Abstract
Active DNA demethylation is crucial for epigenetic control, but the underlying enzymatic mechanisms are incompletely understood. REPRESSOR OF SILENCING 1 (ROS1) is a 5-methylcytosine (5-meC) DNA glycosylase/lyase that initiates DNA demethylation in plants through a base excision repair process. The enzyme binds DNA nonspecifically and slides along the substrate in search of 5-meC. In this work, we have used homology modelling and biochemical analysis to gain insight into the mechanism of target location and recognition by ROS1. We have found that three putative helix-intercalating residues (Q607, R903 and M905) are required for processing of 5-meC:G pairs, but dispensable for excision of mismatched 5-meC. Mutant proteins Q607A, R903A and M905G retain the capacity to process an abasic site opposite G, thus suggesting that all three residues play a critical role in early steps of the base extrusion process and likely contribute to destabilization of 5-meC:G pairs. While R903 and M905 are not essential for DNA binding, mutation of Q607 abrogates stable binding to both methylated and nonmethylated DNA. However, the mutant protein Q607A can form stable complexes with DNA substrates containing blocked ends, which suggests that Q607 intercalates into the helix and inhibits sliding. Altogether, our results suggest that ROS1 uses three predicted helix-invading residues to actively interrogate DNA in search for 5-meC.
INTRODUCTION
DNA methylation at carbon 5 of cytosine (5-meC) is a reversible epigenetic mark essential for cell differentiation and genome defense against transposable elements (1). Active DNA demethylation processes play a pivotal role in shaping methylation patterns, but the underlying mechanisms are still incompletely understood (2,3). In plants, active DNA demethylation is initiated by a family of DNA glycosylases that specifically excise 5-meC and initiate its replacement with unmethylated C in a base excision repair (BER) process (2,4). No 5-meC DNA glycosylases have been unambiguously identified in animals, which may resort to BER of deaminated and/or oxidized derivatives of 5-meC to perform demethylation (5,6).
Plant 5-meC DNA glycosylases are typified by Arabidopsis REPRESSOR OF SILENCING 1 (ROS1) (7–9), and its paralogs DME (DEMETER), DML2 and DML3 (DEMETER-like proteins 2 and 3) (8,10–12). ROS1, DML2 and DML3 counteract excessive methylation at several hundred loci across the genome in vegetative tissues (11–13). DME demethylates the maternal allele of imprinted genes in the endosperm (14), but its basal function appears to be the reactivation of transposons in companion cells to generate short interfering RNAs (siRNAs) that would reinforce transposon silencing in male and female gametes (15,16). ROS1, DML2 and DML3 may also contribute to such reactivation in male gametes (15). All four proteins are bifunctional enzymes with an associate lyase activity that cleaves the phosphodiester backbone at the 5-meC removal site by β,δ-elimination, generating as a major product a single-nucleotide gap flanked by 3′-phosphate and 5′-phosphate termini (8,9,11,12,14). The DNA 3′-phosphatase zinc finger DNA 3′-phosphoesterase (ZDP) functions downstream of ROS1 by removing the blocking 3′-phosphate (17), and the repair protein XRCC1 stimulates 5-meC excision and facilitates 3′-end cleaning and DNA ligation (18).
Plant 5-meC DNA glycosylases belong to the HhH-GPD superfamily (19), but they are distinctively characterized by a bipartite catalytic domain divided by a large insert predicted to have an unstructured conformation (20). They also contain a carboxy-terminal domain of unknown function (8), and a short amino-terminal domain significantly rich in lysine (21). In ROS1, this basic domain mediates strong methylation-independent binding (21) and endows the protein with the capacity to perform facilitated diffusion through random 1D sliding along DNA (22).
ROS1 and its homologs face the challenge to locate and excise a modified, but otherwise nondamaged, correctly paired base. Although no crystal structure is available for any 5-meC DNA glycosylase, the combination of sequence alignment and available structural data of HhH-GPD enzymes allows predicting functional residues that can be tested by site-directed mutagenesis. Most DNA glycosylases use a common base-flipping mechanism to extrude the target lesion from the base stack into a substrate-recognition pocket (23–25). The resulting distortion of the DNA is stabilized by insertion of a bulky intercalating side chain that plugs the vacant space left by the flipped-out nucleotide and a second side chain that wedges between the bases on the opposite strand (25). In a previous study (20), we used homology modeling and biochemical analysis to identify residues important for ROS1 function. Two amino acids predicted to be positioned between the base stack and the recognition pocket (T606 and D611) were found to be essential for catalysis, whereas mutational changes in two aromatic residues presumably located in the substrate-binding pocket (F589 and Y1028) altered the base specificity of the enzyme. Our study also proposed Q607, which is essential for both catalytic activity and stable DNA binding, as a strong candidate for the plug residue that replaces the flipped 5-meC in the base stack (20).
A central question that remains to be answered is how ROS1 locates its target base. In this work, we have built on our previous analysis (20) by investigating in detail the functional role of the putative plug residue Q607 and two other putative helix-intercalating amino acids (R903 and M905) that are predicted to contact the orphan G on the complementary strand. We have found that all three residues are specifically required for excision of 5-meC:G pairs, but dispensable for excision of mismatched 5-meC. We also found evidence that Q607, which is essential for stable methylation-independent DNA binding, slows down ROS1 sliding along DNA. Altogether, our results suggest that ROS1 performs sequential extrusion of every base for extrahelical interrogation while sliding along the DNA in search of 5-meC.
MATERIALS AND METHODS
DNA substrates
Oligonucleotides used as DNA substrates (Supplementary Table S1) were synthesized by Operon or Integrated DNA Technologies (IDT) and purified by polyacrylamide gel electrophoresis (PAGE) before use. Double-stranded DNA substrates were prepared by mixing a 5-μM solution of a 5′-fluorescein-labeled oligonucleotide (upper-strand) with a 10-μM solution of an unlabeled oligomer (lower-strand), heating to 95°C for 5 min and slowly cooling to room temperature. Annealing reactions for the preparation of the 1-nt gapped duplex were carried out at 95°C for 5 min in the presence of a 2-fold molar excess of both unlabeled 5′-phosphorylated oligonucleotide (P30_51) and unlabeled oligonucleotide (CGR) with respect to the 5′-alexa-labeled 3′-phosphorylated oligonucleotide (Al-28P), followed by cooling to room temperature. DNA containing a natural AP site opposite guanine was prepared by incubating a DNA duplex containing a U:G mispair (200 nM) with 2.5 U of Escherichia coli Uracil DNA glycosylase (New England BioLabs) at 30°C for 5 min. Substrates SL1 and SL1-2 contain the same tetraloop obstacle at either one or both molecule ends, respectively. The obstacle is created by a 6-bp DNA helix capped at both termini with tetraloops, which is connected to the DNA substrate via a four-way junction (22). Both substrates were obtained by annealing two oligonucleotides of different lengths, as previously described (22).
Production of ROS1 mutant versions
Site-directed mutagenesis was performed using the Quick-Change II XL kit (Stratagene). The mutations were introduced into the expression vector pET28a (Novagen) containing the full-length wild-type (WT) ROS1 cDNA using specific oligonucleotides (Supplementary Table S2). Mutational changes were confirmed by DNA sequencing, and the constructs were used to transform E. coli BL21 (DE3) dcm− Codon Plus cells (Stratagene). WT and mutant versions were expressed and purified as N-terminal His-tagged proteins, as previously described (21,26).
Enzyme activity assays
Fluorescein-labeled duplex oligonucleotides (20 nM, unless otherwise stated) were incubated at 30°C for the indicated times in a reaction mixture containing 50 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM dithiothreitol (DTT), 0.1 mg/ml bovine serum albumin (BSA) and the indicated amounts of WT ROS1 or mutant variant in a total volume of 50 μl. When reactions included AP endonuclease 1 (APE 1, 5 U; New England BioLabs), EDTA was omitted and 5 mM MgCl2 was added. When measuring AP lyase activity, the reaction mixture was then incubated with 300 mM NaBH4 at 0°C for 30 min and neutralized with 100 mM Tris–HCl pH 7.4. Reactions were stopped by adding 20 mM EDTA, 0.6% sodium dodecyl sulphate and 0.5 mg/ml proteinase K, and the mixtures were incubated at 37°C for 30 min. DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and ethanol precipitated at −20°C in the presence of 0.3 mM NaCl and 16 μg/ml glycogen. Samples were resuspended in 10 μl of 90% formamide and heated at 95°C for 5 min. Reaction products were separated in a 12% denaturing polyacrylamide gel containing 7 M urea. Fluorescein-labeled DNA was visualized in a FLA-5100 imager and analyzed using Multigauge software (Fujifilm).
Kinetic analysis
ROS1 does not exhibit significant turnover in vitro owing to strong product binding (26), and therefore a simple Michaelis–Menten model is inadequate for a correct kinetic analysis of this enzyme. The standard reaction conditions were equimolar (20 nM) enzyme/substrate ratios and incubation at 30°C. Data were fitted to the equation [Product] = Pmax[1 – exp(−kt)] using nonlinear regression analysis and the software Sigmaplot. For each mutant enzyme and substrate, the parameters Pmax (maximum substrate processing within an unlimited period of time), T50 (the time required to reach 50% of the product plateau level, Pmax) and the relative processing efficiency (Erel = Pmax/T50) were determined (27). A representative example of 5-meC DNA glycosylase assay and kinetic analysis is shown in Supplementary Figure S1.
Electrophoretic mobility shift assay
In standard electrophoretic mobility shift assay reactions, increasing amounts of WT ROS1 or mutant variants were incubated with 10 nM fluorescein- or alexa-labeled duplex oligonucleotides, unless otherwise stated. Competition band-shift reactions were performed by preincubating WT ROS1 or mutant variants (130 nM) with 100 nM fluorescein-labeled substrates at 25°C for 5 min and then adding increasing amounts of unlabeled duplex as competitor. DNA binding reactions were carried out at 25°C for 60 min, unless otherwise stated, in 10 mM Tris–HCl, pH 8.0, 1 mM DTT, 10 μg/ml BSA, 1 mM EDTA, in a final volume of 10 μl. Complexes were electrophoresed through 0.2% agarose gels in 1× TAE (40 mM Tris–HCl, pH 8.0, 20 mM acetic acid, 1 mM EDTA). Electrophoresis was carried out in 1× TAE for 40 min at 80 V at room temperature. Fluorescein- or alexa-labeled DNA was visualized in a FLA-5100 imager and analyzed using Multigauge software (Fujifilm).
RESULTS
Identification of putative helix-intercalating residues in ROS1
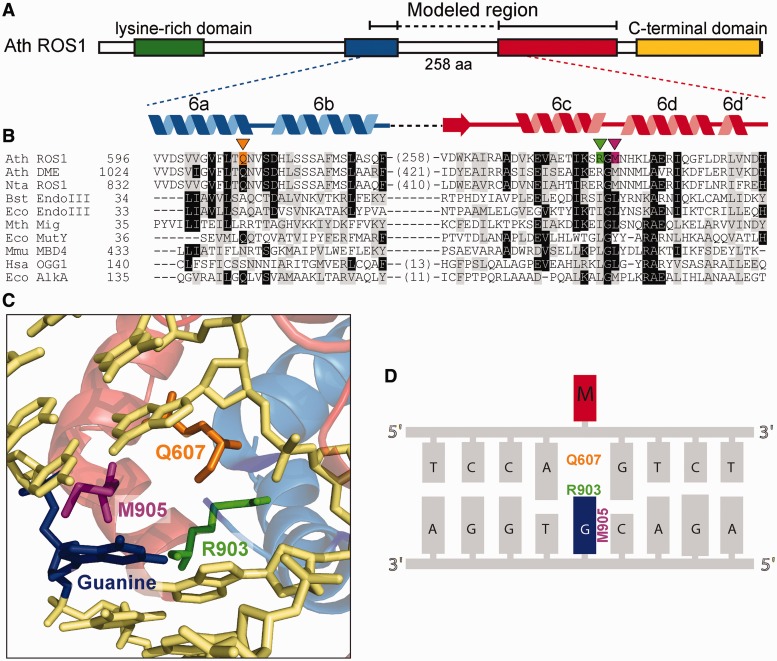
In E. coli Endonuclease III, the side chain of L82 serves as the wedge that intercalates into the DNA duplex and stacks with the G opposite the lesion (estranged G), which donates a hydrogen bond to the backbone carbonyl of I80 (28). The homologous residues of L82 and I80 in ROS1 are M905 and R903, respectively (Figure 1B). The residue R903 is conserved within the ROS1/DME family of DNA demethylases, whereas Q instead of M is observed at the homologous position of M905 in some members of the family (Supplementary Figure S2). The modeled structure of ROS1 (20) predicts that M905 and R903 are positioned close to the G opposite 5-meC (Figure 1C). To test the prediction that M905 and R903 have a role in ROS1 enzymatic activity, we mutated them to Gly (M905G) and Ala (R903A), respectively. In our experiments, we also used the mutant version Q607A, which lacks the putative plug residue (20).
Figure 1.
Identification of putative helix-invading residues in ROS1. (A) Schematic diagram showing conserved regions among members of the ROS1/DME family: a N-terminal lysine-rich region (green), a noncontiguous DNA glycosylase domain distributed over two segments (blue and red) separated by a nonstructured linker region and a highly conserved C-terminal domain (yellow) that is not found in any other protein family. (B) Multiple sequence alignment of part of the DNA glycosylase domain of ROS1/DME proteins and several HhH-GPD superfamily members. ROS1 amino acids analyzed in this work are indicated by inverted triangles and highlighted in orange (Q607), green (R903) or pink (M905). Names of organisms are abbreviated as follows: Ath, Arabidopsis thaliana; Nta, Nicotiana tabacum; Bst, Bacillus stearothermophilus; Eco, Escherichia coli; Mth, Methanobacterium thermoautotrophicum; Mmu, Mus musculus; Hsa, Homo sapiens. Genbank accession numbers are as follows: Ath ROS1: AAP37178; Ath DME: ABC61677; Nta ROS1: BAF52855; Bst EndoIII: 1P59; Eco EndoIII: P20625; Mth Mig: NP_039762; Eco MutY: NP_417436; Mmu MBD4: 1NGN; Hsa OGG1: O15527; Eco AlkA: P04395. (C) Structural model for the DNA glycosylase domain of ROS1 bound to a DNA containing an abasic site. The position of Q607, R903 and M905 residues (colored as in panel B) in relation to the estranged guanine (blue) are shown. The model was generated as described in (20) and the figure was prepared with PyMOL (http://www.pymol.org). (D) Schematic sequence diagram indicating the predicted interactions between mutated amino acids and the orphan guanine (blue) opposite 5-meC (M, in red).
Q607, R903 and M905 are specifically required for processing of 5-meC:G base pairs
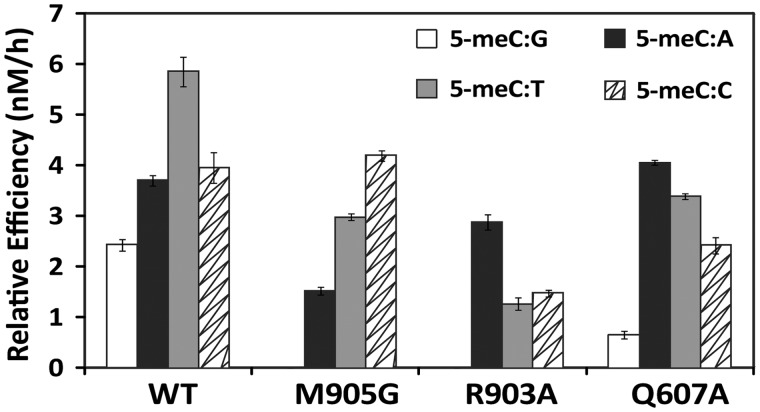
We first examined the ability of WT and mutant proteins M905G, R903A and Q607A to process a 51-mer duplex oligo substrate containing a single 5-meC opposite G, A, T or C (Figure 2). Consistent with our previously reported observations (26), we found that WT ROS1 processed 5-meC with higher efficiency when mispaired with either C, T or A than when paired with G. Mutants M905G and R903A had no detectable base excision activity on a 5-meC:G pair, whereas the Q607A mutant displayed a strongly reduced activity on the same substrate (Figure 2). Interestingly, however, all three mutant proteins retained a significant activity on DNA substrates containing a mismatched 5-meC. In contrast, two catalytically disabled mutants (D611V and T606L) did not show detectable base excision activity on either paired or mispaired 5-meC (data not shown). Therefore, Q607, R903 and M905 are specifically required for efficient excision of 5-meC opposite G.
Figure 2.
Enzymatic activity of WT ROS1 and mutant variants on DNA substrates containing 5-meC opposite different bases. Purified proteins (20 nM) were incubated at 30°C with 51-mer double-stranded oligonucleotide substrates (20 nM) containing 5-meC opposite G, A, T or C, as indicated. Relative processing efficiencies were determined in kinetic assays as described in ‘Materials and Methods’ section. Values are mean ± SE (error bars) from three independent experiments.
These results strongly suggest significant differences in recognition and processing of 5-meC:G pairs and 5-meC mismatches. To explore further such possibility, we examined the temperature dependence of WT ROS1 activity on both types of substrates (Supplementary Figure S3). The catalytic activity on all four DNA substrates increases with temperature from 5 to 30°C, but the temperature dependence is significantly lower for the DNA substrate containing a 5-meC:G pair.
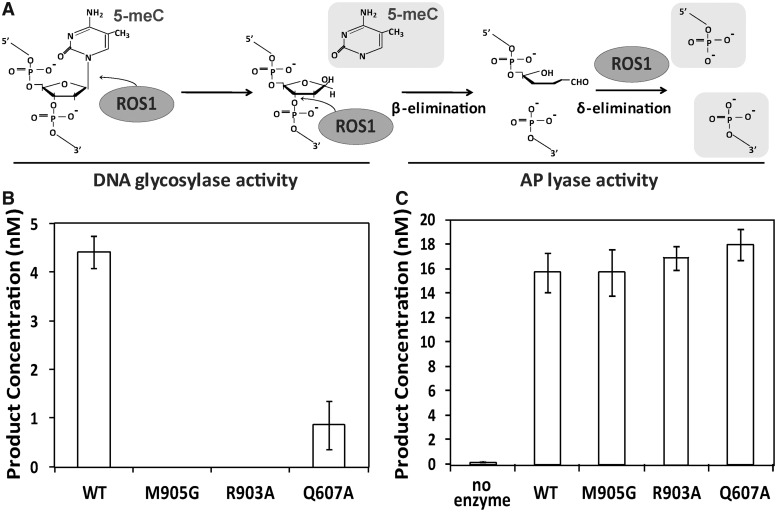
ROS1 is a bifunctional DNA glycosylase/lyase that catalyzes both the release of 5-meC and the cleavage of DNA at the resulting abasic site (8). We therefore asked whether the incapacity of the three mutant proteins to process 5-meC:G pairs is due to a deficiency in DNA glycosylase activity, lyase activity or both (Figure 3). To detect DNA glycosylase activity, we analyzed the reaction products generated by different ROS1 variants after an additional alkaline treatment with NaOH, which cleaves all abasic sites generated by the enzyme and reflects 5-meC excision. We found that R903A and M905G mutant enzymes did not generate detectable incision products, whereas the Q607A variant showed a significantly decreased activity (Figure 3B). Analogous results were obtained when performing reactions in the presence of human APE1 (data not shown). We next tested whether the three variants retained AP lyase activity by incubating the proteins with a 51-mer duplex oligo substrate containing an AP site opposite G. We found that all three mutant variants cleaved the abasic site as efficiently as WT ROS1 (Figure 3C). These results indicate that R903A, M905G and Q607A exhibit a specific defect in catalysis of glycosylic bond cleavage. Because such defect is greatly alleviated when the target base is mispaired, we conclude that Q607, R903 and M905 are critical for destabilization of 5-meC:G base pairs and are likely to perform a key role in extrusion of the target base from DNA.
Figure 3.
Q607A, M905G and R903A mutant proteins lack DNA glycosylase activity on 5-meC:G pairs but retain AP liase activity. (A) Schematic diagram of ROS1 DNA glycosylase/AP lyase activity on 5-meC. ROS1 excises 5-meC as a free base and then cleaves the phosphodiester backbone at the 5-meC removal site by successive β,δ-elimination. (B) DNA glycosylase assay. The generation of incision products was measured by incubating purified WT ROS1 or mutant variants (20 nM) at 30°C for 4 h with a double-stranded oligonucleotide substrate (20 nM) containing a single 5-meC:G pair. After incubation, NaOH (100 nM) was added and samples were immediately transferred to 90°C for 10 min. Products were separated in a 12% denaturing polyacrylamide gel and the amounts of incised oligonucleotide were quantified by fluorescent scanning. (C) AP liase assay. A double-stranded oligonucleotide substrate containing an AP site opposite G (20 nM) was incubated at 30°C for 2 h in the presence of purified WT ROS1 or mutant variants (20 nM). Samples were treated with NaBH4 (300 mM) at 0°C for 30 min to stabilize nonprocessed AP sites and neutralized with 100 mM Tris–HCl, pH 7.4. Products were separated in a 12% denaturing polyacrylamide gel and the amount of incised oligonucleotide was quantified by fluorescent scanning. Values are mean ± SE (error bars) from three independent experiments.
R903 and M905 are dispensable for DNA binding
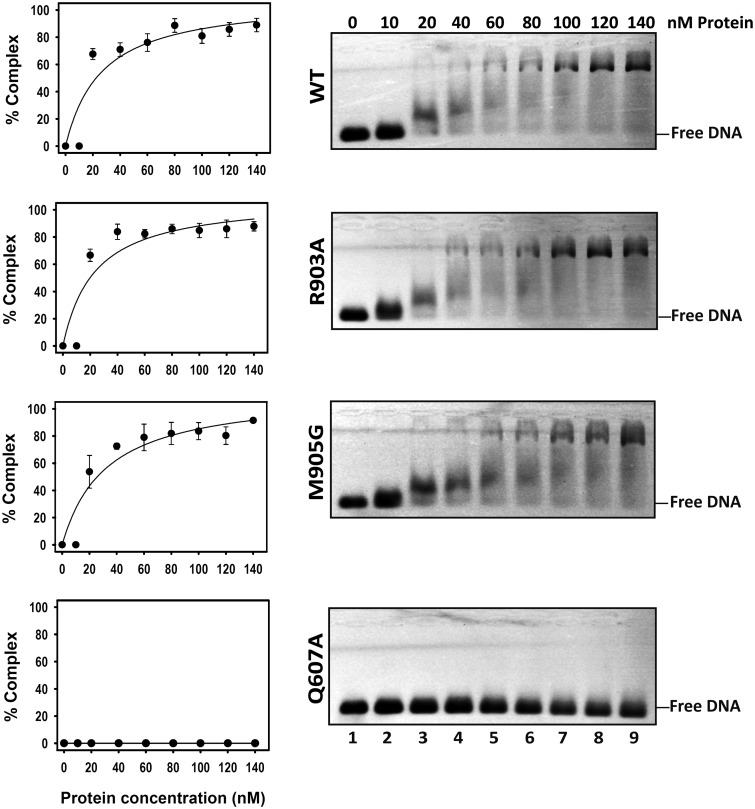
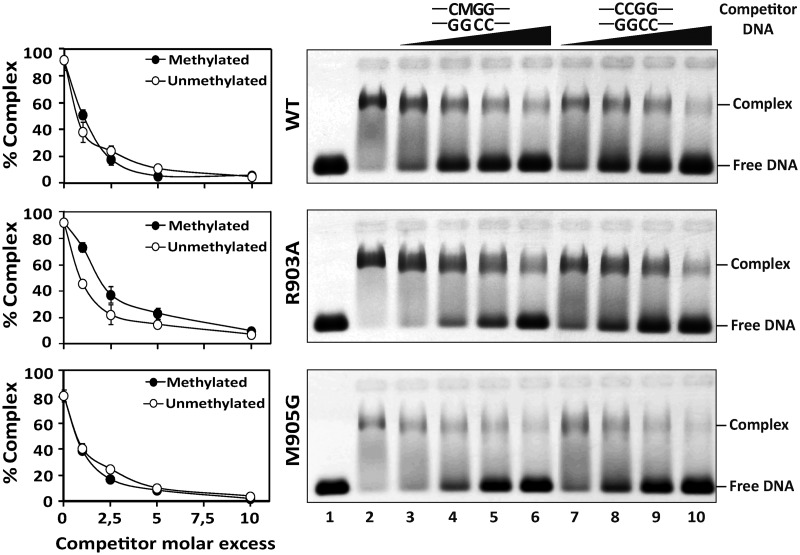
We have previously reported that the Q607A mutant exhibits a drastically reduced DNA binding capacity (20). We therefore decided to examine whether R903 and/or M905 also play a role in substrate binding. We incubated increasing concentrations of WT ROS1, R903A, M905G or Q607A proteins with a labeled DNA substrate containing a 5-meC:G pair (Figure 4). No protein–DNA complexes were detected in binding reactions containing the mutant protein Q607A. In the case of WT ROS1, R903A and M905G proteins, we observed a major band with retarded mobility at high protein concentrations and a minor diffuse band, exhibiting higher mobility that was detectable at low protein concentrations. Analogous results were obtained with nonmethylated DNA (data not shown). These results suggest the formation of protein–DNA complexes containing more than one ROS1 molecule. A similar observation has been previously reported for a mammalian homolog of the DNA glycosylase MutY (29). Quantitation of the total amount of protein–DNA complexes (Figure 4, left) indicates that there are not significant differences in DNA binding capacity between WT ROS1 and either R903A or M905G proteins.
Figure 4.
DNA binding capacity of R903A, M905G and Q607A mutant proteins. Increasing concentrations of purified WT ROS1 or mutant variants were incubated at 25°C for 1 h with 10 nM of fluorescein-labeled 5-meC:G duplex. After nondenaturing gel electrophoresis, gels were scanned to detect fluorescein-labeled DNA. Protein–DNA complexes were identified by their retarded mobility compared with that of free DNA. A representative gel is shown for each protein. Graphs on the left show the percentage of protein–DNA complex versus protein concentration. All bands with slower mobility were used in quantitation of bound protein. Values are mean ± SE (error bars) from three independent experiments.
We next examined in detail the binding affinity of WT ROS1, R903A and M905G through competition experiments with unlabeled oligonucleotides (Figure 5). In agreement with its methylation-independent DNA binding capacity (21), WT ROS1 binding to a methylated DNA probe was reduced with equivalent intensity when incubated with increasing amounts of either nonmethylated or methylated unlabeled competitor DNA (Figure 5, upper panel). We found that the two mutant proteins exhibited dissociation rates not significantly different from those of the WT protein, and both bound methylated and nonmethylated DNA with similar affinity (Figure 5, middle and lower panels). We also found that the two mutant versions do not show any enhanced affinity for DNA substrates containing mismatched 5-meC, or for a single nucleotide gap opposite A, T or C (Supplementary Figure S4). Altogether, these results indicate that R903 and M905 are dispensable for DNA binding.
Figure 5.
Binding of R903A and M905G mutant proteins to methylated and nonmethylated DNA. WT ROS1 (upper panel), R903A (center panel) or M905G (lower panel) proteins (130 nM) were incubated at 25°C for 1 h with 100 nM labeled methylated DNA containing a single 5-meC:G pair, in the presence of increasing amounts (0, 0.1, 0.25, 0.5 and 1.0 μM) of either methylated (lanes 2–6) or nonmethylated (lanes 2 and 7–10) unlabeled competitor DNA. After nondenaturing gel electrophoresis, the gel was scanned to detect fluorescein-labeled DNA. Protein–DNA complexes were identified by their retarded mobility compared with that of free DNA, as indicated. Graphs on the left show the percentage of remaining complex versus competitor molar excess ratios. Values are mean ± SE (error bars) from three independent experiments. M indicates 5-meC.
Q607 inhibits ROS1 sliding along DNA
The results discussed above indicate that residues Q607, R903 and M905 are required for excision of 5-meC opposite G, but only the former is critical for stable DNA binding (20). We have suggested that Q607 is the plug residue used by ROS1 to flip out 5-meC and compensate its extrusion by filling in the vacant space in the DNA base stack (20). Because Q607 is required for stable binding to both methylated and unmethylated DNA (20), we hypothesized that ROS1 performs extrahelical interrogation of unmethylated base pairs by insertion of this residue into the DNA helix.
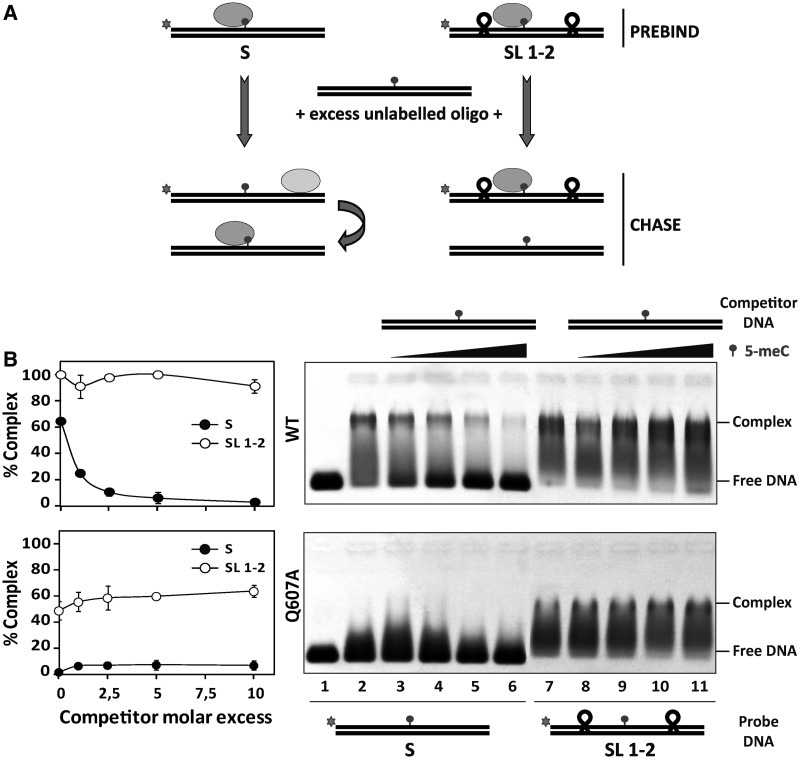
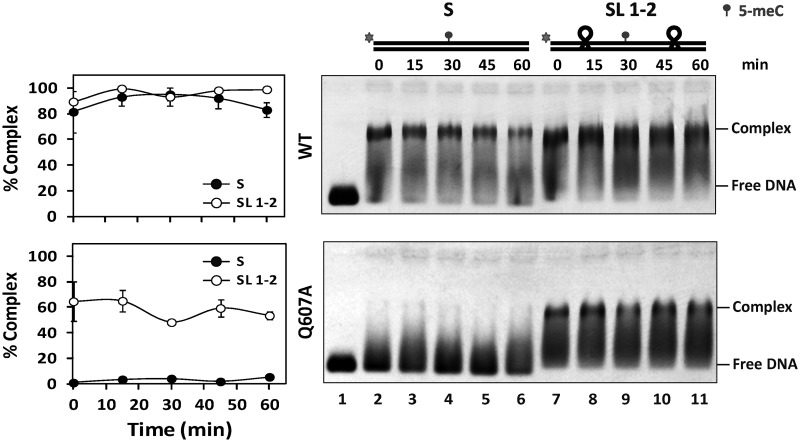
We have recently reported that ROS1 performs sliding on DNA while searching for its target base (22). Therefore, we reasoned that if Q607 plays any role in DNA interrogation, the absence of this residue in the mutant protein should have an effect on DNA sliding. To examine this possibility, we compared the diffusive behavior of WT ROS1 and Q607A on a substrate containing tetraloop obstacles along the DNA surface (Figure 6). We preincubated WT ROS1 or Q607A with labeled substrates S or SL1-2 and then added increasing concentrations of unlabeled S competitor to promote dissociation. Consistent with our previously reported observations (22), we found that WT ROS1 dissociates from substrate S when chased by the competitor, but remains bound to substrate SL1-2 even at high competitor concentrations. As expected, Q607A was unable to bind substrate S. However, the mutant protein was able to form a stable complex with substrate SL1-2, resisting competition with increasing concentrations of unlabeled S (Figure 6). By performing DNA binding measurements at different time points in the absence of competitor, we detected stable complexes of WT ROS1 with both S and SL1-2, whereas Q607A only bound stably to SL1-2 (Figure 7). Furthermore, we found that unblocking one of the substrate ends greatly reduced the capacity of Q607A to form a stable complex with DNA (Supplementary Figure S5). We therefore conclude that the Q607A variant is unable to form a stable complex with a DNA substrate containing free ends, but remains bound to a molecule whose ends are obstructed with tetraloop blocks. Importantly, we found that the stable binding of Q607A to substrate SL1-2 did not have any positive effect on the reduced catalytic activity of the mutant protein (Supplementary Figure S6), thus corroborating the essential role of Q607 for base excision. Altogether, these results indicate that Q607 has an inhibitory effect on ROS1 sliding along DNA, and suggest that the enzyme extrudes unmethylated bases for interrogation by inserting this residue into the base stack.
Figure 6.
The Q607A variant remains bound to a DNA substrate with blocked ends. (A) Schematic diagram showing the experimental setup used to assay for linear diffusion. Proteins were preincubated for 5 min with fluorescein-labeled substrates S or SL1-2, both containing a single 5-meC:G pair, and then chased by addition of unlabeled S competitor. If linear diffusion occurs, dissociation will be faster from labeled substrate S. (B) Gel shift assay showing dissociation of WT ROS1 (upper panel, 130 nM) and the Q607A mutant (lower panel, 130 nM) from fluorescein-labeled substrates S (lanes 2–6, 100 nM) and SL1-2 (lanes 7–11, 100 nM) on addition of increasing amounts (0, 0.1, 0.25, 0.5 and 1.0 μM) of methylated unlabeled competitor S. After nondenaturing gel electrophoresis, the gel was scanned to detect fluorescein-labeled DNA. Protein–DNA complexes were identified by their retarded mobility compared with that of free DNA, as indicated. Graphs on the left show the percentage of remaining complex versus competitor molar excess ratios. Values are mean ± SE (error bars) from three independent experiments.
Figure 7.
Stable binding of Q607A variant to a DNA substrate with blocked ends. Purified WT ROS1 (upper panel, 130 nM) or Q607A mutant variant (lower panel, 130 nM) were incubated at 25°C with 100 nM of fluorescein-labeled substrates S (lanes 2–6) or SL1-2 (lanes 7–11), both containing a single 5-meC:G pair, and the reactions were monitored for 60 min. After nondenaturing gel electrophoresis, the gel was scanned to detect fluorescein-labeled DNA. Protein–DNA complexes were identified by their retarded mobility compared with that of free DNA, as indicated. Graphs on the left show the percentage of protein–DNA complexes at different incubation times. Values are mean ± SE (error bars) from three independent experiments.
DISCUSSION
The conserved base-flipping mechanism used by DNA glycosylases entails the need to stabilize the resulting distortion of DNA. Central to this process is the intercalation of two residues into the base stack: a plug residue that fills the gap left by the flipped-out nucleotide and a wedge residue that intrudes between bases on the opposite strand and interacts with the orphan base. Our homology modeling and functional analysis of ROS1 suggest that Q607 and M905 serve the roles of plug and wedge residues, respectively, with R903 additionally performing a critical function in the stabilization of the orphan G. In this work, we have analyzed the function of these three putative DNA intercalating residues in initiation of DNA demethylation by ROS1 glycosylase.
We have found that Q607A, M905G and R903A variants fail to process 5-meC:G pairs, but remain competent in cleaving an abasic site opposite G. These results demonstrate that mutations in any of these three residues specifically impair early 5-meC recognition events, leaving intact the capacity to perform downstream steps in the base excision cascade. Similar separation of function mutations have been reported in other bifunctional DNA glycosylases. For example, blocking the 8-oxoG recognition pocket of hOGG1 with a bulky amino acid side chain generates a mutant variant unable to flip 8-oxoG nucleotides in DNA, but still able to recognize and cleave abasic sites (30). Interestingly, mutation of two DNA intercalating residues in Fpg (also called MutM) (F114A and M77A) significantly decreases 8-oxoG excision but preserves AP lyase activity (31). Although our results do not exclude the possibility that Q607A, M905G and/or R903A may facilitate AP site hydrolysis following 5-meC excision, they suggest that the major catalytic defect in the three mutant proteins relates to upstream steps of the base extrusion pathway.
WT ROS1 excises 5-meC more efficiently from mismatches (26), which suggests that facile extrusion from the helix plays a critical role in 5-meC excision. The ROS1 model structure predicts that Q607, M905 and R903 are candidates to intercalate in DNA, and therefore they might play a role in destabilization and extrusion of 5-meC opposite G. This hypothesis is supported by the remarkable observation that all three mutant ROS1 proteins retain a significant 5-meC excision activity when the target base is mispaired. Increased base excision activity by disruption of proper base pairing has been previously reported for several glycosylases (32–34). UDG has been proposed to capture spontaneously extruded uracils (35), and displays higher activity when its target base is mispaired with an unnatural adenine analog that lacks Watson–Crick hydrogen-bonding groups (33). DNA glycosylases that use an active base-flipping mechanism, such as Fpg and human AAG, also excise their target bases with higher efficiency when mispaired with nonpolar analogs (32,34), an effect that has been attributed to facilitated base extrusion from the helix. Interestingly, the catalytic activity of human AAG mutated in the plug residue Y162 was partially restored when the base opposite the target was changed to a nonhydrogen bonding partner (34). This result mirrors our observation that variant Q607A retains a significant activity on mismatched 5-meC, thus suggesting that facilitated extrusion of the target base partially bypasses requirement for helix-invading residues.
Insertion of the plug residue into the gap left by the extruded base is likely to play a crucial role in stabilizing the lesion-recognition complex. In fact, it has been consistently observed in different DNA glycosylases that mutation of the plug residue not only curtails catalytic activity, but also abrogates stable binding to DNA substrates (34,36,37). However, there is growing evidence that, in addition to stabilizing the extrahelical state after base flipping, side chains that intercalate into DNA play a critical role during target location and substrate specificity (31,38–40). The results described here strengthen the hypothesis that ROS1 uses Q607 as a plug residue for efficient 5-meC extrusion. In addition, they suggest that ROS1 requires such residue for extrusion of both methylated and unmethylated bases in a DNA scanning process. There are several observations that, taken together with the stable binding of Q607A to a DNA substrate with blocked termini, support the idea that Q607A is not just a DNA binding-deficient mutant, but a faster slider with decreased extrusion rates on both methylated and unmethylated bases. WT ROS1 binds with similar affinity to methylated and nonmethylated DNA, whereas Q607A does not show detectable binding activity on either substrate. However, despite such undetectable binding capacity, Q607A exhibits a significant activity on targets that do not require full extrusion competence, such as mismatched 5-meC and AP sites, and displays excision rates similar to those of the binding-proficient versions M905G and R903A. In contrast, the increased binding of Q607A to a DNA substrate with blocked ends does not result in an increased activity on 5-meC:G pairs (Supplementary Figure S6), which suggests that unstable DNA binding is a derived consequence of a deficiency in a putative plug residue. We therefore propose that the unstable DNA binding exhibited by the Q607A mutant is most likely due to increased sliding along DNA, thus implying that the side chain of Q607 inhibits the scanning rate of ROS1. The role of helix-invading residues in controlling motion of a DNA glycosylase along DNA is not without precedent. In Fpg, which performs intrahelical interrogation (39), mutations of the wedge (F114) and plug (R112) residues increase the diffusion rate (40) and induce strandwise translocation (41), respectively.
Our results shed some light on the possible mechanism used by ROS1 to locate its target base. Three possible strategies have been proposed to explain how DNA glycosylases find their target bases on DNA: (i) passive capture of extrahelical target bases that have spontaneously emerged from the DNA base stack (35), (ii) intrahelical inspection of the relative strength and flexibility of base pairs by inserting one or more residues into the DNA duplex (38,42) and (iii) sequential extrusion of every base out of the DNA helix for extrahelical interrogation (43). Because methylation has been shown to decrease base opening rates (44), it is unlikely that ROS1 passively captures 5-meC residues spontaneously emerged from the DNA base stack at 5-meC:G pairs. By other hand, the inhibitory role of Q607 on ROS1 sliding along DNA suggests that base flipping precedes substrate discrimination, and that the enzyme actively interrogates bases on an extrahelical rather that intrahelical conformation. Intrahelical inspection may be rather inefficient to distinguish between C and 5-meC. Methylation does not induce gross conformational changes in DNA (45) and the 5-methyl group is located on the major groove, whereas the catalytic domain of ROS1, as in all other DNA glycosylases (23), is expected to bind DNA through the minor groove. Intriguingly, both the DNA-binding domain of the methyl-specific endonuclease McrBC from E. coli (46) and the SET- and Ring-associated (SRA) domains of mammalian UHRF1 (47) and Arabidopsis SUVH5 (48) flip out 5-meC from the base stack to achieve discrimination of 5-meC from C. In any case, a full understanding of the search mechanism used by ROS1 must await structural information for this enzyme.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness and the European Regional Development Fund [BFU2010-18838]; Junta de Andalucía, Spain [P07-CVI-02770]; Spanish Ministry of Education, PhD FPU fellowship (to J.T.P.D); Junta de Andalucía, PhD Fellowship (to M.I.P.M.). Funding for open access charge: Spanish Ministry of Economy and Competitiveness and the European Regional Development Fund [BFU2010-18838]; Junta de Andalucía, Spain [P07-CVI-02770].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of their laboratory for helpful discussions and advice.
REFERENCES
- 1.Zilberman D. The evolving functions of DNA methylation. Curr. Opin. Plant Biol. 2008;11:554–559. doi: 10.1016/j.pbi.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Roldan-Arjona T, Ariza RR. In: DNA and RNA modification Enzymes: Comparative Structure, Mechanism, Functions, Cellular Interactions and Evolution. Grosjean H, editor. Austin, TX: Landes Bioscience; 2009. pp. 149–161. [Google Scholar]
- 3.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong Z, Zhu JK. Active DNA demethylation by oxidation and repair. Cell Res. 2011;21:1649–1651. doi: 10.1038/cr.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 8.Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias MI, Ariza RR, Roldan-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl Acad. Sci. USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agius F, Kapoor A, Zhu JK. Role of the arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl Acad. Sci. USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 11.Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc. Natl Acad. Sci. USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- 13.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in arabidopsis. Curr. Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 14.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calarco JP, Borges F, Donoghue MT, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijo JA, Becker JD, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151:194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez-Macías MI, Qian W, Miki D, Pontes O, Liu Y, Tang K, Liu R, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, et al. A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol. Cell. 2012;45:357–370. doi: 10.1016/j.molcel.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Macías MI, Cordoba-Cañero D, Ariza RR, Roldan-Arjona T. The DNA repair protein XRCC1 functions in the plant DNA demethylation pathway by stimulating cytosine methylation (5-meC) excision, gap tailoring, and DNA ligation. J. Biol. Chem. 2013;288:5496–5505. doi: 10.1074/jbc.M112.427617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nash HM, Bruner SD, Scharer OD, Kawate T, Addona TA, Spooner E, Lane WS, Verdine GL. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 20.Ponferrada-Marín MI, Parrilla-Doblas JT, Roldán-Arjona T, Ariza RR. A discontinuous DNA glycosylase domain in a family of enzymes that excise 5-methylcytosine. Nucleic Acids Res. 2011;39:1473–1484. doi: 10.1093/nar/gkq982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponferrada-Marín MI, Martínez-Macías MI, Morales-Ruiz T, Roldán-Arjona T, Ariza RR. Methylation-independent DNA binding modulates specificity of repressor of silencing 1 (ROS1) and facilitates demethylation in long substrates. J. Biol. Chem. 2010;285:23032–23039. doi: 10.1074/jbc.M110.124578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponferrada-Marín MI, Roldán-Arjona T, Ariza RR. Demethylation initiated by ROS1 glycosylase involves random sliding along DNA. Nucleic Acids Res. 2012;40:11554–11562. doi: 10.1093/nar/gks894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat. Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 24.Dalhus B, Laerdahl JK, Backe PH, Bjoras M. DNA base repair—recognition and initiation of catalysis. FEMS Microbiol. Rev. 2009;33:1044–1078. doi: 10.1111/j.1574-6976.2009.00188.x. [DOI] [PubMed] [Google Scholar]
- 25.Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim. Biophys. Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponferrada-Marín MI, Roldán-Arjona T, Ariza RR. ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic Acids Res. 2009;37:4264–4274. doi: 10.1093/nar/gkp390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardeland U, Bentele M, Jiricny J, Schar P. Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. J. Biol. Chem. 2000;275:33449–33456. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- 28.Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pope MA, David SS. DNA damage recognition and repair by the murine MutY homologue. DNA Repair (Amst) 2005;4:91–102. doi: 10.1016/j.dnarep.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Dalhus B, Forsbring M, Helle IH, Vik ES, Forstrom RJ, Backe PH, Alseth I, Bjoras M. Separation-of-function mutants unravel the dual-reaction mode of human 8-oxoguanine DNA glycosylase. Structure. 2011;19:117–127. doi: 10.1016/j.str.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Sung RJ, Zhang M, Qi Y, Verdine GL. Structural and biochemical analysis of DNA helix-invasion by the bacterial 8-oxoguanine DNA glycosylase MutM. J. Biol. Chem. 2013;288:10012–10023. doi: 10.1074/jbc.M112.415612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis AW, Helquist SA, Kool ET, David SS. Probing the requirements for recognition and catalysis in Fpg and MutY with nonpolar adenine isosteres. J. Am. Chem. Soc. 2003;125:16235–16242. doi: 10.1021/ja0374426. [DOI] [PubMed] [Google Scholar]
- 33.Krosky DJ, Song F, Stivers JT. The origins of high-affinity enzyme binding to an extrahelical DNA base. Biochemistry. 2005;44:5949–5959. doi: 10.1021/bi050084u. [DOI] [PubMed] [Google Scholar]
- 34.Vallur AC, Feller JA, Abner CW, Tran RK, Bloom LB. Effects of hydrogen bonding within a damaged base pair on the activity of wild type and DNA-intercalating mutants of human alkyladenine DNA glycosylase. J. Biol. Chem. 2002;277:31673–31678. doi: 10.1074/jbc.M204475200. [DOI] [PubMed] [Google Scholar]
- 35.Cao C, Jiang YL, Stivers JT, Song F. Dynamic opening of DNA during the enzymatic search for a damaged base. Nat. Struct. Mol. Biol. 2004;11:1230–1236. doi: 10.1038/nsmb864. [DOI] [PubMed] [Google Scholar]
- 36.Maiti A, Morgan MT, Drohat AC. Role of two strictly conserved residues in nucleotide flipping and N-glycosylic bond cleavage by human thymine DNA glycosylase. J. Biol. Chem. 2009;284:36680–36688. doi: 10.1074/jbc.M109.062356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 38.Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 39.Qi Y, Spong MC, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature. 2009;462:762–766. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011;39:7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi Y, Nam K, Spong MC, Banerjee A, Sung RJ, Zhang M, Karplus M, Verdine GL. Strandwise translocation of a DNA glycosylase on undamaged DNA. Proc. Natl Acad. Sci. USA. 2012;109:1086–1091. doi: 10.1073/pnas.1111237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi Y, Spong MC, Nam K, Karplus M, Verdine GL. Entrapment and structure of an extrahelical guanine attempting to enter the active site of a bacterial DNA glycosylase, MutM. J. Biol. Chem. 2010;285:1468–1478. doi: 10.1074/jbc.M109.069799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verdine GL, Bruner SD. How do DNA repair proteins locate damaged bases in the genome? Chem. Biol. 1997;4:329–334. doi: 10.1016/s1074-5521(97)90123-x. [DOI] [PubMed] [Google Scholar]
- 44.Mirau PA, Kearns DR. Effect of environment, conformation, sequence and base substituents on the imino proton exchange rates in guanine and inosine-containing DNA, RNA, and DNA-RNA duplexes. J. Mol. Biol. 1984;177:207–227. doi: 10.1016/0022-2836(84)90453-4. [DOI] [PubMed] [Google Scholar]
- 45.Derreumaux S, Chaoui M, Tevanian G, Fermandjian S. Impact of CpG methylation on structure, dynamics and solvation of cAMP DNA responsive element. Nucleic Acids Res. 2001;29:2314–2326. doi: 10.1093/nar/29.11.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sukackaite R, Grazulis S, Tamulaitis G, Siksnys V. The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine. Nucleic Acids Res. 2012;40:7552–7562. doi: 10.1093/nar/gks332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajakumara E, Law JA, Simanshu DK, Voigt P, Johnson LM, Reinberg D, Patel DJ, Jacobsen SE. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 2011;25:137–152. doi: 10.1101/gad.1980311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.