces and structural properties that result in such “multiple network hubs” are unclear, but deserve further study.
The multiparameter nature of our study also enabled the first direct comparison of the rates at which PPIs and PDIs are rewired. We find that TF-in PDIs (TFs that bind a gene’s promoter) are rewired much faster than any other interaction type, while TF-out PDIs (promoters bound by a TF) persist the longest. These observations present an interesting functional contrast between upstream regulatory regions, which are highly plastic and diverge rapidly, and DNA sequence specificities of TFs, which are more stable over evolutionary time.
The respective order of the rates of rewiring for these interaction types suggests that TF sub- and/or neo-functionalization is most easily achieved through changes in expression or the loss or gain of specific PPIs. Consistent with this idea, we find evidence for negative correlation between TF-TF paralog PPIs and TF paralog expression similarity. Specifically, there are significantly fewer than expected paralog pairs that share a high proportion of interacting TFs and that exhibit high co-expression similarity. This has interesting connections to observations reported in yeast, where duplicates resulting from whole-genome duplication events retain more PPI similarity but exhibit high divergence in co-expression and promoter sequence, whereas in genes arising from local duplication events, high co-expression persisted longer while PPI similarity diverged quickly (Guan et al., 2007).
Relationships between evolutionary age and PPI degree have been previously explored in yeast, where older genes tend to engage in more PPIs (Capra et al., 2010; Kim and Marcotte, 2008). We approached the concept of age differently: rather than dating the proteins by the earliest occurrence of a homolog which would group all members of a family into one age category, we used a phylogenetic approach to estimate the time of the most recent duplication producing each individual extant gene. Using this approach, we observed a positive correlation between age and TF-TF PPI degree in the ZF-NHR and ZF-C2H2 families: many high-degree proteins in the TF-TF network were very old in these families (i.e. they had their last duplication in an ancestor very distant from C. elegans). Two models explain this observation. First, the lower connectivity of young NHRs or C2H2 TFs may result from relaxed purifying selection on recently duplicated members of the family (i.e. because the phenotypic effect of mutations in one paralog is masked by the presence of the other paralog). Several previous studies reported evidence for the idea that duplicate genes experience relaxed selective pressure immediately after duplication (Jordan et al., 2004; Lynch and Conery, 2000; Scannell and Wolfe, 2008). This is typically associated with accelerated sequence evolution immediately after the duplication event, which slows down as the two paralogs diverge in function (Scannell and Wolfe, 2008). With respect to our study, young TFs may have lost the majority of their ancestral interactions due to rapid accumulation of mutations, while maintaining a few specialized functions or acquiring new ones, and as a result were retained in the genome. Second, recent work has suggested that there are biases in which genes are “duplicable” due to the fact that some duplications result in harmful imbalances in expression (Papp et al., 2003; Wapinski et al., 2007). Genes with deleterious overexpression phenotypes in yeast are highly enriched for encoding disordered proteins as well as proteins with a large number of PPIs (Vavouri et al., 2009). Thus, the lower PPI degree for young NHRs and C2H2 TFs may simply reflect the fact that the high-degree family members are less duplicable due to deleterious fitness effects upon increased expression. Remarkably, we observed the opposite for the bHLH family, where recently duplicated members exhibit an increased interaction degree. This suggests that different evolutionary pressures have shaped this family. For instance, there may have been a recent gain of interactions through positive selection or a family hub may have undergone several duplication events.
Comprehensive, systems-level networks can be effective guides to gain insights into the molecular determinants that provide interaction specificity. For instance, by combining DNA binding networks with available co-crystal structures, amino acids that dictate the unique recognition of particular DNA sequences could be identified (De Masi et al., 2011; Noyes et al., 2008). The TF networks presented here will provide a fruitful resource for the similar analysis of different TF families (with different DNA binding domains), and different interaction types.
Altogether, our observations indicate that sequence similarity is generally not predictive of interaction profile similarity and illustrate the continued need for the mapping of molecular interaction networks for different species. Our comprehensive multiparameter study provides a framework that can be extended to other types of proteins, other types of networks, as well as other complex organisms, particularly those for which complete clone collections are available that enable the comprehensive delineation of molecular interaction networks.
EXPERIMENTAL PROCEDURES
eY1H and eY2H Assays and Data Collection
eY1H assays have been described previously (Reece-Hoyes et al., 2011b). Briefly, a DNA “bait” is cloned upstream of two reporter genes and each construct is integrated into different sites within the genome of “bait” yeast strain. An array of yeast “prey” strains, that each express a different “prey” TF-Gal4AD fusion, are mated individually to the bait strain, and expression of the reporters are assayed in the resulting diploid yeast. The two reporter genes are LacZ and HIS3; LacZ expression is detected via the conversion of colorless X-gal to a blue compound, while HIS3 expression allows the yeast to grow on media lacking histidine and to overcome the addition of 3-amino-triazole (3AT), a competitive inhibitor of the His3 enzyme used to counter background expression of the reporter. Each interaction is tested four times within each assay and only those that are positive for both reporters at least twice are considered genuine. eY2H assays utilize the same reagents (yeast strains, protein expression constructs, and media) as previous Y2H assays (Walhout et al., 2000), however, they are performed in the same way as eY1H (i.e. the eY2H bait strain is mated with the same TF array, and the same reporters assayed). All eY1H and eY2H screens were processed using the SpotOn software we developed for calling interactions from images of the assay readout plates (Reece-Hoyes et al., 2011b). For data tracking and management, as well as to make the raw and quantified data publicly available, we developed ‘MyBrid’, a web-based tool with which one can retrieve interactions for particular gene(s) or TFs (http://csbio.cs.umn.edu/MyBrid, Diallo et al., in preparation).
Generating Interaction Profile Similarity Scores
For each eY1H and eY2H interaction dataset a binary matrix was generated (i.e. detected interaction indicated by “1”, no interaction indicated by “0”). Similarity in each network was calculated using only the proteins/promoters that reported at least one interaction in that network. A cosine similarity score was generated for every TF-TF pair for each network by comparing their interaction profiles in the relevant dataset (TF-in, TF-out, TF-TF, CF-TF) using the formula:
Calculating Protein Identity
Protein sequences for the longest isoforms of the C. elegans TFs in wTF2.2 (Reece-Hoyes et al., 2011b) were extracted from Wormbase WS200 release (wormbase.org). For most TFs, this isoform matches the cloned version. DNA binding domains (DBDs) were identified using NCBI-CDS (ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), InterProScan (ebi.ac.uk/Tools/pfa/iprscan), or manually. ClustalW (Chenna et al., 2003) was used to align the proteins, and the number of matching residues was counted and divided by the number of residues in the longest of the two proteins. To avoid the complication of assigning similarity scores for each amino acid based on charge, hydrophobicity, etc., we focused on sequence identity rather than similarity. If a TF had multiple DBDs of the same family then the DBD sequences were concatenated in the order they appear in the full-length protein sequence. 22 TFs had multiple DBDs from different families and were not considered for this study, along with another seven TFs for which gene model changes resulted in the removal of their DNA binding domains.
Supplementary Material
Highlights.
Four comprehensive C. elegans transcription factor networks
Extensive and rapid network rewiring during evolution
Dynamic evolutionary pressures for different TF families and different networks
Rich datasets for structural and functional studies
ACKNOWLEDGEMENTS
We thank members of the Walhout and Myers laboratories, J. Dekker, and N. Springer for discussions and critical reading of the manuscript. This work was supported by National Institutes of Health (NIH) grant GM082971 to A.J.M.W. and D.E.H.; NIH grant DK06429 to A.J.M.W.; CIHR grant MOP-111007 to T.R.H.; fellowships from CIHR and CIFAR to M.T.W.; and C.L.M., C.P., J.N., and P.S.M.R. are partially supported by grants from the NIH (HG005084-01A1, HG005853-01) and the National Science Foundation (DBI 0953881).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, three figures and eleven tables.
REFERENCES
- Albert R, Jeong H, Barabasi A-L. Error and attack tolerance of complex networks. Nature. 2000;378:378–381. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interaction Mapping Consortium. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Taubert S, Conine C, Tsuda B, Van Gilst MR, Sequerra R, Doucette-Stam L, Yamamoto KR, Walhout AJM. Functional modularity of nuclear hormone receptors in a C. elegans gene regulatory network. Molecular Systems Biology. 2010;6:367. doi: 10.1038/msb.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burga A, Casanueva MO, Lehner B. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature. 2011;480:250–253. doi: 10.1038/nature10665. [DOI] [PubMed] [Google Scholar]
- Capra JA, Pollard KS, Singh M. Novel genes exhibit distinct patterns of function acquisition and network integration. Genome Biology. 2010;11:R127. doi: 10.1186/gb-2010-11-12-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis CI, Hartl DL, Achaz G. Cis-regulatory and protein evolution in orthologous and duplicate genes. Genome Res. 2004;14:1530–1536. doi: 10.1101/gr.2662504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikina MD, Huttenhower C, Murphy CT, Troyanskaya OG. Global prediction of tissue-specific gene expression and context-dependent gene networks in Caenorhabditis elegans . PLoS Comput Biol. 2009;5:e1000417. doi: 10.1371/journal.pcbi.1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Masi F, Grove CA, Vedenko A, Alibes A, Gisselbrecht SS, Serrano L, Bulyk ML, Walhout AJM. Using a structural and logics systems approach to infer bHLH DNA binding specificity determinants. Nucleic Acids Res. 2011;39:4553–4563. doi: 10.1093/nar/gkr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Dupuy D, Vidal M, Walhout AJM. A Gateway-compatible yeast one-hybrid system. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Mukhopadhyay A, Ao W, Elewa AM, Grove CA, Martinez NJ, Sequerra R, Doucette-Stam L, Reece-Hoyes JS, Hope IA, et al. A gene-centered C. elegans protein-DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Gaudinier A, Zhang L, Reece-Hoyes JS, Taylor-Teeples M, Pu L, Liu Z, Breton G, Pruneda-Paz JL, Kim D, Kay SA, et al. Enhanced Y1H assays to elucidate Arabidopsis gene regulatory networks. Nature Methods. 2011;8:1053–1055. doi: 10.1038/nmeth.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove CA, deMasi F, Barrasa MI, Newburger D, Alkema MJ, Bulyk ML, Walhout AJ. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Dunham MJ, Troyanskaya OG. Functional analysis of gene duplications in Saccharomyces cerevisiae . Genetics. 2007;175:933–943. doi: 10.1534/genetics.106.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Wolfe YI, Koonin EV. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol Biol. 2004;4:22. doi: 10.1186/1471-2148-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WK, Marcotte EM. Age-dependent evolution of the yeast protein interaction network suggests a limited role of gene duplication and divergence. PLos Comp Biol. 2008;4:e1000232. doi: 10.1371/journal.pcbi.1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1154. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- MacNeil LT, Walhout AJM. Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression. Genome Res. 2011;21:645–657. doi: 10.1101/gr.097378.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer; 1970. [Google Scholar]
- Ow MC, Martinez NJ, Olsen P, Silverman S, Barrasa MI, Conradt B, Walhout AJM, Ambros VR. The FLYWCH transcription factors FLH-1, FLH-2 and FLH-3 repress embryonic expression of microRNA genes in C. elegans . Genes Dev. 2008;22:2520–2534. doi: 10.1101/gad.1678808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Barutcu AR, Patton McCord R, Jeong J, Jian L, MacWilliams A, Yang X, Salehi-Ashtiani K, Hill DE, Blackshaw S, et al. Yeast one-hybrid assays for high-throughput human gene regulatory network mapping. Nature Methods. 2011a;8:1050–1052. doi: 10.1038/nmeth.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Deplancke B, Shingles J, Grove CA, Hope IA, Walhout AJM. A compendium of C. elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 2005;6:R110. doi: 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Diallo A, Kent A, Shrestha S, Kadreppa S, Pesyna C, Lajoie B, Dekker J, Myers CL, Walhout AJM. Enhanced yeast one-hybrid (eY1H) assays for high-throughput gene-centered regulatory network mapping. Nature Methods. 2011b;8:1059–1064. doi: 10.1038/nmeth.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D, Wolfe K. A burst of protein sequence evolution and a prolonged period of asymmetric evolution follow gene duplication in yeast. Genome Res. 2008;18:137–147. doi: 10.1101/gr.6341207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis N, Rual JF, Carvunis AR, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi T, Deplancke B, Osato N, Zhu LJ, Barrasa MI, Harrison MM, Horvitz HR, Walhout AJM, Hagstrom K. Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet. 7. 20112011:e1002074. doi: 10.1371/journal.pgen.1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Feizi H, Luo C, Fan SH, Ravasi T, Ideker TG. A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response. Proc Natl Acad Sci U S A. 2008;105:2934–2939. doi: 10.1073/pnas.0708670105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Hall TM. Multiple modes of RNA recognition by zinc finger proteins. Current Opin Struct Biol. 2005;15:367–373. doi: 10.1016/j.sbi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- VanderSluis B, Bellay J, Musso G, Costanzo M, Papp B, Vizeacoumar FJ, Baryshnikova A, Andrews B, Boone C, Myers CL. Genetic interactions reveal the evolutionary trajectories of duplicate genes. Mol Syst Biol. 2010;6:429. doi: 10.1038/msb.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;198:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Vermeirssen V, Barrasa MI, Hidalgo C, Babon JAB, Sequerra R, Doucette-Stam L, Barabasi AL, Walhout AJM. Transcription factor modularity in a gene-centered C. elegans core neuronal protein-DNA interaction network. Genome Res. 2007a;17:1061–1071. doi: 10.1101/gr.6148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V, Deplancke B, Barrasa MI, Reece-Hoyes JS, Arda HE, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Brent M, et al. Matrix and Steiner-triple-system smart pooling assays for high-performance transcription regulatory network mapping. Nat Methods. 2007b;4:659–664. doi: 10.1038/nmeth1063. [DOI] [PubMed] [Google Scholar]
- Wagner A. Asymmetric functional divergence of duplicate genes in yeast. Mol Biol Evol. 2002;19:1760–1768. doi: 10.1093/oxfordjournals.molbev.a003998. [DOI] [PubMed] [Google Scholar]
- Wagner A. Distributed robustness versus redundancy as causes of mutational robustness. BioEssays. 2005;27:176–188. doi: 10.1002/bies.20170. [DOI] [PubMed] [Google Scholar]
- Walhout AJM. Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome Res. 2006;16:1445–1454. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- Walhout AJM. What does biologically meaningful mean? A perspective on gene regulatory network validation. Genome Biol. 2011;12:109. doi: 10.1186/gb-2011-12-4-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJM, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.