Abstract
We have explored the functional relationships between spliceosome and Microprocessor complex activities in a novel class of microRNAs (miRNAs), named Splice site Overlapping (SO) miRNAs, whose pri-miRNA hairpins overlap splice sites. We focused on the evolutionarily conserved SO miR-34b, and we identified two indispensable elements for recognition of its 3′ splice site: a branch point located in the hairpin and a downstream purine-rich exonic splicing enhancer. In minigene systems, splicing inhibition owing to exonic splicing enhancer deletion or AG 3′ss mutation increases miR-34b levels. Moreover, small interfering-mediated silencing of Drosha and/or DGCR8 improves splicing efficiency and abolishes miR-34b production. Thus, the processing of this 3′ SO miRNA is regulated in an antagonistic manner by the Microprocessor and the spliceosome owing to competition between these two machineries for the nascent transcript. We propose that this novel mechanism is commonly used to regulate the relative amount of SO miRNA and messenger RNA produced from primary transcripts.
INTRODUCTION
MicroRNAs (miRNAs) are 21–23-nt long non-coding RNAs that regulate gene expression by affecting translation and/or stability of messenger RNA (mRNAs) (1). Embedded in coding or non-coding genes, the hairpin secondary structure of primary (pri)-miRNAs is initially cropped in the nucleus by Drosha, an RNase III-like enzyme that is part of the Microprocessor Complex (MPC), along with its cofactor DGCR8 (2). The resulting precursor (pre)-miRNA is ∼70 nt long and is exported to the cytoplasm where it is cleaved by Dicer to obtain the final mature form. On the nascent transcript, the MPC-dependent processing of the pri-miRNA hairpin is an important and early regulatory event involved in miRNA biogenesis. Indeed, several proteins interfere with the activity of the MPC, including RNA-binding proteins that either affect components of the MPC (3,4) or directly interact with the pri-miRNA hairpins (5–8).
The splicing reaction allows the maturation of a precursor (pre)-mRNA through the joining of the exonic sequences and the excision of the introns; to correctly identify exons, the splicing machinery recognizes the core cis-acting elements (9,10) that consist of the 5′ and 3′ splice sites (ss) and include the polypyrimidine tract and the branch point (BP) near the 3′ss. Recognition of the exon requires also splicing regulatory elements that are classified, depending on their location and effect on splicing, as exonic/intronic splicing enhancer and exonic/intronic splicing silencers (9,10). These elements are crucial for alternative splicing regulation, a mechanism present in the majority of human genes that enormously increase the transcript diversity through the selection of alternative splice sites (9). The exonic elements are composed of largely degenerated poorly conserved sequences and interact with splicing factors that may have a positive (serine/arginine-rich (SR) proteins) or a negative heterogeneous nuclear ribonucleoproteins (hnRNPs) effect on exon recognition (11).
Several polymerase II (PolII) precursor transcripts are processed co-transcriptionally by the spliceosome and the MPC into spliced mRNAs and miRNAs, respectively. In the case of intronic miRNA hairpins, which represent almost half of miRNAs (12), Drosha cleavage occurs before splicing and does not significantly affect the amount of mRNA (12,13). On the other hand, intronic pri-miRNA hairpins, both in coding or non-coding transcripts, are preferentially located at a distance from splice sites to avoid possible interference between the two processing machineries (12,13). Experiments using minigenes and in vivo analysis indicate that Drosha cleavage at intronic pri-miRNAs can both increase (12,14) and decrease (13,15) the splicing efficiency. pri-miRNA processing is more efficient if hairpins are retained at the sites of transcription (16), and, in some constructs, splicing disruption of both the 5′ss and 3′ss was found to affect miRNA biosynthesis (13). However, for the intronic miR-211, only mutations at the 5′ss were reported to reduce the biogenesis of the miRNA.
miR-34b, along with the related miR-34a and miR-34c, is involved in several physiological and pathological conditions. Originally identified as a tumour suppressor miRNA (17–19), miR-34b is involved in osteoblast proliferation (20,21), pathological cardiac remodelling (22) and Huntington and Parkinson diseases (23,24). miR-34b and miR-34c are part of the same non-coding transcriptional unit on chr11, possibly regulated by a p53-responsive promoter (25). The transcriptional unit is composed of two exons separated by a ∼2 kb long intron. miR-34c is part of the last exon, whereas miR-34b is unexpectedly located on the boundary between intron 1 and exon 2.
In this study, we have identified a peculiar class of miRNAs, including miR-34b, whose hairpins overlap with splice sites and whose biogenesis is regulated by splicing. We have named these Splice site Overlapping (SO)-miRNAs. SO miR-34b overlaps with a non-canonical 3′ss, whose recognition depends on a strong BP and a purine-rich exonic splicing enhancer (ESE). Splicing inhibition by mutation of the 3′ss or the ESE, but not the 5′ss, increases miR-34b biosynthesis, whereas reduction of the Drosha/DGCR8 levels by RNAi knock-down increases splicing efficiency.
MATERIALS AND METHODS
Cell culture, transfections and reverse transcription-PCR analysis
HeLa cell culture and transfection, RNA extraction, reverse transcription (RT)-PCR and quantification of the percentage of splicing were performed as previously described (26). For the analysis of spliced isoforms, pBRA 34b minigenes were amplified with BRC90BsteII for (ctggtgaccaagtttgccagaaaacaccacatcactttaactaatc) and glo800 rev (gctcacagaagccaggaacttgtccagg); pcDNA3pY7 miR-34b constructs were amplified with pY7 ex2 dir (tacaaggcttgtcgaggaggacatc) and miR34b_2505XbaI rev (tatctagaccacgccgacgccgcgct). For co-transfection experiments, HeLa cells were transfected with 500 ng of the minigene construct together with 500 ng of an empty vector or vectors containing the proteins of interest.
Detection of spliced and unspliced miR-34b transcripts in mouse and human tissues
The human total RNA of 20 tissues was purchased from Amsbio, whereas the mouse one was extracted from tissues using TriReagent (Ambion) according to manufacturer’s instructions.
The primers used for the RT-PCRs performed to detect the spliced and unspliced isoforms of human miR-34b transcripts were as follows: 34b_131 for (agtaggcaatgcatcttcatgac) and 34b_521 rev (ccttcgagagaagatgcctg) for the splicing form and 34b_233 for (cttttcaaggcatctgaccc) and 34b_435 rev (aatagtcttcattccattaaca) for the unspliced one. For the mouse spliced isoform, we used mmu_34b_4299 for and mmu_34b_6598 rev (CAATGATAGCTTTGGATGGAAGC), and for the unspliced variant, we used mmu_34b_6397 for (agtagtagaaatagcctccatcc) and mmu_34b_6609 rev (GACAGTTTATGCAATGATAGCT). For the control GAPDH gene, we used GAPDH for (gacagtcagccgcatcttct) and GAPDH rev (ttaaaagcagccctggtgac).
Quantitative RT-PCR
Quantitative (q)RT-PCR to detect the abundance of mouse and human mature miRNAs was performed using the TaqMan® Assay (Applied Biosystems). Both RT and PCR were performed according to manufacturer instructions.
Minigene design
To clone the hairpin structure of miR-34b and its flanking portions (18 bp upstream and 97 bp downstream), we amplified the region of interest by PCR from human genomic DNA. To clone in the pBRA plasmid (27), we used the following set of primers: miR-34b_2310 Xba for (tatctagacagccgcgggtgcccggtgc) and miR34b_2505 Pst rev (tactgcagccacgccgacgccgcgct). To clone the same region in the pcDNA3pY7 construct (28), we used the following oligonucleotides: miR-34b_2310 Xho for (tatactcgagccgcgggtgcccggtgct) and miR34b_2505Xba rev (tatctagaccacgccgacgccgcgct). To generate the mutated constructs, we amplified the sequence of interest with mutated primers built either for direct or overlapping PCR according to the position of the mutation.
Small interfering transfection and western blot
Small interfering (siRNA) transfections were performed in HeLa cells using Oligofectamine Reagent (Invitrogen) as previously described (26). The sense strand of RNAi oligos (Dharmacon) that were used to silence the target genes are as follows: cgaguaggcuucgugacuu (siDrosha) and caucggacaagagugugau (siDGCR8). The siLuc was the siCONTROL Non-Targeting siRNA #2 from Dharmacon. To check for protein silencing, western blot was performed with the following antibodies: antiDGS8 (N-19) from Santa Cruz Biotechnologies, antiDrosha (ab12286) from Abcam and antiTubulin antibody kindly provided by Dr Muro.
Northern blot for small RNAs
To perform northern blot for small RNA, we followed an already described procedure (29). Briefly, we loaded 30 µg of total RNA on a 13.5% (19:1) acrylamide/bisacrylamide, 5 M urea and a denaturing gel. After the run, we performed semi-dry transfer for 1 h at 2 mA/cm2 and ultraviolet-cross-linking of the membrane with 1200 µJ. We used probes against miR-34b (atggcagtggagttagtgattg) and U6 snRNA (atatggaacgcttcacgaatt). We radiolabelled 10 pmol of the probe with γ-32P ATP in the presence of T4 Kinase for 1 h at 37°C. Probe hybridization was performed at 37°C for 2 h (U6snRNA) or O/N (miR-34b).
Bioinformatics analysis and manual annotation of SO pri-miRNAs
The algorithm was designed by Dennis Prickett at CBM, Trieste, Italy. We downloaded the coordinates of human miRNAs (www.mirbase.org) and the coordinates of the spliced expressed sequence tags (ESTs, http://genome.uscs.edu/). The algorithm was designed to compare the position of the 5′ and 3′ ss of the ESTs with the position of 5′ and 3′ ends of the miRNAs. When the distance was <100 bp, the miRNA of interest was printed. Then, through manual annotation, we selected the pri-miRNA hairpins overlapping with splice sites. Donor and acceptor splice site scores were calculated using the Neural Network method available at http://fruitfly.org/seq_tools/splice.html.
RESULTS
Identification of SO pri-miRNAs
To identify pri-miRNAs that overlap with splice sites, we compared the human annotated pri-miRNA hairpin coordinates (from www.mirbase.org) with the position of the splice sites derived from the analysis of the spliced expressed sequence tags (ESTs; extracted from http://genome.uscs.edu/). This analysis, followed by a careful manual annotation to exclude inappropriate intron–exon junctions, identified a group of pri-miRNAs whose predicted hairpins contain a possible splice site. We found 17 pri-miRNA hairpins overlapping with splice sites that accordingly were named SO pri-miRNAs. Eleven SO pri-miRNAs contain a 3′ss, six a 5′ss and eight are evolutionarily conserved among vertebrates (Table 1). Most are located within protein-coding genes, three belong to non-coding transcripts, and two derive from putative open reading frames (ORFs). The splice sites in the hairpins can be located at the base of the hairpin, near the junction between the single-stranded (ss)RNA and the double-stranded (ds)RNA, or in the stems, but none is in the terminal loop. The analysis of the splice site strength, evaluated with a neural network program, showed that most of the splice sites have a good score, but three 3′ss (miR-34b, miR-205 and miR-133a-2) were completely ignored (Table 1). These three non-canonical acceptor sites lack an obvious polypyrimidine tract. Interestingly, these miRNAs are implicated in several physio-pathological conditions, and downstream targets mRNAs have been identified (30,35,39).
Table 1.
Splice site Overlapping miRNAs
| miRNA | chr | miRNA conservation | Transcript | ss strength (neural network) | References |
|---|---|---|---|---|---|
| 3′ SO miRNAs | |||||
| miR-205 | 1 | Y | non-coding | 0 | (30,31,36) |
| miR-943 | 4 | WHSC2 | 0.62 | ||
| miR-936 | 10 | COL17A1 | 0.96 | ||
| miR-1287 | 10 | Y | PYROXD2 | 0.85 | |
| miR-34b | 11 | Y | non-coding | 0 | (17,18,23,32–34 |
| miR-1178 | 12 | CIT | 0.90 | ||
| miR-636 | 17 | SFRS2 | 0.64 | ||
| miR-4315–2 | 17 | Y | PLEKHM1F | 0.67 | |
| miR-4321 | 19 | AMH | 0.58 | ||
| miR-133a-2 | 20 | Y | C20orf166 | 0 | (35,38) |
| miR-1292 | 20 | NOP56 | 0.58 | ||
| 5′ SO miRNAs | |||||
| miR-4260 | 1 | LAMB3 | 0.54 | ||
| miR-761 | 1 | Y | NRD1 | 0.95 | |
| miR-555 | 1 | Y | ASH1L | 0.99 | |
| miR-1204 | 8 | non-coding PVT1 | 0.97 | ||
| miR-611 | 11 | C11orf10 | 0.97 | ||
| miR-638 | 19 | Y | DNM2 | 0.83 | (37,53) |
The non-canonical 3′ss of SO pri-miR-34b is correctly used in vivo and in minigene systems
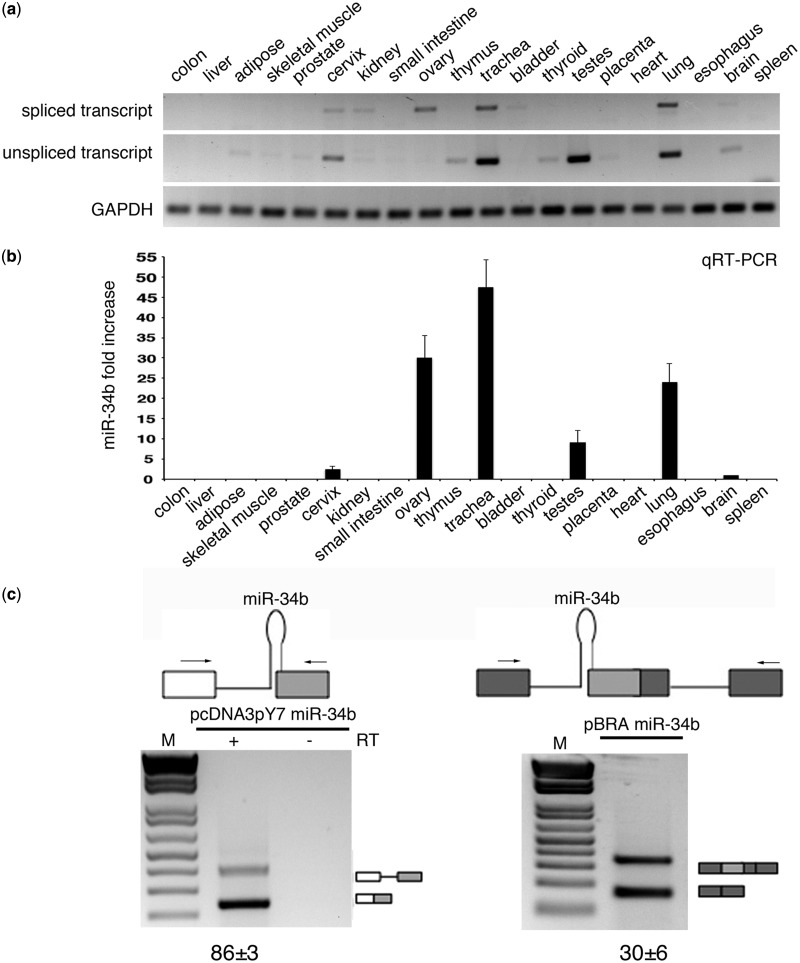
To investigate the dependence of splicing on these peculiar 3′ss that lack a polypyrimidine tract and are embedded in a pri-miRNA hairpin, we focused on pri-miR-34b. pri-miR-34b is part of a non-coding transcript composed of two exons and is located on the distal junction between intron 1 and exon 2 (Supplementary Figure S1). Interestingly, evolutionarily conserved sequences are present at the promoter, at the splice sites, at the pri-miRNAs and at the polyadenylation site (Supplementary Figure S1). The predicted miR-34b hairpin has a typical conserved pri-miRNA secondary structure with three stems (stems A, B and C) and a terminal loop (Figure 1). The AG dinucleotide of the 3′ss is located at the end of the pri-miRNA structure in stem A, four nucleotides above the ssRNA–dsRNA junction (Figure 1). To understand whether the non-canonical acceptor site is effectively used in vivo, we amplified the transcript in a panel of normal human and mouse RNAs with primers located on the exons. The results shown in Figure 2a and Supplementary Figure S2 indicate the presence of the spliced isoform in several human and mouse tissues, and direct sequencing of the bands revealed correct usage of the acceptor site. To understand the role of splicing on miR-34b biosynthesis, we evaluated spliced and unspliced transcripts along with miR-34b abundance in human tissues. In the commercial panel of human total RNAs (Amsbio), miR-34b is significantly expressed in cervix, ovary, trachea, testes and lung (Figure 2b). Interestingly, the relative amount of spliced and unspliced transcripts showed some tissue-specific differences. For example, the spliced and unspliced transcripts are evident in trachea and lung, but the unspliced form is the only product expressed in testes (Figure 2a). Quantitative RT-PCR analysis of the human tissues confirmed this tissue-specific distribution (Supplementary Figure S3). In addition, we observed high levels of the unspliced form also in mouse testes, suggesting that in this tissue production of miR-34b is evolutionarily linked to inefficient intron splicing (Supplementary Figure S2). All together, these data suggest that in some tissues, changes in splicing efficiency might contribute to miR-34b biosynthesis.
Figure 1.
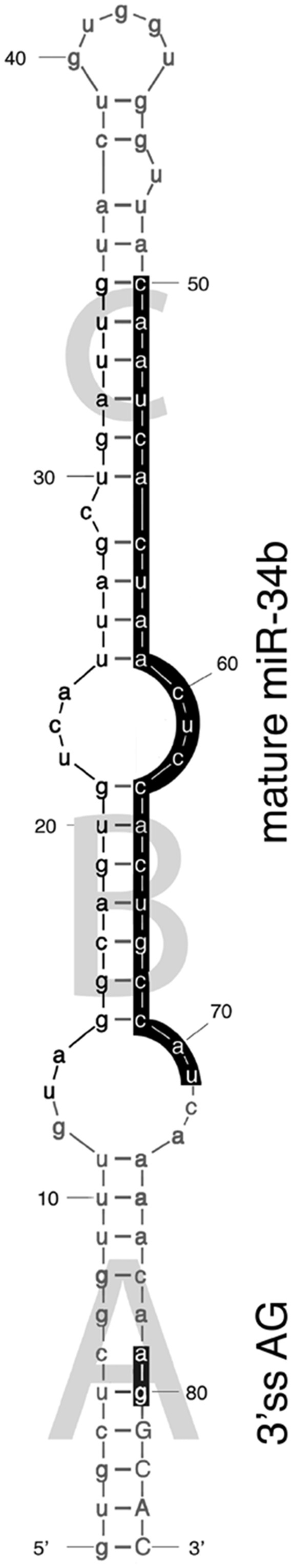
Secondary structure of SO miR-34b hairpin. The secondary structure of SO miR-34b hairpin has been calculated through the mfold web server (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form) with standard parameters. The three stems, named A, B and C, the mature form of miR-34b and the AG dinucleotide of the 3′ss are indicated. Lower and upper cases indicate the intronic and exonic sequences, respectively.
Figure 2.
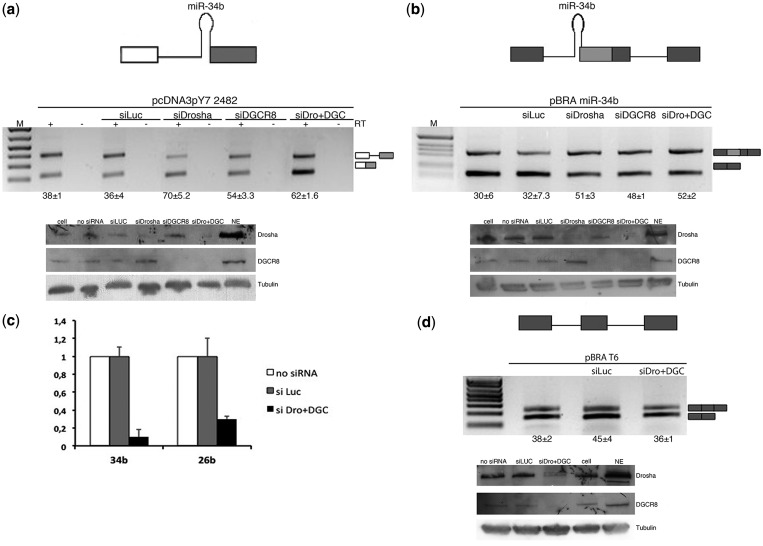
SO pri-miR-34b transcript is spliced in vivo and in minigene systems. (a) RT-PCR profile of miR-34b transcript in 20 human tissues. The upper panels show amplification of the spliced and unspliced transcripts, the lower one amplification of the control gene GAPDH. The identity of the bands was verified through direct sequencing, and quantitative analysis by qRT-PCR is provided in Supplementary Figure S3. (b) qRT-PCR analysis of the mature miR-34b in the different human tissues. Values are normalized for the GAPDH gene. miR-34b abundance in brain is set to 1. (c) Schematic representation of the minigene systems: on the left, pcDNA3pY7 miR-34b and on the right pBRA miR-34b. Thin lines represent introns and miR-34b hairpin. Boxes indicate exons, and, in particular, the light grey box is exon2 of miR-34b. Arrows indicate primers used for PCR analysis. On the bottom, the splicing profile of the two systems after transfection in HeLa cells. The identity of the bands is depicted on the right and was verified through direct sequencing. The numbers under the panel indicate the percentage of splicing (for pcDNA3pY7 miR-34b) or exon inclusion (pBRA miR-34b) ± SD.
To evaluate in more detail how this non-canonical 3′ss is recognized and to identify the minimal sequences required for splicing, we inserted the human pri-miR-34b hairpin structure along with the flanking sequences (18 nt upstream and 97 nt downstream of the ssRNA–dsRNA junctions) in two minigene contexts (Figure 2c): the pcDNA3pY7 minigene (28) in which the 3′ss is before the terminal exon and the pBRA (27) where the 3′ss is part of an alternatively spliced exon. In both minigenes, transfection and RT-PCR analysis in HeLa cells showed correct selection of the acceptor site. In particular, pcDNA3pY7 miR-34b showed two transcripts: the shorter corresponds to splicing of the intron, whereas the longer corresponds to intron retention. In pBRA miR-34b minigene, the upper and the lower bands correspond to exon inclusion and skipping, respectively (Figure 2c). In pcDNA3pY7, miR-34b splicing efficiency was ∼86%, whereas in pBRA, it was ∼30%. Thus, these two minigenes are suitable systems to explore the splicing regulation of this SO pri-miRNA.
Splicing of SO miR-34b requires a BP sequence and a downstream purine-rich ESE
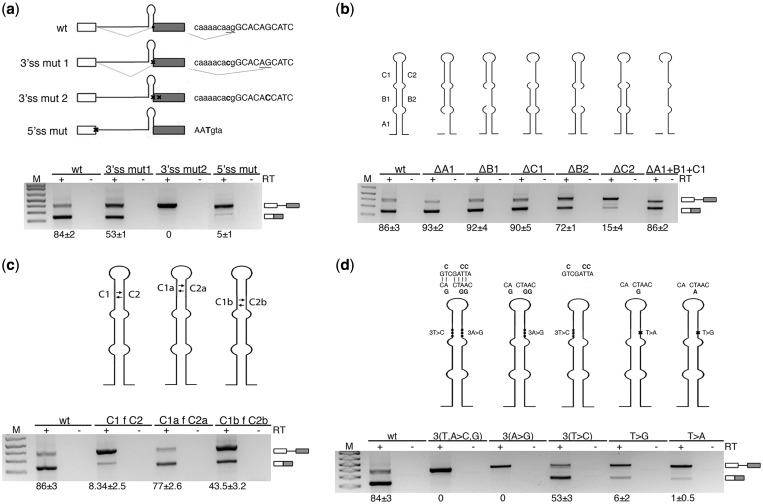
To identify the splicing regulatory elements involved in recognition of the non-canonical SO pri-miR-34b, we performed mutagenesis experiments in the context of the pcDNA3pY7 miR-34b minigene. To evaluate the importance of the AG dinucleotide of the 3′ss, we mutated it to CG, and this activated a 4 bp downstream cryptic acceptor site, located outside the hairpin (3′ss mut 1 in Figure 3a). Mutation of both natural and cryptic sites (3′ss mut 2 in Figure 3a) completely abolished splicing. The disruption of the 5′ss also resulted in complete splicing inhibition, as expected (Figure 3a). To identify the splicing regulatory sequences in the SO pri-miR-34b and explore the possible contribution of the hairpin secondary structure, we performed selective consecutive 5–14-nt long deletions (Figure 3b). Deletions were designed to eliminate the complementary sequences that form the three A, B and C stems of the pri-miRNA secondary structure. Splicing analysis showed that the mutants of the left part of the hairpin, either alone (ΔA1, ΔB1 and ΔC1) or combined (ΔA1+B1+C1), did not affect splicing (Figure 3b). The ΔB2 mutant had only a minor effect. On the contrary, deletion of the 14 bp long C2 (dC2, Figure 3b) or exchange between C2 and C1 (C1fC2, Figure 3c) significantly reduced the splicing efficiency, with only 15% of the intron excised. Fine mapping of the C2 element (Figure 3c) showed that the effect on splicing depends on the C2b portion that affects the ‘CACTAAC’ sequence. It perfectly matches the consensus for BP (YNYURAC, BP underlined) and is also located 18 bp upstream of the 3′ss, in accordance with the optimal conserved distance of BP in acceptor sites (40). Substitution of the three A with G (3A>G) or mutation of the critical T (41) (T>G and T>A) located upstream the A of the BP inhibited splicing (Figure 3d), strongly suggesting that this BP is critical for the processing of the pri-miR-34b transcript.
Figure 3.
Splicing of SO miR-34b requires the AG dinucleotide and a BP sequence. (a) Splicing pattern of pcDNA3pY7 miR-34b mutants that affect the 3′ss and 5′ss after transfection in HeLa cells. Thin lines and boxes represent introns and exons, respectively. The position of miR-34b hairpin is indicated. Splice site mutants are crossed. Lower and upper cases identify intronic and exonic sequences, respectively, and mutated nucleotides are in bold. The results of splice site selection are indicated on the right of each minigene (underlined AG). (b) The deletion mutants of pri-miR-34b hairpin are schematically depicted above each lane. The lower panel shows the splicing pattern of the mutant plasmids after transfection in HeLa cells. (c) Splicing profile of mutants of the C stem of pri-miR-34b. The left and right arms of the C stem are twisted, as indicated in the scheme. (d) Scheme of the BP mutants. Mutants of the left and right arms of miR-34b hairpin are indicated above the pictures. The lower panel indicates the splicing pattern after transfection in HeLa cells. The numbers under each panel indicate the percentage of splicing expressed as mean ± SD of at least three independent experiments.
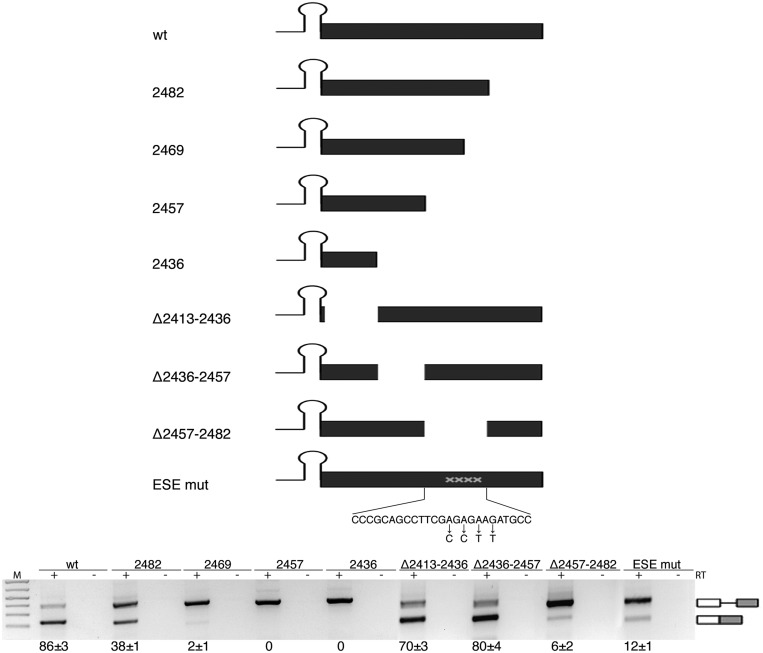
To identify additional splicing regulatory elements, we focused on downstream exonic sequences. In several cases, exonic regulatory sequences act on alternative splicing regulating the 3′ ss recognition (42,43). We prepared minigenes with progressive deletions of the exonic sequences. Minigenes 2482, 2469, 2457 and 2436 (Figure 4) contain progressive deletions of 23, 36, 48 and 69 bp of the exon, respectively. Deletion of the last 23 nt of the exon (mutant 2482) reduced the splicing efficiency to 38%, whereas further deletions completely abolished splicing (mutants 2469, 2457 and 2436; Figure 4). To map the regulatory element involved, we made internal 25 bp deletions (Figure 4), and the results showed that the sequence deleted in mutant Δ2457–2482 was sufficient to abolish splicing. This region contains a 9 bp purine-rich GAGAGAAGA sequence, and substitution of the four adenines into pyrimidines induced nearly complete intron retention (ESEmut, Figure 4).
Figure 4.
Splicing of SO miR-34b requires a purine-rich ESE. Schematic representation of the pcDNA3pY7 miR-34b mutants of the exon. The lines and black box correspond to the intronic miR-34b hairpin and exonic sequences, respectively. The mutated nucleotides of ESEmut are indicated. Lower panel shows the splicing pattern of pcDNA3pY7 miR-34b exonic mutants after transfection in HeLa cells. The identity of the bands is depicted on the right. Numbers below the panel indicate the percentage of splicing expressed as mean±SD of at least three independent experiments.
All together, the experiments with the minigenes indicate that splicing of the non-canonical SO pri-miR-34b acceptor site requires an intronic BP located in the hairpin and a downstream purine-rich ESE. In addition, the absence of any effect on splicing of the combined ΔA1+B1+C1 deletion (Figure 3b) and the results obtained with the flip mutants (Figure 3c and d) indicate that the normal secondary structure of the SO pri-miR-34b is not required for the selection of the acceptor site.
Splicing of SO pri-miR-34b influences miRNA biosynthesis
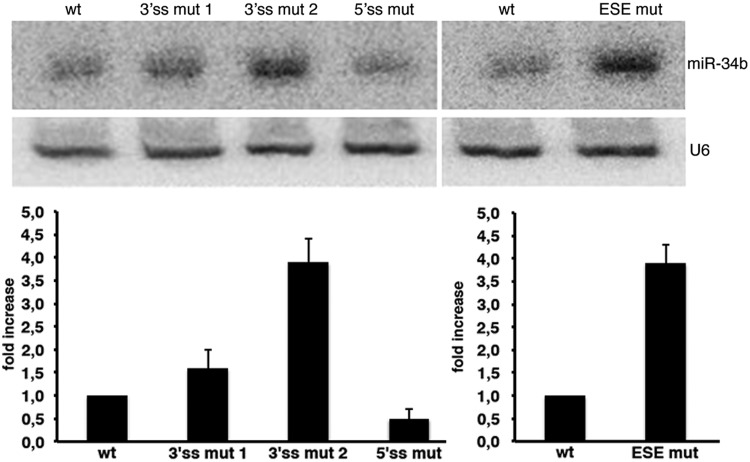
To explore the effect of splicing on miRNA biosynthesis, we analysed the miR-34b derived from processing of the pcDNA3pY7 miR-34b minigenes. We evaluated the wild-type construct, 3′ss mut 1 and 3′ss mut 2 that directly affect the AG dinucleotide (Figure 3a), the 5′ss mutant (Figure 3a) and the ESEmut minigene (Figure 4). In comparison with the wild-type, the amount of mature miR-34b was significantly increased by the mutants that affect 3′ss splicing efficiency (Figure 5). In particular, 3′ss mut 1, which reduced the splicing efficiency to ∼50%, was associated with a ∼1.6-fold increase in miR-34b (Figure 5), whereas the 3′ss mut 2 and the ESEmut, which completely abolished splicing, produced ∼4-fold more miR-34b (Figure 5). On the other hand, splicing inhibition caused by mutation of the 5′ss only slightly reduced the amount of miR-34b (Figure 5). This decrease can be due to the facilitating effect of U1snRNP on Drosha processing, as recently suggested (14). In northern blot analysis, we did not observe any band corresponding to the pre-miRNA intermediate derived from transfection of normal or mutant minigenes. This may indicate that processing of the pre-miRNA by Dicer is efficient and is not a rate-limiting step for its maturation. In addition, we can exclude that the mutants affect miRNA abundance through Dicer-dependent pre-miRNA processing.
Figure 5.
Splicing of SO pri-miR-34b affects miRNA biosynthesis. Northern blot analysis of miR-34b and U6 snRNA. The analysis was performed on the AG dinucleotide mutants, on the 5′ss mutant described in Figure 3 (3′ss mut 1, 3′ss mut 2 and 5′ss mut) and on the mutant of the ESE shown in Figure 4 (ESE mut). Histograms show the fold increase of mature miR-34b normalized to U6. The abundance of miR-34b in the wild-type construct is set to 1.
Silencing of the MPC proteins Drosha and DGCR8 improves SO miR-34b splicing efficiency
As splicing inhibition increases miR-34b biosynthesis, we decided to evaluate whether the MPC-dependent pri-miRNA processing affects the splicing efficiency. To this aim, we silenced Drosha and DGCR8 in HeLa cells that do not express miR-34b, followed by the evaluation of the splicing pattern of transfected minigenes (Figure 6). Western blotting showed almost complete silencing of Drosha and DGCR8 in HeLa cells (Figure 6a and b, lower panels) and a significant reduction of the control endogenous miR-26b (Figure 6c).
Figure 6.
Silencing of Drosha and DGCR8 improves splicing efficiency. (a and b) Splicing pattern of pcDNA3pY7 2482 plasmid (see Figure 4) and pBRA miR-34b (see Figure 2c) after transfection of a control siRNA (siLuc) or silencing of Drosha and DGCR8, alone (siDrosha and siDGCR8) or together (siDro+DGC). Bands identity is indicated on the right. Numbers below the panel indicate the percentage of splicing (a) or exon inclusion (b) ± SD; western blot for Drosha, DGCR8 and tubulin is shown in the bottom panel (c) Quantitative RT-PCR of miR-34b derived from pcDNA3pY7 miR-34b and control endogenous miR-26b, normalized for the GAPDH gene. The abundance of miR-34b and miR-26b in non-treated cells (no siRNA) is set to 1. (d) RT-PCR analysis of pBRA T6 after transfection in HeLa cells and silencing of a control gene (siLuc) or silencing of Drosha and DGCR8 together (siDro+DGC). The percentage of exon inclusion ± SD is indicated below the panel. Western blot for Drosha, DGCR8 and tubulin is shown in the bottom panel.
As the wild-type minigene is nearly completely spliced (pcDNA3pY7 miR-34b) and thus not suitable to appreciate further splicing improvement, we tested Drosha and DGCR8 silencing in pcDNA3pY7 miR-34b 2482 and pBRA miR-34b minigenes. In these two contexts, ∼30–40% of transcripts are spliced in normal conditions (see Figures 4 and 2c, respectively). In pcDNA3pY7 miR-34b 2482, silencing of Drosha and DGCR8 either alone or in combination increased the percentage of splicing from 38 to ∼70% (Figure 6a). Similarly, in the pBRA miR-34b, splicing efficiency increased, raising exon inclusion from 30 to ∼50% (upper band of Figure 6b). On the control pBRAT6 minigene, which does not have the hairpin, we did not observe changes in the splicing pattern (Figure 6d). In addition, we found that Drosha and DGCR8 silencing reduced miR-34b biosynthesis derived from co-transfection of pcDNA3pY7 miR-34b (Figure 6c). As mirtrons (44,45) or 3′-tailed mirtrons (46) do not require the MPC, our results exclude the involvement of these non-canonical pathways in miR-34b biosynthesis.
On the other hand, overexpression of Drosha and DGCR8 in co-transfection experiments reduced the splicing efficiency both in pcDNA3pY7 miR-34b (Supplementary Figure S4a) and pBRA miR-34b (Supplementary Figure S4b) and had no effect on the control pBRA minigene (not shown). Thus, the MPC-dependent pri-miRNA processing interferes with spliceosome and affects the splicing efficiency of SO pri-miR-34b.
DISCUSSION
SO miRNAs are a novel class of miRNAs, characterized by the presence of overlapping pri-miRNA hairpins and splice sites on the nascent transcripts. In SO miR-34b, whose hairpin includes an acceptor site, we have shown that mutations that reduce the 3′ss splicing efficiency increase the mature miRNA form. Conversely, reduction of the Drosha/DGCR8 levels by RNAi knock-down improves splicing efficiency, whereas their overexpression reduces splicing. Thus, in the non-coding SO miR-34b and possibly in the other SO miRNAs, miRNA biosynthesis and production of mature mRNAs are regulated by the competition between the MPC and the spliceosome, owing to the overlap between pri-miRNA hairpins and splice sites. This competition represents a novel mechanism to regulate the level of miRNA biosynthesis through control of pre-mRNA processing.
The biosynthesis of miRNAs can be affected at the level of pri-miRNA processing through changes in the efficiency of MPC-dependent cropping of the nascent transcript (47). Some of the factors that affect cropping, such as Ars2 (3) or FUS (4), lack specificity, as they interact directly with component of the MPC to affect the biosynthesis of multiple miRNAs (4). In some cases, RNA-binding proteins that directly interact with the terminal loop sequence of the pri-miRNA hairpin regulate cropping of miRNAs. For example, LIN-28, SF2/ASF and hnRNPA1 bind to the terminal loop of pri-let-7, pri-miR-18a and pri-miR7, respectively, and affect their processing either acting on the MPC or through changes in the RNA secondary structure (5–7). In SO pri-miRNAs, where the splice sites overlap with the hairpins, changes in the splicing efficiency represent a novel system to directly regulate miRNA biosynthesis. In SO miR-34b, two elements are indispensable for splicing: a consensus BP located in the hairpin and a purine-rich ESE. The interaction of the ESE with RNA-binding protein(s) is the most likely mechanism that regulates splicing of SO miR-34b and consequently its biosynthesis. This ESE contains putative binding sites for SF2/ASF (SF2) and Tra2beta (SFRS10), but neither their overexpression nor silencing affected SO miR-34b splicing (data not shown), suggesting that other splicing factors are involved. Additional experiments are required to identify the mechanism of ESE function and characterize the regulatory splicing factors that specifically affect miR-34b and the other SO miRNAs.
Most of the pre-mRNA processing events occur co-transcriptionally and are functionally coupled through the carboxyl-terminal domain of PolII (48,49), but the precise relationship between splicing and pri-miRNA cropping is not completely understood. In vivo, MPC-dependent cropping of pri-miRNA hairpins located inside introns has been reported to facilitate (12,14) or inhibit (13) splicing. On the other hand, spliceosome assembly was shown to promote cropping (14,16). However, splicing inhibition due to mutations that specifically affect the 5′ss results in a negative effect on production of miRNAs (12,14). The effect of the 5′ss mutation on the production of miR-34b that we observed here is not totally unexpected and consistent with a positive effect of the donor site and U1 snRNP on the MPC, as recently reported (12,14). In this case, a functional donor site, but not the 3′ss, has been shown to be critical for the biosynthesis of the intronic mir-211. Mutations of the 5′ss reduced the production of this miRNA and Drosha recruitment to the miRNA locus (14). Furthermore, intronic plant miRNAs also require a conserved 5′ss for their optimal expression (50). Consistent with the proposed enhancing effect of U1snRNP on the MPC activity (12,14), we also observed that the disruption of the 5′ss reduces the amount of mature miR-34b (Figure 5), in contrast to the mutations that affect splicing of the 3′ss. However, as U1snRNP binding to the first exon can facilitate PolII initiation (51), we cannot exclude a direct effect of the 5′ss mutation on transcription or stability of the nascent transcript. According to Morlando et al. (12), the positive effect on splicing of the MPC is mediated by the carboxyl-terminal domain of the PolII through a tethering mechanism that maintains exonic sequences in place during transcription, whereas cropped intronic fragments are degraded by exonucleases (52). However, in vitro, the two machineries can compete on preformed transcripts (15). In the case of SO miR-34b, and possibly for the other SO miRNAs, when the pri-miRNA hairpins overlap with a splice site, the MPC and the spliceosome act in a mutually exclusive manner to cleave the precursor transcript. The relative proportion of nascent transcripts that are either spliced or cropped in vivo is difficult to assess and might depend on individual SO miRNA features. The analysis of miR-34b in normal human tissues suggests that the spliceosome processes most of the nascent transcripts, as we did not observe a strict reciprocal relationship between the miRNA levels and the amount of spliced RNA. The regulated competition between splicing and the MPC probably affects only a fraction of transcripts in normal conditions, and this explains why in some tissues the levels of mature miR-34b do not correlate with the corresponding amount of spliced transcript (Figure 2a and b). In this case, tissue-specific changes in the splicing efficiency might regulate the competition between the spliceosome and the MPC. Interestingly, several pri-miRNA hairpins do not overlap but are in the proximity of splice sites (within 100 bp) raising the possibility that, in some cases, the position of the miRNA hairpin relative to the intron–exon architecture, along with the presence of intronic splicing regulatory elements, might determine the extent and type of interaction between the MPC and the spliceosome.
The antagonistic effect between the MPC and the spliceosome might have important consequences for SO miRNAs embedded in coding transcripts, which represent a significant proportion of SO miRNAs identified (Table 1). In these cases, the balance between cropping and splicing activities on the nascent transcript might determine how much miRNA is produced at the expense of the mRNA and therefore of protein synthesis. Thus, the coding or non-coding nature of the SO miRNA transcripts targeted by the MPC and the spliceosome might represent an additional level of regulation of gene expression.
Interestingly, several SO miRNAs are associated to physio-pathological conditions. Patients with Parkinson disease or Huntington disease show altered levels of miR-34b (23,24); miR-638 has been correlated with lupus nephritis severity (53); impairment of miR-34b, miR-205 and miR-133a-2 in several types of cancer is a well-established phenomenon (30,35,39). In these cases, the analysis of the splicing pattern of the corresponding transcripts would unveil the unexpected contribution of splicing abnormalities that are at the base of the altered synthesis of miRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana per la Ricerca sul Cancro (AIRC) [10387]. Funding for open access charge: AIRC [10387].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dennis Prickett, CBM, Trieste for the help with the bioinformatics analysis. They also thank Shona Murphy for her support, discussion and critical reading of the manuscript.
REFERENCES
- 1.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 3.Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in. Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morlando M, Dini Modigliani S, Torrelli G, Rosa A, Di Carlo V, Caffarelli E, Bozzoni I. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012;31:4502–4510. doi: 10.1038/emboj.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 6.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. A splicing-independent function of SF2/ASF in microRNA processing. Mol. Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol. Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell. Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 12.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janas MM, Khaled M, Schubert S, Bernstein JG, Golan D, Veguilla RA, Fisher DE, Shomron N, Levy C, Novina CD. Feed-forward microprocessing and splicing activities at a microRNA-containing intron. PLoS Genet. 2011;7:e1002330. doi: 10.1371/journal.pgen.1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kataoka N, Fujita M, Ohno M. Functional association of the Microprocessor complex with the spliceosome. Mol. Cell. Biol. 2009;29:3243–3254. doi: 10.1128/MCB.00360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubo T, Toyooka S, Tsukuda K, Sakaguchi M, Fukazawa T, Soh J, Asano H, Ueno T, Muraoka T, Yamamoto H, et al. Epigenetic silencing of microRNA-34b/c plays an important role in the pathogenesis of malignant pleural mesothelioma. Clin. Cancer Res. 2011;17:4965–4974. doi: 10.1158/1078-0432.CCR-10-3040. [DOI] [PubMed] [Google Scholar]
- 18.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 19.Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH, Hermeking H, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin. Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, Dawson BC, Munivez E, Tao J, Lee BH. miRNA-34c regulates Notch signaling during bone development. Hum. Mol. Genet. 2012;21:2991–3000. doi: 10.1093/hmg/dds129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei J, Shi Y, Zheng L, Zhou B, Inose H, Wang J, Guo XE, Grosschedl R, Karsenty G. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J. Cell. Biol. 2012;197:509–521. doi: 10.1083/jcb.201201057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernardo BC, Gao XM, Winbanks CE, Boey EJ, Tham YK, Kiriazis H, Gregorevic P, Obad S, Kauppinen S, Du XJ, et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc. Natl Acad. Sci. USA. 2012;109:17615–17620. doi: 10.1073/pnas.1206432109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaughwin PM, Ciesla M, Lahiri N, Tabrizi SJ, Brundin P, Bjorkqvist M. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington's disease. Hum. Mol. Genet. 2011;20:2225–2237. doi: 10.1093/hmg/ddr111. [DOI] [PubMed] [Google Scholar]
- 24.Minones-Moyano E, Porta S, Escaramis G, Rabionet R, Iraola S, Kagerbauer B, Espinosa-Parrilla Y, Ferrer I, Estivill X, Marti E. MicroRNA profiling of Parkinson's disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 25.Cha YH, Kim NH, Park C, Lee I, Kim HS, Yook JI. MiRNA-34 intrinsically links p53 tumor suppressor and Wnt signaling. Cell Cycle. 2012;11:1273–1281. doi: 10.4161/cc.19618. [DOI] [PubMed] [Google Scholar]
- 26.Pastor T, Pagani F. Interaction of hnRNPA1/A2 and DAZAP1 with an Alu-derived intronic splicing enhancer regulates ATM aberrant splicing. PLoS One. 2011;6:e23349. doi: 10.1371/journal.pone.0023349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goina E, Skoko N, Pagani F. Binding of DAZAP1 and hnRNPA1/A2 to an exonic splicing silencer in a natural BRCA1 exon 18 mutant. Mol. Cell Biol. 2008;28:3850–3860. doi: 10.1128/MCB.02253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deirdre A, Scadden J, Smith CW. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 1995;14:3236–3246. doi: 10.1002/j.1460-2075.1995.tb07326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan M, Iaconcig A, Muro AF. CPEB2, CPEB3 and CPEB4 are coordinately regulated by miRNAs recognizing conserved binding sites in paralog positions of their 3′-UTRs. Nucleic Acids Res. 2010;38:7698–7710. doi: 10.1093/nar/gkq635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin AY, Zhang XW, Liu L, Yu JP, Li H, Wang SZ, Ren XB, Cao S. MiR-205 in cancer: an angel or a devil? Eur. J. Cell Biol. 2012;92:54–60. doi: 10.1016/j.ejcb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Kim JS, Yu SK, Lee MH, Park MG, Park E, Kim SG, Lee SY, Kim CS, Kim HJ, Chun HS, et al. MicroRNA-205 directly regulates the tumor suppressor, interleukin-24, in human KB oral cancer cells. Mol. Cells. 2013;35:17–24. doi: 10.1007/s10059-013-2154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pigazzi M, Manara E, Baron E, Basso G. miR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer Res. 2009;69:2471–2478. doi: 10.1158/0008-5472.CAN-08-3404. [DOI] [PubMed] [Google Scholar]
- 33.Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai CH, Kao HW, Fang WL, Huang KH, Chan WC, et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int. J. Cancer. 2011;129:2600–2610. doi: 10.1002/ijc.25919. [DOI] [PubMed] [Google Scholar]
- 34.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 35.Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran MN, Choi W, Wszolek MF, Navai N, Lee IL, Nitti G, Wen S, Flores ER, Siefker-Radtke A, Czerniak B, et al. The p63 protein isoform deltaNp63alpha inhibits epithelial-mesenchymal transition in human bladder cancer cells: ROLE OF MIR-205. J. Biol. Chem. 2013;288:3275–3288. doi: 10.1074/jbc.M112.408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li D, Wang Q, Liu C, Duan H, Zeng X, Zhang B, Li X, Zhao J, Tang S, Li Z, et al. Aberrant expression of miR-638 contributes to benzo(a)pyrene-induced human cell transformation. Toxicol. Sci. 2012;125:382–391. doi: 10.1093/toxsci/kfr299. [DOI] [PubMed] [Google Scholar]
- 38.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 40.Chua K, Reed R. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol. Cell Biol. 2001;21:1509–1514. doi: 10.1128/MCB.21.5.1509-1514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kralovicova J, Houngninou-Molango S, Kramer A, Vorechovsky I. Branch site haplotypes that control alternative splicing. Hum. Mol. Genet. 2004;13:3189–3202. doi: 10.1093/hmg/ddh334. [DOI] [PubMed] [Google Scholar]
- 42.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 43.Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 44.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol. Cell. 2010;38:900–907. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 48.Lenasi T, Peterlin BM, Barboric M. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb) J. Biol. Chem. 2011;286:22758–22768. doi: 10.1074/jbc.M111.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montes M, Becerra S, Sanchez-Alvarez M, Sune C. Functional coupling of transcription and splicing. Gene. 2012;501:104–117. doi: 10.1016/j.gene.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 50.Bielewicz D, Kalak M, Kalyna M, Windels D, Barta A, Vazquez F, Szweykowska-Kulinska Z, Jarmolowski A. Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep. 2013;14:622–628. doi: 10.1038/embor.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol. Cell. 2008;29:271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 52.Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol. Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 53.Lu J, Kwan BC, Lai FM, Tam LS, Li EK, Chow KM, Wang G, Li PK, Szeto CC. Glomerular and tubulointerstitial miR-638, miR-198 and miR-146a expression in lupus nephritis. Nephrology (Carlton) 2012;17:346–351. doi: 10.1111/j.1440-1797.2012.01573.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.